Abstract
To identify genes involved in Arabidopsis thaliana petal and stamen organogenesis, we used a gene trap approach to examine the patterns of reporter expression at each stage of flower development of 1765 gene trap lines. In 80 lines, the reporter gene showed petal- and/or stamen-specific expression or lack of expression, or expression in distinct patterns within the petals and/or the stamens, including distinct suborgan domains of expression, such as tissue-specific lines marking epidermis and vasculature, as well as lines demarcating the proximodistal or abaxial/adaxial axes of the organs. Interestingly, reporter gene expression was typically restricted along the proximodistal axis of petals and stamens, indicating the importance of this developmental axis in patterning of gene expression domains in these organs. We identified novel domains of gene expression along the axis marking the midregion of the petals and apical and basal parts of the anthers. Most of the genes tagged in these 80 lines were identified, and their possible functions in petal and/or stamen differentiation are discussed. We also scored the floral phenotypes of the 1765 gene trap lines and recovered two mutants affecting previously uncharacterized genes. In addition to revealing common domains of gene expression, the gene trap lines reported here provide both useful markers and valuable starting points for reverse genetic analyses of the differentiation pathways in petal and stamen development.
INTRODUCTION
Organogenesis consists of coordinated series of cell division and differentiation, which are regulated temporally and spatially to form specific types of tissues and the organ as a whole. In Arabidopsis thaliana flowers, four types of lateral organs arise in concentric rings, or whorls; the sepals, petals, stamens, and carpels are sequentially initiated from the outermost to the innermost whorls (Smyth et al., 1990). The identity of each whorl of organs is specified by a distinct combination of floral homeotic gene functions (Coen and Meyerowitz, 1991; Lohmann and Weigel, 2002). Within an organ, cells in different areas differentiate into diverse tissues depending on further patterning events. Floral organs originate from founder cells in different meristem layers and differentiate to become epidermis, mesophyll, or vascular tissues (Jenik and Irish, 2000). In parallel, each floral organ develops particular axial identities along the proximodistal (base-tip), adaxial/abaxial (toward the shoot apex and away from the shoot apex), and centrolateral (midrib-margin) axes (Hudson, 1999; Bowman et al., 2002; Golz and Hudson, 2002). Here, we initiate studies to examine the specification and orchestration of cell differentiation during organogenesis, using Arabidopsis petals and stamens as models.
The petals are initiated at stage 5 by coordinated cell divisions in the presumptive epidermis (L1 layer) and the presumptive mesophyll and vascular tissues (L2 layer) of the floral meristem (Hill and Lord, 1989; Smyth et al., 1990; Jenik and Irish, 2000; Dinneny et al., 2004). The petals grow more slowly compared with the other organs during the early phases of development. Starting at stage 9, the distal blade and the proximal claw regions become distinct, and changes in cellular morphology begin to be apparent (Smyth et al., 1990; Pyke and Page, 1998). The cells in the blade proliferate extensively at stages 8 to 10, while division of claw cells are more scarce and occur mainly in stages 11 and 12 (Dinneny et al., 2004). The petals expand rapidly after stage 9; the cells at the base mainly elongate in the apical-basal direction, becoming long and narrow, whereas cells in the blade expand more evenly in all directions and become round. The claw cells contain chloroplasts, while the cells in the blade contain leucoplasts and so appear white (Pyke and Page, 1998). Mature petals are reflexed, exposing conical papillae cells on the adaxial surface of the blade (Smyth et al., 1990; Takeda et al., 2004).
The stamen primordia are initiated at stage 5 by cell divisions in the L1, L2, and L3 layers of the floral meristem (Lord et al., 1994; Jenik and Irish, 2000). Each stamen is composed of a filament and an anther, and in Arabidopsis, these structures are first distinguishable at stage 7 (Smyth et al., 1990). The locules, or pollen sacs, are detectable by stage 7 on the adaxial side of the anther, and the sporogenous tissue within the locules is apparent by stage 9 (Sanders et al., 1999; Scott et al., 2004). Meiosis occurs in the anthers of stage 9 flowers, and pollen develops at stages 10 and 11. The filament, which supports the anther, has a simple, radialized structure and contains a single strand of vasculature. Filament cells, which are organized in files, elongate rapidly prior to anthesis. At anthesis, the stomium, which consists of specialized epidermal cells, ruptures to split the lobes of the anther and release pollen (Sanders et al., 1999).
Floral organ development relies on both appropriate lateral organ differentiation, as well as the specification of unique organ identities. It is clear that the floral homeotic genes control organ identity by regulating the appropriate differentiation of organ-specific tissues. However, the floral homeotic genes are expressed largely ubiquitously and throughout the development of the organs they specify (Yanofsky et al., 1990; Jack et al., 1992; Mandel et al., 1992; Goto and Meyerowitz, 1994; Pelaz et al., 2000; Honma and Goto, 2001; Ditta et al., 2004); thus, additional factors are required to regulate specific individual differentiation processes. For instance, the floral homeotic gene AGAMOUS (AG) regulates stamen identity and has been shown to directly activate transcription of SPOROCYTELESS (SPL), which is necessary for formation of the locules and anther walls (Yang et al., 1999; Ito et al., 2004). Constitutive AG expression resulted in ectopic SPL activation and formation of locules, which, however, was restricted to the distal region of lateral lamina of the petals (Ito et al., 2004), indicating that factors providing distal and lateral cues are also involved in SPL regulation. Similarly, in Antirrhinum, MYBML1, a gene required for ventral petal epidermis differentiation, is regulated by the petal identity gene DEFICIENS (Perez-Rodriguez et al., 2005); however, its specific expression pattern also depends on further positional restrictions by DIVARICATA, which specifies ventral petal identity (Almeida et al., 1997). Therefore, a combination of positional signals is required for the correct patterning of gene expression and cell differentiation during organogenesis.
In order to isolate genes important for petal and/or stamen differentiation, we employed a gene trapping strategy. Gene trap lines, in which endogenous proteins are fused to reporter proteins, can be used to visualize expression patterns and functional domains of random genes in the genome (Sundaresan et al., 1995; Bellen, 1999; Springer, 2000). We examined the expression patterns of the β-glucuronidase (GUS) reporter gene in inflorescences of 1765 gene trap lines. We isolated 80 gene trap lines in which specific patterns of GUS expression were observed in the petals and stamens and cloned their gene trap insertion sites. Many of these lines showed distinct suborgan domains of GUS expression, revealing common domains of gene regulation in the Arabidopsis petals and stamens. In particular, a number of lines were recovered whose GUS expression was restricted to specific suborgan domains along the proximodistal axis. The 1765 lines were also examined for unusual floral phenotypes, and eight mutant lines, including two previously uncharacterized mutants, were recovered. Besides providing a collection of gene expression patterns and useful marker lines, the gene trap lines identified in this study define candidate genes involved in petal and stamen identity-specific differentiation pathways.
RESULTS
Gene Trap Screening
Approximately 1765 Trapper gene trap lines were examined for their patterns of reporter expression in inflorescence tissue. To generate these lines, a single copy of the GUS reporter gene flanked by splice acceptor sites and an intron was mobilized to another site in the genome via activity of the Ac-Ds transposon system (Sundaresan et al., 1995; Martienssen, 1998). Approximately 90% of such mobilization events result in single copy insertions (Martienssen, 1998). Lines in which the gene trap insertion landed inside a transcriptional unit generate chimeric proteins consisting of a partial protein product of the transcriptional unit fused to the GUS protein at the C terminus. Therefore, by staining the gene trap lines for GUS enzymatic activity, one can survey gene expression patterns.
For most of the gene trap lines described in this study, genomic regions flanking the gene trap insertion were cloned, and the gene whose expression pattern was most likely reported by the GUS staining was identified (see Methods). As previously observed for the gene trapping system used in this study, insertions were frequently located in the vicinity of the 5′ end of transcriptional units (Parinov et al., 1999; Pan et al., 2005; see Tables 2 to 6). Although the gene trap system was designed to recover insertions within transcription units, in some lines insertions were found outside of transcribed regions (∼50% of lines reported here) or within a transcribed region of an annotated gene but in the wrong orientation (∼25% of lines reported here). Theoretically, no GUS staining should be observed in such a scenario; however, it has been postulated that the GUS gene in the Trapper gene trap lines may contain a minimal promoter and can also act as an enhancer trap (Cocherel et al., 1996). Gene trap insertions within transcribed regions of annotated genes yet in the opposite direction may be tagging the antisense transcripts, since many Arabidopsis genes are also transcribed in the reverse direction (Yamada et al., 2003; Jen et al., 2005). Alternatively, such gene trap inserts could be tagging as yet unannotated genes (Groover et al., 2003). In this report, we focus our discussion on lines with an insertion within a transcription unit or minimal promoter region, since GUS staining in these lines more likely reflects the endogenous gene expression pattern. However, regardless of whether the gene trap lines replicate endogenous gene expression or expression driven by a subset of enhancer sequences, the patterns of expression reported here demarcate a variety of distinct region-specific domains of gene regulation.
Table 2.
Genes Recovered with Known Expression Patterns
| Gene Trap Line | Annotationa | Gene Name | Insertion Siteb | Insertion Orientationc | GUS Staining Pattern | Reference to Expression Pattern |
|---|---|---|---|---|---|---|
| GT7847 | At5g60910 | FRUITFULL | Within | Same | Inflorescence meristem, stage 1 and 2 floral meristems, carpels from stage 7 and on | Gu et al. (1998) |
| GT7953 | At1g70510 | KNAT2 | Within | Same | Inflorescence meristem and young flowers to stage 4; from stage 4, staining is excluded from floral organ primordia, except for the base of carpels and replum | Dockx et al. (1995); Pautot et al. (2001); Byrne et al. (2002) |
| GT8137 | At3g51060 | STYLISH1 | Within | Same | In presumptive floral and sepal primordia, apex of the inflorescence and floral meristems, distal tips of floral organ primordia, and in developing styles and ovules | Kuusk et al. (2002) |
| GT8686 | At1g24260 | SEPALLATA3 | Within | Same | In the 2nd, 3rd, and 4th whorl of flowers in stage 3 and on | Mandel and Yanofsky (1998) |
Genes whose endogenous expression patterns are likely to be reported by the GUS reporter are listed.
Annotated gene closest to the gene trap insertion site identified for the line.
Insertion location respective to the transcribed region of the annotated gene.
Orientation of GUS reporter gene relative to the annotated gene.
Table 3.
Gene Trap Lines with Petal- and/or Stamen-Specific GUS Staining Patterns
| Expression Class | Gene Trap Line | GUS Staining Patterna | Annotationb | Description | Insertion Sitec | Insertion Orientationd |
|---|---|---|---|---|---|---|
| Petal-specific expression | GT7921 | Petal-specific stages 8 to 11e | At5g54880 | Expressed protein | Within | Same |
| GT8132 | Petal-specific stage 6 and on | At1g35560 | Putative TCP family transcription factor | 50 bp upstream | Same | |
| Petal-specific lack of expression | GT8282 | Not in petals, stages 11 and 12e | At3g11340 | Glycosyltransferase family protein | Within | Same |
| GT8309 | Not in petals, stages 9 to 12e | |||||
| GT8454 | Not in expanded petal lamina, stage 13 and one | At3g10060 | Putative FKBP-type peptidyl-prolyl cis-trans isomerase | Within | Same | |
| GT8485 | Not in petals, stages 10 to 14e | At5g14210 | Putative protoporphyrinogen IX oxidase | ∼1 kb upstream | Same | |
| GT8535 | Not in petals, stages 11 and 12e | At5g36710 | Expressed protein | Within | Opposite | |
| GT8607 | Not in petals, all stages | At4g32340 | Putative protein | Within | Opposite | |
| GT8883 | Not in petals, stages 8 to 13e | At1g01170 | Expressed protein | Within | Same | |
| GT9409 | Not in petals, stage 11 and one | At4g16890 | Disease resistance RPP5-like R gene | Within | Same | |
| GT9447 | Not in petals, stage 12 and one | At2g01100 | Expressed protein | Within | Same | |
| Stamen-specific expression | GT7833 | Mainly in anthers, stage 9 and on | At4g00110 | Nucleotide sugar epimerase family protein | ∼1.3 kb upstream | Same |
| GT7848 | Mainly in stomium, stages 7 to 11e | At2g34920 | E3 ligase-like protein | Within | Same | |
| GT7850 | Anther-specific, stage 7 and on | At2g32580 | Expressed protein | Within | Opposite | |
| GT8027 | Mainly in anthers, stage 8 and one | At4g05095 | Putative reverse transcriptase | ∼500 bp upstream | Same | |
| GT8102 | Tapetum-specific, stages 8 to 12 | At5g65870 | AtPSK5: phytosulfokine precursor 5 | ∼400 bp upstream | Opposite | |
| GT8113 | Endothecium-specific, stages 8 to 12 | At5g17800 | MYB family transcription factor MYB56 | 1 kb upstream | Same | |
| GT8115 | Anther-specific, stages 8 to 12e | At1g21000 | Zinc binding protein family protein | Downstream | Same | |
| GT8163 | Anther-specific, stages 8 to 11e | At2g01820 | Putative receptor-like protein kinase | Downstream | Same | |
| GT8335 | Anther-specific, stages 8 to 12 | At4g33355 | Protease inhibitor/lipid transfer protein | 50 bp upstream | Same | |
| GT8362 | Mainly in stamen filaments, stages 8 to 10e | At4g00430 | Probable plasma membrane intrinsic protein PIP1/TMP-C | Within | Same | |
| GT8430 | Anther-specific, stage 8 and on | At3g06430 | PPR-repeat containing protein | 20 bp upstream | Same | |
| GT8435 | Anther-specific, stages 8 to 12e | At5g66200 | Putative protein | Within | Same | |
| GT8473 | Tapetum-specific, stages 8 to 12 | At3g09780 | Putative protein kinase similar to Pto kinase interactor 1 | Downstream | Same | |
| GT8503 | Anther-specific, stages 8 to 14e | At3g11420 | Unknown protein | Within | Opposite | |
| GT8554 | Anther-specific, stages 7 to 11e | At5g65050 | MADS box containing transcription factor AGL31 | ∼2 kb upstream | Opposite | |
| GT8555 | Locule-specific, stages 8 to 12 | At3g10550 | Putative myotubularin | Within | Same | |
| GT8612 | Anther-specific, stage 10 and one | At1g32250 | Putative calmodulin | 100 bp upstream | Same | |
| GT8619 | Anther-specific, stages 9 to 12 | At4g36240 | GATA zinc-finger protein | ∼1.2 kb upstream | Same | |
| GT8623 | Anther-specific, stages 8 to 11 | At5g40960 | Putative protein | Downstream | Opposite | |
| GT8635 | Anther-specific, stage 10 and on | |||||
| GT8749 | Anther-specific, stages 8 to 12 | At3g60970 | ABC transporter family protein | ∼7 kb upstream | Same | |
| GT8761 | Anther-specific, stage 8 and on | At3g10116 | Hypothetical protein | ∼1.3 kb upstream | Opposite | |
| GT8771 | Anther-specific, stages 8 to 13 | |||||
| GT8838 | Anther-specific, stages 8 to 11 | At1g72360 | Putative EREBP/AP2-like transcription factor | 60 bp upstream | Same | |
| GT8854 | Anther-specific, stage 8 and on | At4g24740 | Protein kinase AFC2 | Within | Opposite | |
| GT8884 | Anther-specific, stage 8 and on | At5g57010 | Putative calmodulin binding protein | ∼400 bp upstream | Same | |
| GT8992 | Anther-specific, stages 8 to 12e | |||||
| GT9007 | Anther-specific, stages 8 to 12 | At3g11210 | GDSL-motif lipase/hydrolase family protein 3 | Within | Same | |
| GT9013 | Anther-specific, stages 8-12 | At5g66270 | Expressed protein | Within | Same | |
| GT9098 | Anther-specific, stage 8 and on | At5g26675 | FLAP endonuclease-like protein | ∼1.2 kb upstream | Opposite | |
| GT9099 | Anther-specific, stages 8 to 12 | At3g06433 | Pseudogene; hypothetical protein | 180 bp upstream | Same | |
| GT9197 | Anther-specific, stage 8 and on | At4g02110 | BRCT domain-containing protein | Within | Opposite | |
| GT9207 | Anther-specific, stages 8-12 | At2g10950 | BSD domain-containing protein | Within | Same | |
| GT9280 | Anther-specific, stage 8 and on | At3g15510 | NAC-domain/NAM family protein | Within | Same | |
| GT9315 | Filament-specific, stages 7 to 9e | At3g50870 | GATA3-like transcription factor HANABA TARANU | Within | Opposite | |
| GT9316 | Anther-specific, stage 8 and on | At3g12630 | AN1-like zinc-finger family protein | Within | Same | |
| GT9389 | Anther-specific, stages 8 to 14e | At4g03560 | Two-pore calcium channel TPC1 | Within | Same | |
| GT9551 | Anther-specific, stages 8 to 11 | At2g03580 | Hypothetical protein | Within | Opposite | |
| GT9606 | Mainly in anthers, stage 6 and one | At3g14230 | Putative EREBP/AP2-like transcription factor | Within | Same | |
| GT9639 | Anther-specific, stages 8 to 11 | At3g06680 | Ribosomal protein L29 | Downstream | Opposite | |
| GT9609 | Anther-specific, stages 8 to 12e | At1g76170 | Hypothetical protein | Downstream | Same | |
| GT9643 | Anther-specific, stages 8 to 12 | |||||
| Stamen-specific lack of expression | GT8763 | In vascular tissues of sepals, petals, and carpelse | At4g13260 | Flavin-containing monooxygenase YUCCA2 | ∼4 kb downstream | Opposite |
| GT9167 | Not expressed in anthers stages 7 to 11e | At2g02070 | Zinc-finger protein | Downstream | Same | |
| Petal- and stamen-specific expression | GT6545 | Petal- and stamen-specific, stages 9 to 12 | At5g10320 | Expressed protein | Within | Same |
| GT7991 | Petal- and stamen-specific, stages 12 and 13e | At3g47350 | Putative 11 β-hydroxysteroid dehydrogenase | ∼2.4 kb upstream | Opposite | |
| GT8007 | Petal- and stamen-specific, stages 8 to 11e | At3g10700 | Galactokinase-like protein | Within | Same | |
| GT8103 | Petal- and stamen-specific, stages 11 and 12e | At5g08180 | Nuclear high mobility protein 2-like protein | 150 bp upstream | Same | |
| GT8121 | In petals, stamens, and tip of sepals, stages 6 to 12e | At4g38620 | MYB family transcription factor | Downstream | Opposite | |
| GT8311 | Specific to petal and stamen vascular tissues | At5g01370 | Hypothetical protein | ∼600 bp upstream | Same | |
| GT8400 | Petal- and stamen-specific, stage 11 and on | At1g63140 | Putative O-methyltransferase 1 | ∼300 bp upstream | Same | |
| GT8472 | Petal- and stamen-specific, stages 9 to 11e | At3g12370 | 50S ribosomal protein L10-like protein | Within | Same | |
| GT8503 | Mainly in petals and stamens | At3g11420 | Unknown protein | Within | Opposite | |
| GT8517 | Mainly in petals and stamens, stage 11e | At2g43680 | SF16-like protein with calmodulin binding domains | Within | Same | |
| GT8869 | Petal- and stamen-specific, stages 9 and 10e | At4g00150 | Scarecrow-like transcription factor SCL6 | Downstream | Opposite | |
| GT8990 | Petal- and stamen-specific, stages 12 and 13e | At5g04200 | Metacaspase-like protein AMC9 | 90 bp upstream | Same | |
| Petal- and stamen-specific lack of expression | GT8066 | Not in petals or stamens, stage 10 and one | At1g54270 | Eukaryotic translation initiation factor | 150 bp upstream | Same |
| GT8096 | Not in petals or stamens, stage 10 and one | At2g01110 | Thylakoid membrane formation protein cpTatC | Within | Same | |
| GT8378 | Vascular-specific in sepals and carpels | At2g39700 | Putative expansin protein EXP4 | ∼3.5 kb upstream | Same | |
| GT8465 | Not in petals or stamens, stage 9 and one | At2g45200 | Putative cis-Golgi SNARE protein | Downstream | Opposite |
Genes whose endogenous expression patterns are likely to be reported by the GUS reporter are listed in bold.
Patterns of GUS expression in petals and stamens. Stages according to Smyth et al. (1990).
Annotated gene closest to the gene trap insertion.
Insertion location respective to the transcribed region of the annotated gene.
Orientation of GUS reporter gene relative to the annotated gene.
Different expression patterns observed at other stages.
Table 4.
Gene Trap Lines with Tissue-Specific Staining Patterns
| Expression Class | Gene Trap Line | GUS Staining Patterna | Annotationb | Description | Insertion Sitec | Insertion Orientationd |
|---|---|---|---|---|---|---|
| Epidermal-specific | GT7038 | Epidermal-specific in petals and stamense | At3g01500 | Carbonic anhydrase chloroplast precursor CN1 | Within | Same |
| GT7912 | Predominantly in abaxial epidermise | At5g57800 | Lipid transfer protein WAX2 | Within | Same | |
| GT8121 | Epidermal-specific in developing petals and stamense | At4g38620 | MYB family transcription factor | Downstream | Opposite | |
| GT8252 | Epidermal-specific in petals and stamense | At1g62440 | Protein similar to tomato disease resistance protein | Within | Opposite | |
| Anther cell type–specific | GT7848 | Mainly in stomiume | At2g34920 | Expressed protein | Within | Same |
| GT8102 | Tapetum-specific | At5g65870 | AtPSK5: phytosulfokine precursor 5 | ∼400 bp upstream | Opposite | |
| GT8113 | Endothecium and middle layer cell-specific | At5g17800 | MYB family transcription factor MYB56 | 1 kb upstream | Same | |
| GT8473 | Tapetum-specific | At3g09780 | Putative protein kinase similar to Pto kinase interactor 1 | Downstream | Same | |
| Vascular-specific | GT8311 | Vascular-specific in petals and stamens | At5g01370 | Hypothetical protein | ∼650 bp upstream | Same |
| GT8378 | Vascular-specific in sepals and carpelse | At2g39700 | Putative expansin protein EXP4 | 3.5 kb upstream | Same | |
| GT8450 | Vascular-specific in stamens and carpels at later stagese | At2g33860 | ARF3/ETT | 1 kb upstream | Opposite | |
| GT8763 | Vascular-specific in sepals, petals, and carpels; distal vascular-specific in mature organse | At4g13260 | Flavin monooxygenase YUCCA2 | 4.5 kb downstream | Opposite |
Genes whose endogenous expression patterns are likely to be reported by the GUS reporter are listed in bold.
Patterns of GUS expression in petals and stamens. Stages according to Smyth et al. (1990).
Annotated gene closest to the gene trap insertion.
Insertion location respective to the transcribed region of the annotated gene.
Orientation of GUS reporter gene relative to the annotated gene.
Also expressed in organs other than petals and/or stamens.
Table 5.
Gene Trap Lines Showing Abaxial or Adaxial Staining Patterns
| Expression Class | Gene Trap Line | GUS Staining Patterna | Annotationb | Description | Insertion Sitec | Insertion Orientationd |
|---|---|---|---|---|---|---|
| Abaxial | GT7885 | Largely abaxial | At2g01110 | Thylakoid membrane formation protein cpTatC | ∼300 bp upstream | Same |
| GT7912 | Predominantly in abaxial epidermis | At5g57800 | Lipid transfer protein WAX2 | Within | Same | |
| Adaxial | GT7969 | Adaxial in floral organ primordia; guard cell–specific expression in mature sepals | At3g09730 | Hypothetical protein | Within | Same |
| GT9206 | Adaxial in floral organ primordia at first two to four stages after organ initiation | At3g60390 | PHD-finger containing HD-ZIP protein HAT3 | Within | Same |
Patterns of GUS expression in petals and stamens. Stages according to Smyth et al. (1990).
Annotated gene closest to the gene trap insertion.
Insertion location respective to the transcribed region of the annotated gene.
Orientation of GUS reporter gene relative to the annotated gene.
Table 6.
Gene Trap Lines Expressed in Specific Proximodistal Domains
| Expression Class | Gene Trap Line | GUS Staining Patterna | Annotationb | Description | Insertion Sitec | Insertion Orientationd |
|---|---|---|---|---|---|---|
| Petal midregion | GT7299 | Strong in midregion of petal, stages 9 and 10 | At3g56220 | Expressed protein with actin interaction domain | Within | Same |
| GT8398 | Strong in midregion of petal, stages 9 and 10 | At3g12930 | IojAP-like protein | ∼1.2 kb upstream | Opposite | |
| Anther-filament junction | GT7953 | Tip of filament, stages 10 and 11 | At1g70510 | Class I KNOX transcription factor KNAT2 | Within | Same |
| GT8400 | Anther-filament junction, stage 11 and on | At1g63140 | Putative O-methyltransferase 1 | ∼300 bp upstream | Same | |
| GT8450 | Anther-filament junction or tip of filament, stage 13 and on | At2g33860 | ARF3/ETT | 1 kb upstream | Opposite | |
| GT8990 | Anther-filament junction, stage 12 and on | At5g04200 | Metacaspase-like protein AMC9 | 90 bp upstream | Same | |
| Anther-proximal or distal | GT7850 | Throughout anthers, stages 7 to 12; proximal region of anthers, stage 13 and on | At2g32580 | Expressed protein | Within | Opposite |
| GT7991 | Throughout anthers, stages 8 to 10; anther-proximal, stages 11 and 12; anther-distal, stage 13 and on | At3g47350 | Putative 11 β-hydroxysteroid dehydrogenase | ∼2.4 kb upstream | Opposite | |
| GT9280 | Throughout anthers, stages 8 to 12; distal region of anthers, stage 13 and on | At3g15510 | NAC-domain/NAM family protein | Within | Same | |
| GT9316 | Throughout anthers, stages 8 to 12; distal region of anthers, stage 13 and on | At3g12630 | AN1-like zinc-finger protein | Within | Same |
Genes whose endogenous expression patterns are likely to be reported by the GUS reporter are listed in bold.
Patterns of GUS expression in petals and stamens. Stages according to Smyth et al. (1990).
Annotated gene closest to the gene trap insertion.
Insertion location respective to the transcribed region of the annotated gene.
Orientation of GUS reporter gene relative to the annotated gene.
A primary screen was conducted, in which inflorescence tissue from each line was treated with two different GUS staining solutions containing either 0 or 2 mM ferri/ferrocyanide (FeCN). Two concentrations of FeCN were used to explore the range of staining patterns for each line, since FeCN prevents diffusion of the GUS stain, yet also interferes with GUS enzymatic activity (Springer, 2000). The stained tissues were cleared and dissected, and spatial patterns of GUS staining were examined for organ and tissue specificity in young (up to stage 9) and maturing (stage 9 and beyond) flowers (stages according to Smyth et al., 1990).
Approximately 300 of these 1765 lines showed staining in floral organs (see Supplemental Table 1 online), and it can be estimated that ∼50% of Arabidopsis genes are expressed to some extent in flowers, given that expression patterns of ∼600 genes were effectively examined in our screen. This estimate is based on the probability of the GUS gene being positioned in the same orientation as the target gene (50%) and the frequency at which these insertion events occur in transcribed regions (∼70%) (Parinov et al., 1999; Pan et al., 2005). Our estimate is similar to estimates from microarray-based surveys of gene expression, in which ∼65% of annotated genes were found to be expressed in floral tissues (Yamada et al., 2003). Although organ type–specific staining patterns were observed in flowers older than stage 9, no whorl-specific expression was found in young flowers in the primary screen, presumably due to overstaining and/or stain diffusion. Therefore, 144 lines showing either petal and/or stamen-specific staining or lack of staining in older flowers and/or any staining in young flowers were reevaluated in a secondary screen using more controlled conditions.
In the secondary screen, tissues were treated with and without 5 mM FeCN, and the staining reaction for each line was quenched when the GUS signal first became obvious. Dissected inflorescences were examined, and GUS staining patterns in stage 1 to 14 flowers were recorded at each stage. As expected, staining was generally more limited and specific in the treatment with FeCN; however, in some lines, staining was abolished in the presence of FeCN. Thus, patterns in both conditions were considered in evaluating each line, and the patterns we describe in this report reflect qualitative aspects of gene expression (Table 1). Table 2 summarizes the GUS staining patterns observed in four lines, in which the gene trap insertions were found in genes with known expression patterns. In these four cases, GUS staining patterns were essentially identical to the previously reported highly specific expression patterns of the corresponding genes (Figure 1), suggesting that the secondary screen conditions were successful in recapitulating endogenous gene expression patterns.
Table 1.
Frequency of Gene Trap Lines Showing Petal- and/or Stamen-Specific Staining Patterns
| GUS Staining Pattern | No. of Lines Showing the Staining Pattern | Estimated Percentage of Arabidopsis Genes with Similar Expression Patterna | ||
|---|---|---|---|---|
| Petal-specific | Expression | 2 | 0.3 | |
| Lack of expression | 9 | 1.5 | ||
| Stamen-specific | Expression | Filament-specific | 2 | 0.3 |
| Anther-specific | 39 | 6.5 | ||
| Total | 41 | 6.8 | ||
| Lack of expression | 2 | 0.3 | ||
| Petal- and stamen-specific | Expression | 12 | 2.0 | |
| Lack of expression | 4 | 0.6 | ||
Based on the estimate that ∼600 genes (2.5% of genes in the Arabidopsis genome) were examined in this screen.
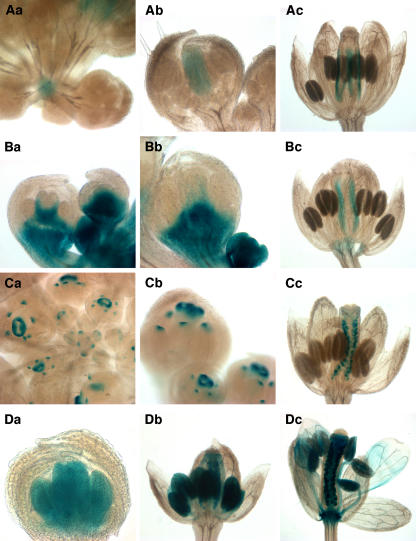
Figure 1.
Expression Patterns of Gene Trap Lines Tagging Genes with Known Expression Patterns.
(A) GT7847: GUS expression was observed in the inflorescence meristem and young floral buds (a) and in carpels at later stages (b and c).
(B) GT7953: GUS expression was detected throughout young flowers, but became excluded from the floral organ primordia (a and b). In older flowers, expression was predominantly in the replum (c).
(C) GT8137: GUS expression was evident in young flowers (a) and at the distal tips of the floral organs (a and b). In later stages, expression was localized to developing ovules (c).
(D) GT8686: GUS expression was detected in the petal, stamen, and carpel primordia at stage 7 (a) and at later stages (b and c).
Gene Trap Lines Showing Staining Patterns Specific to Petals and/or Stamens
Table 3 lists 71 gene trap lines that showed staining or lack of staining specific to petals and/or stamens. Genes expressed in such patterns are likely to function in pathways unique to petal and/or stamen development. As noted, some lines showed petal- and/or stamen-specific patterns exclusively, while others had dynamic staining patterns that change over developmental time and exhibited petal- and/or stamen-specific patterns only in some stages of differentiation.
We recovered relatively few lines that expressed GUS in a petal-specific manner. Only GT7921 and GT8132 had petal-specific staining patterns (Table 3, Figures 2A and 2B), suggesting that a small number of genes (estimated to be 0.3% of genes in the genome; see Table 1) are expressed exclusively in petals. This observation correlates with results from previous work. When an enhancer trap collection was examined for floral expression patterns in Arabidopsis, very few petal-specific lines were recovered (Campisi et al., 1999). Microarray-based genome surveys also identified a small number of putative petal-specific genes in Arabidopsis (estimated at 0.06% of the genome in Wellmer et al., 2004 and 0.2% in Zik and Irish, 2003), supporting the idea that the petal is a rather simple organ that requires relatively few specific gene functions.
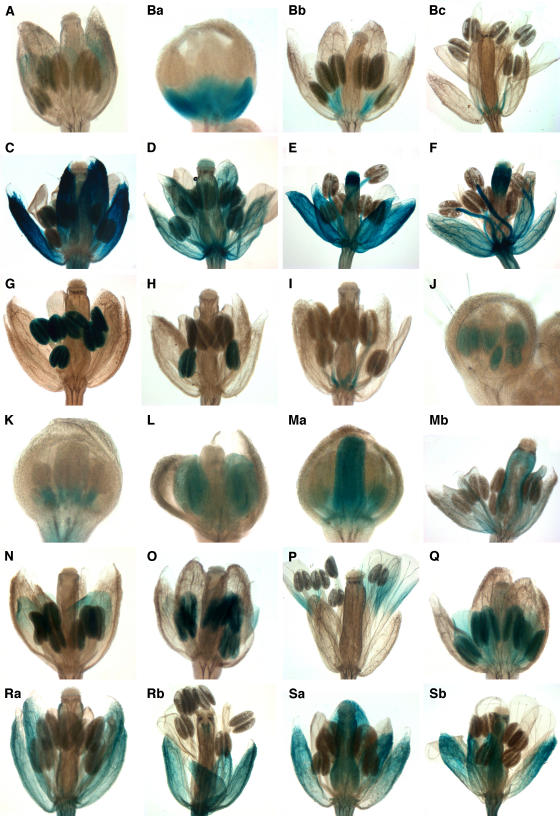
Figure 2.
Gene Trap Lines Showing Petal- and/or Stamen-Specific Expression Patterns.
(A) and (B) Petal-specific expression.
(A) GT7921 flower at stage 11.
(B) GT8132 flowers at stage 7 (a), stage 10 (b), and stage 14 (c).
(C) to (F) Petal-specific lack of expression.
(C) GT8454 flower at stage 12.
(D) GT8485 flower at stage 13.
(E) GT8883 flower at stage 11.
(F) GT9447 flower at stage 13.
(G) to (L) Stamen-specific expression.
(G) GT7833 flower at stage11, expression mainly in anthers.
(H) GT8335 flower with anther-specific expression at stage 11.
(I) GT8362 flower at stage 11 with expression confined to the filaments.
(J) GT8555 flower at stages 9 and 10.
(K) GT9315 flower at stage 8 with filament-specific expression.
(L) GT9551 anther specific expression at stage 8.
(M) Stamen-specific lack of expression. GT9167 flowers at stage 8 (a) and stage 11 (b).
(N) to (Q) Petal- and stamen-specific expression.
(N) GT6545 flower at stage 11.
(O) GT8007 flower at stage 11.
(P) GT8400 flower at stage 14.
(Q) GT8472 flower at stage 11.
(R) and (S) Petal- and stamen-specific lack of expression.
(R) GT8066 flowers at stage 11 (a) and stage 14 (b).
(S) GT8096 flowers at stage 11 (a) and stage 12 (b).
In GT8132, a gene trap insertion was found 50 bp upstream of the transcription start site of a gene encoding a putative TCP-class transcription factor. Functional characterization of this gene has not yet been conducted; however, it belongs to the PCF-like TCP gene family. PCF proteins were first isolated in rice (Oryza sativa) as trans-acting factors that bind to the promoter regions of PCNA, a marker of dividing cells, and are thought to regulate cell proliferation (Kosugi and Ohashi, 1997, 2002). GT8132 shows GUS expression shortly after petal initiation until maturity, and its expression becomes restricted to the claws at later stages of development.
Interestingly, our screen identified a larger number of lines that specifically lacked expression in petals, and ∼1.5% of genes in the genome are estimated to have such a pattern of expression (Tables 1 and 3, Figures 2C to 2F). This observation suggests that suppression may be a more common mode of gene regulation than activation in the petals. Genes that are specifically downregulated in petals include a disease resistance RPP5-like resistance (R) gene (GT9409). This R gene activates both salicylic acid–dependent and -independent disease resistance pathways, and constitutive activity of this gene causes dwarfing and curly leaf morphology (Stokes et al., 2002; Zhang et al., 2003). Salicylic acid has been postulated to inhibit cell growth by affecting ion uptake (Raskin, 1992), and specific downregulation of this gene may be important for petal growth control.
In contrast with petals, many genes are expressed specifically in the stamens (∼6% of genes in the genome), especially in the anthers (Tables 1 and 3, Figures 2G to 2J). Studies in several angiosperm species also resulted in the isolation of numerous anther-specific genes (Kamalay and Goldberg, 1984; Koltunow et al., 1990; Nacken et al., 1991; Scott et al., 1991; Rubinelli et al., 1998; Amagai et al., 2003; Zik and Irish, 2003; Wellmer et al., 2004). This likely reflects the fact that the anther is a highly specialized tissue system, containing a variety of cell types necessary for pollen generation and dispersal (Koltunow et al., 1990; Scott et al., 1991, 2004; Irish, 1999; Sanders et al., 1999).
Several transcription factors were identified as anther specific in this screen (Table 3) and are good candidates for regulators of anther differentiation. A NAC-domain containing transcription factor tagged in GT9280, At3g15510, had been identified as stamen specific (Wellmer et al., 2004). It is a close relative of the NAC-LIKE ACTIVATED BY AP3/PI gene (Ooka et al., 2003), for which decreased levels of expression result in defects in stamen elongation and anther dehiscence (Sablowski and Meyerowitz, 1998). Two AP2/EREBP-class transcription factors (GT8838 and GT9606) belong to the small subfamily of B-2 class ethylene response factor-like proteins in Arabidopsis (Sakuma et al., 2002), suggesting that ethylene plays a role in anther development.
Calcium signaling may be distinctively important in anther differentiation. We have identified three calcium signaling-related genes that showed an overlapping pattern of GUS staining in anthers (Table 3): a putative calmodulin (GT8612), a putative calmodulin binding protein (GT8884), and a calcium channel (GT9389). Previous work in tobacco (Nicotiana tabacum) has shown that a calcium/calmodulin-dependent protein kinase was also expressed specifically in differentiating anthers, and calcium oscillations have been postulated to regulate synchronization of anther differentiation (Poovaiah et al., 1999).
Only two lines, GT8362 and GT9315, showed filament-specific GUS expression patterns (Tables 1 and 3, Figures 2I and 2K). This low number of specific genes (∼0.3% of genes in the genome) probably reflects the general functional roles of the filaments, which are to transport water and nutrients to the anthers and elongate prior to pollination (Goldberg et al., 1993; Scott et al., 1991, 2004). A plasma membrane intrinsic protein (PIP) is expressed in the filament during the rapid elongation phase before anthesis (GT8362; Figure 2I). The gene, TMP-C/PIP1e/PIP1;4, belongs to the PIP1 class of the aquaporin gene family (Weig et al., 1997; Johansson et al., 2000; Johanson et al., 2001). Aquaporins are passive water channels and often function in cell enlargement; thus, PIP1;4 may be involved in the elongation of filament cells.
Several lines showed staining restricted to both petals and stamens (Figures 2N to 2Q) or to sepals and carpels (Figures 2R and 2S). The trapped genes in these lines are likely to function in pathways shared in petal and stamen differentiation. One common characteristic of petals and stamens in Arabidopsis is that they are the only aerial organs that are largely nonphotosynthetic. Thus, genes involved in photosynthetic pathways are expected to be downregulated in these organs. Consistent with this assumption, expression of the gene encoding chloroplast TatC protein, which translocates proteins to the chloroplast and is necessary for chloroplast function (Motohashi et al., 2001), was largely absent in petals and stamens (GT8096; Table 3, Figure 2S).
Gene Trap Lines Marking Various Tissue Types in Petals and/or Stamens
Some gene trap lines showed expression only in certain tissues within the petals and/or stamens (Table 4, Figures 3A to 3J). Some of these lines showed GUS activity in other organs as well, while others were largely specific to petals and stamens (Table 3).
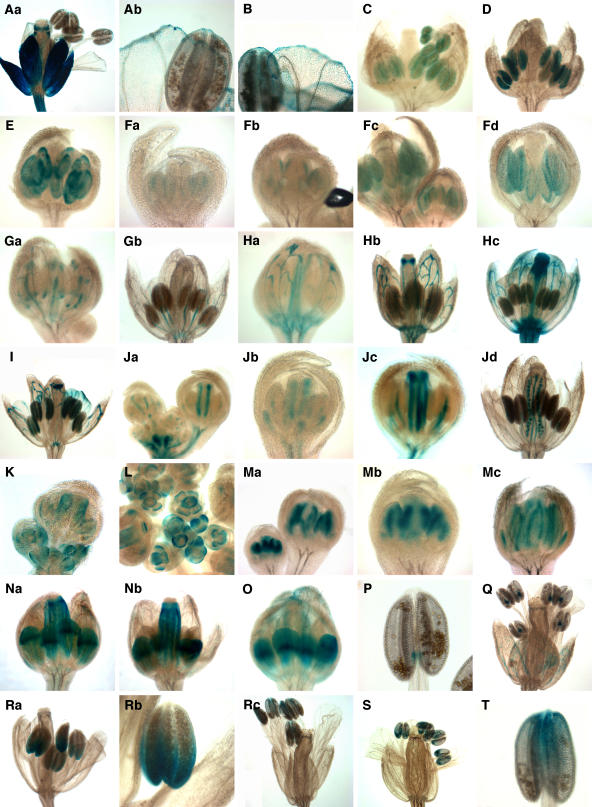
Figure 3.
Tissue- or Regional-Specific GUS Expression Patterns.
(A) and (B) Epidermal-specific lines.
(A) GT7038 flower at stage 13 (a) and stage 13 petals and stamens (b).
(B) GT8252 flower at stage 13 petals and stamens.
(C) to (F) Anther cell type–specific lines.
(C) GT8102 flower at stage 9 showing tapetum expression.
(D) GT8473 flower at stage 10 with tapetum expression.
(E) GT8113 flower at stage 9 with endothecium and middle layer cell expression.
(F) GT7848 flowers at stage 7 (a), stage 8 (b), stage 9 (c), and stage 10 (d) showing stomium-specific expression.
(G) to (J) Vascular-specific lines.
(G) GT8311 flowers with petal and stamen vascular expression at stages 9 (a) and 11 (b).
(H) GT8378 flowers with sepal and carpel vascular-specific expression at stage 9 (a), stage 10 (b), and stage 11 (c).
(I) GT8763 flowers at stage 11 with distal vascular expression in sepals, petals, and carpels.
(J) GT8450 inflorescence meristem and cluster of stages 1 to 7 flowers (a) and individual flowers at stage 7 (b), stage 9 (c), and stage 11 (d).
(K) to (M) Abaxial/adaxial-specific lines.
(K) Abaxial expression in GT7885 flowers from stages 1 to 10.
(L) GT7912 inflorescence showing flowers with abaxial expression at stages 4 to 10.
(M) Adaxial expression in GT9206 flowers at stages 6 and 8 (a), stage 7 (b), and stage 10 (c).
(N) to (T) Lines marking distinct proximodistal domains.
(N) GT7299 flowers with stronger expression in midregion of petal at stage 10 (a) and stage 11 (b).
(O) GT8398 flower with stronger expression in midregion of petal at stage 10.
(P) Expression at junction of anther and filament in GT8400 stage 13 stamen.
(Q) GT8990 flower with expression in connectives at stage 13.
(R) GT7991 stage 11 flower (a) and anther showing GUS expression in the proximal region (b) and stage 14 flower with expression in the distal region of the anthers (c).
(S) GT7850 stage 13 flower showing expression in the proximal region of the anthers.
(T) GT9280 stage 14 anther with expression in the distal region.
The epidermis of the petals and stamens was specifically stained in four lines (Table 4). GT7038, in which a carbonic anhydrase CN1 is trapped, showed epidermal-specific GUS staining in the petals and stamens, although broader staining patterns were observed in sepals and carpels (Figure 3A). CN1 is thought to be transported into chloroplasts and hydrates carbon oxide into bicarbonate, stabilizing carbon availability to ribulose-1,5-bis-phosphate carboxylase/oxygenase (Fett and Coleman, 1994). Its roles in photosynthetic carbon fixation are consistent with the more widespread and stronger staining in sepals and carpels. Given the absence of chloroplasts in petal lamina and stamen anther epidermis, it is unclear as to the role of CN1 in these tissues.
Four lines marking specific cell types in the anther were recovered (Table 4). In GT8102 and GT8473, GUS staining was limited to the tapetum cells (Figures 3C and 3D), while GUS staining in GT8113 was restricted to the endothecium and perhaps also the middle layer cells (Figure 3E). GT7848 showed GUS staining patterns exclusively in areas where the stomium forms and differentiates (Figure 3F). Since expression was detectable at stage 7, before stomium cells are apparent (Sanders et al., 1999), the gene trapped in this line, encoding an E3 ligase-like protein, may play roles in stomium specification as well as differentiation.
GUS staining was confined to vascular tissues in four lines (Table 4). Interestingly, these lines exhibited different organ specificities. GT8311 showed vascular staining in petals and stamens (Figure 3G), whereas staining was observed only in sepals and carpels in GT8378 (Figure 3H). In GT8763, vascular tissues in all floral organs but stamens were stained (Figure 3I). GT8450 has GUS activity in flower buds and in all floral organ primordia, but expression persisted only in stamen and carpel vasculature (Figure 3J). The insert in GT8450 is in the promoter of the ETTIN (ETT) gene, and the GUS expression pattern recapitulates vascular aspects of the expression pattern of the endogenous ETT gene (Sessions et al., 1997). Together, these staining patterns suggest that vascular differentiation and/or function are to some extent distinctive in the different floral organs, although there has been little documentation of such dissimilarities. Vascular patterns vary among the different floral organs (Sessions and Zambryski, 1995; Christensen et al., 2000; Semiarti et al., 2001); thus, vascular patterning genes perhaps are differentially expressed depending on the organ type. In line with this, ETT has been shown to specifically pattern carpel vasculature (Sessions and Zambryski, 1995; Sessions et al., 1997).
Gene Trap Lines Demarcating Abaxial-Adaxial and Proximodistal Domains
Abaxial/adaxial-specific staining was observed in four lines (Table 5). GT7885 and GT7912 showed a stronger GUS signal on the abaxial side of all floral organs (Figures 3K and 3L). The gene trap insertion in GT7912 was located in the WAX2 gene, which is homologous to the maize Glossy1 gene (Chen et al., 2003). The GUS staining pattern, which is epidermal in addition to being abaxial, correlates with the gene's function in cuticle differentiation. WAX2 is important for the maintenance of organ separation on the abaxial side of floral organs, since its loss-of-function mutant phenotype in flowers consists of postgenital organ fusion along the abaxial side (Chen et al., 2003).
Conversely, GT7969 and GT9206 exhibited adaxial-specific staining (Table 5, Figure 3M). The corresponding insertions in these lines were located within the transcriptional units of a hypothetical protein and a PHD finger-containing HD-ZIP gene HAT3, respectively. HAT3 has been reported to be the most abundantly expressed in roots, and only low levels of expression were detected in floral tissues (Schindler et al., 1993). GUS staining in GT9206 was highly specific and only detected on the adaxial side of floral primordia during a few stages subsequent to initiation of the organs. The adaxial GUS expression in the stamens became further restricted to the locules as the anthers progressed through differentiation (Figure 3Mc).
More commonly, asymmetrical staining patterns along the proximodistal axis were recovered (Table 6, Figures 2 and 3). GUS staining in differentiating petals in most lines was generally restricted to either blade or claw (for example, Figures 2A, 2B, and 2N to 2P). Two lines, GT7299 and GT8398, showed particularly strong GUS expression in the midregion of petals, starting at stage 9, when the proximal and distal parts of the petals first become distinct (Figures 3N and 3O; Smyth et al., 1990). This region corresponds to the border between the blade and claw and is the area where bending occurs when petals mature. Genes expressed in this middle region could play roles in establishing boundaries between distal and proximal halves of petals; alternatively, such genes may act in cellular events leading to petal bending. The gene trapped in GT7299 encodes a protein with an actin-interacting domain and perhaps is involved in cytoskeleton changes responsible for organ curvature. Cytoskeletal rearrangements have been shown to be important for bending of other plant organs (Nick et al., 1990; Fischer and Schopfer, 1998).
Asymmetric staining was also frequently observed along the proximodistal axis in stamens (Tables 1 and 6). GUS staining in stamens was usually restricted either to the anther or the filament (for example, Figures 2C to 2Q and 3C to 3F). In addition, four lines exhibited staining restricted to the connective at the junction of the filament and anther (Figures 3P and 3Q). The connective is rich in transmitting tissues for efficient nutrient transport from the single vascular strand in the filament to the anthers (Sanders et al., 1999). After dehiscence at stage 12, anthers start to senesce, and connective cells degenerate at stage 13 (Sanders et al., 1999). GT8990 has a gene trap insert just before the transcription initiation site of a gene encoding a caspase-like protein (Table 3), which likely functions in triggering programmed cell death (Watanabe and Lam, 2004, 2005). In GT8860 flowers after pollination, strong GUS activity was observed in the connectives, which could reflect the programmed onset of degeneration (Figure 3Q).
The anthers themselves also displayed distinct proximodistal domains of gene expression. Four lines had GUS staining patterns restricted to the apical or basal half of the mature anthers (Table 6, Figures 3R to 3T). In GT7991 flowers, GUS staining in anthers shifted from the proximal half to the distal half as anthers differentiated (Figure 3R). Although such gene expression patterns suggest that the apical and basal parts of the anther are distinctive, little is known about differences between the two regions within the anther.
Gene Trap Lines Exhibiting Visible Phenotypes
Gene trapping is not only a means to identify patterns of gene expression but also a tool to generate mutations by disrupting gene function. In addition to assessing the reporter expression patterns, we scored the 1765 gene trap lines for visible mutant phenotypes in flowers (Table 7).
Table 7.
Floral Mutants Recovered in Phenotypic Screen
| Gene Trap Line | Annotationa | Description | Insertion Siteb | Insertion Orientationc | GUS Staining | Homozygous Mutant Phenotype |
|---|---|---|---|---|---|---|
| GT7847 | At5g60910 | MADS transcription factor FRUITFULL | Within | Same | See Table 2 | Short, indehiscent carpels |
| GT7912 | At5g57800 | Lipid transfer protein WAX2 | Within | Same | See Tables 4 and 5 | Shiny, brighter stem and silique surface; shorter internodes in inflorescences; smaller petals |
| GT8096 | At2g01110 | Thylakoid membrane formation protein cpTatC | Within | Same | See Table 3 | Albino seedlings; seedling lethal |
| GT8686 | At1g24260 | MADS transcription factor SEPALLATA3/AGL9 | Within | Same | See Table 2 | Slower growing greener petals that eventually grow larger than the wild type; petals have stomata; smaller, slower developing silique |
| GT8860 | At5g35770 | STERILE APETALA | Within | Opposite | No | Stunted inflorescences; small floral organs; carpelloid sepals; smaller, narrower petals; underdeveloped stamens |
| GT9315 | At3g50870 | GATA3-like transcription factor HANABA TARANU | Within | Opposite | See Table 3 | Fewer petals and stamens, carpelloid sepals; sterile gynoecium |
| GT9356 | At2g25210 | 60S ribosomal protein L39 | Within | Opposite | No | Rough deeper green leaves; excess branching; inflorescence terminates early with a mass of flower-like structures; phenotype less severe in later arising flowers |
| GT9411 | At1g16380 | Putative cation/hydrogen exchanger CHX1 | Within | Opposite | No | Dwarfed, paler green plants; smaller flowers missing some organs, especially in the 2nd and 3rd whorls; bent carpels; sterile |
Genes whose endogenous expression patterns are likely to be reported by the GUS reporter are listed in bold.
Patterns of GUS expression in petals and stamens. Stages according to Smyth et al. (1990).
Annotated gene closest to the gene trap insertion.
Insertion location respective to the transcribed region of the annotated gene.
Insertions in GT7847, GT8096, GT8860, and GT8686 were found in the previously characterized genes FRUITFULL, cpTatC, STERILE APETALA (SAP), and SEPALLATA3 (SEP3), respectively, and lines homozygous for the gene trap insertions exhibited mutant phenotypes identical to those reported in the literature (Table 7, Figures 4B and 4D to 4F) (Gu et al., 1998; Byzova et al., 1999; Motohashi et al., 1999, 2001; Budziszewski et al., 2001; Pelaz et al., 2001). The gene trap insertions in these lines were located inside transcriptional units of the corresponding genes and probably completely impaired the function of the gene products.
Figure 4.
Phenotypes of Mutants Recovered in This Screen.
(A) to (C) and (E) to (I) An overview of inflorescence is shown in the left panels and ethanol-cleared flower in right panels.
(A) Wild type (Landsberg erecta).
(B) GT7847/ful shows stunted carpels with a short nondehiscent silique.
(C) GT7912/wax2 flowers have a shinier stem and carpel surface.
(D) GT8096/cpTatC: phenotypically normal heterozygous (left) and albino homozygous (right) seedlings.
(E) GT8686/sep3 flowers have short siliques and wider and longer petals.
(F) GT8860/sap mutants display compact flowers lacking most of the petals and stamens.
(G) GT9315/han flowers are generally sterile and lack most petals and stamens.
(H) GT9356 disrupts a gene encoding a 60S ribosomal protein L39 and displays inflorescences with flower-like structures (early arising flowers shown).
(I) GT9411 has an insertion in the CHX1 gene resulting in a dwarf, pale green inflorescence with flowers containing deformed and fewer floral organs.
On the other hand, GT7912 and GT9315, disrupting the WAX2 and HANABA TARANU (HAN) genes, respectively, exhibited weaker phenotypes than corresponding loss-of-function mutations. GT7912 plants have deeper green, shinier organs (especially carpels), as well as smaller siliques, but do not show any postgenital organ fusion as seen in wax2 knockout mutants (Figure 4C) (Chen et al., 2003). Since the gene trap insertion is in the last exon of WAX2, the WAX2-GUS chimeric proteins probably retained some native function in GT7912. Severe han flowers are sterile and have fused sepals and fewer organs in all whorls (Zhao et al., 2004); by contrast, GT9315 flowers are weakly fertile and mainly lack petals and stamens (Figure 4G), and fused sepals with stigmatic tissues were observed only on later arising flowers. Since the GUS gene in GT9315 is inserted in the end of the last exon of HAN in the opposite orientation, HAN transcripts are presumably truncated but still retain some protein function.
Two previously uncharacterized genes were trapped in GT9356 and GT9411, which display pleiotropic mutant phenotypes. GT9356, which had an insert in the gene encoding a 60S ribosomal protein L39, exhibited rough surfaces, twisted leaves, and excess branching. The inflorescences terminated early after giving rise to a disorganized mass of flower-like structures (Figure 4H). This GT9356 phenotype was ameliorated in later arising flowers that formed more normal floral organs and were slightly fertile. GT9411 plants contained a gene trap insertion in a cation/proton antiporter gene, were dwarf and pale green, and made curly leaves. Their flowers had smaller, deformed floral organs, especially in the second and third whorls, and were largely sterile (Figure 4I).
DISCUSSION
Gene Trap Lines Define Axial and Tissue-Specific Domains in Petals and Stamens
By assaying for reporter gene expression in 1765 gene trap lines, expression patterns of ∼600 genes were examined in this study. We have recovered 80 gene trap lines showing GUS staining patterns with various organ or suborgan specificities in the petals and stamens. Among these, 71 lines had GUS expression upregulated or downregulated specifically in petals and stamens (Table 3). Genes with such expression patterns probably play roles in petal- and stamen-specific differentiation and are good candidates for being targets of APETALA3 (AP3) or PISTILLATA (PI), floral homeotic genes that specify petal and stamen identities (Bowman et al., 1989; Hill and Lord, 1989; Jack et al., 1992; Goto and Meyerowitz, 1994). In contrast with AP3 and PI, both of which are expressed throughout the petals and stamens until late stages of development, the expression patterns of these candidate target genes are more dynamic and restricted to small suborgan domains at specific stages of development (Table 3, Figure 2). Thus, genes acting in petal- and/or stamen-specific differentiation are likely to be regulated by factors restricting their spatial and temporal domains of expression, in addition to the floral homeotic gene products.
One common subdomain of expression that we recovered was specific expression in petal and stamen epidermis. Epidermal specification depends on two HD-GL2–type homeodomain-containing transcription factors that are expressed in the L1 layer of the shoot apical meristem and the L1 progenitor cells in differentiating epidermis (Lu et al., 1996; Abe et al., 2003). These transcription factors bind to the L1-box, a promoter element that drives epidermal expression, and are considered as the key regulators of epidermal patterning (Abe et al., 2001, 2003). The genes annotated in the four gene trap lines marking the petal and stamen epidermis (Table 4) contain at least one L1-box in the promoter, intron, or exon (data not shown); thus, they could be directly regulated by these HD-GL2–type transcription factors.
We also recovered four lines expressed in an abaxial/adaxial-specific fashion. In Arabidopsis lateral organs, adaxial regulators belonging to the class III HD-ZIP transcription factors are expressed in the adaxial domains (McConnell and Barton, 1998; Zhong and Ye, 1999; McConnell et al., 2001), while abaxial fate is promoted by the abaxially expressed YABBY zinc-finger proteins and KANADI GARP-type transcription factors (Eshed et al., 1999; Siegfried et al., 1999; Kerstetter et al., 2001). The counteracting effects of these regulators specify and maintain the abaxial and adaxial domains within lateral organs, and such specification is thought to be necessary for lateral organ outgrowth (Eshed et al., 2001; Kumaran et al., 2002). However, how these transcriptional domains are translated into cell type–specific abaxial and adaxial patterns of differentiation are unknown. Adaxially or abaxially expressed genes, such as the WAX2 and HAT3 genes (this work; Kidner and Martienssen, 2004), are good candidates for being such downstream effectors of the axial polarity (Table 6). The relative paucity of such abaxial-adaxial restricted and organ-specific patterns of expression recovered in our screen, despite the abundant developmental asymmetry along this axis, could reflect the fact that abaxial-adaxial polarity may be a relatively general patterning process common to all lateral organs. Consistent with this view, all asymmetric expression patterns we observed along the abaxial-adaxial axis did not show any organ specificity (Table 5, Figures 3K to 3M).
To our surprise, domains of gene expression in the petals and stamens were far more commonly restricted along the proximodistal axis, suggesting the central role of this developmental axis in patterning domains of gene regulation. GUS staining in the petals and stamens was usually confined either to the distal or proximal parts, blade and claw in the petals, and anther and filament in the stamens, respectively. We also recovered two lines, GT7299 and GT8398, that displayed particularly strong expression in the midregion of petals (Figures 3N and 3O). Similarly, GUS reporter activity in stamens in some lines was to the connectives at the junction of the anther and filament (Figures 3P and 3Q).
Together, these observations suggest that patterning along the proximodistal axis of these lateral organs depends on subdivision into three regions. Likewise, Arabidopsis carpels consist of three proximodistal domains: stigma and style (distal), ovary (middle), and gynophore (proximal) (Dinneny and Yanofsky, 2005). Auxin has been suggested to act in establishing these axial domains; when auxin flow was disrupted, the ovary domain was shortened (Bennett et al., 1995; Nemhauser et al., 2000). ETT and SPATULA (SPT) genes are thought to translate axial information into morphological domains. In carpel primordia, ETT is expressed in the presumptive ovary and establishes the middle domain, while in later stages of carpel development, it is expressed mainly in vasculature and functions in ovary differentiation (GT8450; Table 4, Figure 3J) (Sessions et al., 1997; Nemhauser et al., 2000). ETT encodes an auxin response factor and functions largely via repression of SPT, which is expressed in the distal region and promotes distal tissue formation (Sessions et al., 1997; Alvarez and Smyth, 1999; Nemhauser et al., 2000; Heisler et al., 2001).
The plethora of proximodistal gene expression patterns perhaps emanates from the importance of the proximodistal axis for cellular differentiation not only spatially but also temporally. Analysis of maize (Zea mays) leaves and mutations altered in establishment of three domains along the proximodistal axis showed that proximal cells differentiate more slowly than distal regions and that this differential developmental schedule is important for setting domain boundaries (Freeling, 1992; Muehlbauer et al., 1997). Ectopic expression of class I KNOX genes results in proximal differentiation in the distal areas of leaves (Smith et al., 1992; Schneeberger et al., 1995; Chuck et al., 1996; Hareven et al., 1996; Chen et al., 1997). These leaf phenotypes can be interpreted as delayed maturation of distal cells by expression of KNOX genes, which are normally expressed in meristems and prevent differentiation, causing such cells to acquire proximal leaf characteristics (Freeling, 1992). Consistent with this idea, we generally observed distal-specific expression in somewhat earlier stages (starting at stage 7) than proximal-specific expression (starting at stage 9) (for example, Table 3, Figure 2).
A few regulators of proximodistal patterning have been identified in Arabidopsis petals and stamens. A YABBY-class transcription factor FILAMENTOUS FLOWERS is expressed in the distal tip and abaxial side of stamen primordia and is required for normal anther differentiation (Komaki et al., 1988; Sawa et al., 1999; Siegfried et al., 1999). We have recovered three anther-specific putative transcription factors, which are good candidate regulators of distal differentiation in the stamens (Table 3). In petals, the distally expressed JAGGED (JAG) gene, encoding a putative transcription factor containing C2H2-type zinc-finger, has been shown to regulate later-stage cell divisions in the blade and is a good candidate for regulating blade-specific genes (Dinneny et al., 2004). The BLADE ON PETIOLE1 and 2 genes encoding putative transcriptional cofactors were recently shown to downregulate JAG expression in the proximal regions of floral organs (Hepworth et al., 2005; Norberg et al., 2005). We have identified a TCP-class transcription factor that is expressed in the claw region of the petals (GT8132; Table 3), which also may be important in controlling proximal fates in these organs.
Isolation of Candidate Genes Involved in Petal and Stamen Organogenesis
We identified genes with distinct expression patterns in the petals and stamens, which are likely to act in organ- or tissue-specific differentiation pathways. Comparison of our findings to previous large-scale screens for genes likely involved in petal or stamen development highlights the complementary nature of our gene trap strategy and microarray-based methods (see Supplemental Table 3 online).
The stamen-specific genes annotated in GT8102, GT8612, and GT9280 had been recovered by microarray screen for genes that are involved in stamen identity specification or that are expressed predominantly in late-stage stamens (Wellmer et al., 2004; Schmid et al., 2005). We also identified similar classes of genes to those that have been recovered in differential display or microarray analyses aimed at identifying stamen-specific/enriched gene functions, such as genes encoding lipid transfer proteins, GDSL-type lipases, and calcium signaling proteins (Koltunow et al., 1990; Amagai et al., 2003; Zik and Irish, 2003; Wellmer et al., 2004).
We have recovered a higher proportion (12% = 4/33 genes in bold in Table 3; also see Supplemental Table 3 online) of transcriptional regulators compared with previous screens for genes preferentially expressed in the petals and/or stamens (0 to 5.5%) (Amagai et al., 2003; Zik and Irish, 2003; Bey et al., 2004; Wellmer et al., 2004). Since a gene trap screen examines gene expression qualitatively (i.e., patterns), rather than quantitatively (i.e., levels), this strategy likely preferentially recovers genes that tend to be expressed in spatiotemporally restricted patterns, such as transcription factors (Birnbaum et al., 2003; Gong et al., 2004; Gray et al., 2004). This qualitative nature of gene trap screening is also effective in identification of dynamically expressed genes, whose expression is critical for identity-specific organ development (Lee et al., 1997; Samach et al., 1999; Durfee et al., 2003; Laufs et al., 2003).
The differences in the sets of genes obtained by various microarray studies and our gene trap screen (see Supplemental Table 3 online) likely reflect the methods used for identifying such genes. Microarray analyses measure mRNA expression levels, while the expression signals in gene trapping come from reporter fusion proteins, which can reflect translational or posttranslational regulation of gene expression patterns. Furthermore, microarray analyses generally depend on amplifying and averaging signal intensities across tissues, organs, or stages, which can serve to mask expression level differences. The direct visual assessment of reporter gene expression in our gene trap screen provides a sensitive means of detecting such subtle stage- or tissue-specific differences.
Identification of Gene Functions Required for Petal and Stamen Development
The gene trap strategy can also serve as an effective tool to analyze gene function, since gene trap insertions can disrupt genes and result in a mutant phenotype. Among the 1765 gene trap lines, we recovered eight lines with visible mutant phenotypes in the flowers. These included several genes that have previously been identified by forward mutagenesis approaches, such as FUL (Figure 4B), SAP (Figure 4F), and HAN (Figure 4G), all of which are required for petal and stamen development. Mutants with pleiotropic defects were also recovered in our forward genetic screen. Among other phenotypes, GT9356 and GT9411 develop abnormal petals and stamens; thus, the corresponding genes are necessary for proper petal and stamen differentiation (Table 7, Figures 4H and 4I).
Gene trapping can also be used as a reverse genetic approach, which can overcome the difficulty of identifying subtle, pleiotropic, or lethal phenotypes. For instance, we recovered the cpTatC gene (GT8990) based on its downregulated expression in the petals and stamens (Table 3); homozygous mutants produce a seedling lethal phenotype that has been described previously (Motohashi et al., 2001). Furthermore, this strategy is particularly advantageous for characterization of essential genes, since it often results in partial protein function, as seen in GT7912 and GT9316 (Figures 4C and 4G), resulting in weak alleles. By examining segregating populations, we identified the subtle sep3 phenotype of GT8686, in which the petals grow larger (Figure 4E) and contain stomata; similar phenotypes were described in a reverse genetic characterization of SEP3 (Pelaz et al., 2001). The strengths of reverse genetic approaches are particularly useful in the identification of mutants impaired in organ differentiation, since differentiation genes likely play multiple roles in a variety of developmental processes (thus leading to lethality or pleiotropy) or affect only specific sets of specialized cells (thus leading to subtle phenotypes). The results reported here demonstrate the utility of such an approach, and continued analyses of the genes identified here should provide further insight into the mechanisms regulating organ-specific differentiation.
METHODS
Gene Trap Screen
The 1765 gene trap lines (see Supplemental Table 1 online) were screened in a primary screen. Twenty to forty segregating plants per line (F3 or F4 generation) were grown on soil, 20 to 30 inflorescences per line were harvested, and GUS activity was assayed in whole mounts. Tissues were vacuum infiltrated for 10 min and incubated at 37°C for 48 to 58 h in a GUS staining solution containing 10 mM sodium phosphate buffer, pH 7, 10 mM EDTA, pH 8, 0.1% Triton X-100, 100 μg/mL chloroamphenicol, 0.5 mg/mL X-glucuronic acid, and 0 or 2 mM FeCN. After termination of staining, tissues were cleared with 70% ethanol. Stained inflorescences were dissected, mounted in 50 to 70% glycerol, and examined using a Zeiss Axiophot microscope. Staining patterns were recorded as weak, medium, or strong for each organ type and major tissue types.
The 144 lines (see Supplemental Table 1 online) were chosen based on the staining patterns observed in the primary screen and reassessed in a more controlled secondary screen. Each line was grown under kanamycin selection (50 μg/mL), and 10 plants per line were transferred to soil. Twenty to forty inflorescences per line were harvested and assayed for GUS activity. The GUS staining solution used in the secondary screen varied from the one used in the primary screen in the concentrations of X-glucuronic acid (2 mg/mL) and concentration of FeCN (0 or 5 mM). Tissues were incubated at 37°C and examined every 4 to 8 h, and staining was terminated when strong staining was detected. Staining duration ranged from 12 to 48 h. Dissected whole-mount flowers were examined, and staining patterns were recorded graphically for each stage of flower development (Smyth et al., 1990). Images were captured using an AxioVision digital camera (Zeiss) and assembled with Adobe Photoshop (Adobe Systems).
Gene Trap Insertion Site Identification
The flanking DNA sequences of gene trap insertions were first identified by thermal asymmetric interlaced PCR (Liu and Whittier, 1995; Tsugeki et al., 1996) following the protocol described at http://genetrap.cshl.org/traps.html. For some gene trap lines, we modified the thermal asymmetric interlaced PCR protocol or used a suppression PCR method instead (Siebert et al., 1995; Balzergue et al., 2001). Multiple PCR reactions were performed with a primer series nested inside the gene trap insertion: Ds5-1, Ds5-3, and Ds5-4 for the 5′ end, Ds3-1, Ds3-2, and Ds3-4 for the 3′ end, and degenerate AD1, AD2, AD3, and AD5 primers were used (Liu and Whittier, 1995; Tsugeki et al., 1996; http://genetrap.cshl.org/traps.html). The PCR reactions were performed basically as described (http://genetrap.cshl.org/traps.html; Balzergue et al., 2001); however, the annealing temperatures were changed to 62°C and 60°C in reactions with Ds5-1/Ds3-1 and Ds5-3/Ds3-2, respectively. Specific PCR products were identified on agarose gels and sequenced after cloning into the pCR-TOPO vector (Invitrogen) using TOPO-FW (5′-GTGTGATGGATATCTGCAG-3′) or TOPO-RV (5′-CTCGGATCCACTAGTAAC-3′) primers. We confirmed these insertion sites either by multiple independent cloning of the same site or PCR with a gene-specific primer. The corresponding Arabidopsis thaliana genomic region was identified using BLAST software. The gene trap lines are available on request at http://genetrap.cshl.org.
Accession Numbers
Arabidopsis Genome Initiative locus identification numbers are provided in Tables 2 to 7. All flanking sequences have been deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/), and the accession numbers for flanking sequences and gene-specific primers are listed in Supplemental Table 2 online.
Supplementary Material
Acknowledgments
We thank R. Shen, Q. Tang, and U. Ramu for generating the gene trap lines, the Cold Spring Harbor Genetrap Consortium, as well as individual users from the research community for supporting the Trapper gene trap collection. We thank Tim Mulligan for plant care and the Gene Trap crew and E. Chae for assistance in planting and harvesting during the primary screen. We also thank our colleagues for helpful comments on the manuscript. This work was supported by a grant from the National Science Foundation (NSF-IBN 0212222) to V.F.I.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Robert Martienssen (martiens@cshl.org) and Vivian F. Irish (vivian.irish@yale.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.033985.
References
- Abe, M., Katsumata, H., Komeda, Y., and Takahashi, T. (2003). Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130, 635–643. [DOI] [PubMed] [Google Scholar]
- Abe, M., Takahashi, T., and Komeda, Y. (2001). Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 26, 487–494. [DOI] [PubMed] [Google Scholar]
- Almeida, J., Rocheta, M., and Galego, L. (1997). Genetic control of flower shape in Antirrhinum majus. Development 124, 1387–1392. [DOI] [PubMed] [Google Scholar]
- Alvarez, J., and Smyth, D.R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126, 2377–2386. [DOI] [PubMed] [Google Scholar]
- Amagai, M., Ariizumi, T., Endo, M., Hatakeyama, K., Kuwata, C., Shibata, D., Toriyama, K., and Watanabe, M. (2003). Identification of anther-specific genes in a cruciferous model plant, Arabidopsis thaliana, by using a combination of Arabidopsis macroarray and mRNA derived from Brassica oleracea. Sex. Plant Reprod. 15, 213–220. [Google Scholar]
- Balzergue, S., et al. (2001). Improved PCR-walking for large-scale isolation of plant T-DNA borders. Biotechniques 30, 496–498, 502, 504. [DOI] [PubMed] [Google Scholar]
- Bellen, H.J. (1999). Ten years of enhancer detection: Lessons from the fly. Plant Cell 11, 2271–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, J., Alvarez, J., Bossinger, G., and Smyth, D.R. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8, 505–520. [Google Scholar]
- Bey, M., Stuber, K., Fellenberg, K., Schwarz-Sommer, Z., Sommer, H., Saedler, H., and Zachgo, S. (2004). Characterization of Antirrhinum petal development and identification of target genes of the class B MADS box gene DEFICIENS. Plant Cell 16, 3197–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J.W., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. (2003). A gene expression map of the Arabidopsis root. Science 302, 1956–1960. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Eshed, Y., and Baum, S.F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18, 134–141. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budziszewski, G.J., et al. (2001). Arabidopsis genes essential for seedling viability: Isolation of insertional mutants and molecular cloning. Genetics 159, 1765–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, M.E., Simorowski, J., and Martienssen, R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129, 1957–1965. [DOI] [PubMed] [Google Scholar]
- Byzova, M.V., Franken, J., Aarts, M.G., de Almeida-Engler, J., Engler, G., Mariani, C., Van Lookeren Campagne, M.M., and Angenent, G.C. (1999). Arabidopsis STERILE APETALA, a multifunctional gene regulating inflorescence, flower, and ovule development. Genes Dev. 13, 1002–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi, L., Yang, Y., Yi, Y., Heilig, E., Herman, B., Cassista, A.J., Allen, D.W., Xiang, H., and Jack, T. (1999). Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 17, 699–707. [DOI] [PubMed] [Google Scholar]
- Chen, J.J., Janssen, B.J., Williams, A., and Sinha, N. (1997). A gene fusion at a homeobox locus: Alterations in leaf shape and implications for morphological evolution. Plant Cell 9, 1289–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Goodwin, S.M., Boroff, V.L., Liu, X., and Jenks, M.A. (2003). Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15, 1170–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S.K., Dagenais, N., Chory, J., and Weigel, D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100, 469–478. [DOI] [PubMed] [Google Scholar]
- Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8, 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocherel, S., Perez, P., Degroote, F., Genestier, S., and Picard, G. (1996). A promoter identified in the 3′ end of the Ac transposon can be activated by cis-acting elements in transgenic Arabidopsis lines. Plant Mol. Biol. 30, 539–551. [DOI] [PubMed] [Google Scholar]
- Coen, E.S., and Meyerowitz, E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Dinneny, J.R., Yadegari, R., Fischer, R.L., Yanofsky, M.F., and Weigel, D. (2004). The role of JAGGED in shaping lateral organs. Development 131, 1101–1110. [DOI] [PubMed] [Google Scholar]
- Dinneny, J.R., and Yanofsky, M.F. (2005). Drawing lines and borders: How the dehiscent fruit of Arabidopsis is patterned. Bioessays 27, 42–49. [DOI] [PubMed] [Google Scholar]
- Ditta, G., Pinyopich, A., Robles, P., Pelaz, S., and Yanofsky, M.F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14, 1935–1940. [DOI] [PubMed] [Google Scholar]
- Dockx, J., Quaedvlieg, N., Keultjes, G., Kock, P., Weisbeek, P., and Smeekens, S. (1995). The homeobox gene ATK1 of Arabidopsis thaliana is expressed in the shoot apex of the seedling and in flowers and inflorescence stems of mature plants. Plant Mol. Biol. 28, 723–737. [DOI] [PubMed] [Google Scholar]
- Durfee, T., Roe, J.L., Sessions, R.A., Inouye, C., Serikawa, K., Feldmann, K.A., Weigel, D., and Zambryski, P.C. (2003). The F-box-containing protein UFO and AGAMOUS participate in antagonistic pathways governing early petal development in Arabidopsis. Proc. Natl. Acad. Sci. USA 100, 8571–8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99, 199–209. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., Perea, J.V., and Bowman, J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251–1260. [DOI] [PubMed] [Google Scholar]
- Fett, J.P., and Coleman, J.R. (1994). Characterization and expression of two cDNAs encoding carbonic anhydrase in Arabidopsis thaliana. Plant Physiol. 105, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, K., and Schopfer, P. (1998). Physical strain-mediated microtubule reorientation in the epidermis of gravitropically or phototropically stimulated maize coleoptiles. Plant J. 15, 119–123. [DOI] [PubMed] [Google Scholar]
- Freeling, M. (1992). A conceptual framework for maize leaf development. Dev. Biol. 153, 44–58. [DOI] [PubMed] [Google Scholar]
- Goldberg, R.B., Beals, T.P., and Sanders, P.M. (1993). Anther development: Basic principles and practical applications. Plant Cell 5, 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz, J.F., and Hudson, A. (2002). Signaling in plant lateral organ development. Plant Cell 14 (suppl.), S277–S288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, W., et al. (2004). Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 135, 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, K., and Meyerowitz, E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Gray, P.A., et al. (2004). Mouse brain organization revealed through direct genome-scale TF expression analysis. Science 306, 2255–2257. [DOI] [PubMed] [Google Scholar]
- Groover, A.T., Fontana, J.R., Arroyo, J.M., Yordan, C., McCombie, W.R., and Martienssen, R.A. (2003). Secretion trap tagging of secreted and membrane-spanning proteins using Arabidopsis gene traps. Plant Physiol. 132, 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Q., Ferrandiz, C., Yanofsky, M.F., and Martienssen, R. (1998). The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125, 1509–1517. [DOI] [PubMed] [Google Scholar]
- Hareven, D., Gutfinger, T., Parnis, A., Eshed, Y., and Lifschitz, E. (1996). The making of a compound leaf: Genetic manipulation of leaf architecture in tomato. Cell 84, 735–744. [DOI] [PubMed] [Google Scholar]
- Heisler, M.G., Atkinson, A., Bylstra, Y.H., Walsh, R., and Smyth, D.R. (2001). SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Hepworth, S.R., Zhang, Y., McKim, S., Li, X., and Haughn, G.W. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17, 1434–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, J.P., and Lord, E.M. (1989). Floral development in Arabidopsis thaliana: A comparison of the wild type and the homeotic pistillata mutant. Can. J. Bot. 67, 2922–2936. [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. [DOI] [PubMed] [Google Scholar]
- Hudson, A. (1999). Axioms and axes in leaf formation? Curr. Opin. Plant Biol. 2, 56–60. [DOI] [PubMed] [Google Scholar]
- Irish, V.F. (1999). Petal and stamen development. Curr. Top. Dev. Biol. 41, 133–161. [DOI] [PubMed] [Google Scholar]
- Ito, T., Wellmer, F., Yu, H., Das, P., Ito, N., Alves-Ferreira, M., Riechmann, J.L., and Meyerowitz, E.M. (2004). The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430, 356–360. [DOI] [PubMed] [Google Scholar]
- Jack, T., Brockman, L.L., and Meyerowitz, E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. [DOI] [PubMed] [Google Scholar]
- Jen, C.H., Michalopoulos, I., Westhead, D.R., and Meyer, P. (2005). Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol. 6, R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik, P.D., and Irish, V.F. (2000). Regulation of cell proliferation patterns by homeotic genes during Arabidopsis floral development. Development 127, 1267–1276. [DOI] [PubMed] [Google Scholar]
- Johanson, U., Karlsson, M., Johansson, I., Gustavsson, S., Sjovall, S., Fraysse, L., Weig, A.R., and Kjellbom, P. (2001). The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 126, 1358–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, I., Karlsson, M., Johanson, U., Larsson, C., and Kjellbom, P. (2000). The role of aquaporins in cellular and whole plant water balance. Biochim. Biophys. Acta 1465, 324–342. [DOI] [PubMed] [Google Scholar]
- Kamalay, J.C., and Goldberg, R.B. (1984). Organ-specific nuclear RNAs in tobacco. Proc. Natl. Acad. Sci. USA 81, 2801–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter, R.A., Bollman, K., Taylor, R.A., Bomblies, K., and Poethig, R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411, 706–709. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428, 81–84. [DOI] [PubMed] [Google Scholar]
- Koltunow, A.M., Truettner, J., Cox, K.H., Wallroth, M., and Goldberg, R.B. (1990). Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2, 1201–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki, K., Okada, E., Nishino, E., and Shimura, Y. (1988). Isolation and characterization of novel mutants of Arabidopsis thaliana defective in flower development. Development 104, 195–203. [Google Scholar]
- Kosugi, S., and Ohashi, Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9, 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348. [DOI] [PubMed] [Google Scholar]
- Kumaran, M.K., Bowman, J.L., and Sundaresan, V. (2002). YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 14, 2761–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusk, S., Sohlberg, J.J., Long, J.A., Fridborg, I., and Sundberg, E. (2002). STY1 and STY2 promote the formation of apical tissues during Arabidopsis gynoecium development. Development 129, 4707–4717. [DOI] [PubMed] [Google Scholar]
- Laufs, P., Coen, E., Kronenberger, J., Traas, J., and Doonan, J. (2003). Separable roles of UFO during floral development revealed by conditional restoration of gene function. Development 130, 785–796. [DOI] [PubMed] [Google Scholar]
- Lee, I., Wolfe, D.S., Nilsson, O., and Weigel, D. (1997). A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr. Biol. 7, 95–104. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., and Whittier, R.F. (1995). Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25, 674–681. [DOI] [PubMed] [Google Scholar]
- Lohmann, J.U., and Weigel, D. (2002). Building beauty: The genetic control of floral patterning. Dev. Cell 2, 135–142. [DOI] [PubMed] [Google Scholar]
- Lord, E.M., Crone, W., and Hill, J.P. (1994). Timing of events during flower organogenesis: Arabidopsis as a model system. Curr. Top. Dev. Biol. 29, 325–356. [DOI] [PubMed] [Google Scholar]
- Lu, P., Porat, R., Nadeau, J.A., and O'Neill, S.D. (1996). Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8, 2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel, M.A., Gustafson-Brown, C., Savidge, B., and Yanofsky, M.F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1998). The Arabidopsis AGL9 MADS box gene is expressed in young flower primordia. Sex. Plant Reprod. 11, 22–28. [Google Scholar]
- Martienssen, R.A. (1998). Functional genomics: Probing plant gene function and expression with transposons. Proc. Natl. Acad. Sci. USA 95, 2021–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. [DOI] [PubMed] [Google Scholar]
- Motohashi, R., Nagata, N., Ito, T., Takahashi, S., Hobo, T., Yoshida, S., and Shinozaki, K. (2001). An essential role of a TatC homologue of a delta pH-dependent protein transporter in thylakoid membrane formation during chloroplast development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98, 10499–10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlbauer, G.J., Fowler, J.E., and Freeling, M. (1997). Sectors expressing the homeobox gene liguleless3 implicate a time-dependent mechanism for cell fate acquisition along the proximal-distal axis of the maize leaf. Development 124, 5097–5106. [DOI] [PubMed] [Google Scholar]
- Nacken, W.K., Huijser, P., Beltran, J.P., Saedler, H., and Sommer, H. (1991). Molecular characterization of two stamen-specific genes, tap1 and fil1, that are expressed in the wild type, but not in the deficiens mutant of Antirrhinum majus. Mol. Gen. Genet. 229, 129–136. [DOI] [PubMed] [Google Scholar]
- Nemhauser, J.L., Feldman, L.J., and Zambryski, P.C. (2000). Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127, 3877–3888. [DOI] [PubMed] [Google Scholar]
- Nick, P., Bergfeld, R., Schafer, E., and Schopfer, P. (1990). Unilateral reorientation of microtubules at the outer epidermal wall during photo- and gravitropic curvature of maize coleoptiles and sunflower hypocotyls. Planta 181, 162–168. [DOI] [PubMed] [Google Scholar]
- Norberg, M., Holmlund, M., and Nilsson, O. (2005). The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 132, 2203–2213. [DOI] [PubMed] [Google Scholar]
- Ooka, H., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247. [DOI] [PubMed] [Google Scholar]
- Pan, X., Li, Y., and Stein, L. (2005). Site preferences of insertional mutagenesis agents in Arabidopsis. Plant Physiol. 137, 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov, S., Sevugan, M., Ye, D., Yang, W.C., Kumaran, M., and Sundaresan, V. (1999). Analysis of flanking sequences from dissociation insertion lines: A database for reverse genetics in Arabidopsis. Plant Cell 11, 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautot, V., Dockx, J., Hamant, O., Kronenberger, J., Grandjean, O., Jublot, D., and Traas, J. (2001). KNAT2: Evidence for a link between knotted-like genes and carpel development. Plant Cell 13, 1719–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Gustafson-Brown, C., Kohalmi, S.E., Crosby, W.L., and Yanofsky, M.F. (2001). APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 26, 385–394. [DOI] [PubMed] [Google Scholar]
- Perez-Rodriguez, M., Jaffe, F.W., Butelli, E., Glover, B.J., and Martin, C. (2005). Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Antirrhinum majus flowers. Development 132, 359–370. [DOI] [PubMed] [Google Scholar]
- Poovaiah, B.W., Xia, M., Liu, Z., Wang, W., Yang, T., Sathyanarayanan, P.V., and Franceschi, V.R. (1999). Developmental regulation of the gene for chimeric calcium/calmodulin-dependent protein kinase in anthers. Planta 209, 161–171. [DOI] [PubMed] [Google Scholar]
- Pyke, K.A., and Page, A.M. (1998). Plastid ontogeny during petal development in Arabidopsis. Plant Physiol. 116, 797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin, I. (1992). Role of salicylic acid in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 439–463. [Google Scholar]
- Rubinelli, P., Hu, Y., and Ma, H. (1998). Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Mol. Biol. 37, 607–619. [DOI] [PubMed] [Google Scholar]
- Sablowski, R.W., and Meyerowitz, E.M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92, 93–103. [DOI] [PubMed] [Google Scholar]
- Sakuma, Y., Liu, Q., Dubouzet, J.G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. [DOI] [PubMed] [Google Scholar]
- Samach, A., Klenz, J.E., Kohalmi, S.E., Risseeuw, E., Haughn, G.W., and Crosby, W.L. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20, 433–445. [DOI] [PubMed] [Google Scholar]
- Sanders, P., Bui, A., Weterings, K., McIntire, K., Hsu, Y., Lee, P., Truong, M., Beals, T., and Goldberg, R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11, 297–322. [Google Scholar]
- Sawa, S., Ito, T., Shimura, Y., and Okada, K. (1999). FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems. Plant Cell 11, 69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler, U., Beckmann, H., and Cashmore, A.R. (1993). HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 4, 137–150. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Schneeberger, R.G., Becraft, P.W., Hake, S., and Freeling, M. (1995). Ectopic expression of the knox homeo box gene rough sheath1 alters cell fate in the maize leaf. Genes Dev. 9, 2292–2304. [DOI] [PubMed] [Google Scholar]
- Scott, R., Dagless, E., Hodge, R., Paul, W., Soufleri, I., and Draper, J. (1991). Patterns of gene expression in developing anthers of Brassica napus. Plant Mol. Biol. 17, 195–207. [DOI] [PubMed] [Google Scholar]
- Scott, R.J., Spielman, M., and Dickinson, H.G. (2004). Stamen structure and function. Plant Cell 16 (suppl.), S46–S60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128, 1771–1783. [DOI] [PubMed] [Google Scholar]
- Sessions, A., Nemhauser, J.L., McColl, A., Roe, J.L., Feldmann, K.A., and Zambryski, P.C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124, 4481–4491. [DOI] [PubMed] [Google Scholar]
- Sessions, R.A., and Zambryski, P.C. (1995). Arabidopsis gynoecium structure in the wild and in ettin mutants. Development 121, 1519–1532. [DOI] [PubMed] [Google Scholar]
- Siebert, P.D., Chenchik, A., Kellogg, D.E., Lukyanov, K.A., and Lukyanov, S.A. (1995). An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23, 1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128. [DOI] [PubMed] [Google Scholar]
- Smith, L.G., Greene, B., Veit, B., and Hake, S. (1992). A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116, 21–30. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, P.S. (2000). Gene traps: Tools for plant development and genomics. Plant Cell 12, 1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes, T.L., Kunkel, B.N., and Richards, E.J. (2002). Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 16, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Takeda, S., Matsumoto, N., and Okada, K. (2004). RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 131, 425–434. [DOI] [PubMed] [Google Scholar]
- Tsugeki, R., Kochieva, E.Z., and Fedoroff, N.V. (1996). A transposon insertion in the Arabidopsis SSR16 gene causes an embryo-defective lethal mutation. Plant J. 10, 479–489. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., and Lam, E. (2004). Recent advance in the study of caspase-like proteases and Bax inhibitor-1 in plants: Their possible roles as regulator of programmed cell death. Mol. Plant Pathol. 5, 65–70. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., and Lam, E. (2005). Two Arabidopsis metacaspases AtMCP1b and AtMCP2b are arginine/lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. J. Biol. Chem. 280, 14691–14699. [DOI] [PubMed] [Google Scholar]
- Weig, A., Deswarte, C., and Chrispeels, M.J. (1997). The major intrinsic protein family of Arabidopsis has 23 members that form three distinct groups with functional aquaporins in each group. Plant Physiol. 114, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer, F., Riechmann, J.L., Alves-Ferreira, M., and Meyerowitz, E.M. (2004). Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16, 1314–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K., et al. (2003). Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302, 842–846. [DOI] [PubMed] [Google Scholar]
- Yang, W.C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13, 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, J.L., Drews, G.N., Feldmann, K.A., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Goritschnig, S., Dong, X., and Li, X. (2003). A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15, 2636–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Medrano, L., Ohashi, K., Fletcher, J.C., Yu, H., Sakai, H., and Meyerowitz, E.M. (2004). HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 16, 2586–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zik, M., and Irish, V.F. (2003). Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell 15, 207–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.