Abstract
SWITCH/SUCROSE NONFERMENTING (SWI/SNF) chromatin-remodeling complexes mediate ATP-dependent alterations of DNA–histone contacts. The minimal functional core of conserved SWI/SNF complexes consists of a SWI2/SNF2 ATPase, SNF5, SWP73, and a pair of SWI3 subunits. Because of early duplication of the SWI3 gene family in plants, Arabidopsis thaliana encodes four SWI3-like proteins that show remarkable functional diversification. Whereas ATSWI3A and ATSWI3B form homodimers and heterodimers and interact with BSH/SNF5, ATSWI3C, and the flowering regulator FCA, ATSWI3D can only bind ATSWI3B in yeast two-hybrid assays. Mutations of ATSWI3A and ATSWI3B arrest embryo development at the globular stage. By a possible imprinting effect, the atswi3b mutations result in death for approximately half of both macrospores and microspores. Mutations in ATSWI3C cause semidwarf stature, inhibition of root elongation, leaf curling, aberrant stamen development, and reduced fertility. Plants carrying atswi3d mutations display severe dwarfism, alterations in the number and development of flower organs, and complete male and female sterility. These data indicate that, by possible contribution to the combinatorial assembly of different SWI/SNF complexes, the ATSWI3 proteins perform nonredundant regulatory functions that affect embryogenesis and both the vegetative and reproductive phases of plant development.
INTRODUCTION
Chromatin-remodeling complexes (CRCs), which mediate ATP-dependent alterations of DNA–histone contacts, provide essential links between signaling pathways and the chromatin-based control of transcription, replication, repair, and recombination (Klochendler-Yeivin et al., 2002; Martens and Winston, 2003; Roberts and Orkin, 2004). Based on characteristics of their SUCROSE NONFERMENTING2 (SNF2) family ATPase subunits, the ATP-dependent CRCs are classified into SWITCH2 (SWI2)/SNF2, IMITATION SWITCH (ISWI), Mi-2/Chromodomain-Helicase-DNA binding protein (Mi-2/CHD), and INO80 subfamilies (Becker, 2002; Brzeski et al., 2003). SWI/SNF-like complexes studied in yeast, Drosophila, and mammals constitute at least nine subunits. The evolutionarily conserved core SWI/SNF subunits, corresponding to homologs of yeast Swi2p/Snf2p ATPase, Snf5p, Swi3p, and Swp73p proteins, are essential and sufficient to remodel chromatin in vitro (Phelan et al., 1999; Sudarsanam and Winston, 2000).
In yeast, two ATP-dependent CRCs, SWI/SNF and Remodel the Structure of Chromatin (RSC) (carrying Swi2p/Snf2p and Sth1p ATPase, respectively), contribute to both transcriptional activation and repression (Sudarsanam and Winston, 2000). Mutations in genes encoding SWI/SNF subunits cause defects in mating-type switch and sucrose fermentation and affect the transcription of ∼5% of yeast genes. The RSC complex influences many more genes and is essential for viability and progression through mitosis (Cairns et al., 1996; Ng et al., 2002). In Drosophila, a single ATPase (Brahma) is found in two different SWI/SNF- and RSC-related complexes (BAP and PBAP) that show different subunit composition and chromosomal distribution (Mohrmann et al., 2004). As core subunits, these complexes carry the trithorax group (trxG) Brahma (ATPase), Snr1 (Snf5p), and Moira (Swi3p) proteins that are required for the maintenance of homeotic gene expression patterns affecting oogenesis, embryogenesis, and segmentation (Crosby et al., 1999). Human prototypes of mammalian SWI/SNF-like complexes, BAF (SWI/SNF-BAP type) and PBAF (RSC/PBAP type), contain either the Brahma or BRG1 ATPase in complex with the INI1 (Snf5p), BAF170, and BAF155 (Swi3p type) core subunits. In various mammalian SWI/SNF complexes, these core subunits are combined with different regulatory subunits, including histone deacetylase and retinoblastoma tumor-suppressor Rb binding proteins (RbAP48). BRG1 and human Brahma bind directly to Rb, regulating cell cycle progression. Mutations affecting core components of SWI/SNF complexes lead to tumorigenesis in somatic tissues of mice and humans, indicating their roles in tumor suppression (Roberts and Orkin, 2004). Compared with late effects of Rb deficiency during embryo development, mutations that inactivate BRG1 (but not human Brahma), INI1 (Snf5p), and SRG3 (BAF155/SWI3) result in early lethality during the blastocyte stage (Bultman et al., 2000; Klochendler-Yeivin et al., 2000; Guidi et al., 2001; Kim et al., 2001).
Although no ATP-dependent CRCs have been characterized to date in plants, comparative genome analyses indicate that plants encode a remarkably high number of potential CRC ATPase subunits (Reyes et al., 2002; Wagner, 2003). From 42 putative Arabidopsis thaliana SNF2-like ATPases (see the Plant Chromatin Database at http://chromdb.org), 4 belong to the canonical SWI2/SNF2 subfamily (Verbsky and Richards, 2001). However, only one of these (Arabidopsis BRAHMA [At2g46020]) carries a C-terminal region resembling a bromodomain, a hallmark for binding acetylated Lys residues of histone tails (Hudson et al., 2000; Brzeski et al., 2003). Genetic studies suggest both positive and negative regulatory roles for members of the Arabidopsis SNF2 gene family. Silencing of Arabidopsis BRAHMA results in reduced fertility, curly leaves, homeotic transformations during flower development, and photoperiod-independent early flowering by derepression of CONSTANS (CO), FLOWERING LOCUS T (FT), and SUPPRESSION OF OVEREXPRESSION OF CONSTANS1 (SOC1) (Farrona et al., 2004). SPLAYED, which encodes a SNF2p/Sth1p-like ATPase with an AT-hook motif (Wagner and Meyerowitz, 2002), is required together with LEAFY (LFY) and UNUSUAL FLORAL ORGANS (UFO) for homeotic class B gene expression but also acts as a repressor of floral transition under noninductive conditions. PHOTOPERIOD-INDEPENDENT EARLY FLOWERING1 (PIE1), encoding an ISWI-type ATPase (Noh and Amasino, 2003), stimulates FRIGIDA-dependent expression of the floral suppressor FLOWERING LOCUS C (FLC). The pie1 mutation causes early flowering under short days independent of FLC and acts as a suppressor of petal defects caused by deficiency of the CURLY LEAF Polycomb (PcG) factor, a repressor of AGAMOUS (Goodrich et al., 1997). The CHD3-type ATPase PICKLE carries two copies of a chromodomain for potential recognition of methyl-lysines in histone tails and acts as a repressor of LEAFY COTYLEDON1, a key activator of embryonic development (Ogas et al., 1999). Mutations of INO80 affect the transcription of >100 genes and reduce the frequency of homologous recombination (Fritsch et al., 2004). A more distantly related member of the SNF2 family, DECREASE IN DNA METHYLATION1 (DDM1) (Jeddeloh et al., 1999; Brzeski and Jerzmanowski, 2003), is required for the maintenance of heterochromatin DNA CpG and histone H3K9 methylation. DDM1 also controls the zygotic stability of parent-of-origin imprinting effects caused by mutations of the MEDEA, FERTILIZATION-INDEPENDENT SEED2 (FIS2), and FERTILIZATION INDEPENDENT ENDOSPERM genes, which code for PcG repressors of fertilization-independent endosperm development (Luo et al., 2000; Gendrel et al., 2002). On the other hand, DRM1 (a member of the RAD54/ATRX SNF2-like ATPase subfamily [Kanno et al., 2004]) is involved in the maintenance of RNA-directed non-CpG (i.e., CpNpG and CpNpN) methylation, whereas MORPHEUS MOLECULE1 (a protein carrying part of a SNF2-like ATPase domain [Amedeo et al., 2000]) is required for transcriptional gene silencing.
In contrast to the multiplicity of SNF2-like ATPase proteins, Arabidopsis has only one gene, BUSHY (BSH) (Brzeski et al., 1999), coding for a structural and functional homolog of SNF5. Unlike yeast and mammals, however, Arabidopsis contains as many as four genes encoding SWI3 homologs (Sarnowski et al., 2002). Because the BSH and ATSWI3 proteins are predicted to form various SWI/SNF complexes with different SWI2/SNF2 subunits, mutations that affect the BSH and SWI3 genes are expected to cause defects that are more severe than or overlapping with those that result from mutations of various SWI2/SNF2 subunits. In fact, partial silencing of BSH in seedlings using an antisense approach led to complex pleiotropic defects, including infertility and reduction of apical dominance (Brzeski et al., 1999), whereas similar silencing of ATSWI3B/CHB2 resulted in dwarfism, delayed flowering, and abnormal seedling and leaf development (Zhou et al., 2003). The multiplicity of ATSWI3 genes and the finding that ATSWI3B interacts with FCA, a putative RNA binding protein that regulates flowering time, supports the hypothesis that Arabidopsis may use specialized SWI/SNF complexes for various chromatin-based regulatory functions (Sarnowski et al., 2002). Here, we describe the characterization of knockout mutations in all four Arabidopsis ATSWI3 genes, which indicates that, compared with yeast and animals, the members of the Arabidopsis SWI3 family have undergone considerable functional diversification. Mutations in ATSWI3A and ATSWI3B cause similar blocks of embryo development at the early globular stage. However, unlike atswi3a, the atswi3b mutations result in aberrant segregation of progeny with arrested ovules, which suggests a possible role for ATSWI3B in imprinting. By contrast, mutations in ATSWI3C and ATSWI3D do not prevent embryonic development but cause characteristic alterations in the development of vegetative and reproductive organs as well as an early-flowering phenotype that is characterized by a reduction in the number of rosette leaves under noninductive conditions.
RESULTS
Classification of Plant SWI3 Proteins
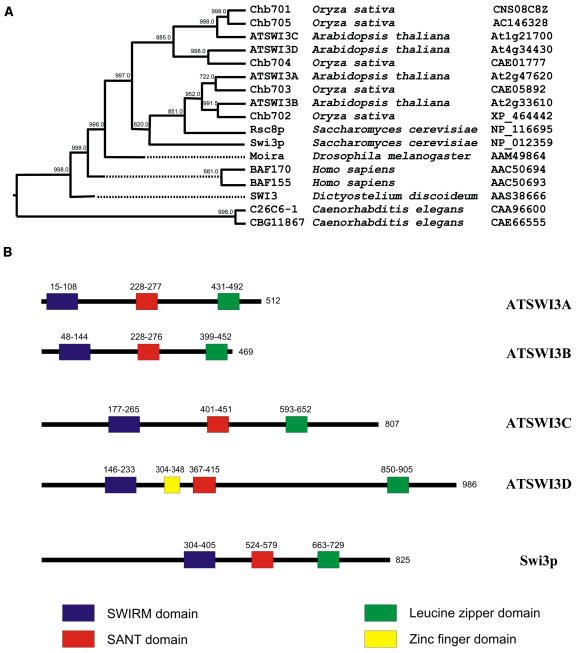
Analysis of the SWI3 superfamily shows that plants have the largest number of nonallelic SWI3 isoforms among multicellular organisms (see the Plant Chromatin Database at http://chromdb.org). Phylogenetic analysis indicates that the sequenced Arabidopsis and rice (Oryza sativa) genomes encode two distinct SWI3 subfamilies (Figure 1A; see Supplemental Table 1 online). The A/B family is composed of two closely related branches, one with the Arabidopsis ATSWI3A and rice Chb703 proteins, and another with ATSWI3B and Chb702. The C/D family branches include ATSWI3C and the related rice proteins Chb701 and Chb705 as well as ATSWI3D with its closest rice homolog, Chb704. The topology of the phylogeny tree predicts that the appearance of four SWI3 subfamilies preceded the separation of monocotyledonous and dicotyledonous plant species during evolution. Compared with the situation in animals and humans, which have either one (Drosophila) or two (Caenorhabditis elegans, mammals, and humans) SWI3 variants, an early duplication of plant SWI3 families suggests a potential functional diversification between their members. In contrast with a previous report by Zhou et al. (2003), comparative analysis of conserved domains using the Smith–Waterman algorithm (Smith and Waterman, 1981) as well as a search of the PFAM_LS database of global domain hidden Markov models (Bateman et al., 1999) showed that all characteristic SWI3 domains, including a SWIRM domain (Swi3/Rsc8/Moira), a SANT domain, and a Leu zipper (Crosby et al., 1999), are present in all four Arabidopsis SWI3 proteins (Figure 1B). However, the length and sequence of interdomain regions vary significantly among members of the A/B and C/D subfamilies.
Figure 1.
Phylogenetic Tree and Domain Organization of Arabidopsis SWI3-Type Proteins.
(A) A consensus maximum parsimony tree from phylogenetic analysis of SWI3-type proteins was constructed. The names of SWI3 proteins from different species along with the accession numbers of the corresponding sequences are listed. Numbers on each branch represent the corresponding bootstrap probability values obtained in 1000 replications.
(B) Protein domains in yeast and Arabidopsis SWI3 proteins were identified using the PFAM-LS database of global domain hidden Markov models (Bateman et al., 1999) and the EMBOSS implementation of the HMMER package (Rice et al., 2000).
Interactions between Arabidopsis SWI3 Proteins in the Yeast Two-Hybrid System
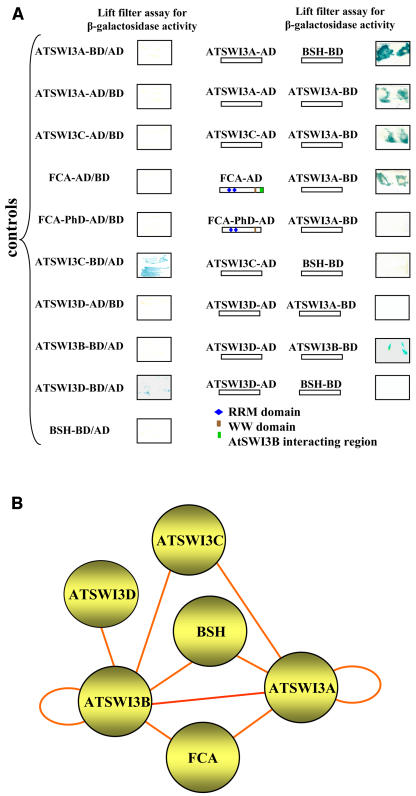
In our previous studies, we demonstrated that ATSWI3B can form homodimers as well as heterodimers with ATSWI3A, ATSWI3C, and BSH/SNF5 in the yeast two-hybrid system (Sarnowski et al., 2002). In addition, we found that ATSWI3B could recruit proteins carrying RNA binding RRM motifs, including FCA, a positive regulator of flower transition (Macknight et al., 2002). By assaying protein interactions of other ATSWI3 family members (Figure 2A), we observed that ATSWI3A can also form homodimers as well as heterodimers with ATSWI3C. In addition to binding ATSWI3B, ATSWI3A also showed interaction with BSH/SNF5 and with the C-terminal region of FCA, which also mediates the interaction with ATSWI3B (Sarnowski et al., 2002). ATSWI3A and ATSWI3B thus recognized similar partners in the two-hybrid assays. However, we failed to detect an interaction between ATSWI3C and BSH. Therefore, it is possible that ATSWI3C associates with BSH only by forming a heterodimer with either ATSWI3A or ATSWI3B. By contrast, ATSWI3A and ATSWI3B may occur in complex with BSH as either a homodimer or a heterodimer. ATSWI3D recognized only ATSWI3B as a binding partner and failed to interact with BSH. This finding suggested that ATSWI3D may recruit BSH only in a heterodimeric complex with AtSWI3B (Figure 2B). Finally, when tethered to DNA by fusion with the Gal4 DNA binding domain, both ATSWI3C and ATSWI3D were capable of activating the transcription of the lacZ reporter gene, which was not observed with the ATSWI3A and AtSWI3B baits in yeast.
Figure 2.
Yeast Two-Hybrid Assays with Arabidopsis SWI3 Proteins.
(A) β-Galactosidase filter lift assays of pair-wise interactions between ATSWI3A and BSH, ATSWI3C and ATSWI3A, ATSWI3A and FCA, ATSWI3C and BSH, ATSWI3D and ATSWI3A, and ATSWI3B and BSH. Blue indicates protein interactions. Control self-activation assays are shown at left in yeast strains carrying ATSWI3A, ATSWI3B, ATSWI3C, ATSWI3D, and FCA bait (BD, fusion with the Gal4 DNA binding domain) or prey (AD, fusion with the Gal4 activation domain) constructs in combination with empty bait or prey vectors, respectively. Note that the ATSWI3C-BD and ATSWI3D-BD baits show self-activation in yeast. Small geometric shapes indicate the domain organization of FCA. The FCA-AD prey encodes full-length FCA, whereas FCA-PhD-AD codes for a C-terminally truncated form of FCA lacking the ATSWI3B-interacting region.
(B) Model for combinatorial interactions between ATSWI3, BSH, and FCA proteins based on the interaction screens in (A) and earlier results obtained by Sarnowski et al. (2002). Circles indicate the homodimerization capability of ATSWI3A and ATSWI3B.
Identification of T-DNA Insertion Mutations in Genes Encoding ATSWI3 Homologs
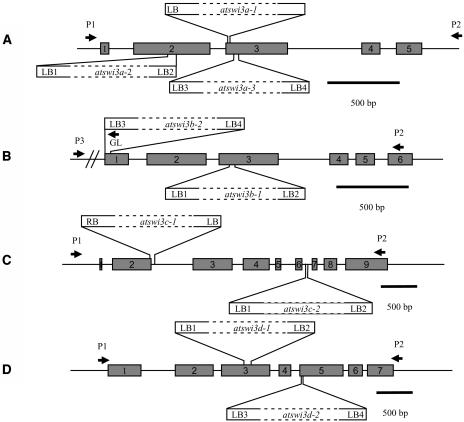
Arabidopsis lines carrying T-DNA insertions in the ATSWI3 genes were identified by PCR screening of different insertion mutant populations (Figure 3). Three atswi3a mutant alleles were found in the SALK collection (Alonso et al., 2003). The atswi3a-1 allele carries a single T-DNA tag (with an intact left border junction and a deletion of 125 bp at the right border; see Methods) in the third exon, replacing a target site deletion of 35 bp. The other mutant alleles, atswi3a-2 and atswi3a-3, contain inverted T-DNA repeats with left border junctions at their termini in the second and third exons, respectively. The T-DNA insertions generated deletions of 54 and 44 bp in the atswi3a-2 and atswi3a-3 alleles, respectively (Figure 3A). In the ATSWI3B gene, two T-DNA tags were identified. The atswi3b-1 allele was isolated by PCR screening of our collection (Ríos et al., 2002), whereas atswi3b-2 was found in the GABI-Kat flanking sequence tag database (Rosso et al., 2003). Both atswi3b-1 and atswi3b-2 carry inverted T-DNA repeats with left border junctions at their termini in the third and first exons, respectively (Figure 3B). The T-DNA integration events were accompanied by deletions of 37 and 47 bp in atswi3b-1 and atswi3b-2, respectively. In addition, insertion of the T-DNA in the first exon added an in-frame stop codon 21 bp downstream of the ATG in atswi3b-2.
Figure 3.
Schemes of the Positions, Orientation, and Organization of T-DNA Insertions in the ATSWI3A, ATSWI3B, ATSWI3C, and ATSWI3D Genes.
The positions of exons (gray boxes with numbers) and introns are indicated. The boundaries of T-DNA inserts (white boxes) marked as LB or RB refer to the junction sequences described in Methods. PCR primers used to identify genotypes of insertion mutants are labeled by arrowheads.
(A) ATSWI3A.
(B) ATSWI3B.
(C) ATSWI3C.
(D) ATSWI3D.
Two T-DNA insertions inactivating ATSWI3C were identified in our collection. The atswi3c-1 allele harbors a single-copy T-DNA insertion within a target site deletion of 42 bp in the third exon (Figure 3C). The atswi3c-2 allele contains an inverted T-DNA repeat with left border junctions facing the boundaries of a deletion of 31 bp in the sixth intron. Although the latter tag is located in an intron, atswi3c-1 and atswi3c-2 confer identical mutant phenotypes. In the atswi3d-1 and atswi3d-2 mutant alleles derived from the SALK lines and our collection, we found inverted T-DNA repeats with left border junctions at their termini, which defined the break points of deletions of 24 and 8 bp in the third and fifth exons of the ATSWI3D gene, respectively (Figure 3D). All T-DNA–tagged mutant lines identified by PCR screening were backcrossed with the wild type, verified by DNA gel blot hybridization and also by segregation analysis of the T-DNA–encoded antibiotic resistance genes (except for the SALK lines, which all carried silenced selectable markers; see Methods).
Mutations in ATSWI3A Affect Early Embryo Development
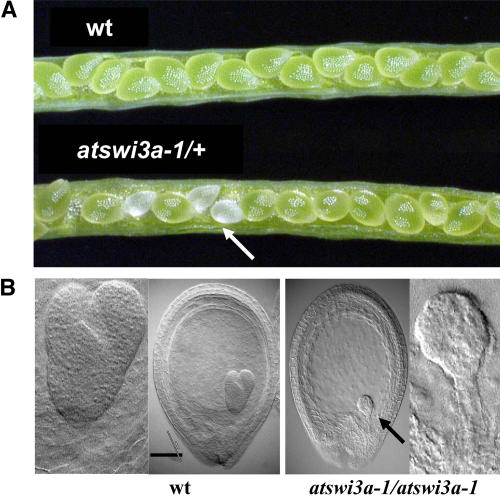
In the segregating SALK mutant lines, we failed to identify homozygous atswi3a mutants. Hence, for each T-DNA insertion allele, we examined the progeny of 30 PCR-genotyped atswi3a/+ plants after self-pollination. Young fruits of all atswi3a/+ lines contained ∼25% white translucent seeds that degenerated into collapsed, brown aborted seeds during maturation (Figure 4A; see Methods). This observation suggested that independent of the position of T-DNA tags in the ATSWI3A gene, all three insertion mutant alleles resulted in a similar, recessive embryo-lethal phenotype. To confirm this conclusion, we compared the developmental status of embryos in green wild-type seeds and white aborted seeds 4 d after fertilization (Figure 4B). Compared with wild-type seeds, which carried embryos in the heart stage, all white seeds showed delayed development and carried embryos arrested in the late globular stage. Although cell divisions appeared to occur normally in the suspensor, the embryo proper displayed aberrant globular shape and degeneration, indicating that ATSWI3A function is essential for proper early embryo development.
Figure 4.
T-DNA Insertions in the ATSWI3A Gene Cause Embryo Lethality.
(A) Open siliques of a wild-type control plant and an atswi3a-1/+ plant showing segregation of green wild-type and white aborting seeds (arrow).
(B) Nomarski images of the same age wild-type and mutant (atswi3a-1/atswi3a-1) seeds, and enlarged sections showing a normal heart-stage embryo in the wild type and an aberrant globular-stage embryo in the mutant. Bar = 100 μm.
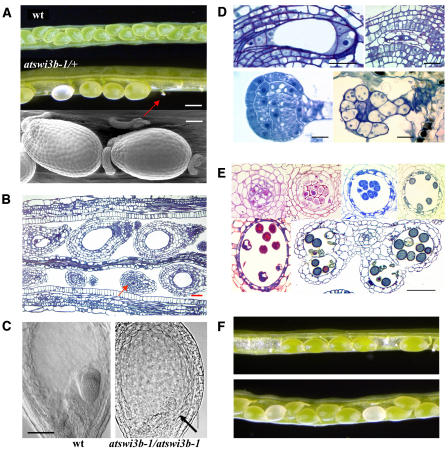
Mutations of ATSWI3B Result in Early Embryo Lethality and Gametophytic Defects Causing Distorted Segregation
By PCR screening, we also failed to identify homozygous atswi3b mutants. Therefore, we examined the segregation of hygromycin and sulfadiazine resistance markers of T-DNA tags of atswi3b-1 and atswi3b-2 alleles, respectively. All M2 lines that carried single T-DNA tags showed a segregation ratio of 2:1 (resistant:sensitive) progeny (see Methods). PCR genotyping of 50 M3 plants confirmed that the resistant lines were heterozygous for the mutant atswi3b alleles. Thus, the segregation data suggested that the atswi3b mutations caused recessive embryo lethality. However, inspection of young siliques of atswi3b-1/+ and atswi3b-2/+ plants revealed an unusual segregation of three distinct phenotypes (Figures 5A and 5B). In addition to green wild-type seeds, representing 40 to 41% of progeny, the fruits contained 48 to 49% arrested ovules and 10 to 11% white aborting seeds, which turned brown and collapsed within 6 to 7 d after fertilization (Table 1). Inspection of embryos 4 d after pollination using Nomarski optics showed that compared with wild-type seeds, which carried late-heart-stage embryos, the white aborting seeds were delayed in development and contained aberrant globular embryos (Figure 5C). A more detailed cytological analysis showed that in white seeds, the endosperm was not cellularized, which is a characteristic trait for early stages of seed development. The embryos displayed abnormal cell shapes and division patterns in the embryo proper and suspensor and remained arrested in the early globular stage (Figure 5D).
Figure 5.
Mutations of the ATSWI3B Gene Cause Embryo Lethality and Non-Mendelian Segregation of Arrested Ovules Showing a Defect in Megasporogenesis.
(A) Open siliques of wild-type control and atswi3b-1/+ mutant plants. A red arrow marks an arrested embryo resulting from maternal imprinting of the atswi3b-1 mutation. Bar = 0.5 mm. The bottom panel shows a scanning electron microscopic image of wild-type seeds and an arrested ovule. Bar = 100 μm.
(B) Longitudinal cross section of an atswi3b-1/+ silique 4 d after pollination showing the structures of aborting (top row, first embryo from left) and wild-type (top row, second embryo from left) seeds and arrested ovules (bottom row, red arrow). Bar = 100 μm.
(C) Nomarski images of a heart-stage embryo in the wild type and an aberrant globular embryo (black arrow) in atswi3b-1/atswi3b-1 mutant seeds. Bar = 100 μm.
(D) Top, the cross section at left shows the embryo sac of an unfertilized wild-type ovule with a typical arrangement of synergids and egg cell at the micropylar pole and antipodal cells at the chalazal pole. The cross section at right shows an arrested ovule, which contains no embryo sac and displays a centrally entrapped vacuolated macrospore. Bottom, cross sections of globular embryos from a wild-type seed (left) and a white aborting seed (right). In the white aborting atswi3b-2 seed, the defective globular embryo shows aberrant cell divisions in the suspensor, unusual shape of the hypophysis cell, and degeneration of internal cells in the embryo proper. Bars = 50 μm.
(E) Stamen cross sections showing the process of microsporogenesis in the atswi3b-2/+ mutant. Top, images from left to right show the normal division of a pollen mother cell, formation of the tetrads, resolution of callose layers connecting the tetrads, and release of the tetrads. Bottom, after separation of the tetrads, ∼50% of microspores fail to divide and undergo vacuolization, senescence, and degeneration. The cross section of an anther shows lobules containing a mixture of wild-type and degenerated spores. Bar = 50 μm.
(F) The white aborting seed phenotype linked to the atswi3b-1 locus and the arrested ovule phenotype, resulting from atswi3b-1–mediated imprinting, segregate as unlinked traits.
Table 1.
Seed Composition of F1 Siliques Obtained by Self-Pollination and Crosses between atswi3b/+ and Wild-Type Plants
| Female × Male | Wild-Type Seeds (%) | Wild-TypeSegregation (R:Sa) | White Aborted Seeds (%) | Arrested Ovules (%) | No. |
|---|---|---|---|---|---|
| atswi3b-1/+ × atswi3b-1/+ | 41 | 2:1 | 11 | 48 | 2000 |
| atswi3b-1/+ × wild type | 46 | 1:1 | <1 | 53 | 494 |
| Wild type × atswi3b-1/+ | 87 | 1:1 | <1 | 12 | 2000 |
| atswi3b-2/+ × atswi3b-2/+ | 40 | 2:1 | 11 | 49 | 2000 |
| atswi3b-2/+ × wild type | 45 | 1:1 | <1 | 55 | 752 |
| Wild type × atswi3b-2/+ | 80 | 1:1 | <1 | 20 | 1668 |
| Wild type × wild type | 98 | – | <1 | 1 | 1301 |
| atswi3b-1/+ (second backcross) | 39 | 2:1 | 11 | 50 | 1047 |
| atswi3b-2/+ (second backcross) | 38 | 2:1 | 11 | 51 | 1024 |
Both atswi3b mutants were backcrossed twice with the wild type.
R, resistant; S, sensitive.
To characterize the defect leading to the formation of arrested ovules, we examined the process of megasporogenesis before fertilization in atswi3b/+ plants. Although apparently normal megaspores were detected in all ovules after meiosis, ∼50% of ovules failed to form embryo sacs; thus, the megaspores did not undergo further divisions but remained arrested in a central position (Figure 5D). As the number of wild-type plus white seeds and that of arrested ovules yielded a segregation ratio of ∼1:1, whereas the T-DNA–tagged atswi3b and wild-type alleles segregated at a ratio of 2:1, the arrested ovule phenotype appeared to be independent of the atswi3b mutations. Therefore, we conclude that the recessive atswi3b mutations resulted in the early embryo lethality seen in white aborting seeds, although the ratio between wild-type and white seeds (∼3.6:1) deviated somewhat from the expected 3:1 ratio. Thus, the phenotypes of the atswi3a and atswi3b mutations appeared to be very similar, although the atswi3b mutations led to earlier arrest of globular embryos and aberrant cell division patterns in the suspensor.
To study the inheritance of the arrested ovule phenotype, we performed recurrent reciprocal crosses between heterozygous atswi3b/+ and wild-type plants (Table 1). When using the wild type as the male pollen donor, we observed an ∼1:1 segregation of green seeds and arrested ovules in the hybrid fruits. This segregation ratio suggested female gametophytic lethality (i.e., predicting no female transmission of the mutant atswi3b alleles). However, germination of wild-type F1 seeds found in the hybrid siliques revealed a 1:1 segregation of T-DNA–tagged atswi3b and wild-type alleles, clearly excluding this possibility. Inspection of 5060 ovules in the F2 progeny of self-pollinated atswi3b/+ F1 plants revealed a segregation of 37.4% ± 0.9% green wild-type seeds and 11.5% ± 0.4% white mutant seeds as well as 51.1% ± 0.9% arrested ovules. In hybrid siliques of the reciprocal cross obtained by pollination of the wild type with either atswi3b-1/+ or atswi3b-2/+ pollen donors, we observed 82 to 87% wild-type seeds, but also a consistent appearance of ∼12 to 17% unfertilized ovules. Viable progeny from these reciprocal crosses also showed a 1:1 segregation of T-DNA–tagged mutant and wild-type alleles, indicating successful male transmission of atswi3b alleles. Upon self-pollination of reciprocal atswi3b/+ F1 hybrids, we scored 4252 F2 progeny, which consisted of 36.5% ± 0.8% wild-type seeds and 11.5% ± 0.6% white mutant seeds as well as 52.0% ± 0.7% arrested ovules. After two backcrosses with the wild type, the heterozygous atswi3b-1/+ and atswi3b-2/+ families showed similar segregation ratios of arrested ovule and aborting seed phenotypes as the original heterozygous M2 mutant families, indicating stable inheritance of both traits.
The appearance of unfertilized ovules in wild-type carpels pollinated by atswi3b/+ males suggested partial male sterility, which implied that the atswi3b mutations could also influence male gametogenesis in addition to affecting macrosporogenesis. Examination of microsporogenesis in atswi3b/+ plants showed that the pollen mother cells could undergo meiosis, producing callose-separated tetrads and later fully separated vacuolated microspores (Figure 5E). However, only approximately half of the microspores (245 of 472 examined) entered into further division, whereas the rest remained vacuolated and exhibited senescence (i.e., lipid deposition, organelle degradation; data not shown) or collapsed. Thus, the observed severe defects in microsporogenesis provided a plausible explanation for the occurrence of unfertilized ovules in fruits derived from the cross of wild-type females with atswi3b/+ males. As the atswi3b mutations were transmitted by the female, we could also exclude the possibility that potential imprinting of the wild-type ATSWI3B male allele caused a defect immediately after fertilization or later during embryo development. However, the strikingly different results of reciprocal crosses, which contradicted Mendel's first law on the uniformity of F1 hybrids, suggested that the potential haploinsufficiency of ATSWI3B might lead to the imprinting of some genes essential for mitotic divisions of macrospores or microspores, or both. The fact that the atswi3b alleles showed similar male and female transmission suggested that, if there were any imprinted genes, they were probably unlinked at the ATSWI3B locus. We examined 196 M3 offspring of a self-pollinated atswi3b/+ plant. From these, 65 AtSWI3B/AtSWI3B lines produced only wild-type seeds, 126 atswi3b/+ lines segregated wild-type and white seeds together with arrested ovules, 4 atswi3b/+ lines had only wild-type and white aborted seeds, and 1 ATSWI3B/ATSWI3B line segregated only arrested ovules. After crossing the last line with a wild-type male, the fruits of F1 hybrids contained 30 to 40% aborted ovules in addition to wild-type seeds. Thus, it appeared that the locus (or loci) conferring the arrested-ovule phenotype was genetically separable from the atswi3b locus (Figure 5F). This observation, of course, raised the possibility that the aborted-ovule phenotype was caused by an independent mutation and not by an atswi3b-dependent imprinting effect. However, the observed aberrant inheritance of the arrested-ovule phenotype, which did not fit Mendelian predictions, its transmission into the F2 progeny obtained by recurrent reciprocal backcrosses with the wild type, and its occurrence in two independent atswi3b mutant lines suggested instead that the arrested-ovule phenotype is an epigenetic trait, which may be further targeted by mapping experiments to verify a potential role of ATSWI3B in imprinting.
Inactivation of ATSWI3C Causes Alterations in Leaf, Root, and Flower Development
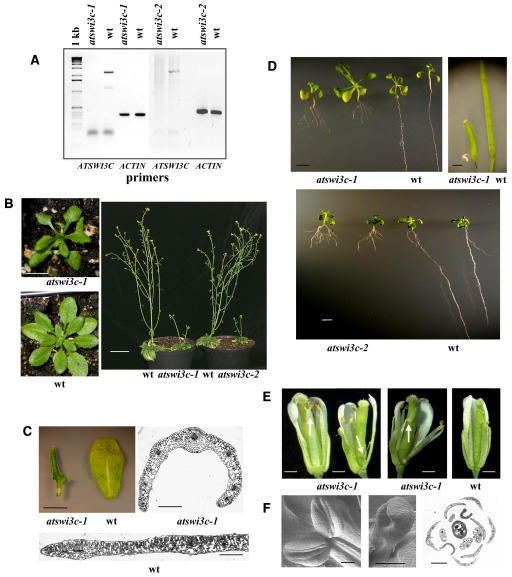
In contrast to atswi3a and atswi3b, PCR screening identified viable plants homozygous for the atswi3c-1 and atswi3c-2 mutations. RT-PCR analysis revealed an absence of wild-type ATSWI3C transcript in the homozygous mutants, indicating that both atswi3c alleles represented null mutations (Figure 6A). Plants carrying the atswi3c-1 and atswi3c-2 alleles showed identical phenotypic alterations, including delayed development, semidwarf growth habit, and formation of aberrant rosettes (Figure 6B). The rosette and cauline leaves were severely twisted along the proximodistal (base-to-tip) axis, which led to a downward curvature of the lateral edges toward the abaxial leaf surface. Cross sections of leaves showed an increased protrusion of mesophyll cell files around the vascular bundles (Figure 6C). This phenotype resembled those caused by the CURLY LEAF (Polycomb repressor of AGAMOUS) and pie (ISWI-type SWI2/SNF2 ATPase) mutations (Kim et al., 1998; Noh and Amasino, 2003; Katz et al., 2004) and by silencing of the SWI2/SNF2-like gene BRAHMA (Farrona et al., 2004). The root system of homozygous atswi3c seedlings was greatly reduced, because the elongation of primary roots was inhibited and the seedlings developed several side roots and secondary root branches (Figure 6D).
Figure 6.
Developmental Defects Caused by the atswi3c Mutations.
(A) RT-PCR assay of the wild type and homozygous atswi3c-1 and atswi3c-2 mutant lines with ATSWI3C and control ACTIN2 primers (see Methods). The absence of PCR product with the gene-specific primer indicates that the atswi3c alleles correspond to null mutations. The size marker is a 1-kb DNA ladder.
(B) Left, rosette phenotypes of 3-week-old atswi3c-1/atswi3c-1 mutant and wild-type plants. Bar = 1 cm. Right, size comparison of 91-d-old wild-type and atswi3c-1 (left pot) and wild-type and atswi3c-2 (right pot) plants grown under short days. Bar = 5 cm.
(C) Cauline leaves of homozygous atswi3c-1 and wild-type plants are shown at top left. Bar = 1 cm. Cross sections of atswi3c-1 mutant (top right) and wild-type (bottom) leaves indicate the enlargement of mesophyll cell files around the vascular bundles in the mutant. Bars = 500 μm.
(D) Left, compared with the wild type, the elongation of the primary root is inhibited in atswi3c-1 (top) and atswi3c-2 (bottom) plants, which display enhanced side root initiation and root branching. Bars = 1 cm. Top right, comparison of mutant and wild-type siliques indicates that the atswi3c-1 mutation inhibits normal fruit development and elongation. Bar = 1 mm.
(E) Left, in whorl C, flowers of the atswi3c-1 mutant contain at least one fused twin stamen and one or two additional staminoid filaments (arrow). Middle and right, compared with the wild type, stamens in atswi3c-1 flowers occasionally carry petalloid tissues in place of anthers (arrow). Bars = 0.25 mm.
(F) Left and middle, scanning electron microscope images of twin anthers (left) and an arrested stamen primordium (middle) from atswi3c-1 flowers. Right, cross section of a mutant flower showing four petals and sepals surrounding three normal and two aberrant stamens and the carpel in the center. Bars = 100 μm.
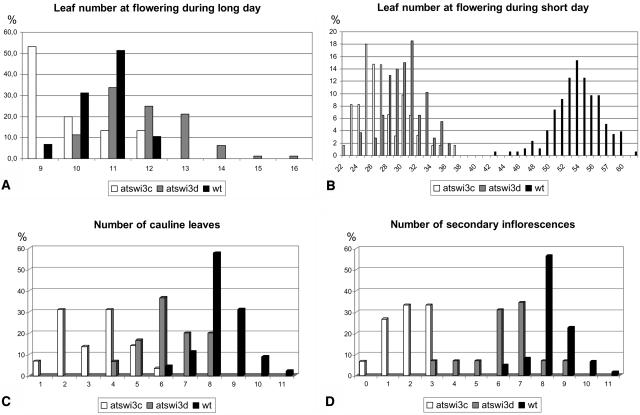
When grown in soil under inductive long-day (LD) conditions (16 h light/8 h dark), the atswi3c mutants flowered 24 to 25 d after planting, with a mean leaf number between 9 and 10, whereas wild-type plants segregating in the same pots flowered at days 20 and 21, with a mean leaf number of 11 (Figure 7A). Under noninductive short-day (SD) conditions (8 h light/16 h dark), the atswi3c mutants flowered between 65 and 75 d after planting, with a mean leaf number of 27, whereas the wild type flowered between 58 and 67 d after planting, with a mean leaf number of 54 (Figure 7B). As in other phenotypic traits, plants carrying the atswi3c-1 and atswi3c-2 alleles showed no difference in flowering time behavior. Based on leaf number, both atswi3c mutants were early flowering during SD conditions and slightly early during LD conditions. In addition to reduced leaf number, both atswi3c mutants had reduced numbers of cauline leaves on the primary inflorescence stem (mean three to four versus eight in the wild type; Figure 7C) and fewer secondary inflorescences (mean two versus eight in the wild type; Figure 7D).
Figure 7.
Number of Rosette Leaves under Long and Short Days, and Number of Cauline Leaves and Secondary Inflorescence Stems under Short Days, in the atswi3c and atswi3d Mutants.
(A) To score the number of rosette leaves under long days, segregating families of atswi3c-1 (n = 104), atswi3c-2 (n = 66), atswi3d-1 (n = 100), and atswi3d-2 (n = 329) were germinated in soil. The data were combined for presentation of both atswi3c and both atswi3d alleles, as pairwise analysis revealed identical flowering time behavior for both.
(B) Leaf number under the SD condition was scored in 61 atswi3c, 108 atswi3d, and 176 wild-type plants.
(C) The number of cauline leaves was counted on primary inflorescence stems of 29 atswi3c, 30 atswi3d, and 45 wild-type plants.
(D) The number of secondary inflorescences was scored using 30 atswi3c, 29 atswi3d, and 65 wild-type plants.
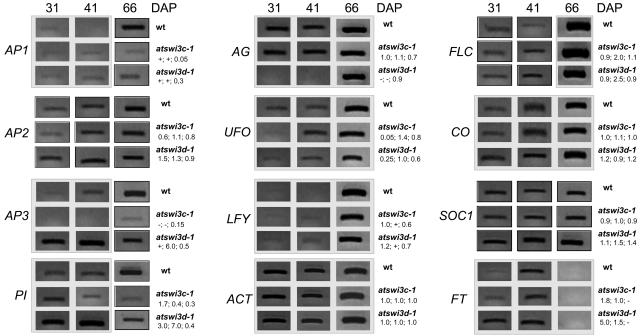
To monitor the developmental regulation of flowering time integrator and floral homeotic genes (for recent reviews, see Ferrario et al., 2004; He and Amasino, 2004) during SD conditions, a series of semiquantitative RT-PCR assays was performed. RNA samples were prepared from whole atswi3c-1 mutant and wild-type seedlings, excluding roots, at days 15, 31, and 41 after planting, as well as from tips of emerging inflorescences on the first day of flowering (i.e., day 66, overlapping between the periods of onset of flowering in the atswi3c mutants and the wild type). The RT-PCR assays with all RNA samples were performed using three different PCR cycle numbers and ACTIN2 as an internal control. Although FT and FLC transcript levels were 1.8- and 2-fold higher in mutant than in wild-type seedlings at days 31 and 41, respectively, no dramatic change in the regulation of flowering time integrator genes was observed in the atswi3c-1 mutant during SD conditions (Figure 8). RT-PCR analyses with plants grown under LD conditions yielded similar data, except that compared with the wild type, FT transcript levels were reduced by 30 to 50% in samples collected from atswi3c-1 seedlings 15 d after planting and from emerging inflorescence tips on the first day of flowering (data not shown). In contrast to the wild type, no APETALA3 (AP3) transcript was detected in vegetative tissues of the atswi3c-1 mutant, whereas PISTILLATA (PI) transcript levels showed a reduction before flowering. In inflorescence tips carrying the differentiating floral meristems, transcript levels of the floral homeotic genes AP1, AP3, and PI were significantly lower in the atswi3c-1 mutant than in the wild type under short days (Figure 8). In addition to these genes controlling the development of sepals, petals, and stamens, AP2 transcript levels also showed a reduction in inflorescence tips of LD-grown mutant plants (data not shown). In association with these transcriptional changes, the atswi3c mutants displayed a 40 to 50% size reduction of flowers, which was especially severe under short days and low humidity and more pronounced in basipetal flowers, as described in BRAHMA-silenced plants by Farrona et al. (2004). However, the size of mature flowers under LD conditions was comparable to that of the wild type.
Figure 8.
RT-PCR Analysis of Transcript Levels of Flowering Time Integrator and Floral Homeotic Genes during Development of the Wild Type, atswi3c-1, and atswi3d-1 during SD Conditions.
Samples were taken at 31 and 41 d after planting (DAP) by collecting whole seedlings, except for roots, whereas samples collected at day 66 correspond to inflorescence tips on the first day of flowering. Numbers under mutant names indicate (from left to right) signal intensities compared with RT-PCR determined with control wild-type RNA samples collected at days 31, 41, and 66. Black framed images show RT-PCR products obtained with 22 cycles of PCR, whereas unframed images correspond to cDNA products amplified by 30 cycles.
More than 80% of mutant flowers contained a fused pair of stamens with degenerated twin anthers. The anthers displayed greatly reduced dehiscences and frequent disjunction between the pair of locules. Occasionally, the anthers were replaced by sepalloid tissues. In addition, approximately one-third of the flowers contained one or two staminoid filaments or arrested stamen primordia (corresponding to a reduction in the number of stamens; Figures 6E and 6F). The atswi3c mutants showed greatly reduced fertility. Although the pollen grains appeared to be normal, during maturation the majority of them remained entrapped and degenerated in the anthers. The siliques contained only a few, often degenerating seeds. Even after manual fertilization with wild-type pollen, fruit development remained abnormal and few viable seeds were obtained. The atswi3c mutants showed distorted segregation of the T-DNA–encoded hygromycin resistance marker. Self-fertilization of the atswi3c-1 mutant, for example, yielded a 2.5:0.4:1.1 segregation of resistant wild-type, mutant, and sensitive offspring, indicating a reduction in the homozygous mutant class (homogeneity χ2 = 0.378, P = 10−7). As no aborting seeds were observed in the siliques of self-pollinated atswi3c/+ plants, the segregation data suggested a possible decrease in either male transmission or the viability of atswi3c gametes. However, because homozygous mutant seeds could be recovered, the development of mutant embryos in atswi3c/+ plants appeared to be unaffected. By contrast, in homozygous atswi3c mutants, the recovery of seed progeny was dramatically compromised, which indicated that the atswi3c mutations caused pleiotropic defects in sporophytic organ development, but unlike atswi3a and atswi3b, they did not result in embryo lethality.
Mutations in ATSWI3D Affect the Number and Development of Leaves and Flower Organs
Like the atswi3c mutants, plants homozygous for the atswi3d-1 and atswi3d-2 mutations were viable and could readily be identified by PCR screening. RT-PCR analysis confirmed a lack of wild-type ATSWI3D RNA in the homozygous lines (Figure 9A), indicating that both atswi3d alleles represented null mutations. Unlike the atswi3c mutants, homozygous mutant seedlings could not be distinguished from wild-type seedlings soon after germination, as they showed normal leaf shape and root elongation. However, plants grown in soil for 15 to 20 d displayed slower development and dwarfism (Figures 9B and 9C). Downward curling of the tips of rosette leaves provided a useful trait for the identification of homozygous mutants (Figure 9C).
Figure 9.
Developmental Alterations Caused by the atswi3d Mutations.
(A) RT-PCR assay of the wild type and homozygous atswi3d-1 and atswi3d-2 mutant lines with ATSWI3D and control ACTIN2 primers (see Methods). The size marker is a 1-kb DNA ladder.
(B) Size comparison of wild-type and dwarf atswi3d-1 (left pot) and wild-type and atswi3d-2 (right pot) plants grown for 91 d under SD conditions. Bar = 5 cm.
(C) Rosettes of atswi3d-1 mutant and wild-type plants grown for 21 d in soil. Note the downward bending of mutant rosette leaves. Bars = 1 cm.
(D) Flowers of homozygous atswi3d-1 and wild-type plants (see description in text). Bar = 0.5 mm.
(E) Scanning electron microscopic images of aberrant carpel development and occasional occurrence of fused twin stamens detected in atswi3d-1 mutant flowers. Bars = 200 μm.
The flowering time behavior of both atswi3d mutants was tested under exactly the same conditions described above for the atswi3c mutants. Both atswi3d mutants flowered under long days, with a mean leaf number of 11, similar to the wild type but 4 to 5 d later (Figure 7A). Under noninductive SD conditions, the atswi3d mutants flowered with a mean leaf number of 34 (54 in the wild type; Figure 7B) between days 60 and 75 after planting (the wild type flowered between days 58 and 67). The atswi3d mutations also reduced the number of cauline leaves on the primary inflorescence (mean between six and seven; eight in the wild type) and the number of secondary inflorescences (mean between six and seven; eight in the wild type) (Figures 7C and 7D). Based on leaf number, the atswi3d mutants were classified as early flowering during short days but showed a less severe flowering time phenotype compared with the atswi3c mutants.
The developmental regulation of flowering time integrator and floral homeotic genes was monitored in the atswi3d-1 mutant by RT-PCR assays as described above for atswi3c-1. Compared with the wild type, the transcript levels of FT and FLC were 5- and 2.5-fold higher at days 31 and 41, respectively, after planting in atswi3d-1 seedlings under SD conditions. Although FT and FLC RNA levels also showed some differences at day 15 and later in mutant and wild-type plants grown under LD conditions (data not shown), the changes detected by RT-PCR did not correlate with the observed early-flowering phenotype and with the comparable transcription of CO and SOC1 genes in mutant and wild-type plants grown under SD and LD conditions. Intriguingly, transcript levels of AP3 and PI were threefold to sevenfold greater, whereas no AG RNA was detected at days 31 and 41 after planting in atswi3d-1 seedlings compared with the wild type. However, in the tips of inflorescences, AG appeared to be transcribed normally, whereas transcript levels of AP1, AP3, PI, and UFO were reduced twofold or more compared with the wild type. As seen in the atswi3c-1 mutant, in addition to these genes, AP2 also showed reduced transcript levels in the atswi3d-1 mutant under LD conditions (data not shown).
A general downregulation of the transcription of floral homeotic genes, except LFY, correlated with severe developmental defects of atswi3d flowers. The sizes of sepals, petals, and stamens of mature mutant flowers were 50 to 60% of wild-type sizes (Figure 9D). The first two whorls of mutant flowers contained five sepals and a variable number of partially developed petals, some of which appeared as aberrant sepal-like filaments. The atswi3d mutants occasionally produced fused twin stamens, like the atswi3c mutants, but with a lower frequency (Figure 9E). The stamens carried small degenerated anthers that failed to produce pollen. The carpels were distorted and highly degenerated; their lower section appeared as an extension of the inflorescence stem. Because of a complete lack of fertilization, the homozygous mutant plants produced no seeds and did not carry normally developed siliques. Despite some similarities, the developmental defects caused by the atswi3d mutations thus appeared to be much more severe than and distinct from those observed in the atswi3c mutants.
DISCUSSION
The SWI/SNF ABC: Arabidopsis SWI3 Family Members and Their Potential Interactions
Compared with yeast, Caenorhabditis, and mammals, the number of genes encoding SWI3 homologs is duplicated from two to four in Arabidopsis. Although the genome annotation is still incomplete, rice probably contains even more, at least five SWI3 genes. The Arabidopsis and rice SWI3 proteins can be classified in two families, the branching of which probably preceded the separation of monocotyledonous and dicotyledonous species. ATSWI3A and ATSWI3B are related to yeast Rsc8p both in size (57, 52, and 63 kD, respectively) and domain organization. Both ATSWI3A and ATSWI3B carry characteristic SWIRM (Swi3/Rsc8/Moira), Leu zipper, and SANT domains, the latter with predicted DNA binding activity to the Myb binding consensus AAC(G/T)G. Members of the second family, ATSWI3C and ATSWI3D, more closely resemble yeast Swi3p and Drosophila Moira, as they are larger (88 and 108 kD), but they nonetheless share all conserved domains with ATSWI3A and ATSWI3B. ATSWI3D is the only member of the ATSWI3 family that contains an additional ZnF-ZZ (CxxCxxxC) domain. The functional significance of SWI3 domains is indicated by site-specific mutagenesis studies in yeast and Drosophila. Deletion of the SANT domain results in defective SWI/SNF complexes in yeast (Boyer et al., 2002), whereas the removal of the C-terminal Leu zipper domain decreases the ability of Moira to self-associate in vitro (Crosby et al., 1999).
In support of sequence-based classification, members of the A/B and C/D ATSWI3 families show different protein interaction properties. ATSWI3A and ATSWI3B are both capable of homodimerization and can form heterodimers with each other. Both ATSWI3A and ATSWI3B can bind ATSWI3C as well as BSH/SNF5 and the flowering regulator FCA protein in yeast two-hybrid assays. By contrast, ATSWI3C and ATSWI3D fail to interact with BSH/SNF5, and ATSWI3D can only bind ATSWI3B. This suggests that ATSWI3D can only recruit BSH/SNF5 through ATSWI3B, whereas ATSWI3C may be tethered to BSH/SNF5 through either ATSWI3A or ATSWI3B. The SNF5 subunit, which is known to coordinate the assembly and nucleosome-remodeling activities of SWI/SNF complexes (Geng et al., 2001), occurs as a single isoform in all species analyzed, including Arabidopsis. Our data suggest that the ATSWI3 subunits could interact with BSH/SNF5 in at least six (AA, BB, AB, AC, BC, and BD) different combinations and form core SWI/SNF complexes with one of the two known Arabidopsis SWP73 homologs (Brzeski et al., 2003) and one specific SNF2-type ATPase subunit.
In the absence of biochemical data, it is impossible to predict which of the 42 potential SNF2-like ATPase homologs will occur in SWI/SNF complexes with BSH/SNF5 and the ATSWI3 proteins. The ATPases found in SWI/SNF complexes of other species all carry characteristic SNF2_N (i.e., a variant of the typical DEXD/H domain) and bromodomain motifs. The bromodomain is a conserved C-terminal motif of ∼110 amino acids that is implicated in the recognition of acetylated Lys residues of core histone tails (Hudson et al., 2000). In Arabidopsis, BRAHMA is the only SNF2/BRAHMA homolog that carries a C-terminal bromodomain, which may enable the SWI/SNF complexes to stimulate the activation of transcription in synergy with histone acetyltranferases.
Recently, BRAHMA was found to interact with ATSWI3C in the two-hybrid system (Farrona et al., 2004). As a canonical ATPase subunit, BRAHMA may occur in SWI/SNF complexes carrying ATSWI3C bound to either ATSWI3A or ATSWI3B and BSH. Silencing of BRAHMA by an RNA interference approach was found to cause an overall size reduction of vegetative and reproductive organs, resulting in curly leaves, short sepals and stamens, reduced fertility, and homeotic transformations between whorls B and C. Downregulation of BRAHMA resulted in precocious flowering under both inductive and noninductive conditions and upregulated the transcript levels of CO, FT, and SOC1 (Farrona et al., 2004). Our studies show that T-DNA knockout mutations of ATSWI3C cause similar morphological alterations to BRAHMA silencing. In addition, the atswi3c null mutants display reduced numbers of rosette leaves (i.e., ∼50% of wild type), especially under SD conditions, and can thus be classified as early-flowering mutants, although they actually flower 4 to 5 d later than the wild type under inductive long days and ∼8 to 10 d later during short days. In the experiments described by Farrona et al. (2004), plants expressing BRAHMA RNA interference flowered after 28 ± 3.6 d during short days (10 h light/14 h dark), whereas wild-type plants bolted after 59 ± 3.4 d. Farrona et al. (2004) compared RNA samples prepared from whole seedlings collected at days 10, 13, and 16 from silenced and wild-type plants and detected significantly higher levels of CO, FT, and SOC1 transcript in plants carrying the BRAHMA RNA interference construct. In our experiments, wild-type plants flowered between days 58 and 67, and samples were collected at days 15, 31, 41, and 66 after planting However, in samples harvested at day 15, only extremely low CO, SOC1, and FT transcript levels were found in both wild-type and atswi3c (and atswi3d) plants, which could only be detected with a large number of PCR cycles. Therefore, we relied instead on a comparison of transcript levels at later time points of development (Figure 7). Our results suggest that the early-flowering phenotype of atswi3c (and atswi3d) mutants did not result from a significant upregulation of genes acting in the photoperiod-sensitive flowering pathway. Nonetheless, more rigorous analysis should address this question (e.g., by monitoring the activity of FT, SOC1, and CO genes using promoter-luciferase fusions in the mutants throughout the full time period between planting and bolting). In addition, testing potential interactions with the autonomous and vernalization-dependent flowering pathways (for review, see He and Amasino, 2004; Henderson and Dean, 2004) should reveal whether the reduced leaf number of atswi3c (and atswi3d) mutants simply reflects defective sporophytic development or is a consequence of the altered expression of genes involved in the regulation of flowering time (Boss et al., 2004). However, it is also conceivable that in the atswi3c mutants, BRAHMA, defined as a repressor of CO (and thereby FT and SOC1), remains capable of delaying flowering. This suggests that BRAHMA may either act as a repressor independent of SWI/SNF complexes or occur in alternative SWI/SNF complexes together with BSH and ATSWI3A or ATSWI3B (or possibly with ATSWI3D). Further studies should clarify whether BRAHMA can interact with other members of the ATSWI3 family and whether FCA (a negative regulator of FLC) and CO (a positive regulator of photoperiod-dependent flowering) would mediate the opposite changes in flowering time caused by the atswi3c mutations and the silencing of BRAHMA.
The interaction of FCA with ATSWI3A and ATSWI3B also suggests that these SWI/SNF core subunits could be targeted by FCA to its partner FY, which is implicated in the control of splicing and 3′ polyadenylation of the FCA transcripts (for review, see Simpson et al., 2004). A recent finding indicates that the human SNF2 ATPase hLodestar/huF2 can interact with CDC5L, a key pre-mRNA splicing factor (Leonard et al., 2003). This suggests that it will be necessary to examine whether the atswi3c mutations affect the splicing and polyadenylation of FCA pre-mRNA.
Through the bromodomains of BRAHMA, the ATSWI3C-containing SWI/SNF complexes may be targeted to acetylated histone cores in transcriptionally active chromatin. The fact that BRAHMA silencing results in precocious flowering suggests that the BRAHMA SWI/SNF complex could act as a repressor by recruiting histone deacetylases to the active chromatin. In fact, mutations that cause late flowering by inactivating the FLOWERING LOCUS D (FLD) gene in the autonomous flowering pathway were recently shown to result in the hyperacetylation of histone H4 in FLC chromatin. FLD encodes a protein showing a close relationship to a subunit of human HISTONE DEACETYLASE1/2 complexes (He et al., 2003). A second gene in the autonomous flowering pathway, FVE, encodes an Rb binding protein, MSI4, that is also required for the downregulation of FLC via histone H3 deacetylation (Ausin et al., 2004). Therefore, it will be important to examine how the atswi3c mutations affect the histone acetylation status of ATSWI3C-regulated genes.
Inactivation of ATSWI3C and ATSWI3D Causes Distinct Defects in the Development of Vegetative and Reproductive Organs
In addition to reducing leaf number at flowering, mutations in ATSWI3C cause characteristic defects in vegetative organ development, which include the formation of curly leaves and the inhibition of primary root elongation, leading to enhanced root branching. The atswi3c mutations also result in a high degree of male and female sterility as well as the appearance of fused twin stamens and petalloid bracts in whorl B. These developmental alterations are accompanied by a significant decrease in AP1, PI, and AP3 transcript levels in the inflorescence tips of atswi3c mutants grown under both SD and LD conditions. The phenotype of atswi3d mutants clearly differs from that of atswi3c mutants, although both mutations affect flower development and leaf number at flowering. The atswi3d mutations do not result in the typical curly-leaf phenotype and inhibition of root elongation, but they cause severe developmental defects in all floral whorls. The mutant flowers contain five sepals in whorl A and defective organs in whorl B, including shorter and degenerated petals and petalloid filaments. Because of severe deficiencies in anther and carpel development, the homozygous mutants are sterile. The atswi3d mutations result in a reduction of expression of AP1, AP3, PI, and UFO genes in inflorescence tips, but they also alter the expression levels of AP3, PI, and AG in sporophytic tissues before flowering (Figure 7). Thus, in addition to ATSWI3C, ATSWI3D also appears to be an important regulator of floral organ identity and development. The pleiotropic phenotype of atswi3d mutations shows no resemblance to known mutations of SNF2-like ATPases. Thus, better understanding of ATSWI3D function requires further genetic dissection of the interactions between ATSWI3D and the Arabidopsis homologs of SWI2/SNF2 ATPases.
ATSWI3A and ATSWI3B Are Essential for Embryonic Development
Our protein interaction studies suggested that ATSWI3A or ATSWI3B, together with BSH/SNF5, could represent a selective core of CRCs, which may recruit ATSWI3C or ATSWI3D as well as different SNF2-like ATPases to regulate specific chromatin-associated functions. This model suggests two different testable predictions. The first is that ATSWI3A and ATSWI3B are functionally equivalent and therefore redundant. Hence, in the absence of ATSWI3A, the equivalent ATSWI3B CRC subunit could recruit all other SWI/SNF components to perform the same functions. By contrast, the second prediction is that ATSWI3A and ATSWI3B perform specific and possibly essential functions. Thus, the functions of ATSWI3 subunit combinations could be specific to different regulatory pathways. In this study, we found that both atswi3a and atswi3b mutations result in lethality by arresting embryo development during the globular (blastocyte) stage. This observation indicated that the ATSWI3A and ATSWI3B genes perform essential and nonredundant functions required for early development. However, whether similar phenotypes of atswi3a and atswi3b mutations imply that a SWI3 complex (AB or AA or BB) is required only for embryogenesis is a question that should be answered by examining the effects of these mutations in somatic cells (i.e., using genetic mosaics). Inhibition of ATSWI3B expression by an antisense approach was reported to cause leaf curling, dwarfism, and delayed flowering, suggesting that ATSWI3B also performs important regulatory functions in sporophytic tissues (Zhou et al., 2003). However, it is unclear whether this antisense approach was specific for ATSWI3B or also influenced the expression of other ATSWI3 homologs. The latter possibility is suggested by phenotypic similarities between ATSWI3B-silenced plants and the atswi3c and atswi3d insertion mutants.
The phenotypes of Arabidopsis atswi3a and atswi3b mutations are comparable to those of knockouts in the mouse Srg3 and Caenorhabditis psa1 SWI3 homologs. These and other mutations, affecting the mouse SWI/SNF subunits Brg1 and SNF5/Ini1, all result in early embryonic death. Heterozygous Brg1+/− and SNF5/Ini1+/− animals show predisposition to tumor formation, indicating that SWI/SNF complexes found in direct association with Rb (the retinoblastoma tumor suppressor) play a key role in controlling cell cycle progression (Kim et al., 2001; Cui et al., 2004; Roberts and Orkin, 2004). Null mutations inactivating the Arabidopsis Rb-related RBR1 gene result in embryonic defects before or during the globular stage. The rbr1 mutation does not prevent normal megagametogenesis and allows the formation of a normal embryo sac. However, in emasculated rbr1/+ flowers, ∼25% of the ovules show fertilization-independent cell division, producing supernumerary nuclei at the micropylar pole. The rbr1 allele is not transmitted maternally and cannot be rescued by paternal RBR1, as a result of a general absence of expression of some paternal genes in the embryos until the late globular stage. The rbr mutant alleles also show greatly reduced paternal transmission and result in the formation of degenerated pollen (Ebel et al., 2004).
In contrast with the rbr1 mutation, the atswi3a and atswi3b mutations do not prevent female transmission and can be rescued in the female by the paternal wild-type allele. The effects of these mutations on embryo development differ slightly, because the atswi3b mutation results in cell divisions with aberrant polarity in the suspensor, which is not seen in the atswi3a mutants. A remarkable effect of the atswi3b mutation is that after pollination of atswi3b/+ plants with the wild type, ∼50% of the progeny are represented by arrested ovules, and the viable offspring show a 1:1 transmission of the atswi3b allele. We found that the arrested-ovule phenotype is caused by the absence of embryo sac formation and the lack of mitotic divisions of the megaspore. This arrested-ovule phenotype is observed in two different insertion mutants, segregates independently of the atswi3b alleles, and shows a non-Mendelian inheritance, suggesting atswi3b-dependent imprinting of a yet unknown locus. An experimental difficulty was revealed by the reciprocal cross, in which we pollinated wild-type females with atswi3b/+ males. Namely, these crosses consistently produced unfertilized ovules, the phenotype of which could not be easily distinguished by visual screening from that of arrested ovules resulting from imprinting. Therefore, we examined the effect of the atswi3b mutation on the process of microsporogenesis and found that ∼50% of microspores underwent degeneration after the formation and separation of tetrads. Yet, the wild-type progeny from the reciprocal crosses segregated the atswi3a and wild-type alleles at a 1:1 ratio. Thus, degeneration of ∼50% of microspores in males and 50% of macrospores in females could only be attributed to a novel imprinting effect caused by the atswi3b mutation. As no similar imprinting effect was observed in the atswi3a mutants, this trait appeared to be specific for the atswi3b mutant alleles.
Novel Aspects in the Organization of Plant SWI/SNF-Type Complexes
The data discussed above demonstrate that the SWI3 proteins are of similar importance for critical developmental functions in both animals and plants. However, there may be considerable differences between the two kingdoms in the details of the mechanisms in which these proteins are involved. The plant SWI3-type proteins have clearly evolved into forms that are functionally much more diversified than their animal counterparts. All SWI/SNF complexes characterized to date in other eukaryotes carry bromodomain-type SWI2/SNF2 ATPases as central catalytic subunits. We have shown that members of the Arabidopsis SWI3 A/B subfamily are required for embryo viability, similar to their mammalian homologs. Yet, our preliminary data indicate that a null mutation of the only Arabidopsis bromodomain-type SWI2/SNF2 ATPase, BRAHMA, does not cause embryo lethality (K. Brzeska and M. Prymakowska-Bosak, personal communication). This would indicate that BRAHMA may not be part of SWI/SNF complexes that carry only the essential ATSWI3A or ATSWI3B subunits. Rather, BRAHMA may be part of an ATSWI3C-based SWI/SNF complex, as suggested by phenotypic similarities between atswi3c mutants and BRAHMA-silenced plants. Because the bromodomain is thought to increase the processivity of SWI/SNF-mediated nucleosome remodeling through its affinity for acetylated histones, the lack of a bromodomain could greatly affect the interactions between ATP-dependent remodeling complexes and epigenetic chromatin marks. Why would a canonical SWI/SNF complex carrying the only known Arabidopsis bromodomain ATPase be excluded from the control of essential functions in plants? Is the acetylated histone binding function of plant SWI2/SNF2 ATPases taken over by other unknown domains or associated with noncore subunits? The answers to these questions require further identification and characterization of the components of different SWI3-containing CRCs in plants.
METHODS
Plant Material and Growth Conditions
The atswi3a-1 (SALK_035320), atswi3a-2 (SALK_065548), atswi3a-3 (SALK_068234), and atswi3d-1 (SALK_100310) mutant alleles were identified in Arabidopsis thaliana seed populations received from the SALK collection (Alonso et al., 2003). Using a PCR-based screening strategy, the atswi3b-1 (Koncz_2208), atswi3c-1 (Koncz_27320), atswi3c-2 (Koncz_3737), and atswi3d-2 (Koncz_14259) mutant alleles were identified in our T-DNA mutant collection (Ríos et al., 2002). The atswi3b-2 (GABI_302G08) mutant allele was obtained from the GABI-Kat T-DNA mutant population (Rosso et al., 2003). For germination, seeds were surface-sterilized with 5% calcium hypochlorite containing 0.01% (v/v) Tween 20, rinsed three times with sterile water, and plated on Murashige and Skoog (MS) seed germination medium containing 0.5% sucrose as described (Koncz et al., 1994). Segregation of antibiotic resistance markers encoded by the T-DNA tags was assayed by growing seedlings in MS medium containing 15 mg/L hygromycin (our lines), 50 mg/L kanamycin (SALK lines), or 12 mg/L sulfadiazine (4-amino-N-[2-pyrimidinyl]benzene-sulfonamide-Na; GABI-Kat lines).
Segregation analysis of the SALK lines was performed with a limited number (30 to 40) of seedlings by PCR as described by Ríos et al. (2002). From each identified heterozygous atswi3a line, the genotypes of 30 offspring obtained after self-pollination were determined by PCR. In addition, 20 to 30 siliques were opened on each atswi3a/+ plant to determine the segregation ratio of wild-type seeds (wt) and aborted seeds (as). The observed segregation ratios were as follows: atswi3a-1/+, 2.95wt:1as (χ2 = 0.0896, P = 0.8); atswi3a-2, 3.02wt:1as (χ2 = 3.85 × 10−3, P = 0.95); and atswi3a-3, 3.008wt:1as (χ2 = 1.85 × 10−4, P = 0.97). The segregation ratios of antibiotic-resistant (R) versus antibiotic-sensitive (S) progeny of atswi3b mutants were as follows: atswi3b-1, 2.1R:1S (χ2 = 0.55, P = 0.5); and atswi3b-2, 1.77R:1S (χ2 = 1.689, P = 0.22). The original GABI_302G08 line carrying the atswi3b-2 allele contained two independently segregating T-DNA insertions, which were separated by repeated outcrosses with the wild type. The segregation of wild-type, aborted-seed, and arrested-ovule traits of self-pollinated atswi3b/+ mutants is shown in Table 1. The segregation ratios of hygromycin-resistant to hygromycin-sensitive progeny of the atswi3c and atswi3d alleles were as follows: atswi3c-1, 2.645R:1S (χ2 = 2.01, P = 0.16); atswi3c-2, 3.307R:1S (χ2 = 0.0952, P = 0.76); and atswi3d-2, 2.75R:1S (χ2 = 4.2 × 10−2, P = 0.84). PCR screening for the atswi3d-1 allele indicated a segregation of 0.9:2.07:0.98 (wt:atswi3c-2/+:atswi3d-2/atswi3d-2; χ2 = 0.163, P = 0.9). The χ2 and P values indicate deviations from the expected 3:1, 2:1, or 1:2:1 segregation ratios and suggest a potential reduction of male or female transmission.
Analysis of T-DNA Insert Junctions in the Insertion Mutant Lines
We screened for T-DNA insertions in all ATSWI3 genes using PCR with pairs of gene-specific and T-DNA end–specific primers. The gene-specific primers were used in subsequent PCRs to classify the heterozygous and homozygous mutant lines. After isolation of the PCR-amplified T-DNA–plant DNA junction fragments from the screening gels, their sequences were determined. In the sequences provided for the T-DNA junctions of each mutant allele below, uppercase letters mark plant sequences and lowercase letters mark T-DNA sequences. The atswi3a-1 allele was identified using combinations of gene-specific FSSB1 (5′-CCGTGTGGTTTGGATTTGGCGATTG-3′) and FSSB2 (5′-CGCGTGTGAATTAGTAGAGAGACCCA-3′) primers with the T-DNA end primers LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′) and TDNAUSALK (5′-GATGAGACCTGCTGCGTAAG-3′). In atswi3a-1, the left border junction of the T-DNA (LB in Figure 3) was 5′-GGCAGGGTATAttgtggtgtaaacaaa-3′. The right border junction carried a deletion of 125 bp in the T-DNA, and the junction with the truncated T-DNA end was 5′-tttaaactatcagtgtttaaacACTATGGGGAGAACAA-3′. The T-DNA insertions in atswi3a-2 and atswi3a-3 mutants were confirmed by PCR using the gene-specific primers FSSB1 and FSSB2 and the T-DNA–specific primer LBb1. In the atswi3a-2 mutant allele, the left borders (LB1 and LB2) of an inverted T-DNA repeat were linked to plant DNA (Figure 3). The sequence of the LB1 junction was 5′-CTCCTGCTGGAAcaaattgacgctt-3′, whereas that of the LB2 junction was 5′-gcgtcaacctcgtTTGAGGAAAGAG-3′. The atswi3a-3 mutant allele similarly carried an insert of inverted T-DNA repeat, the ends of which were designated LB3 and LB4. The sequence corresponding to LB3 was 5′-GCGCTAGAATCTGTGacacaaattgacgctt-3′ and that of LB4 was 5′-ccatgtgtaatttgttTACCAAGAGTAG-3′.
T-DNA tags in the ATSWI3B gene were identified as described above using the gene-specific primers FSSA1 (5′-CTTCTCCGGCGAAGTTGCGTTAGTT-3′) and FSSA2 (5′-CTCCAATTGTTTCCGGCTTCTCTCCAT-3′) together with the T-DNA–specific primer FISH1 (5′-CTGGGAATGGCGAAATCAAGGCATC-3′) for our mutant collection. The T-DNA insertion in the atswi3b-2 allele was identified using the gene-specific primers FSSA1 and FSSA2 together with the T-DNA–specific primer for the GABI-Kat collection, GABI-LEFT (5′-TCTCCATATTGACCATCATACTCATTGC-3′). The junctions of an inverted T-DNA repeat in the atswi3b-1 mutant allele (Figure 3) were LB1 (5′-AGTCAAAGCCAGgatatattcaat-3′) and LB2 (5′-ctcattgctcatTGTTATGCACTA-3′). The atswi3b-2 mutant allele carried a similar inverted T-DNA repeat, the junctions of which were LB3 (5′-GAGAGTCATGGCtaccgaaaaattgataaaatga-3′) and LB4 (5′-aatatatcctgaCACTCCCTCTCT-3′). The positions of the ATG codon of AtSWI3B and an in-frame stop codon in the T-DNA sequence are underlined.
To identify T-DNA tags in the ATSWI3C gene, we used the gene-specific primers FSSC1 (5′-AATGCGCGACGGTCGACTAATGTATC-3′) and FSSC2 (5′-AGCCTGAACCTGTGGAAGACCTAAC-3′) in combination with the T-DNA left border primer FISH1 and the RB primer FISH2 (5′-CAGTCATAGCCGAATAGCCTCTCCA-3′). The junction sequences of the atswi3c-1 allele (Figure 3) were as follows: right border junction (5′-AGATTCTCCTCTtattttatttttatt-3′) and left border junction (5′-tccttatgtaggataataaTGGTGCCACA-3′). The junction sequences of the inverted T-DNA tag in the atswi3c-2 mutant allele were LB1 (5′-ATAAATTCTCTGTAGTatatattcaattg-3′) and LB2 (5′-aattgaatatatcCTGAAACTTCTT-3′).
To identify the T-DNA insertion in the ATSWI3D gene, PCR screening was performed with combinations of the gene-specific primers WTDU (5′-GGCGTTGGTAGTGGGAAGTGGA-3′) and WTDL (5′-TCTGGTTCTGGAACTTCTTTCA-3′) with the T-DNA end primer LBb1 for the SALK collection. To screen our insertion mutant collection, we used the gene-specific primers AtSWI3D1 (5′-AGAAGCGAAATTGGGGGATTGGGAATAG-3′) and AtSWI3D2 (5′-AGGTTGAAGCCGCAAAAGGTGGGATAAT-3′) in combination with the T-DNA end primer HOOK1 (5′-CTACACTGAATTGGTAGCTCAAACTGTC-3′). The insertions in both atswi3d-1 and atswi3d-2 alleles represented inverted T-DNA repeats facing the plant DNA with their left borders. The junction sequences of the T-DNA insert in atswi3d-1 were LB1 (5′-CACCTGAAGAGGacaacttaataaca-3′) and LB2 (5′-aagcgtcaatttgttattAAGATGAAACTATG-3′), whereas in atswi3d-2 they were LB3 (5′-CAGAAGAGGTCaatatggatcagc-3′) and LB4 (5′-gacatgaagccatTTCAGATGATAG-3′).
Assays of Flowering Time during SD and LD Conditions
Segregating families of atswi3c-1/+, atswi3c-2/+, atswi3d-1/+, and atswi3d-2/+ mutant lines were planted into soil and grown either under SD conditions using an 8-h light/16-h dark cycle or under LD conditions with a 16-h light/8-h dark cycle. During the day, temperature was a constant 23°C with 60% humidity, whereas the night temperature was 20°C with 70% humidity. The intensity of irradiance was 200 μE·m−2·s−1. Samples for RNA isolation were collected under LD conditions at midday (8 h after dawn) from seedlings at day 15 after sowing and from inflorescence tips on the day of appearance of the first visible flower bud. Under SD conditions, samples were taken 6 h after dawn at days 15, 31, and 41 from seedlings and on the first day of flowering (i.e., day 66, which overlapped in the flowering time schedules of the atswi3c and atswi3d mutants and the wild type).
Analysis of mRNA Levels in the atswi3c and atswi3d Mutants
Total RNA was isolated from 100 mg of tissue of atswi3c, atswi3d, and wild-type plants using the RNeasy plant mini kit (Qiagen). One microgram of total RNA was reverse-transcribed using the Transcriptor First Strand cDNA synthesis kit (Roche) according to the manufacturer's instructions. Equal aliquots from each RT reaction were used as template to amplify the ATSWI3C and ATSWI3D cDNAs shown in Figures 7A and 9A. Amplification of full-length ATSWI3C cDNA was performed with the gene-specific primers SWI3C1 (5′-TCCCCCGGGGCCAGCTTCTGAAGAT-3′) and SWI3C2 (5′-ACGCGTCGACTAGTTTAAGCCTAAGCCGGA-3′) using denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 2 min, and elongation at 72°C for 10 min. Amplification of full-length ATSWI3D cDNA was performed with the primers SWI3D1 (5′-ATGGAGGAAAAACGACGCGATT-3′) and SWI3D2 (5′-CTAAACCGAAGAACATTGTCTG-3′) using denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 4 min, and extension at 72°C for 10 min.
For RT-PCR analysis of flowering time integrator and floral homeotic genes, cDNA templates were prepared from two independently isolated sets of mRNAs. PCR was performed after denaturation at 95°C for 5 min using 22, 30, or 35 cycles of amplification (95°C for 30 s, 56°C for 30 s, and 68°C for 1 min) followed by an elongation step at 68°C for 5 min. The following pairs of gene-specific primers were used: AP1-1 (5′-acaagtgacattctcgaaaagaaga-3′) and AP1-2 (5′-tggactcgtacataagttggtttttc-3′); AP2-1 (5′-cgattatgatgatgacttgaaacag-3′) and AP2-2 (5′-cttgacttttatgcttcgaattagc-3′); AP3-1 (5′-atgagtatatcagccctaacaccac-3′) and AP3-2 (5′-gagtcgtaatctcctccattgtcta-3′); PI-1 (5′-gatgattgattactgttgtccttcc-3′) and PI-2 (5′-atcatgatctctcatcatcattcct-3′); AG-1 (5′-agaaagcttacgagctctctgttct-3′) and AG-2 (5′-cgttatgcaaatcaacttctctttt-3′); FLC-1 (5′-atcaagcgaattgagaacaaaagta-3′) and FLC-2 (5′-attctcaacaagcttcaacatgagt-3′); FT-1 (5′-atagtaagcagagttgttggagacg-3′) and FT-2 (5′-tgtcgaaacaatataaacacgacac-3′); CO-1 (5′-tgaatacagtcaacaccaacaaaac-3′) and CO-2 (5′-tgtcttctcaaatttccttgtcttc-3′); SOC1-1 (5′-agtgactttctccaaaagaaggaat-3′) and SOC1-2 (5′-ctgctcaatttgttccttaaacact-3′); UFO-1 (5′-tctctcttttgcttatatcccttca-3′) and UFO-2 (5′-cgctaaaagggctatagttcataca-3′); LFY-1 (5′-gtccgtacggtatacgtttctacac-3′) and LFY-2 (5′-gttgcttcttcatctttccttgac-3′); and Actin2 control ACT1 (5′-agagattcagatgcccagaagtcttgtcc-3′) and ACT2 (5′-aacgattcctggacctgcctcatcatactc-3′).
Cytological Techniques
Immature seeds were removed from green siliques at various times after pollination with a dissecting microscope and cleared in a small amount of Hoyer's solution (30 mL of water, 100 g of chloral hydrate, 7.5 g of gum arabic, and 5 mL of glycerin) on a glass slide. Mutant seeds usually cleared in 4 to 12 h, depending on their developmental stage, and were examined with a compound microscope equipped with Nomarski optics. To prepare cross sections of leaves and flowers, regular segments of 2 to 3 mm were cut from the same position of leaves and flower buds. The sections were fixed for 2 h in 2% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2, dehydrated using a graded series of ethanol and propylene oxide, and embedded in Spurr resin (Spurr, 1969). Alternatively, the sections were fixed in 4% glutaraldehyde in 50 mM PIPES buffer, pH 7.1, washed with the buffer three times for 15 min, and postfixed in 1% osmium tetroxide for 24 h in the same buffer. After washing and dehydration in acetone, the samples were infiltrated and embedded in Spurr medium. Semithin sections of 1 to 2 μm were cut using a Leica RM2065 microtome or an LKB ultramicrotome, stained with 0.1% toluidine blue in 1% borax or by basic fuchsine in combination with toluidine blue or methylene blue, as described by Farrás et al. (2001), and observed with a Leica Aristoplan light microscope using a Hitachi HV-20 camera for image recording. For scanning electron microscopy, the samples were processed with an electron microscope cryopreparation system (CT1500; Oxford Instruments) and examined with a Zeiss DSM 940 microscope.
Protein Interaction Assays in the Yeast Two-Hybrid System
Plasmids pGADSWI3A, pGADSWI3C, pGBSH, pGADFCA, and pFCATRUNCATED used in yeast two-hybrid assays were described previously (Sarnowski et al., 2002). Plasmids pGBTSWI3A and pGBTSWI3C were obtained by cloning full-length ATSWI3A and ATSWI3C cDNAs from pGADSWI3A and pGADSWI3C, respectively, in pGBT9 (DNA-BD) using SmaI and BamHI. A full-length ATSWI3D cDNA was amplified using PCR with primers SWI3D1 and SWI3D2 (see above) and cloned into the TOPO 2.1 cloning vector (Invitrogen). Upon sequencing, the ATSWI3D cDNA was moved by EcoRI into the vectors pGBT9 and pGAD424, yielding pGBTSWI3D and pGADSWI3D, respectively. Plasmid pCL1 was used as a positive control (Sarnowski et al., 2002). Yeast strain Y190 was transformed with the following plasmid pairs: pGBSH-pGADSWI3A, pGBSH-pGADSWI3C, pGBTSWI3A-pGADSWI3A, pGBTSWI3A-pGADSWI3C, pGBTSWI3A-pGADFCA and pGBTSWI3A-pFCATRUNCATED (FCA-PhD-AD), pGBTSWI3A-pGADSWI3D, pGBTSWI3B-pGADSWI3D, pGBSH-pGADSWI3D, as well as with the control plasmids pGBSH, pGBTSWI3A, pGADSWI3A, pGBTSWI3C, pGADSWI3C, pFCATRUNCATED, pGADFCA, pGBTSWI3D, pGADSWI3D, and pCL1 in combination with either pGBT9 or pGAD424. All transformants were grown in selection medium containing 100 mM 3-amino-1,2,4-triazole, and the level of β-galactosidase activity was monitored by the replica filter lift method described in the Clontech yeast protocol handbook.
Sequence Homology Searches and Phylogenetic Tree Analysis
Arabidopsis and rice (Oryza sativa) SWI3 homologs were identified using the National Center for Biotechnology Information tBLASTN search facility and the Oryza database (http://riceblast.dna.affrc.go.jp) and were compared with entries in the Plant Chromatin Database (http://chromdb.org). Multiple alignments were generated using the ClustalW program package at the Pasteur Institute (http://bioweb.pasteur.fr/), including all correction and statistical control facilities for improving the sensitivity, weighting, and position-specific gap penalties. The multiple alignments were used for phylogenetic analyses. The tree consensus was generated either by the quartet puzzling maximum likelihood method (Strimmer and von Haeseler, 1996) or by generating a consensus maximum parsimony tree using the PHYLIP package (Felsenstein, 1989). The bootstrap analysis was based on 1000 replications.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY081570 (ATSWI3A), NM_128921 (ATSWI3B), AY091026 (ATSWI3C), NM_202953 (ATSWI3D), NM_112639 (BSH), and Z82989 (FCA).
Supplementary Material
Acknowledgments
We thank Sabine Schäfer, Andrea Lossow, Christine Gerdes, and Ingrid Reintsch for their help in the insertion mutant screens. The Polish laboratory was supported by the Center of Excellence for Multiscale Biomolecular Modeling, Bioinformatics, and Applications. T.J.S. was supported by a Federation of European Biochemical Societies Short-Term Fellowship (2004). S.S. was supported by the British Council and Polish Ministerstwo Nauki i Informatyzacji Young Scientists Programme. This work was supported by Howard Hughes Medical Institute Grant 55000312 and Polish Committee for Scientific Research Grant PBZ-039/PO4/2001 to A.J., by European Union Grants QLK-5-200101871 and QLK5-CT-2002-00841 and Deutsche Forschungsgemeinshaft Grant AFGN Ko 1438/9-1 to C.K., and by a Marie Curie Fellowship (EU HPMF-CT-2000-00597) to G.R.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Tomasz Sarnowski (tsarn@poczta.ibb.waw.pl) and Gabino Ríos (gabino.rios@uv.es).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031203.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Amedeo, P., Habu, Y., Afsar, K., Mittelstein Scheid, O., and Paszkowski, J. (2000). Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405, 203–206. [DOI] [PubMed] [Google Scholar]
- Ausin, I., Alonso-Blanco, C., Jarillo, J.A., Ruiz-Garcia, L., and Martinez-Zapater, J. (2004). Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 36, 162–166. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Durbin, R., Eddy, S.R., Finn, R.D., and Sonnhammer, E.L.L. (1999). Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 27, 260–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, P.B. (2002). Nucleosome sliding: facts and fiction. EMBO J. 21, 4749–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss, P.K., Bastow, R.M., Mylne, J., and Dean, C. (2004). Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell 16, S18–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer, L.A., Langer, M.R., Crowley, K.A., Tan, S., Denu, J.M., and Peterson, C.L. (2002). Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 10, 935–942. [DOI] [PubMed] [Google Scholar]
- Brzeski, J., Dyczkowski, J., Kaczanowski, S., Zielenkiewicz, P., and Jerzmanowski, A. (2003). Plant chromatin: Learning from similarities and differences. Adv. Bot. Res. 40, 107–142. [Google Scholar]
- Brzeski, J., and Jerzmanowski, A. (2003). Deficient in DNA Methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J. Biol. Chem. 278, 823–828. [DOI] [PubMed] [Google Scholar]
- Brzeski, J., Podstolski, W., Olczak, K., and Jerzmanowski, A. (1999). Identification and analysis of the Arabidopsis thaliana BSH gene, a member of the SNF5 gene family. Nucleic Acids Res. 27, 2393–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman, S., Gebuhr, T., Yee, D., La Mantia, C., Nicholson, J., Gilliam, A., Randazzo, F., Metzger, D., Chambon, P., Crabtree, G., and Magnuson, T. (2000). A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6, 1287–1295. [DOI] [PubMed] [Google Scholar]
- Cairns, B.R., Lorch, Y., Li, Y., Zhang, M., Lacomis, L., Erdjument-Bromage, H., Tempst, P., Du, J., Laurent, B., and Kornberg, R.D. (1996). RSC, an essential, abundant chromatin-remodeling complex. Cell 87, 1249–1260. [DOI] [PubMed] [Google Scholar]
- Crosby, M.A., Miller, C., Alon, T., Watson, K.L., Verrijzer, C.P., Goldman-Levi, R., and Zak, N.B. (1999). The trithorax group gene moira encodes a brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol. Cell. Biol. 19, 1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, M., Fay, D.S., and Han, M. (2004). lin35/Rb cooperates with the SWI/SNF complex to control Caenorhabditis elegans larval development. Genetics 167, 1177–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel, C., Mariconti, L., and Gruissem, W. (2004). Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429, 776–780. [DOI] [PubMed] [Google Scholar]
- Farrás, R., Ferrando, A., Jásik, J., Ökrész, L., Tiburcio, A., Salchert, K., del Pozo, C., Schell, J., and Koncz, C. (2001). SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 20, 2742–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona, S., Hurtado, L., Bowman, J.L., and Reyes, J.C. (2004). The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131, 4965–4975. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1989). Phylip: Phylogeny Inference Package (Version 3.2). Cladistics 5, 164–166. [Google Scholar]
- Ferrario, S., Immink, R.G., and Angenent, G.C. (2004). Conservation and diversity in flower land. Curr. Opin. Plant Biol. 7, 84–91. [DOI] [PubMed] [Google Scholar]
- Fritsch, O., Benvenuto, G., Bowler, C., Molinier, J., and Hohn, B. (2004). The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol. Cell 16, 479–485. [DOI] [PubMed] [Google Scholar]
- Gendrel, A.-V., Lippman, Z., Yordan, C., Colot, V., and Martienssen, R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297, 1871–1873. [DOI] [PubMed] [Google Scholar]
- Geng, F., Cao, Y., and Laurent, B.C. (2001). Essential roles of Snf5p in Snf-Swi chromatin remodeling in vivo. Mol. Cell. Biol. 21, 4311–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, J., Puangsomlee, P., Martin, M., Long, M., Meyerowitz, A.M., and Coupland, G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51. [DOI] [PubMed] [Google Scholar]
- Guidi, C.J., Sands, A.T., Zambrowicz, B.P., Turner, T.K., Demers, D.A., Webster, W., Smith, T.W., Imbalzano, A.N., and Jones, S.N. (2001). Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell. Biol. 21, 3598–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., and Amasino, R.M. (2004). Role of chromatin modification in flowering-time control. Trends Plant Sci. 10, 30–35. [DOI] [PubMed] [Google Scholar]
- He, Y., Michaels, S.D., and Amasino, R.M. (2003). Regulation of flowering time by histone acetylation in Arabidopsis. Science 302, 1751–1754. [DOI] [PubMed] [Google Scholar]
- Henderson, I.R., and Dean, C. (2004). Control of Arabidopsis flowering: The chill before the bloom. Development 131, 3829–3838. [DOI] [PubMed] [Google Scholar]
- Hudson, B.P., Martinez-Yamout, M.A., Dyson, H.J., and Wright, P.E. (2000). Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J. Mol. Biol. 304, 355–370. [DOI] [PubMed] [Google Scholar]
- Jeddeloh, J.A., Stokes, T.L., and Richards, E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22, 94–97. [DOI] [PubMed] [Google Scholar]
- Kanno, T., Mette, F.M., Kreil, D.P., Aufsatz, W., Matzke, M., and Matzke, A.J.M. (2004). Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 14, 801–805. [DOI] [PubMed] [Google Scholar]
- Katz, A., Oliva, M., Mosquna, A., and Ohad, N. (2004). FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 37, 707–719. [DOI] [PubMed] [Google Scholar]
- Kim, G.-T., Tsukaya, H., and Uchimiya, H. (1998). The CURLY LEAF gene controls both division and elongation of cells during the expansion of the leaf blade in Arabidopsis thaliana. Planta 206, 175–183. [DOI] [PubMed] [Google Scholar]
- Kim, J.K., Huh, S.O., Choi, H., Lee, K.S., Shin, D., Lee, C., Nam, J.S., Kim, H., Chung, H., Lee, H.W., Park, S.D., and Seong, R.H. (2001). Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol. Cell. Biol. 21, 7787–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klochendler-Yeivin, A., Fiette, L., Barra, J., Muchardt, C., Babinet, C., and Yaniv, M. (2000). The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 1, 500–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klochendler-Yeivin, A., Muchardt, C., and Yaniv, M. (2002). SWI/SNF chromatin remodeling and cancer. Curr. Opin. Genet. Dev. 12, 73–79. [DOI] [PubMed] [Google Scholar]
- Koncz, C., Martini, N., Szabados, L., Hrouda, M., Bachmair, A., and Schell, J. (1994). Specialized vectors for gene tagging and expression studies. In Plant Molecular Biology Manual, Vol. B2, S. Gelvin and B. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic), pp. 1–22.
- Leonard, D., Ajuh, P., Lamond, A.I., and Legerski, R.J. (2003). hLodestar/huF2 interacts with CDC5L and is involved in pre-mRNA splicing. Biochem. Biophys. Res. Commun. 308, 793–801. [DOI] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Dennis, E.S., Peacock, J.W., and Chaudhury, A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97, 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macknight, R., Duroux, M., Laurie, R., Dijkwel, P., Simpson, G., and Dean, C. (2002). Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14, 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens, J.A., and Winston, F. (2003). Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13, 136–142. [DOI] [PubMed] [Google Scholar]
- Mohrmann, L., Langenberg, K., Krijgsveld, J., Kal, A.J., Heck, A.J.R., and Verrijzer, C.P. (2004). Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol. Cell. Biol. 24, 3077–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, H.H., Robert, F., Young, R.A., and Struhl, K. (2002). Genome-wide location and regulated recruitment of the RSC nucleosome remodelling complex. Genes Dev. 16, 806–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh, Y.S., and Amasino, R.M. (2003). PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15, 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas, J., Kaufmann, S., Henderson, J., and Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan, M.L., Sif, S., Narlikar, G.J., and Kingston, R.E. (1999). Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3, 247–253. [DOI] [PubMed] [Google Scholar]
- Reyes, J.C., Hennig, L., and Gruissem, W. (2002). Chromatin-remodeling and memory factors: Novel regulators of plant development. Plant Physiol. 130, 1090–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, P., Longden, I., and Bleasby, A. (2000). EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277. [DOI] [PubMed] [Google Scholar]
- Ríos, G., et al. (2002). Rapid identification of Arabidopsis insertion mutants by nonradioactive detection of T-DNA tagged genes. Plant J. 32, 243–253. [DOI] [PubMed] [Google Scholar]
- Roberts, C.W.M., and Orkin, S.H. (2004). The SWI/SNF complex: Chromatin and cancer. Nat. Rev. Cancer 4, 133–142. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53, 247–259. [DOI] [PubMed] [Google Scholar]
- Sarnowski, T.J., Swiezewski, S., Pawlikowska, K., Kaczanowski, S., and Jerzmanowski, A. (2002). AtSWI3B, an Arabidopsis homolog of SWI3, a core subunit of yeast Swi/Snf chromatin remodeling complex, interacts with FCA, a regulator of flowering time. Nucleic Acids Res. 30, 3412–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, G.G., Quesada, V., Henderson, I.R., Dijkwel, P.P., Macknight, R., and Dean, C. (2004). RNA processing and Arabidopsis flowering time control. Biochem. Soc. Trans. 32, 565–566. [DOI] [PubMed] [Google Scholar]
- Smith, T.F., and Waterman, M.S. (1981). Identification of common molecular subsequences. J. Mol. Biol. 147, 195–197. [DOI] [PubMed] [Google Scholar]
- Spurr, A.R. (1969). A low viscosity embedding medium for electron microscopy. J. Ultrastruct. Res. 26, 31–43. [DOI] [PubMed] [Google Scholar]
- Strimmer, K., and von Haeseler, A. (1996). Quartet puzzling: A quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13, 964–969. [Google Scholar]
- Sudarsanam, P., and Winston, F. (2000). The Swi/Snf family of nucleosome-remodeling complexes and transcriptional control. Trends Genet. 16, 345–351. [DOI] [PubMed] [Google Scholar]
- Verbsky, M.L., and Richards, E.J. (2001). Chromatin remodeling in plants. Curr. Opin. Plant Biol. 4, 494–500. [DOI] [PubMed] [Google Scholar]
- Wagner, D. (2003). Chromatin regulation of plant development. Curr. Opin. Plant Biol. 6, 20–28. [DOI] [PubMed] [Google Scholar]
- Wagner, D., and Meyerowitz, E.M. (2002). SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr. Biol. 12, 85–94. [DOI] [PubMed] [Google Scholar]
- Zhou, C., Miki, B., and Wu, K. (2003). CHB2, a member of the SWI3 gene family, is a global regulator in Arabidopsis. Plant Mol. Biol. 52, 1125–1134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.