Abstract
In this study, we characterize the evolutionarily conserved TOUGH (TGH) protein as a novel regulator required for Arabidopsis thaliana development. We initially identified TGH as a yeast two-hybrid system interactor of the transcription initiation factor TATA-box binding protein 2. TGH has apparent orthologs in all eukaryotic model organisms with the exception of the budding yeast Saccharomyces cerevisiae. TGH contains domains with strong similarity to G-patch and SWAP domains, protein domains that are characteristic of RNA binding and processing proteins. Furthermore, TGH colocalizes with the splicing regulator SRp34 to subnuclear particles. We therefore propose that TGH plays a role in RNA binding or processing. Arabidopsis tgh mutants display developmental defects, including reduced plant height, polycotyly, and reduced vascularization. We found TGH expression to be increased in the amp1-1 mutant, which is similar to tgh mutants with respect to polycotyly and defects in vascular development. Interestingly, we observed a strong genetic interaction between TGH and AMP1 in that tgh-1 amp1-1 double mutants are extremely dwarfed and severely affected in plant development in general and vascular development in particular when compared with the single mutants.
INTRODUCTION
Throughout their life cycle, plants produce new cells that subsequently differentiate to give rise to new cell types and organs. Cell differentiation and organ formation are closely linked to the activities of the phytohormones auxin and cytokinin. Applications of specific concentrations of auxin and cytokinin can stimulate the formation of undifferentiated callus from differentiated tissue and conversely the formation of differentiated shoot and root tissue from undifferentiated callus (Murashige and Skoog, 1962). Within the plant, the proper distribution of auxin and cytokinin as well as the activities of hormone-specific signal transduction pathways appear to be the main determinants of the effects of these hormones during differentiation (Murashige and Skoog, 1962; Reinhardt, 2003).
Cotyledons and leaves of dicotyledonous plants have an interconnected vascular network that transports essential nutrients throughout the plant. The differentiation of the vascular system requires proper auxin transport and response (Mattsson et al., 1999, 2003). Auxin is produced in the margins of cotyledons and leaves and from there transported inwards, resulting in auxin accumulation within specific cells. Most but not all phenomena associated with vascular differentiation can be explained by the canalization hypothesis, which suggests that these auxin-accumulating cells subsequently differentiate and connect to give rise to the vascular networks of cotyledons and leaves (Sachs, 1991; Reinhardt, 2003). The role of auxin transport and signaling in vascular development is supported by physiological experiments as well as by several Arabidopsis thaliana mutants defective in proteins that presumably participate in the transport of auxin (e.g., GNOM and PIN-FORMED1) or in the transcriptional regulation of auxin-induced gene expression (e.g., BODENLOS, MONOPTEROS, and AUXIN-RESISTANT6) (Gälweiler et al., 1998; Hardtke and Berleth, 1998; Mattsson et al., 1999, 2003; Steinmann et al., 1999; Christensen et al., 2000; Hobbie et al., 2000; Carland et al., 2002; Hamann et al., 2002; Hellmann et al., 2003; Willemsen et al., 2003). In addition, mutants of the cell cycle–regulated HOBBIT gene, which encodes a protein with homology to the CDC27 subunit of the anaphase promoting complex, display reduced auxin responses, suggesting that cell cycle regulation, auxin response, and cell differentiation may be tightly interconnected processes (Willemsen et al., 1998; Blilou et al., 2002).
In addition to the proteins and mutants described above, several other mutants defective in vascular development have been described that can at present not be linked to auxin transport or auxin signaling. The gene ALTERED MERISTEM PATTERNING1 (AMP1) was isolated in multiple mutant screens (Jürgens et al., 1991; Chaudhury et al., 1993; Hou et al., 1993; Conway and Poethig, 1997). The amp1 mutant has elevated cytokinin levels, expresses higher levels of the cytokinin-induced cell cycle regulator cyclin D3 (CYCD3), and has an enlarged meristem (Chaudhury et al., 1993; Riou-Khamlichi et al., 1999). Increased CYCD3 expression, cell cycle activity, and enlarged meristem size may be the cause for the supernumerary cotyledons and the faster vegetative growth observed in the amp1 mutant. Interestingly, amp1 mutants fail to form a proper vascular system in leaves, and it may be suggested that increased cell cycle activity in the amp1 mutant prevents proper leaf vascularization (Conway and Poethig, 1997).
To specify the set of available proteins in specific cells or cell types, eukaryotes control gene expression at the transcriptional level or posttranscriptionally by pre-mRNA processing and alternative splicing (Smith and Valcarcel, 2000; Orphanides and Reinberg, 2002; Proudfoot et al., 2002). Pre-mRNA processing involves the removal of introns by the spliceosome, capping of the mRNA 5′-end, polyadenylation of the mRNA 3′-end, and transport of the mature mRNAs to the cytoplasm (Orphanides and Reinberg, 2002). Alternative splicing, the alternative removal of exons or introns by differential selection of 5′ and 3′ splice sites, permits the generation of different polypeptides from one pre-mRNA (Blencowe et al., 1999; Graveley, 2000). Thereby, alternative splicing not only enhances the number of different proteins encoded from a limited number of genes but it also represents an important regulatory mechanism during gene expression. RNA binding proteins such as the Ser Arg (SR) proteins have not only been implicated in RNA processing but also in transcriptional activation and elongation, suggesting that transcription and pre-mRNA processing are tightly linked (Fong and Zhou, 2001; Orphanides and Reinberg, 2002; Proudfoot et al., 2002).
In plants, pre-mRNA processing, alternative splicing, and other RNA-directed protein activities are required for various processes, including floral transition, floral patterning, vegetative phase change, and signal transduction (Chen and Cheng, 2004). However, little is known about the role of RNA binding and processing proteins when compared with the large set of proteins with a proposed role in these processes (Lorkovic and Barta, 2002). Here, we describe the previously uncharacterized and evolutionarily conserved TOUGH (TGH) protein, which we initially isolated as a yeast two-hybrid interactor of TATA binding protein 2 (TBP2). TGH contains two conserved protein domains found in proteins with a role in RNA binding and RNA processing, and TGH localizes with the Arabidopsis splicing regulator SRp34 to specific subnuclear particles. Both observations are indicative for a role of TGH in RNA binding or processing. Arabidopsis tgh mutants are defective in vascular patterning, and TGH expression is increased in the amp1-1 mutant. Interestingly, TGH interacts genetically with AMP1, and our data suggest that both proteins control growth in general and vascular patterning in particular.
RESULTS
TGH Is an Evolutionarily Conserved Protein
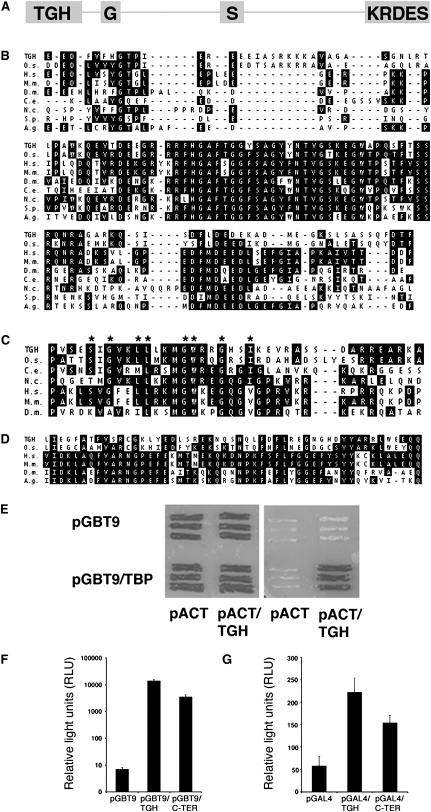
TGH (At5g23080) is a previously uncharacterized protein from Arabidopsis. The TGH open reading frame is composed of 16 exons and encodes a 931–amino acid protein with a calculated molecular mass of 104.9 kD (Figure 1A). BLASTP searches using full-length TGH protein reveal that TGH is not related to any other Arabidopsis protein. However, proteins clearly related to TGH are present in rice (Oryza sativa) and in many eukaryotic model organisms, including all model animal species and the fission yeast Schizosaccharomyces pombe, although not in the budding yeast Saccharomyces cerevisiae (Figures 1B and 1C). Since, with the exception of budding yeast, each organism subjected to analysis was found to contain exactly one protein with obvious similarity to TGH, we assume that these proteins arose from one common ancestor and that they are TGH orthologs. The TGH orthologs share highest sequence similarity within their N termini, and these similarities range from 69% sequence identity (83% similarity) between Arabidopsis and rice TGH to 52% identity (72% similarity) between Arabidopsis and Caenorhabditis elegans TGH, the most distantly related TGH identified in our analyses (Figure 1B). Since the N-terminal domain is specific to Arabidopsis TGH and its orthologs, we named it the TGH domain (Figures 1A and 1B).
Figure 1.
The TGH Protein Is Evolutionarily Conserved and May Interact with TBP2.
(A) Schematic representation of the TGH protein. Gray boxes indicate the conserved TGH (TGH), the G-patch (G), and the SWAP (S) domains (drawn to scale). The KRDES domain of Arabidopsis TGH is enriched in basic and acidic amino acids as well as in Ser residues.
(B) Protein sequence alignment of the highly conserved N termini of TGH (amino acids 5 to 123) and its orthologs from rice (O.s.; amino acids 6 to 124), human (H.s.; amino acids 9 to 116), mouse (M.m.; amino acids 9 to 116), Drosophila (D.m.; amino acids 3 to 111), C. elegans (C.e.; amino acids 5 to 111), Neurospora crassa (N.c.; amino acids 18 to 131), S. pombe (S.p.; amino acids 18 to 119), and Anopheles gambiae (A.g.; amino acids 3 to 112). Identical conserved amino acids are shaded.
(C) Protein sequence alignment of the G-patch domains of TGH orthologs from Arabidopsis (TGH; amino acids 157 to 197), rice (O.s.; amino acids 158 to 197), C. elegans (C.e.; amino acids 143 to 191), N. crassa (N.c.; amino acids 146 to 197), human (H.s.; amino acids 150 to 199), mouse (M.m.; amino acids 150 to 199), and Drosophila (D.m.; amino acids 151 to 200). Accession numbers are listed in Methods. Identical conserved amino acids are shaded. Conserved Gly residues of the G-patch are indicated by asterisks.
(D) Protein sequence alignment of the SWAP domains of TGH from Arabidopsis (TGH; amino acids 405 to 455) and rice (O.s., amino acids 408 to 458) and SWAP domain containing proteins from human (H.s.; amino acids 15 to 65), mouse (M.m.; amino acids 15 to 65), Drosophila (D.m.; amino acids 14 to 63), and A. gambiae (A.g.; amino acids 36 to 85). Accession numbers are listed in Methods. Identical amino acids are shaded. Conserved residues of the SWAP domain are indicated by asterisks. Sequence alignments in (B) to (D) were performed using the Jotun-Hein algorithm (gap penalty 11, gap length penalty 3, Ktuple 2).
(E) The yeast two-hybrid interaction between TBP and TGH visualized by differential growth on full media (cotransformation control, left panel) and on selective media (interaction experiment, right panel). The respective empty vector controls with pGBT9 and pACT fail to grow on selective media.
(F) and (G) Protein fusions of the GAL4 DNA binding domain to TGH or to the TGH KRDES domain activate transcription from a GAL4-reponsive LACZ reporter in yeast (F) or a luciferase reporter construct in transiently transformed tobacco mesophyll protoplasts (G).
In addition to the highly conserved and specific TGH domain, the TGH orthologs contain two recognizable conserved protein domains, a G-patch and a Suppressor-of-white-apricot (SWAP) domain (Figures 1A, 1C, and 1D) (Denhez and Lafyatis, 1994; Spikes et al., 1994; Aravind and Koonin, 1999). The G-patch is defined by a series of conserved Gly residues, and G-patches are exclusively found in proteins with a predicted or known role in RNA binding or RNA processing (Aravind and Koonin, 1999) (Figure 1C). The SWAP domain is a conserved domain with a presumed function in RNA binding that was first identified in the splicing regulator SWAP from Drosophila melanogaster (Denhez and Lafyatis, 1994; Spikes et al., 1994) (Figure 1D). The G-patch and the SWAP domains have unknown biochemical properties or functions. Nevertheless, these domains have only been found together in proteins with a role in RNA binding and RNA processing, inviting the hypothesis that SWAP and G-patch domain-containing proteins and therefore also TGH may play a role in these processes (Aravind and Koonin, 1999). With regard to the G-patch, we also noted that several conserved residues of the G-patch are not conserved in Arabidopsis and rice TGH but in their animal counterparts (Figure 1C). It can therefore not be ruled out that the G-patch has lost or changed functionality during plant evolution.
The G-patch domain is often accompanied by the repetitive RS (Arg, Ser) and RGG (Arg, Gly, Gly) sequences (Blencowe et al., 1999; Graveley, 2000). RS and RGG sequences are found in splicing regulators, such as SR proteins, and have been proposed to mediate protein–RNA or protein–protein interactions (Aravind and Koonin, 1999). While canonical RS and RGG sequences are absent from TGH, the domain comprising the C-terminal 170 amino acids of Arabidopsis TGH is significantly enriched in basic (53; 31%) and acidic (36; 21%) amino acids and in Ser residues (36; 21%). This domain was therefore designated KRDES (Lys, Arg, Asp, Glu, Ser) domain (Figure 1A; data not shown). The KRDES domain seems to be unique to Arabidopsis TGH.
TGH May Interact with TATA Binding Protein
Our interest in TGH was initially triggered by its identification as an interactor of the general transcription factor TBP2. We found that the TGH full-length protein and the TGH C-terminal 245 amino acids, including the KRDES domain, interact with Arabidopsis TBP2 (At1g55520) in the yeast two-hybrid system (Figure 1E). TBPs recognize the TATA-box in eukaryotic gene promoters, where they initiate the assembly of other general transcription factors to form the preinitiation complex, a prerequisite for RNA polymerase II binding (Orphanides and Reinberg, 2002; Proudfoot et al., 2002). TBPs interact with transcriptional coactivators, such as the TBP-associated factors (TAFs), which in turn serve as interaction platforms for transcription activators (Tansey and Herr, 1997; Wu and Chiang, 2001). To make an allusion to the TBP-interacting TAF proteins, the hitherto uncharacterized TBP-interacting protein At5g23080 was designated TGH.
Numerous studies show that the interactions between TBP and transcriptional regulators, such as the TAFs, are essential for transcriptional activation (Dynlacht et al., 1991; Goodrich and Tjian, 1994; Lieberman and Berk, 1994; Tjian and Maniatis, 1994; Verrijzer and Tjian, 1996). We therefore examined whether TGH functions as a transcriptional regulator. To this end, we fused the full-length TGH protein as well as its C terminus including the KRDES domain to the DNA binding domain of the GAL4 transcription factor. We found that the TGH full-length protein as well as the TGH C terminus can activate gene expression from a GAL4-responsive reporter in yeast and in transiently transformed tobacco (Nicotiana tabacum) mesophyll protoplasts (Figures 1F and 1G). It could therefore be envisioned that TGH regulates gene expression as a transcriptional activator or as a regulator of another process that occurs cotranscriptionally and that possibly requires TBP2 interaction.
Subcellular Localization of TGH
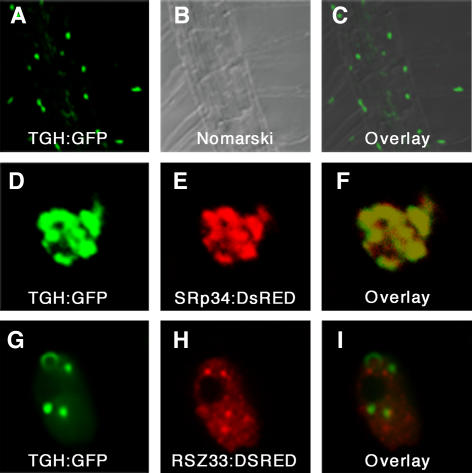
The observed yeast two-hybrid interaction with TBP2 and the presence of several predicted nuclear targeting sequences within the KRDES domain (RKKR, RRRKR, KKRRR, and RRESSREKRSSHKKHS) are suggestive of a nuclear localization (Dingwall and Laskey, 1991). To study the subcellular localization of TGH, we generated transgenic Arabidopsis lines expressing TGH:green fluorescent protein (GFP) fusions. Transgenic plants transformed with construct TGH:TGH:GFP express the genomic TGH gene fragment fused to GFP under control of a 1-kb TGH promoter fragment. Transgenic plants transformed with 35S:TGH:GFP express the TGH cDNA fused to GFP under control of the constitutive 35S promoter of Cauliflower mosaic virus. Both constructs produce a functional TGH:GFP fusion protein that is able to rescue the mutant phenotype (Figures 2M to 2P; see Supplemental Table 1 online). In transgenic lines transformed with either construct, we found TGH:GFP to accumulate exclusively in the nucleus of all cells amenable to fluorescence microscopy (Figures 3A to 3C). When we analyzed TGH:GFP localization in transiently transformed Arabidopsis protoplasts, we noticed that the fusion protein is present in subnuclear particles that varied in size and shape in different cells examined, regardless of the TGH:GFP construct used (Figures 3D and 3G). To study the identity of these subnuclear particles, we conducted colocalization experiments between the TGH:GFP constructs and SRp34:DsRED as well as between the TGH:GFP constructs and RSZ33:DsRED. SRp34 is one of two proteins present in Arabidopsis closely related to human SPLICING FACTOR2/ALTERNATIVE SPLICING FACTOR, which is required for pre-mRNA processing (Lopato et al., 1999). RSZ33 is a plant-specific protein with a predicted role in RNA binding and processing (Lopato et al., 2002; Lorkovic et al., 2004b). Both SRp34 and RSZ33 are known to localize to distinct particles within the nucleus. As observed with the TGH:GFP contructs, SRp34:DSRED and RSZ33:DsRED proteins also accumulate in subnuclear particles that had different sizes and shapes in different cells examined. Regardless of these differences in size and shape, our studies revealed colocalization of the TGH:GFP constructs with SRp34:DsRED in all of the cells examined (Figures 3D to 3F) but not of the TGH:GFP constructs with RSZ33:DsRED (Figures 3G to 3I). We therefore propose that the TGH protein functions close to the Arabidopsis splicing regulator SRp34.
Figure 2.
Mutations in the TGH Gene Cause Severe Growth Defects.
(A) Genomic organization of the TGH gene. Gray boxes indicate exons, and lines indicate introns (drawn to scale). The T-DNA insertion positions of tgh-1 and tgh-2 are indicated by arrowheads.
(B) Result of PCR genotyping of segregants from a tgh-1/TGH and a tgh-2/TGH population. The presence or absence of a band indicates the presence or absence of the gene indicated on the left side of the panel. Only plants homozygous for the T-DNA insertions show the mutant phenotype.
(C) to (E) Three-week-old tgh-1 (D) and tgh-2 (E) mutants are dwarfed compared with the wild type (C). Bars = 3 cm.
(F) tgh-1 mutant plants (right) have an increased number of lateral shoots and are sterile (inset). Bar = 5 cm.
(G) and (H) Partially dissected wild-type (G) and tgh-1 mutant (H) flower showing reduced stamen length and sterility in the tgh mutants.
(I) Reduced hypocotyl elongation and apical hook formation in 5-d-old dark-grown wild-type (left) and tgh-1 mutant (right) seedlings.
(K) and (L) tgh mutants frequently develop three cotyledons. (K), the wild type; (L), tgh-1 mutant.
(M) to (P) The TGH:GFP fusion constructs complement the tgh-1 mutant phenotype. Phenotype of an 18-d-old wild-type plant (M) and tgh-1 mutants containing no TGH:GFP transgene (N), containing the TGH:TGH:GFP transgene (O), and the 35S:TGH:GFP transgene (P).
Figure 3.
TGH:GFP and SRp34:DsRED Colocalize to Subnuclear Particles.
(A) to (C) 35S:TGH:GFP and TGH:TGH:GFP accumulate in the nuclei of root cells of transgenic Arabidopsis seedlings. Confocal fluorescence image (A), Nomarski image (B), and merged image (C) of a root expressing 35S:TGH:GFP.
(D) to (I) Confocal images of Arabidopsis protoplast nuclei expressing 35S:TGH:GFP ([D] and [G]) and SRp34:DsRED (E) or RSZ33:DsRED (H). 35S:TGH:GFP and SRp34:DsRED ([D] to [F]) but not 35S:TGH:GFP and RSZ33:DsRED ([G] to [I]) colocalize in subnuclear particles.
TGH Is Required for Proper Arabidopsis Development
To gain insight into the biological function of TGH, we examined two TGH T-DNA insertion mutants, namely SALK_053445 (tgh-1) and GABI_774H04 (tgh-2) (Figure 2A). The T-DNA insertions are located at the end of intron 11 and at the beginning of exon 12, respectively (Figure 2A). Plants homozygous for the TGH T-DNA insertions were isolated by PCR-based genotyping from a large segregating population (Figures 2B to 2E). Plants homozygous for both TGH gene insertions displayed identical phenotypes that clearly distinguished them from wild-type and hemizygous segregants (Figures 2B to 2E). In general terms, adult tgh mutant plants have a reduced stature and significantly smaller lanceolate leaves (Figures 2C to 2F). Elongation defects could also be observed in the anthers of tgh mutant flowers, which fail to elongate and produce pollen (Figures 2G and 2H). At the seedling stage, tgh mutant seedlings are recognizable by developmental defects, including triple cotyledons, altered cotyledon shape, and reduced elongation growth in dark-grown seedlings (Figures 2I to 2L; data not shown). The triple cotyledon phenotype is, however, not fully penetrant (e.g., only 7 of 362 progeny seedlings of a TGH/tgh-1 parent line displayed the triple cotyledon phenotype corresponding to a penetrance of 7.7%). Due to the almost complete infertility of tgh mutants, all subsequent analyses were performed with segregants of heterozygous TGH/tgh-1 and TGH/tgh-2 plants.
TGH Is Required for the Initiation of Vascular Development
Altered cotyledon number as observed in the tgh mutants is frequently associated with defects in cotyledon and leaf vascular initiation (Chaudhury et al., 1993; Hardtke and Berleth, 1998; Hamann et al., 1999; Hobbie et al., 2000; Mattsson et al., 2003). We therefore examined vascularization in cotyledons and leaves of tgh-1 and tgh-2 mutants and found that both tgh mutant alleles form an incomplete vascular system (Figure 4). While wild-type cotyledons typically have a simple interconnected vascular system, tgh mutant cotyledons have an imperfect vascular system with differentiated but frequently unconnected vascular strands (Figures 4A and 4D). The defects in vascularization are also detectable in leaves where second and third order vascular strands often remain unconnected in tgh mutants, while they are almost always interconnected in the wild type (Figures 4B and 4E). While the vascular network is altered in the tgh mutants, no obvious defects were observed with respect to vascular strand positions and vascular strand alignment.
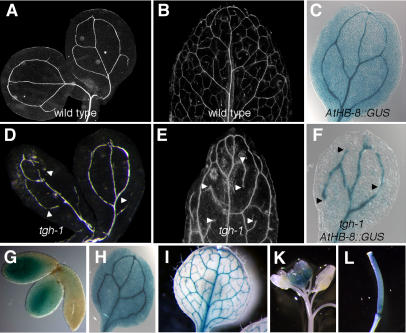
Figure 4.
Vascularization Defects in the tgh Mutants.
(A) and (B) Cleared cotyledons and leaf of wild-type plants.
(C) GUS staining of AtHB-8:GUS in cotyledons of 5-d-old Arabidopsis seedlings clearly delineates the vascular system.
(D) and (E) Cleared cotyledons and leaf of the tgh-1 mutant reveal defects in vascularization. Arrowheads mark unconnected vascular strands.
(F) GUS staining of AtHB-8:GUS in the cotyledons of tgh-1 seedlings delineates the interrupted vascular system observed in these mutants. Arrowheads mark unconnected vascular strands.
(G) to (L) GUS expression in embryos (G), cotyledon (H), leaf (I), young inflorescence (K), and the pistil (L) of transgenic lines expressing the reporter GUS under control of a 1021-bp TGH promoter fragment.
The morphological examination of cleared tissue only allows the visualization of differentiated vascular tissue but not the visualization of provascular cells. The expression of the Arabidopsis gene HOMEOBOX GENE 8 (AtHB-8) as monitored with the AtHB-8:β-glucuronidase (GUS) transgene is strong in provascular cells and reduced after vascularization is complete (Baima et al., 1995). To differentiate between defects at the level of vascular initiation and vascular differentiation, we examined the expression of AtHB-8:GUS in the tgh-1 mutant. We found that GUS staining in tgh-1 mutant embryonic and mature cotyledons delineates the interrupted vascularization pattern observed in the clearing sections (Figures 4C and 4F). Taken together, this shows that tgh mutants have defects in the initiation of vascular development as determined by the expression of the AtHB-8:GUS reporter construct.
To examine whether TGH gene expression can be correlated with its apparent role in vascular development, we generated transgenic lines that express the GUS reporter under control of a 1021-bp TGH promoter fragment. The same promoter fragment had been used in the TGH:TGH:GFP expression construct, which was able to rescue the tgh mutant phenotype. Analysis of TGH expression by virtue of GUS reporter activity allowed us to reproducibly detect strong GUS expression in the cotyledons of embryos, in the vasculature of cotyledons and leaves, in young meristematic tissue, in trichomes, and in the pistil (Figures 4G to 4L). Hence, in this analysis TGH expression in the vascular system can be correlated with its role in vascular development.
Auxin Response Is Not Altered in the tgh Mutant
Vascular differentiation requires auxin transport and auxin signal transduction (Mattsson et al., 2003; Fukuda, 2004). The importance of auxin transport for vascularization is supported by the observation that Arabidopsis mutants of auxin transport proteins (e.g., pin-formed1 [pin1] and pinoid) or of proteins involved in the proper localization of auxin transport proteins (e.g., gnom) fail to form a proper vascular system (Gälweiler et al., 1998; Steinmann et al., 1999; Christensen et al., 2000). The importance of auxin signal transduction for vascular development is suggested by the discontinuous vascular system observed in the Arabidopsis mutants monopteros (mp), bodenlos (bdl), and auxin-resistant 6 (axr6) (Hardtke and Berleth, 1998; Hobbie et al., 2000; Hamann et al., 2002). The current model of auxin signal transduction predicts that these mutants fail to express a subset of auxin-induced genes required for proper vascular development since they lack the transcription activator MP or are unable to degrade the repressor BDL. To examine a possible role for TGH in auxin response, we investigated the possibility that TGH participates in auxin-induced gene expression. To this end, we compared the expression of the three auxin-induced genes GH3, IAA19, and SAUR10 by reverse transcription PCR in tgh-1 mutant and in wild-type seedlings that had been subjected to a 2-h treatment with 5 and 50 μM of the synthetic auxin 2,4-D (Figure 5A). In these analyses, we detected different levels of auxin-induced gene expression for all three genes; however, we failed to detect any obvious differences between the wild type and the tgh-1 mutant (Figure 5A). These data suggest that TGH is not required for auxin-induced gene expression.
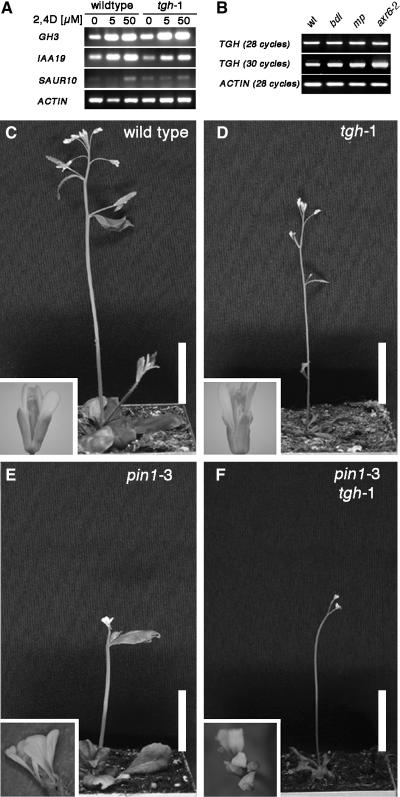
Figure 5.
Auxin Response and Auxin Transport Are Not Affected in the tgh-1 Mutant.
(A) Auxin-induced gene expression of the genes GH3, IAA19, and SAUR10 is not impaired in tgh-1 mutants compared with the wild type. Three-week-old plants were treated for 2 h with the synthetic auxin 2,4-D and subsequently analyzed by reverse transcription followed by 28 PCR amplification cycles.
(B) TGH gene expression is obviously unaltered in 7-d-old bdl, mp, and axr6-2 mutant seedlings as determined by RT-PCR analysis. ACTIN served as an input control for all experiments. Number of PCR amplification cycles is indicated in parentheses.
(C) to (F) Mutants defective in TGH and PIN1 show additive phenotypes. Shown are 4-week-old wild-type (C), tgh-1 (D), pin1-3 (E), and pin1-3 tgh-1 double mutant (F) plants as well as flowers (insets). Bars = 1.5 cm.
Next, we examined whether TGH is a downstream target of the gene expression system that is composed of MP, BDL, and AXR6 (Hardtke and Berleth, 1998; Hamann et al., 2002; Hellmann et al., 2003). To this end, we tested whether TGH expression is altered in a loss-of-function allele of the transcriptional activator MP identified in the Salk T-DNA collection, as well as in the bdl and axr6-2 mutants that fail to degrade BDL in response to auxin (Hardtke and Berleth, 1998; Hamann et al., 2002; Hellmann et al., 2003; D. Weijers and G. Jürgens, unpublished data). When we examined TGH gene expression by RT-PCR, we failed to detect any obvious differences in any of the mutants examined when compared with the wild type (Figure 5B). We therefore suggest that TGH acts independently of the BDL, MP, and AXR6 proteins and conclude furthermore that TGH is not a component of the auxin response pathway.
Additionally, we examined whether TGH is required for proper auxin transport. Mutants of the putative auxin efflux carrier PIN1 fail to form a proper vascular system (Gälweiler et al., 1998). We therefore investigated the genetic interaction of TGH and PIN1 in a pin1-3 tgh-1 double mutant. Since this double mutant displayed the additive phenotype of the respective single mutants with respect to plant size, leaf shape, and flower morphology, we conclude that TGH and PIN1 regulate vascular development by independent mechanisms (Figures 5C to 5F).
Genetic Interaction of TGH and AMP1
Arabidopsis AMP1 has been identified in several mutant screens (Jürgens et al., 1991; Chaudhury et al., 1993; Hou et al., 1993; Conway and Poethig, 1997). amp1 mutants fail to form proper cotyledon and leaf vasculature and frequently produce multiple cotyledons (Chaudhury et al., 1993; Conway and Poethig, 1997). Furthermore, amp1 mutants are constitutively photomorphogenic, grow fast, and flower early (Chaudhury et al., 1993). At least some of the amp1 mutant phenotypes may be explained by the increased cytokinin levels measured in the amp1 mutant resulting in the increased expression of the cell cycle regulator CYCD3 (Chaudhury et al., 1993; Riou-Khamlichi et al., 1999). AMP1 has homology to N-acetyl-α-linked acidic dipeptidases, and based on this homology, AMP1 has been proposed to participate in the processing of small acidic peptides and folate polyglutamate (Helliwell et al., 2001). However, the postulated biochemical role of AMP1 in protein processing as well as its role as a developmental regulator has not been defined yet.
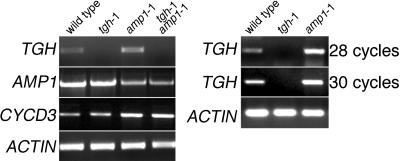
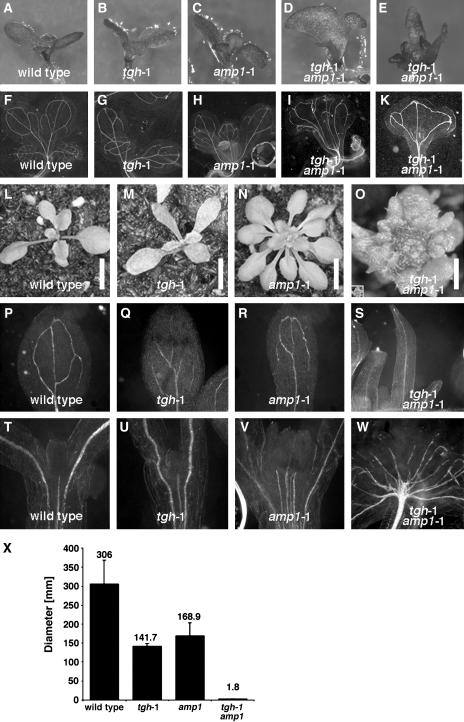
To examine a possible link between TGH and AMP1 function, we investigated TGH gene expression in the amp1-1 mutant using RT-PCR. amp1-1 is an ethyl methanesulfonate–induced loss-of-function allele with a base change mutation leading to an early stop codon mutation (Helliwell et al., 2001). In our studies, we consistently observed reduced AMP1 expression in the amp1-1 mutant allele, possibly a result of the destabilization of the mutant transcript (Figure 6). Consistent with previous reports, we found CYCD3 expression to be elevated in the amp1-1 mutant, indicative of increased cell cycle activity in the mutant (Riou-Khamlichi et al., 1999). Interestingly, our TGH gene expression analyses also revealed increased TGH expression in the amp1-1 mutant compared with the wild type (Figure 6). We therefore went on to examine the genetic interaction between AMP1 and TGH in amp1-1 tgh-1 double mutants. In the progeny of amp1-1 tgh-1/TGH lines, we identified plants with a new phenotype that was not observed in the parental lines or in the single mutants. In seedlings, this phenotype is characterized by dramatic alterations of cotyledon shape, such as fused cotyledons and the impairment of leaf and shoot growth and differentiation (Figures 7A to 7E, 7L, and 7M). In 2-week-old adult plants, the rosette diameter of these mutants was reduced at least 10-fold (Figures 7N, 7O, inset in 7O, and 7X). By genotyping and gene expression analysis, we could show that these plants represented the amp1-1 tgh-1 double mutant segregants of the amp1-1 tgh-1/TGH lines, and we therefore propose a genetic interaction between TGH and AMP1.
Figure 6.
TGH Expression Is Elevated in the amp1-1 Mutant.
Gene expression analysis of TGH, AMP1, and the G1 cell cycle phase marker CYCD3 in 3-week-old wild-type, tgh-1, amp1-1, and tgh-1 amp1-1 double mutants (28 PCR amplification cycles). TGH expression was reproducibly found to be elevated in the amp1-1 mutant. Two additional independent experiments are shown in the right panel for comparison. ACTIN expression (28 cycles) was used as a control for all experiments.
Figure 7.
Genetic Interaction between TGH and AMP1.
(A) to (S) Development of 7-d-old seedlings ([A] to [E]) and 2-week-old adult plants ([L] to [O]) is severely affected in the tgh-1 amp1-1 double mutants. Genotypes are as indicated in the panel. All pictures are taken at comparable magnification, except (O), which is 10 times magnified. See the inset in (O) for the original size of the adult tgh-1 amp1-1 mutant in scale with the single mutants shown in (L) and (M). Growth impairment of the double mutant is reflected at the level of vascular development in seedlings ([F] to [K]) and cotyledons ([P] to [S]). Bars = 5 mm in (L) to (N) and 1 mm in (O).
(T) to (W) Vascularization in the cotyledons and leaves of cleared 10-d-old dark-grown seedlings.
(X) Quantitative analysis of rosette diameters of adult wild-type plants, single mutants, and the tgh-1 amp1-1 double mutant (n ≥ 10; bars and error bars represent means and standard deviations).
Since amp1-1 and tgh-1 single mutants have defects in vascular initiation, we also examined vascularization in these mutants. We found that vascular initiation is significantly more disturbed in the amp1-1 tgh-1 double mutants than in the respective single mutants (Figures 7F to 7K, 7S, and 7W). For example, in cotyledons and leaves of the amp1-1 tgh-1 double mutants, often only the primary vascular strand but no secondary or tertiary strands differentiated (Figures 7P to 7W). Furthermore, the lack of vascularization in these double mutant plants is accompanied by an inability to form normal leaf blades. We therefore propose that TGH and AMP1 are required for the proper differentiation of the vascular network and normal leaf growth.
The amp1-1 mutant has increased cytokinin levels and increased levels of the cytokinin-inducible G1 cell cycle regulator CYCD3 (Riou-Khamlichi et al., 1999) (Figure 6). Since CYCD3 expression has been proposed to be a direct consequence of the amp1-1 mutant's increased cytokinin levels, we reasoned that the observed amp1-1 tgh-1 double mutant phenotype may be a result of the combined effects of increased cytokinin levels and loss of TGH gene function. We therefore examined the effects of cytokinin treatment on the tgh-1 mutant seedlings. However, by treating tgh-1 mutants with different cytokinins and different cytokinin concentrations, we failed to induce phenotypes in the tgh-1 mutant comparable to those observed in the amp1-1 tgh-1 double mutant (data not shown). At the same time, we found CYCD3 expression to be elevated to the same extent in both the amp1-1 and the amp1-1 tgh-1 double mutant, suggesting that TGH does not influence CYCD3 expression (Figure 6). We therefore conclude that the strong amp1-1 tgh-1 double mutant phenotype is not the direct result of cytokinin overproduction or CYCD3 overexpression in the amp1-1 tgh-1 double mutants.
DISCUSSION
In this report, we describe the previously uncharacterized TGH protein and its role in Arabidopsis growth and development. TGH is an evolutionarily conserved regulator with a G-patch and a SWAP domain, and both domains are exclusively found in RNA binding and processing proteins (Figure 1) (Denhez and Lafyatis, 1994; Spikes et al., 1994; Aravind and Koonin, 1999). TGH colocalizes with the SR protein SRp34 to subnuclear particles and the enrichment of Ser and Arg residues in the KRDES domain of TGH is reminiscent of the Arg- and Ser-rich RS domains found in SR proteins (Figures 1 and 2). SR proteins play key roles in constitutive and alternative splicing either as essential splicing factors or as specific regulators that act during different stages of spliceosome assembly (Blencowe et al., 1999; Graveley, 2000). The only other data available on TGH function besides the studies reported here come from a Drosophila interactome study where the Drosophila TGH ortholog CG8833 was found to interact with CG6843 (Giot et al., 2003). CG6843 is an uncharacterized SR protein, which in turn interacts with splicing factors such as SWAP and SRp54. Taken together, the different findings strongly suggest that TGH functions as an RNA binding and processing protein that may act close to the splicing machinery. Another point that indirectly supports this hypothesis is the absence of a TGH ortholog from the budding yeast S. cerevisiae, an organism where splicing is not an important regulatory mechanism (Figure 1). On the other side, two large-scale purifications of spliceosomes have been reported, and none of these reports identified TGH orthologs as components of the spliceosome (Rappsilber et al., 2002; Zhou et al., 2002). It is therefore unlikely that TGH is an integral spliceosome subunit.
In our study, we observed a yeast two-hybrid interaction between TGH and TBP2, and we observed that TGH promotes gene expression when tethered to the promoter of a reporter gene (Figures 1E to 1G). Transcription and pre-mRNA processing are known to be intimately related processes, and alternative splicing can be determined by the identity of the promoter driving gene expression (Smith and Valcarcel, 2000; Fong and Zhou, 2001; Proudfoot et al., 2002). Multiple splicing regulators have been reported to interact with the C-terminal domain (CTD) of RNA polymerase II, and it is thought that CTD serves as an assembly platform for and a regulator of transcription and pre-mRNA processing machines (Maniatis and Reed, 2002). Interestingly, in addition to the clear sequence homologies described in this report, TGH has also limited similarity to the CTD-interacting protein 6, an interactor of the C terminus of RNA polymerase II (data not shown). It may therefore be hypothesized that TGH is an RNA binding or processing protein that functions cotranscriptionally.
Our developmental analysis reveals that TGH is essential for proper plant development. Two independent mutant alleles show identical phenotypes, including reduced elongation growth, tricotyly, reduced vascularization, and reduced pollen formation (Figures 2 and 4). Altered cotyledon number in combination with reduced vascularization has been observed in a limited number of mutants that are defective in auxin transport and response as well as in mutants defective in AMP1, a putative N-acetyl-α-linked acidic dipeptidase (Helliwell et al., 2001). In our studies, we failed to establish a link between TGH function and the auxin transport and auxin response pathways (Figure 5). By contrast, however, we observed TGH gene expression to be elevated in an amp1 mutant. This observation may suggest that a TGH expressing cell type is more abundant in the amp1 mutant or that AMP1 represses TGH gene expression in the wild type (Figure 6). The latter explanation is unlikely based on the proposed biochemical function of AMP1 as a putative N-acetyl-α-linked acidic dipeptidase with a presumed role in protein or peptide processing. On the other side, amp1 mutants have increased cell cycle activity and increased meristem size (Riou-Khamlichi et al., 1999). Since TGH:GUS data suggest that TGH is expressed in meristematic and young tissue, the increased TGH gene expression observed by RT-PCR may be a consequence of the amp1 mutant's increased meristem size.
TGH shows a strong genetic interaction with AMP1 (Figure 7). Double mutants defective in both gene functions initiate but fail to differentiate leaves. It may be postulated that the impaired and strongly reduced differentiation of leaves in the double mutant restricts proper vascularization, so that impaired cotyledon and leaf development may be the primary cause for reduced vascularization. Conversely, since both mutants are defective in vascularization and since this effect is strongly enhanced in the double mutant, it may also be postulated that reduced vascularization is the biological cause for the dramatic double mutant phenotype.
Although the strong genetic interaction between TGH and AMP1 is very intriguing, the interaction of their gene products can at present not be explained based on the proposed biochemical function of TGH and AMP1. While we postulate TGH to act in RNA binding or processing, AMP1 is a putative N-acetyl-α-linked acidic dipeptidase with a presumed role in protein or peptide processing (Helliwell et al., 2001). To date, no substrates for AMP1's proposed peptidase activity have been identified nor has AMP1 been localized to any subcellular compartment. We can therefore at present not provide an explanation for the strong genetic interaction observed between the two genes based on the nature of their gene products.
In plants, RNA binding and processing proteins have so far only been implicated in floral transition and patterning, vegetative phase change, as well as abscisic acid signaling, stress responses, and circadian rhythms (Chen and Cheng, 2004). The putative RNA binding and processing protein TGH with its proposed role in vascularization adds a new biological function to this list. However, based on the large numbers of proteins with domains indicative for a role in RNA binding and processing and based on the essential role of these proteins in gene expression, it can be expected that there are many novel developmental functions for these proteins that await to be identified (Lorkovic and Barta, 2002).
METHODS
Biological Material
The TGH T-DNA insertion lines SALK_053445 (Columbia background) and GABI_774H04 (Columbia background) were identified in the SIGNAL database (http://signal.salk.edu/cgi-bin/tdnaexpress) and obtained from the Nottingham Arabidopsis Stock Centre (NASC) and from GABI-KAT at the Max-Planck Institute for Plant Breeding Research (Cologne, Germany), respectively (Alonso et al., 2003; Rosso et al., 2003). To test for the presence of the TGH wild-type gene and the TGH T-DNA insertion, segregants of SALK_053445 were PCR genotyped using combinations of primers flanking the T-DNA insertion site, TGHFW 5′-ATGTTGAGGATGAAGATGTCTATGC-3′ and TGHRV 5′-AGATGAAGCTGATTTGGTGAATGTG-3′, or TGHFW and the T-DNA specific primer LBb1 5′-GCGTGGACCGCTTGCTGCAACT-3′. Segregants of GABI_774H04 were genotyped with TGHFW and TGHRV or TGHRV in combination with the T-DNA–specific GABI primer 5′-ATATTGACCATCATACTCATTGC-3′.
The reporter line AtHB-8:GUS was obtained from Giorgio Morelli (Rome, Italy) and crossed to tgh-1/TGH lines. Tissue-specific GUS expression was analyzed in TGH wild-type and mutant segregants of the F2 and F3 progeny as described elsewhere (Baima et al., 1995; Weigel and Glazebrook, 2002). The mp allele was derived from the T-DNA insertion line SALK_023812 and was a gift from D. Weijers (Tübingen, Germany). bdl and axr6-2 alleles were previously described (Hamann et al., 1999; Hobbie et al., 2000; Hellmann et al., 2003). pin1-3/PIN1 mutant seed were a gift from N. Geldner (Tübingen University, Germany). pin1-3/PIN1 were identified by a ScaI cleaved-amplified polymorphic sequence marker and then crossed with tgh-1/TGH. The resulting F1 plants were genotyped to identify pin1-3/PIN1 tgh-1/TGH lines. The F2 progeny of these lines was then used for phenotype and genotype analyses. amp1-1 (Columbia background) mutant seed were obtained from NASC and crossed to tgh-1/TGH. amp1-1/amp1-1 tgh-1/TGH plants were identified among the F2 progeny based on the amp1-1 phenotype and the genotype of the TGH locus. The F3 progeny of these lines was subjected to phenotype analyses. Phenotypes were linked to the segregating genotypes using PCR analysis as described above. When different ecotypes were combined for genetic crosses, segregating wild-type plants were used as wild-type control.
Subcellular Localization Studies
Constructs expressing TGH:GFP fusions were prepared using the Gateway system (Invitrogen). To generate 35S:TGH:GFP, the TGH gene fragment was amplified by PCR using the primers 5′-attB1-TCATGGGGTCAGACGAGGAAGATTTCGTGTTTC-3′ and 5′-attB2-CGTCTCGTCGCCTCTTCTTCTCCCGCCTTGAC-5′ and introduced into pDONR 201. One entry clone was fully sequenced and inserted into the vector 35S-GW-GFP(Kan), a gift from F. Turck (Max-Planck Institute for Plant Breeding Research, Cologne, Germany). To generate TGH:TGH:GFP, a 6.6-kb TGH genomic fragment was amplified by PCR using the primers 5′-attB1-CATGCTCAGGAGCAATCGTCCGTTTATC-3′ and attB2-CGTCTCGTCGCCTCTTCTTCTCCCGCCTTGAC-5′. The fragment was introduced into pDONR 201, and one clone was fully sequenced and then inserted into the vector pGWB8 (a gift from T. Nakagawa, Shimane University, Japan). Twenty stable transgenic lines expressing each TGH:GFP fusion construct were generated and propagated in Arabidopsis thaliana ecotype Columbia (Clough and Bent, 1998). For transient expression, the 35S:TGH:GFP and TGH:TGH:GFP constructs were introduced alone or in combination with the constructs SRp34:DsRED and RSZ33:DsRED (a gift from Z. Lorkovic and A. Barta, Vienna Biocentre, Austria) into Arabidopsis var Columbia suspension culture cells using established procedures (Negrutiu et al., 1987; Lorkovic et al., 2004a). The DsRED constructs express the respective fusion proteins from the 35S promoter of Cauliflower mosaic virus. Protein fluorescence was analyzed using a Leica TCS SP2 confocal microscope.
Tissue Clearing
Tissue of 7-d-old Arabidopsis seedlings and 3-week-old Arabidopsis plants was fixed for 12 h in an ethanol:acetic acid (6:1) mixture followed by two 30-min washes with 100% ethanol and one wash with 70% ethanol. Tissues were finally cleared with chloral hydrate:glycerol:water (8 g:1 mL:2 mL) for several hours as previously described (Weigel and Glazebrook, 2002).
Yeast Two-Hybrid System
A previously described yeast two-hybrid system cDNA library was used to perform a library screen with pGBT9/pTBP expressing potato (Solanum tuberosum) TBP (Holdsworth et al., 1992; Kim et al., 1997; Schwechheimer and Deng, 2002). To this end, full-length pTBP was cloned in frame by ligating a blunt-ended BamHI fragment to the blunt-ended SalI site of the pGBT9 vector. The screen resulted in the isolation of pACT/2-8/5 expressing the C-terminal 245 amino acids of TGH, including its KRDES domain. pACT/TGH was obtained by cloning a BamHI linked full-length TGH cDNA fragment into pACT2. The interaction between full-length TGH and Arabidopsis TBP2 was then confirmed in the yeast two-hybrid system with pGBT9/AtTBP expressing the Arabidopsis TBP2 ortholog (At1g55520) cloned in an identical manner to construct pGBT9/pTBP.
Isolation of TGH Full-Length cDNA Clones
A size-fractionated Arabidopsis cDNA library provided by NASC was screened for full-length cDNAs using the TGH cDNA fragment isolated in the yeast two-hybrid system as a probe. Sequence analyses of several full-length cDNAs confirmed the exon-intron predictions for the TGH gene found in the databases and revealed that polyadenylation of TGH mRNAs occurs at two alternative positions 76 and 285 base pairs after the stop codon.
Transactivation Studies
pGBT9/TGH containing the full-length TGH gene and pGBT9/KRDES containing the TBP2-interacting TGH fragment originally identified in the yeast two-hybrid system library screen were tested for their ability to transactivate expression of the LacZ reporter gene in the yeast strain HF7C using Galacto-Star (Tropix). The TGH coding sequence and the KRDES fragment present in the original yeast two-hybrid clone were subcloned into the vector pGAL4 and tested as previously described for their ability to transactivate transcription from UAS4xGALLUC in tobacco (Nicotiana tabacum) mesophyll protoplasts (Negrutiu et al., 1987; Schwechheimer et al., 1998).
The TGH Promoter:GUS Fusion Construct
A 1021-bp TGH promoter fragment was amplified using 5′-attB1-CATGCTCAGGAGCAATCGTCCGTTTATC-3′ and 5′-attB2-TGTCTTCACCACCGAGACCGAGAGGAGCAG-3′. The fragment was introduced into pDONR 201, fully sequenced, and subsequently cloned into the Gateway compatible pGWB3 vector (a gift from T. Nakagawa). Twenty transgenic lines were established harboring the TGH:GUS transgene. Seedlings and plants were stained for GUS expression using standard procedures (Weigel and Glazebrook, 2002).
Gene Expression Analysis
Gene expression analyses were performed by RT-PCR. RNA was extracted from plant material using the RNeasy method (Qiagen). Two micrograms of total RNA were used for the reverse transcription reaction with an oligo(dT) primer and Platinum reverse transcriptase as previously described (Invitrogen) (Frohman et al., 1988). TGH gene expression was monitored using the gene-specific primers TGHFW and TGHRV, primers identical to those used for TGH genotyping. Gene expression of auxin-induced genes was monitored using GH3-FW 5′-ATGGAGGAGTCGTTGAACTCTGTG-3′ and GH3-RV 5′-AAGCTCCATTATTGGCGTGAAACTC-3′ for GH3 (At2g23170); IAA19-FW 5′-GTGATGTACCTTGGGGGATGTTTC-3′ and IAA19-RV 5′-AATGAACCAGCTCCTTGCTTCTTG-3′ for IAA19 (At3g15540); SAUR10-FW 5′-CGAAGTCGGTACATCGTTCCTATC-3′ and SAUR10-RV 5′-CATGGAGATAAGAGACCTGAAGAAGA-3′ for SAUR10 (At2g18010). Gene expression of AMP1 and CYCD3 was determined using the primers AMP1-FW 5′-ATGTCACAACCTCTCAC-3′ and AMP1-RV 5′-TCATGTGAAACCTCCTT-3′ for AMP1 and CYCD3-FW 5′-ATGGCTTTAGAAGAGGAGGAAGA-3′ and CYCD3-RV 5′-TTAGCGAGGACTACTACTAAGCAC-3′ for CYCD3 (At3g50070). ACTIN-FW 5′-ATTCAGATGCCCAGAAGTCTTGTTC-3′ and ACTIN-RV 5′-GCAAGTGCTGTGATTTCTTTGCTCA-3′ for ACTIN (At3g18780) were used as a loading control in all cases. All RT-PCR reactions were repeated using independent RNA preparations and reverse transcription reactions. Transcripts were amplified using 28 or 30 amplification cycles as indicated in the text. Each experiment was repeated at least twice with independent RNA preparations.
Accession Numbers
TGH sequence data from this article have been deposited with the GenBank data library under accession number AAR99647. TOUGH (TGH) is registered as a gene class symbol at www.arabidopsis.org. Arabidopsis Genome Initiative locus identifiers for the Arabidopsis genes used in this article are as follows: TGH (At5g23080), AtTBP2 (At1g55520), GH3 (At2g23170), IAA19 (At3g15540), SAUR10 (At2g18010), CYCD3 (At3g50070), and ACTIN (At3g18780). Accessions of the TGH orthologs listed in Figures 1B and 1C are as follows: rice (BAD37703.1), human (NP_060495.2), mouse (NP_080457.1), Drosophila (NP_648669.1), C. elegans (CAA83621.1), N. crassa (EAA31042.1), S. pombe (NP_593626.1), and A. gambiae (NP_593626.1). Accessions of the SWAP domain containing proteins listed in Figure 1D are as follows: rice (BAD37703.1), human (AAN77183.1), mouse (NP_613051.2), Drosophila (AAN77184.1), A. gambiae (EAA11741.1).
Supplementary Material
Acknowledgments
We thank Dolf Weijers and Melina Zourelidou for helpful comments on the manuscript, Eike Rademacher for assistance during the initial characterization of the tgh-1 mutant, Catarina Brancato for Arabidopsis protoplast transformations, colleagues in Tübingen for providing mutant seed, the Salk Institute Genomic Analysis Laboratory and NASC for generating and providing the sequence-indexed Arabidopsis T-DNA insertion mutant SALK_053445, B. Weisshaar and the GABI-KAT program (Max-Planck Institute for Plant Breeding Research) for the T-DNA mutant GABI774H04, Z.J. Lorkovic and A. Barta for providing the constructs SRp34:DsRED and RSZ33:DsRED, and Giovanni Spena for AtHB8:GUS seeds. The work in our laboratory was supported by funds from Tübingen University and by the Deutsche Forschungsgemeinschaft (SCHW 751/4).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Claus Schwechheimer (claus.schwechheimer@zmbp.uni-tuebingen.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031302.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 633–657. [DOI] [PubMed] [Google Scholar]
- Aravind, L., and Koonin, E.V. (1999). G-patch: A new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 24, 342–344. [DOI] [PubMed] [Google Scholar]
- Baima, S., Nobili, F., Sessa, G., Lucchetti, S., Ruberti, I., and Morelli, G. (1995). The expression of the AtHB-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121, 4171–4182. [DOI] [PubMed] [Google Scholar]
- Blencowe, B.J., Bowmann, J.A.L., McCracken, S., and Rosonina, E. (1999). SR-related proteins and the processig of messenger RNA precursors. Biochem. Cell Biol. 77, 277–291. [PubMed] [Google Scholar]
- Blilou, I., Fugier, F., Folmer, S., Serralbo, O., Willemsen, V., Wolkenfeldt, H., Eloy, N.B., Ferreira, P.C.G., Weisbeek, P.J., and Scheres, B. (2002). The Arabidopsis HOBBIT gene encodes a CDC27 homolog that links the plant cell cycle to progression of cell differentiation. Genes Dev. 16, 2566–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland, F.M., Fujioka, S., Takatsuto, S., Yoshida, S., and Nelson, T. (2002). The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 14, 2045–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, A.M., Letham, S., Craig, S., and Dennis, E.S. (1993). amp1: A mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J. 4, 907–916. [Google Scholar]
- Chen, X., and Cheng, Y. (2004). Posttranscriptional control of plant development. Curr. Opin. Plant Biol. 7, 20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S.K., Dagenais, N., Chory, J., and Weigel, D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100, 469–478. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Conway, L.J., and Poethig, R.S. (1997). Mutations of Arabidopsis thaliana that transform leaves into cotyledons. Proc. Natl. Acad. Sci. USA 94, 10209–10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhez, F., and Lafyatis, R. (1994). Conservation of regulated alternative splicing and identification of functional domains in vertebrate homologs to the Drosophila splicing regulator, suppressor-of-white-apricot. J. Biol. Chem. 269, 16170–16179. [PubMed] [Google Scholar]
- Dingwall, C., and Laskey, R.A. (1991). Nuclear targeting sequences: A consensus? Trends Biochem. Sci. 16, 478–481. [DOI] [PubMed] [Google Scholar]
- Dynlacht, B.D., Hoey, T., and Tjian, R. (1991). Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell 66, 563–576. [DOI] [PubMed] [Google Scholar]
- Fong, Y.W., and Zhou, Q. (2001). Stimulatory effect of splicing factors on transcriptional elongation. Nature 414, 929–933. [DOI] [PubMed] [Google Scholar]
- Frohman, M.A., Dush, M.K., and Martin, G.R. (1988). Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, H. (2004). Signals that control plant vascular cell differentiation. Nat. Rev. Mol. Cell Biol. 5, 379–391. [DOI] [PubMed] [Google Scholar]
- Gälweiler, L., Guan, C., Müller, A.J., Wismann, E., Medgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Development 282, 2226–2230. [DOI] [PubMed] [Google Scholar]
- Giot, L., et al. (2003). A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736. [DOI] [PubMed] [Google Scholar]
- Goodrich, J.A., and Tjian, R. (1994). TBP-TAF complexes: Selectivity factors for eukaryotic transcription. Curr. Biol. 6, 403–409. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R. (2000). Sorting out the complexity of SR protein functions. RNA 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann, T., Benkova, E., Bäurle, I., Kientz, M., and Jürgens, G. (2002). The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 16, 1610–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann, T., Mayer, U., and Jürgens, G. (1999). The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126, 1387–1395. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, C.A., Chin-Atkins, A.N., Wilson, I.W., Chapple, R., Dennis, E.S., and Chaudhury, A. (2001). The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell 13, 2115–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann, H., Hobbie, L., Chapman, A., Dharmasiri, S., Dharmasiri, N., del Pozo, C., Reinhardt, D., and Estelle, M. (2003). Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis. EMBO J. 22, 3314–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie, L., McGovern, M., Hurwitz, L.R., Pierro, A., Liu, N.Y., Bandyopadhyay, A., and Estelle, M. (2000). The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 127, 23–32. [DOI] [PubMed] [Google Scholar]
- Holdsworth, M.J., Grierson, C., Schuch, W., and Bevan, M. (1992). DNA-binding properties of cloned TATA-binding protein from potato tubers. Plant Mol. Biol. 19, 455–464. [DOI] [PubMed] [Google Scholar]
- Hou, Y., von Arnim, A.G., and Deng, X.-W. (1993). A new class of Arabidopsis constitutive photomorphogenic genes involved in regulating cotyledon development. Plant Cell 5, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens, G., Mayer, U., Torres Ruiz, R.A., Berleth, T., and Miséra, S. (1991). Genetic analysis of pattern formation in the Arabidopsis embryo. Dev. Suppl. 1, 27–38. [Google Scholar]
- Kim, J., Harter, K., and Theologis, A. (1997). Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman, P.M., and Berk, A.J. (1994). A mechanism for TAFs in transcriptional activation: Activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 8, 995–1006. [DOI] [PubMed] [Google Scholar]
- Lopato, S., Forstner, C., Kalyna, M., Hilscher, J., Langhammer, U., Indrapichate, K., Lorkovic, Z.J., and Barta, A. (2002). Network of interactions of a novel plant-specific Arg/Ser-rich protein, atRSZ33, with atSC35-like splicing factors. J. Biol. Chem. 277, 39989–39998. [DOI] [PubMed] [Google Scholar]
- Lopato, S., Kalyna, M., Dorner, S., Kobayashi, R., Krainer, A.R., and Barta, A. (1999). atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 13, 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic, Z.J., and Barta, A. (2002). Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30, 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic, Z.J., Hilscher, J., and Barta, A. (2004. a). Use of fluorescent protein tags to study nuclear organization of the spliceosomal machinery in transiently transformed living plant cells. Mol. Biol. Cell 15, 3233–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic, Z.J., Lopato, S., Pexa, M., Lehner, R., and Barta, A. (2004. b). Interactions of Arabidopsis RS domain containing cyclophilins with SR proteins and U1 and U11 small nuclear ribonucleoprotein-specific proteins suggest their involvement in pre-mRNA splicing. J. Biol. Chem. 279, 33890–33898. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., and Reed, J. (2002). An extensive network of coupling among gene expression machines. Nature 416, 499–506. [DOI] [PubMed] [Google Scholar]
- Mattsson, J., Ckurshumova, W., and Berleth, T. (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131, 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, J., Sung, Z.R., and Berleth, T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126, 2979–2991. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Plant Physiol. 15, 473–497. [Google Scholar]
- Negrutiu, I., Shillito, R., Potrykus, I., and Sala, F. (1987). Hybrid genes in the analysis of transformation conditions. I. Setting up a simple method for direct gene transfer in plant protoplasts. Plant Mol. Biol. 8, 363–373. [DOI] [PubMed] [Google Scholar]
- Orphanides, G., and Reinberg, D. (2002). A unified theory of gene expression. Cell 108, 439–451. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. (2002). Integrating mRNA processing with transcription. Cell 108, 501–512. [DOI] [PubMed] [Google Scholar]
- Rappsilber, J., Ryder, U., Lamond, A.I., and Mann, M. (2002). Large-scale proteomic analysis of the human spliceosome. Genome Res. 12, 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D. (2003). Vascular patterning: More than just auxin? Curr. Biol. 13, R485–R487. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi, C., Huntley, R., Jacqmard, A., and Murray, J.A.H. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283, 1541–1544. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y.-F., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53, 247–259. [DOI] [PubMed] [Google Scholar]
- Sachs, T. (1991). Cell polarity and tissue patterning in plants. Dev. Suppl. 1, 83–93. [Google Scholar]
- Schwechheimer, C., and Deng, X.W. (2002). Studying protein-protein interactions with the yeast two-hybrid system. In Molecular Plant Biology, P.M. Gilmartin and C. Bowler, eds (Oxford: Oxford University Press), pp. 173–198.
- Schwechheimer, C., Smith, C., and Bevan, M. (1998). The activities of acidic and glutamine-rich transcriptional activation domains in plant cells: Design of modular transcription factors for high-level expression. Plant Mol. Biol. 36, 195–204. [DOI] [PubMed] [Google Scholar]
- Smith, C.W.J., and Valcarcel, J. (2000). Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem. Sci. 25, 381–388. [DOI] [PubMed] [Google Scholar]
- Spikes, D.A., Kramer, J., Bingham, P.M., and van Doren, K. (1994). SWAP pre-mRNA splicing regulators are a novel, ancient protein family sharing a highly conserved sequence motif with the prp21 family of constitutive splicing proteins. Nucleic Acids Res. 22, 4510–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Moangold, S., Jackson, C.L., Paris, S., Gälweiler, L., Palme, K., and Jürgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286, 316–318. [DOI] [PubMed] [Google Scholar]
- Tansey, W.P., and Herr, W. (1997). TAFs: Guilt by association? Cell 88, 729–732. [DOI] [PubMed] [Google Scholar]
- Tjian, R., and Maniatis, T. (1994). Transcriptional activation: A complex puzzle with few easy pieces. Cell 77, 5–8. [DOI] [PubMed] [Google Scholar]
- Verrijzer, C.P., and Tjian, R. (1996). TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem. Sci. 21, 338–342. [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Willemsen, V., Friml, J., Grebe, M., van den Toorn, A., Palme, K., and Scheres, B. (2003). Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15, 612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen, V., Wolkenfelt, H., de Vrieze, G., Weisbeek, P.J., and Scheres, B. (1998). The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development 125, 521–531. [DOI] [PubMed] [Google Scholar]
- Wu, S.Y., and Chiang, C.M. (2001). TATA-binding protein-associated factors enhance the recruitment of RNA polymerase II by transcriptional activators. J. Biol. Chem. 276, 34235–34243. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., Licklider, L.J., Gygi, S.P., and Reed, R. (2002). Comprehensive proteomics analysis of the human spliceosome. Nature 419, 182–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.