Abstract
In mammals, electron-transfer flavoprotein:ubiquinone oxidoreductase (ETFQO) and electron-transfer flavoprotein (ETF) are functionally associated, and ETF accepts electrons from at least nine mitochondrial matrix flavoprotein dehydrogenases and transfers them to ubiquinone in the inner mitochondrial membrane. In addition, the mammalian ETF/ETFQO system plays a key role in β-oxidation of fatty acids and catabolism of amino acids and choline. By contrast, nothing is known of the function of ETF and ETFQO in plants. Sequence analysis of the unique Arabidopsis thaliana homologue of ETFQO revealed high similarity to the mammalian ETFQO protein. Moreover, green fluorescent protein cellular localization experiments suggested a mitochondrial location for this protein. RNA gel blot analysis revealed that Arabidopsis ETFQO transcripts accumulated in long-term dark-treated leaves. Analysis of three independent insertional mutants of Arabidopsis ETFQO revealed a dramatic reduction in their ability to withstand extended darkness, resulting in senescence and death within 10 d after transfer, whereas wild-type plants remained viable for at least 15 d. Metabolite profiling of dark-treated leaves of the wild type and mutants revealed a dramatic decline in sugar levels. In contrast with the wild type, the mutants demonstrated a significant accumulation of several amino acids, an intermediate of Leu catabolism, and, strikingly, high-level accumulation of phytanoyl-CoA. These data demonstrate the involvement of a mitochondrial protein, ETFQO, in the catabolism of Leu and potentially of other amino acids in higher plants and also imply a novel role for this protein in the chlorophyll degradation pathway activated during dark-induced senescence and sugar starvation.
INTRODUCTION
In mammals, the nuclear-encoded mitochondrial protein, electron-transfer flavoprotein:ubiquinone oxidoreductase (ETFQO), which is associated with the inner mitochondrial membrane, accepts electrons from the electron-transfer flavoprotein (ETF) localized in the mitochondrial matrix and reduces ubiquinone (Ruzicka and Beinert, 1977; Beckmann and Frerman, 1985, 1987). ETF is the physiological electron acceptor for at least nine mitochondrial matrix flavoprotein dehydrogenases; hence, the ETF/ETFQO system can be thought of as a branch of the electron transport system with multiple input sites from seven acyl-CoA dehydrogenases and two N-methyl dehydrogenases (Frerman, 1988; Frerman and Goodman, 2001). These dehydrogenases include four chain length–specific acyl-CoA dehydrogenases involved in straight-chain fatty acid β-oxidation: isovaleryl CoA dehydrogenase, 2-methyl branched-chain acyl-CoA dehydrogenase, and glutaryl-CoA dehydrogenase, which play a role in the oxidation of amino acids; and sarcosine and dimethylglycine dehydrogenases, which are involved in choline metabolism (Frerman, 1988; Frerman and Goodman, 2001). The mammalian mitochondrial proteins, ETF and ETFQO, are essential for the catabolism of fatty acids, several amino acids, and choline and are important in supplying mitochondria with respiratory substrates auxiliary to those derived from sucrose. In humans, mutations in either ETF or ETFQO results in the fatal genetic disease type II Glutaric acidemia (multiple acyl-CoA dehydrogenase dysfunctional disease) (Frerman and Goodman, 2001).
In the higher plant Arabidopsis thaliana, both α- and β-subunits of ETF (the corresponding genes are At1g50940 and At5g43430, respectively) were identified in mitochondria by the use of gel-based or liquid chromatography tandem mass spectrometry mitochondrial proteomic analysis (Heazlewood et al., 2004). However, the ETFQO protein has not been identified in plant mitochondria, although a unique homologue of the ETFQO gene (At2g43400) has been identified in the Arabidopsis genome (Arabidopsis Genome Initiative, 2000).
One of the functions of the ETF/ETFQO system in mammals is to allow respiration of substrates other than glucose. In plants, sucrose availability can be restricted during extended darkness (Graham et al., 1992); however, plants have apparently evolved a range of metabolic responses that allow them to survive for extended periods during dark-induced sucrose starvation. Such metabolic responses lead to the mobilization of alternate cellular components, which compensate for decreases in primary respiratory substrate and maintain important biochemical processes, thus allowing survival in adverse conditions (Aubert et al., 1996). Genome-wide responses to sucrose starvation have been recently highlighted by several microarray experiments, where it was shown that a range of genes involved in metabolic processes, such as lipid metabolism, chlorophyll degradation, protein degradation, and amino acid catabolism, are upregulated, whereas genes encoding components of pathways such as photosynthesis, carbon fixation, ribosome biogenesis, amino acid biosynthesis, glycolysis, and many others are downregulated (Contento et al., 2004; Lin and Wu, 2004; Thimm et al., 2004; Buchanan-Wollaston et al., 2005).
In this study, we have investigated the role of ETFQO in Arabidopsis during sucrose starvation induced by extended dark treatment and have focused on the characterization of Arabidopsis T-DNA knockouts of the gene encoding ETFQO at the molecular, physiological, and metabolite levels. We have confirmed the mitochondrial location of the protein by means of green fluorescent protein (GFP) localization experiments and have performed expression analysis of ETFQO in Arabidopsis seedlings following transfer to extended dark periods. Finally, we have studied the consequences of these gene mutations on plant viability following periods of extended darkness (up to 15 d) in addition to studying their impact on cellular metabolism under these conditions. The data are discussed in the context of current models of mitochondrial metabolism in plant tissues.
RESULTS
Arabidopsis ETFQO Is a Unique Homologue of Human ETFQO and Is Localized in Mitochondria
We have identified a homologue of ETFQO in the Arabidopsis genome, designated as ETFQO (At2g43400), that encodes a protein with 70% similarity to human ETFQO (accession number Q16134). The mammalian ETFQO is a unique integral membrane protein containing one equivalent of flavin adenine dinucleotide (FAD) and a [4Fe-4S]2+1+ cluster (Goodman et al., 1994), characteristics shared with the Arabidopsis ETFQO. The N-terminal sequence of the Arabidopsis ETFQO protein has characteristics consistent with that of a mitochondrial transit peptide and is predicted to target ETFQO to the mitochondrion by the three targeting prediction programs Predotar (http://genoplante-info.infobiogen.fr/predotar/), MitoProt II (Claros and Vincens, 1996), and PSORT (Nakai and Horton, 1999), consistent with the location of mammalian ETFQO in mitochondria. Alignment of the amino acid sequence of Arabidopsis ETFQO to homologues from bacteria to human shows a high overall similarity, including a putative FAD binding domain and [4Fe-4S]2+1+ cluster domain (see Supplemental Figure 1 online). Taken together, the above observations give strong support to our claim that Arabidopsis ETFQO is a functional homologue of ETFQO from other organisms.
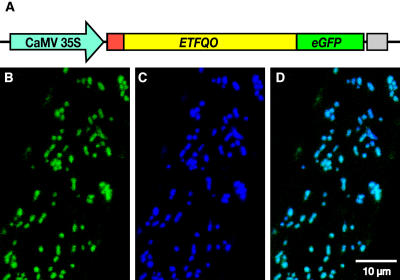
In order to confirm the in vivo subcellular localization of Arabidopsis ETFQO, a full-length cDNA fragment was fused to the cDNA of GFP (Figure 1A). The cDNA fusion construct was subcloned downstream of the cauliflower mosaic virus (CaMV) 35S RNA promoter and transformed into Arabidopsis. Five-day-old seedlings of stable transformants were incubated with MitoTracker Orange for specific staining of mitochondria. Inspection of the seedlings transformed with the Pro35S:ETFQO:GFP construct by confocal microscopy revealed that the GFP fluorescence within root cells was restricted to numerous spherical or elongated bodies, which correspond well with mitochondrial morphology (Figures 1B to 1D). The colocalization of the MitoTracker Orange dye and the GFP signal in root cells confirmed mitochondrial targeting of the ETFQO:GFP fusion protein (Figures 1B to 1D). These data clearly demonstrate the mitochondrial targeting of the ETFQO:GFP fusion protein in vivo and thus establish the mitochondrial localization of the ETFQO in Arabidopsis.
Figure 1.
Mitochondrial Localization of an ETFQO:eGFP Fusion Protein in Arabidopsis Seedlings.
(A) The construct carrying the gene encoding ETFQO:eGFP fusion protein. The ETFQO cDNA encoding the full-length coding sequence except stop codon (ETFQO, yellow box), which includes a sequence for the predicted mitochondrial target peptide (red box), is cloned upstream of the eGFP reading frame (eGFP, green box). Expression of the resulting fusion protein is controlled by the CaMV 35S promoter (blue box) and the NOS terminator (gray box).
(B) to (D) Images taken from Arabidopsis root cells expressing the ETFQO:eGFP fusion protein. (B), (C), and (D) are GFP signal (green), MitoTracker Orange signal (blue), and merged image of (C) and (D), respectively.
ETFQO Is Induced under Sucrose-Starved Conditions
The ETFQO gene is represented on the Affymetrix ATH1 Genome Array, and analysis of microarray data stored in the GENEVESTIGATOR database (Zimmermann et al., 2004) revealed that ETFQO transcripts are expressed constitutively at a low level in all plant tissues and developmental states, including leaf senescence (Buchanan-Wollaston et al., 2005); however, intriguingly, it was highly expressed in dark-induced senescent leaves (Lin and Wu, 2004), senescent cell cultures (Buchanan-Wollaston et al., 2005), and sucrose-starved cell cultures (Contento et al., 2004). RNA gel blot analysis confirmed that ETFQO expression was not significantly induced during normal developmental senescence (data not shown).
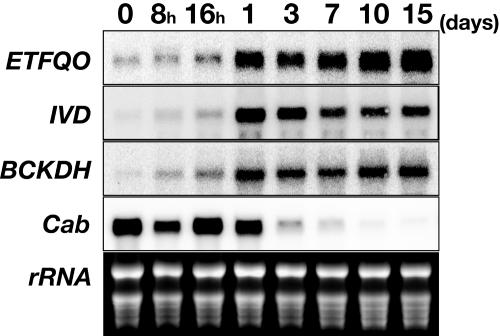
To further investigate regulation of ETFQO, transcript levels were measured in 4-week-old Arabidopsis leaves following transfer to darkness by RNA gel blot analysis. During dark adaptation, the transcripts from a light-regulated photosynthetic gene, Cab4, gradually declined (Figure 2) (Fujiki et al., 2001), whereas the ETFQO transcripts accumulated to very high levels (Figure 2). As a positive control, the transcript levels of the isovaleryl-CoA dehydrogenase gene (IVD) and the E1α subunit of branched-chain α-keto dehydrogenase gene (BCKDH), which have previously been shown to be induced by sucrose starvation (Daschner et al., 2001; Fujiki et al., 2001), were analyzed. The IVD and BCKDH transcripts also accumulated in the dark-adapted leaves (Figure 2), suggesting that expression of the ETFQO gene is also induced by sucrose starvation following prolonged darkness.
Figure 2.
Accumulation of Transcripts from the ETFQO in Dark-Treated Arabidopsis Leaves.
RNA was isolated from leaves harvested at indicated times after the onset of dark treatment. As control, we examined the expression of the IVD gene, that of BCKDH E1α subunit gene, and that of the chlorophyll a/b binding protein gene (Cab). Equal loading was confirmed by comparing ethidium bromide staining of rRNA bands.
Isolation of T-DNA Insertion Mutants of Arabidopsis ETFQO
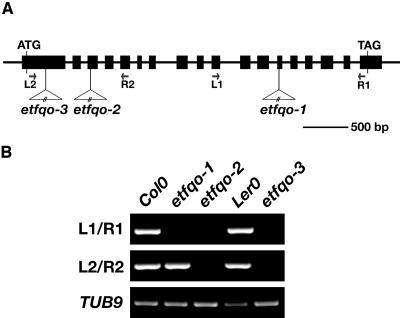
To investigate the in vivo functions of the ETFQO protein, three independent Arabidopsis lines containing T-DNA or dissociation (Ds) transposon element insertional mutations of ETFQO were isolated. Segregation of the encoded resistance markers of all three insertion elements were in good agreement with the 3:1 (resistant:susceptible) ratio, suggesting insertion at a single Mendelian locus. Homozygous lines for each mutant were confirmed by genomic PCR and designated etfqo-1, etfqo-2, and etfqo-3, respectively (Figure 3A), and the respective sites of insertion of these mutants were confirmed by sequencing PCR products amplified from each mutant.
Figure 3.
Identification of etfqo-1, etfqo-2, and etfqo-3 Mutants.
(A) Genomic structure of ETFQO loci. Arrows represent positions of primers used for genotype and RT-PCR analyses of wild-type and mutant lines; closed boxes indicate exons. In etfqo-1 and etfqo-2, the T-DNA is inserted in exon 4 and exon 12, respectively. In etfqo-3, Ds transposon element is inserted in exon 1.
(B) RT-PCR analysis on total RNA from the wild type, Col-0, Ler-0, and the mutant lines etfqo-1, etfqo-2, and etfqo-3, with primer sets indicated on the left and the positions of each primer in ETFQO genomic loci represented in (A).
As described above, ETFQO contains a putative [4Fe-4S]2+1+ cluster domain near its C terminus that is likely to be required for electron flow from flavin to ubiquinone. In all three mutant lines, the insertional element is positioned upstream of the putative [4Fe-4S]2+1+ cluster, and it would be anticipated that this would severely compromise its electron transfer capacity and so result in a complete loss of functional protein. RT-PCR using primer pairs designated to span the T-DNA insertion site of each mutant locus was used to investigate ETFQO transcription. The Arabidopsis β-tubulin gene was used as a control to demonstrate the integrity of the RNA preparation. ETFQO mRNAs were detected in the wild type (Columbia-0 [Col-0] and Landsberg erecta [Ler-0]) using the primer sets L1/R1 and L2/R2. In etfqo-1, ETFQO transcripts were detected using the L2/R2 primer set; however, no amplification products were observed for the transcripts using the L1/R1 primer set. In etfqo-2 and etfqo-3, amplification products were not detected using either the L1/R1 or L2/R2 primer set (Figure 3B). These results confirmed that transcripts spanning the T-DNA insertion site, and downstream of the T-DNA insertion, are absent in all these mutant lines.
Phenotype of etfqo Mutant Lines
After the molecular identity of the T-DNA insertional mutants was established, they were grown in soil under long-day conditions alongside the corresponding wild-type controls. Under these conditions, there was no visible phenotype in the mutants during the early stage of vegetative growth; however, in the reproductive stage, etfqo-1 and etfqo-2 plants often produced shorter siliques with a lower number of seeds than the corresponding wild type, whereas etfqo-3 had siliques with length and number of seeds similar to the wild type (see Supplemental Table 1 online). This difference might be due to the fact that the etfqo-3 mutant is in a different wild-type strain background (Ler-0) from etfqo-1 and etfqo-2 (Col-0). From crosses using wild-type pollen as the male parent and etfqo-1 and etfqo-2 as female parents, shorter siliques with lower seed set were produced, whereas crosses using pollen from etfqo-1 and etfqo-2 to create crosses with wild-type plants as female parent produced normal siliques with seed number similar to wild-type siliques, suggesting an effect of the ETFQO defect on the development of female gametophyte cells (see Supplemental Table 2 online).
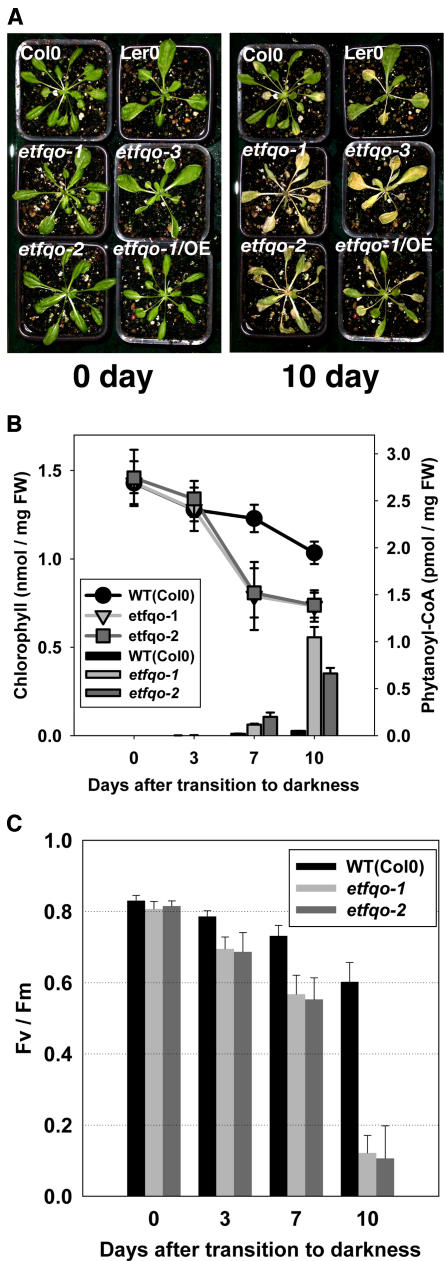
When 5-week-old etfqo mutants grown under short-day conditions were transferred to dark conditions, a dramatic phenotype became apparent. All of the homozygous mutant lines started to wilt and show signs of senescence after 10 d of continuous darkness and were apparently dead after 15 d of continuous darkness, whereas both wild-type ecotypes are still alive after 15 d of continuous darkness and show only limited signs of senescence (Figure 4A). To further investigate this apparent accelerated senescence in the etfqo mutants, we measured two parameters related to the function of chloroplasts, chlorophyll content and photochemical efficiency (maximum variable fluorescence/maximum yield of fluorescence [Fv/Fm]), as diagnostics for leaf senescence (Oh et al., 1996). During extended dark conditions, chlorophyll content declined more rapidly in etfqo mutants than in the wild type (Figure 4B), and the decline of chlorophyll content was accompanied by a decrease in the photochemical efficiency of photosystem II (PSII) (Fv/Fm) (Figure 4C). This result indicates that the process of senescence is more rapid in etfqo mutants than wild-type plants during extended dark conditions.
Figure 4.
Phenotype of etfqo Mutants in Extended Dark Treatment.
(A) Photos of 4-week, short-day grown Arabidopsis plants after further growth for 0 and 10 d in extended darkness. The plant designated as etfqo-1/OE is the etfqo-1 line transformed with the CaMV 35S promoter–driven ETFQO ORF. The leaves of etfqo mutants, etfqo-1, etfqo-2, and etfqo-3, were yellowed and dehydrated following 10 d of growth in darkness compared with the wild-type control, Col-0 and Ler-0, and etfqo-1/OE.
(B) Chlorophyll (line graph) and phytanoyl-CoA (bar graph) content of 9th or 10th leaves of 4-week, short-day grown Arabidopsis plants of wild type (Col-0), etfqo-1, and etfqo-2 after further growth for 0, 3, 7, and 10 d in extended dark treatment. Values are means ± sd for six independent samplings. FW, fresh weight.
(C) Fv/Fm, maximum quantum yield of PSII electron transport (maximum variable fluorescence/maximum yield of fluorescence) of 9th or 10th leaves of 4-week, short-day grown Arabidopsis plant of the wild type (Col-0), etfqo-1, and etfqo-2 after further growth for 0, 3, 7, and 10 d in extended darkness. Values are means ± sd for six independent samplings.
In order to confirm that the phenotypes of the mutant plants were caused by mutations in ETFQO, a CaMV 35S promoter–driven ETFQO open reading frame (ORF) was introduced into the etfqo-1 mutant (designated as etfqo-1/OE line) and shown to completely reverse the partial sterility and prevent the accelerated senescence phenotype observed in the etfqo-1 mutant line (Figure 4A; see Supplemental Table 1 online).
Leu Catabolism Is Blocked in etfqo Mutants
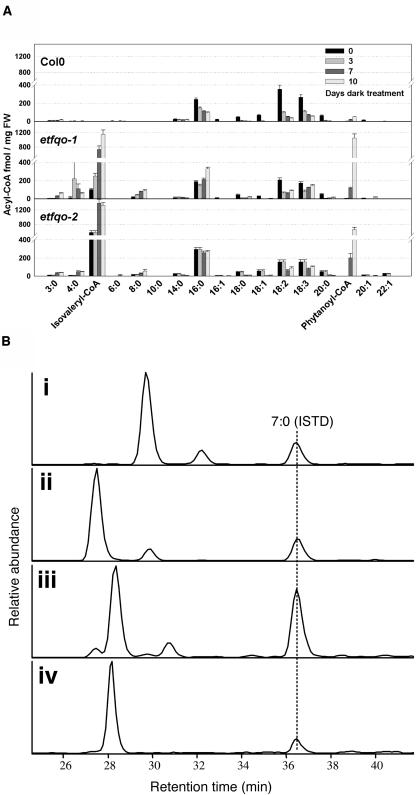
In mammals, defects in ETFQO result in a functional deficiency of mitochondrial flavoprotein dehydrogenases and accumulation of their substrates (Frerman and Goodman, 2001). To gain further insight into the role of the ETFQO protein in Arabidopsis, we used the method of Larson and Graham (2001) to analyze changes in acyl-CoAs in the wild type and etfqo mutants during the extended dark treatment. The results shown in Figure 5 demonstrate a dramatic increase in the amount of five-carbon acyl-CoA in the etfqo-1 and etfqo-2 mutants compared with the wild type during the extended dark period. There were four candidates in Arabidopsis for the five-carbon acyl-CoA compounds: isovaleryl-CoA (3-methyl-butyryl-CoA), 2-methyl-butyryl-CoA, valeryl-CoA, and acetoacetyl-CoA. However, the retention time of synthetic acetoacetyl-etheno-CoA was earlier than the five-carbon acyl-CoA peak (data not shown), eliminating acetoacetyl-CoA as a candidate for the peak. Furthermore, given the improved resolution among peaks in the short-chain acyl-CoA region by HPLC using the Hypercarb porous graphitic carbon column, we could successfully discriminate this five-carbon acyl-CoA peak from valeryl-CoA and 2-methyl-butyryl-CoA, and identified this peak as isovaleryl-CoA (Figure 5B). Isovaleryl-CoA is an intermediate of the Leu catabolic pathway, being subsequently dehydrogenated to 3-methylcrotonyl-CoA by isovaleryl-CoA dehydrogenase. In animals, ETF is the physiological electron acceptor of isovaleryl-CoA dehydrogenase (Ikeda and Tanaka, 1983). These results suggest a similar interaction of isovaleryl-CoA dehydrogenase, ETF, and ETFQO in Arabidopsis.
Figure 5.
Accumulation of Isovaleryl-CoA in etfqo-1 and etfqo-2 Mutants in Extended Dark Treatment.
(A) Acyl-CoA profiles of the wild type (Col-0), etfqo-1, and etfqo-2 during the extended dark treatment. Values are means ± se for six samples of 10 mg each, derivatized to their acyl-etheno-CoA esters, separated by HPLC, and detected fluorometrically. FW, fresh weight.
(B) Identification of the increased short-chain acyl-CoA peak in etfqo-1. Short-chain acyl-CoAs were chemically synthesized from their respective fatty acids, derivatized to their acyl-etheno-CoA esters, separated by HPLC on a Hypercarb porous graphitic carbon column, and detected by MS. The 5:0 acyl-etheno-CoA peaks were identified by their characteristic parent and daughter ion mass-to-charge (m/z) ratios of 876 and 452, respectively. Peaks were aligned against added heptanoyl-etheno-CoA (7:0) internal standard (ISTD; m/z 904). Shown are the extracted ion chromatograms for m/z values of 876 and 904. i, Valeryl-etheno-CoA; ii, 2-methyl-butyryl-etheno-CoA; iii, isovaleryl (3-methyl butyryl)-etheno-CoA; iv, etfqo-1 extract at 10 d in extended darkness.
Involvement of ETFQO in Phytol Degradation
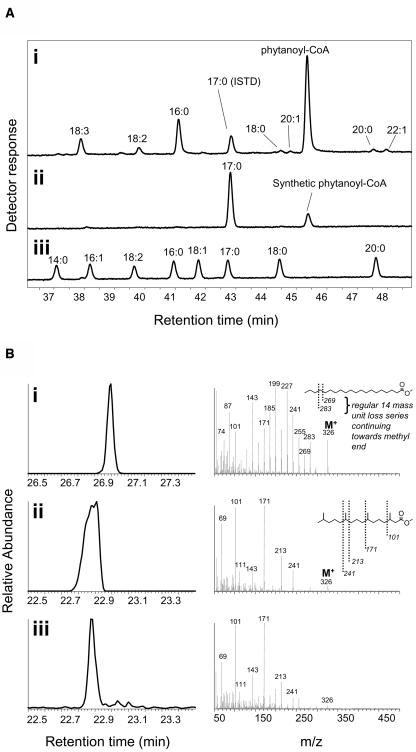
Acyl-CoA profiles from both etfqo-1 and etfqo-2 mutants from the 7- and 10-d extended dark treatments also showed dramatically increased content of a compound that had the same mass as the straight-chain eicosanoyl (20:0)-CoA, but which had a different retention time from the 20:0-CoA peak in the standard mixture. The retention time of this unknown acyl-CoA peak from the etfqo-1 mutant was identical to synthetic phytanoyl-CoA (Figure 6A), and both shared the same molecular mass and diagnostic acyl-CoA–specific fragmentation pattern when analyzed by liquid chromatography–mass spectrometry (LC-MS) (data not shown). Unfortunately, LC-MS fragmentation did not provide any information on the branching pattern of the acyl chain and so could not distinguish 20:0-CoA from phytanoyl-CoA. Therefore, in order to further confirm the identity of this unknown peak, it was purified in bulk, transmethylated, and analyzed by gas chromatography–mass spectrometry (GC-MS). The mass spectrum of the transmethylated peak was distinguishable from that of eicosanoic acid methyl ester but matched well with that of phytanic acid methyl ester, which displayed an electron-impact fragmentation spectrum expected for methyl-branched acyl chains (Figure 6B). From these results, the accumulated unknown peak near 20:0 acyl-CoA was unequivocally identified as phytanoyl-CoA. During extended dark treatment, etfqo-1 and etfqo-2 mutants contained dramatically increased concentrations of phytanoyl-CoA relative to the wild type (Figures 4B and 6A). However, analysis of leaf lipid fatty acids by GC showed no measurable increase in phytanic acid during the extended dark treatment (data not shown). In humans, phytanoyl-CoA is a known breakdown product of phytol, the hydrophobic tail of chlorophylls a and b. That said, relatively little is currently known about the exact mechanism of phytol or chlorophyll degradation in plants (Matile et al., 1999; Hortensteiner, 2004); however, our data suggest that Arabidopsis ETFQO is involved in phytol degradation.
Figure 6.
Identification of Phytanoyl-CoA Peak.
(A) Comparison of phytanoyl-CoA peak in etfqo-1 with standards. HPLC profiles of acyl-etheno-CoA derivatives are shown for etfqo-1 extract at 10 d in extended darkness (i), phytanoyl-CoA synthesized from phytanic acid by Pseudomonas acyl-CoA synthetase (ii), and the calibration standard mix (iii).
(B) Confirmation of phytanoyl-CoA identity by GC-MS. Collected fractions of the putative phytanoyl-CoA peak from HPLC runs of derivatized etfqo-1 extracts at 10 d in extended darkness were subjected to alkaline transmethylation to form the methyl ester. Left panels show GC-MS chromatograms; right panels show mass spectra for the corresponding peaks. i, eicosanoic acid methyl ester; ii, synthetic phytanic acid methyl ester; iii, transmethylated peak from etfqo-1.
Metabolite Contents of ETFQO Mutants during the Extended Dark Condition
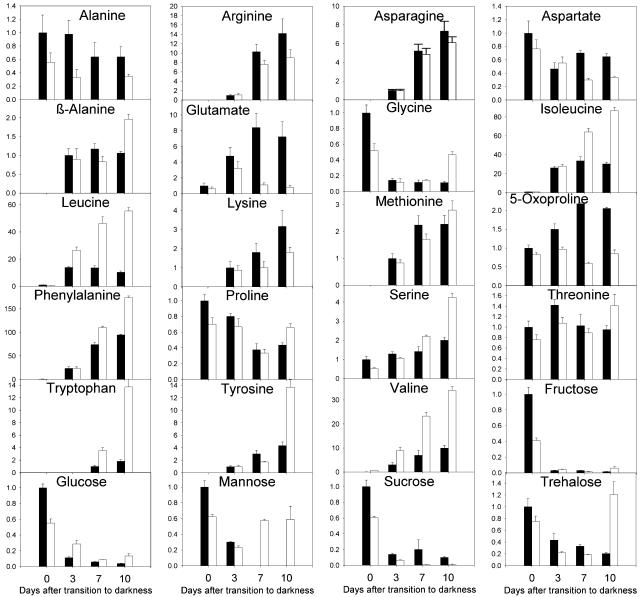
Further characterization of the accelerated senescence phenotype in etfqo mutants was performed using a nonbiased GC-MS metabolic profiling protocol allowing the simultaneous determination of >100 compounds (Roessner et al., 2001) As was expected, the extended dark treatment led to a rapid decline in sucrose and other sugar contents in both the wild type and etfqo-1 (Figure 7). Most free amino acids increased significantly in both the wild type and etfqo-1, including, Leu, Isle, Val, Asn, Arg, Trp, and Phe, suggesting that protein degradation was elevated under these conditions (Figure 7). The significantly elevated levels of several amino acids, especially Leu and Trp, in the etfqo mutants suggest involvement of ETFQO in their degradation pathways. Analysis of tricarboxy acid (TCA) cycle intermediates showed a dramatic increase in succinate, fumarate, and malate in etfqo-1 at 7 and 10 d dark treatment (see Supplemental Figure 2 online). While these accumulations are striking, the exact mechanism underlying this phenomenon cannot be elucidated from the results in this study. There are two possible explanations for the progressive accumulation of these metabolites. First, it is conceivable that the TCA cycle is progressively upregulated in the mutant, during the course of the extended darkness, in an attempt to compensate for the reduced availability of respiratory substrate. Secondly, the accumulation of TCA cycle intermediates may be a consequence of a general downregulation of biosynthesis that would be anticipated under conditions of carbon starvation. While we favor the second hypothesis, further experimentation, including direct measurements of fluxes within the constituent pathways, will ultimately be required in order to fully comprehend these data. Interestingly, Glu and pyroglutamic acid concentrations declined in etfqo-1 at 7 to 10 d, whereas they increased in the wild type (Figure 7; see Supplemental Figure 2 online). It has been suggested, in sucrose starvation experiments, that amino acid catabolism is activated and supplies respiratory substrates to the TCA cycle (Contento et al., 2004); however, in etfqo-1, one or more amino acid catabolic pathways appear to be blocked. One possible consequence of this block could be an overactivation of Glu catabolism in order to compensate for the shortfall of respiratory substrate. Furthermore, accumulation of uracil and ribose in etfqo-1 in the 10-d dark treatment suggests that RNA degradation was activated in etfqo mutants at this stage, and this is most probably associated with the process of cell death (data not shown). The above observations were all made by analyzing data on the basis of specific metabolites; however, interesting features of the data also emerge when the entire metabolite profiles are considered. While the majority of the metabolite levels, in the etfqo-1 mutant, exhibit monophasic behavior with respect to time in continuous darkness, a subset of metabolites, such as Gln, Gly, oxyproline, Pro, mannose and trehalose, exhibit biphasic behavior. It is tempting to speculate that monophasic responses are observed for the metabolites more intimately related to the reactions catalyzed by ETFQO, while the biphasic responses are probably secondary effects arising from the extreme stress that the etfqo mutants are under following longer periods of darkness. However, further experimentation will be required to determine whether this is indeed the case. Future work should also include characterization of mannose and trehalose metabolism in these lines, since both of these carbohydrates increase following extensive periods in continuous darkness despite initially decreasing. It is important to note that although the data presented here were solely determined from etfqo-1, essentially the same results were obtained from analysis of a second mutant (etfqo-2).
Figure 7.
Relative Levels of Amino Acids and Sugars in the Wild Type (Black Bars) and etfqo-1 Mutant (White Bars) during Extended Dark Conditions.
The y axis values represent metabolite level relative to the wild type. Data were normalized to mean response calculated for the 0-d dark-treated leaves of the wild type (in case no response was detected at 0 d, normalization was done against 3-d dark-treated leaves of the wild type). Values presented are mean ± se of determinations on six individual plants.
DISCUSSION
The essential roles in central metabolism of mammalian ETF and ETFQO have been well established; however, we have no knowledge of the role of their homologues in higher plants. ETF and ETFQO homologues were identified in the Arabidopsis genome and also in the genomes and EST databases of many other plant species, including, rice (Oryza sativa), tomato (Lycopersicon esculentum), barley (Hordeum vulgare), and wheat (Triticum aestivum) (Plant Genome Database; http://www.plantgdb.org/). This suggests that ETF and ETFQO are widely expressed in higher plants and potentially play an important role in plant metabolism. In this article, we describe an investigation of the functional importance of ETFQO in Arabidopsis and its role during dark-induced senescence.
In mammals, the reducing equivalents from various flavoprotein dehydrogenases are transferred via ETF to ETFQO, which is localized in the inner mitochondrial membrane, and the accepted electrons are further passed from ETFQO into the electron transport chain via coenzyme Q to the CoQH2-cytochrome c oxidoreductase (Complex III) (Parker and Engel, 2000). The deduced amino acid sequence of Arabidopsis ETFQO showed high overall similarity to that of human ETFQO, including its two putative functional domains, the FAD binding domain, and the [4Fe-4S]2+1+ cluster domain (see Supplemental Figure 1 online) and a mitochondrial targeting peptide at the N terminus. We confirmed the mitochondrial localization of Arabidopsis ETFQO by colocalization of MitoTracker Orange with a fusion to GFP (Figure 1). In Arabidopsis, α- and β-subunits of ETF have been found in mitochondria (Heazlewood et al., 2004), and their amino acid sequences also show high similarity to the both subunits of the human ETF (data not shown). Hence, it is quite feasible that the basic function of ETFQO in electron transfer to coenzyme Q in the mitochondrial respiratory chain is conserved between plants and animals.
ETFQO Is Induced and Essential for Viability in Sucrose-Starved Condition
Analysis of microarray data in public databases and RNA gel blot analyses confirmed that ETFQO was constitutively expressed in all tissues and stages of development in Arabidopsis plants and was upregulated following transfer of mature plants to extended periods of darkness over a period of 15 d (Figure 2). During experimentally induced sucrose starvation, there is a marked decrease in expression of many genes involved in carbohydrate breakdown, glycolysis, the TCA cycle, mitochondrial electron transport, and ATP synthesis (Buchanan-Wollaston et al., 2005). This is associated with a decrease in respiratory energy metabolism and increased expression of a number of genes involved in proteolysis, amino acid catabolism, and fatty acid degradation, suggesting the induction of salvage mechanisms that release amino acids and fatty acids from structural proteins and lipids as an alternative source of respiratory substrate to maintain viability of carbohydrate-starved cells (Contento et al., 2004; Lin and Wu, 2004; Thimm et al., 2004; Buchanan-Wollaston et al., 2005). The increased expression of ETFQO in dark-adapted plants (Figure 2) suggests a central role for ETFQO in supplying mitochondria with an alternative source of respiratory substrates from proteolysis and/or lipid degradation. The observation that the etfqo mutants senesce and die before wild-type plants under extended dark conditions (Figure 4A) taken together with their altered metabolic profiles provides further evidence that ETFQO plays a key role in supplying alternative respiratory substrates. However, it is possible that the accelerated senescence and early death of the etfqo mutants (Figure 4) could be, at least in part, due to the hyperaccumulation of toxic metabolites, since both branched-chain amino acids and α-keto acids are known to be cytotoxic compounds that can induce apoptosis in mammals (Eden and Benvenisty, 1999; Ogier de Baulny and Saudubray, 2002). Thus, our results suggest that ETFQO plays an essential role during sucrose starvation in plants by facilitating the use of alternative respiratory pathways either by supplying alternate substrates, by promoting the metabolism of toxic products thereof, or both.
Contribution of ETFQO to the Reproductive Development of Arabidopsis
The ecotype-specific reduced seed set and silique size of etfqo mutants (see Supplemental Table 1 online) suggest an additional role for ETFQO in Arabidopsis. Numerous studies on cytoplasmic male sterility have revealed the important role of the mitochondrion during pollen development (Hanson and Bentolila, 2004). In this case, however, the reproductive phenotype was not associated with pollen quality (see Supplemental Table 2 online). Thus, the mitochondrion also appears to play an important role in female gametophyte development as previously shown (Skinner et al., 2001; Christensen et al., 2002).
Involvement of ETFQO in Leu Catabolism
In Arabidopsis, isovaleryl-CoA dehydrogenase is localized in plant mitochondria (Daschner et al., 2001; Taylor et al., 2004), and our metabolite profiling data (Figure 5) demonstrated a marked accumulation of the isovaleryl-CoA dehydrogenase substrate, isovaleryl-CoA, in etfqo mutants, suggesting dysfunction of isovaleryl-CoA dehydrogenase in these mutants. These results further support the existence of the electron transfer cascade from isovaleryl-CoA dehydrogenase to ETF, ETF to ETFQO, and ETFQO to ubiquinone in the electron transport chain of plant mitochondria as reported in mammals (Ikeda and Tanaka, 1983), although the direct interaction of these four components has yet to be confirmed.
The recent proteomic study on Arabidopsis mitochondria by Taylor et al. (2004) revealed that the enzymes necessary for Leu catabolism are all localized within plant mitochondria, confirming the findings of 14C-Leu tracer experiments on isolated pea (Pisum sativum) mitochondria (Anderson et al., 1998). In this study, our demonstration of a marked accumulation of isovaleryl-CoA, as a result of mutation in the mitochondrial protein ETFQO, provides further support for the mitochondrial location of Leu catabolism in Arabidopsis. By contrast, we did not detect an accumulation of the intermediates of either Ile or Val catabolism, 2-methyl-butyryl-CoA, and isobutyryl-CoA, respectively, in etfqo mutants during the extended dark treatment, although it has been shown in mammals that ETF/ETFQO is also involved in dehydrogenation of 2-methyl-butyryl-CoA and isobutyryl-CoA (Ikeda et al., 1983). Our data imply that the mitochondrial ETF/ETFQO is less involved in Ile and Val catabolism in Arabidopsis than it is in Leu catabolism. However, it is important to note that these amino acids and several others accumulate in the etfqo mutants relative to the wild type (Figure 7). It is not clear why CoA intermediates for Ile and Val were not detected. It is possible that they are not as actively degraded as Leu; thus, intermediates are below the level of detection. Alternatively, the CoA intermediates could be actively and specifically deacylated to free fatty acids, but short chain acyl-CoA thioesterases that would catalyze this process have not been described in plants. Finally it is possible that the breakdown of Val and Ile may not be dependent on the ETFQO, and the dehydrogenation/oxidation step could be localized to the peroxisomes, as is the case for straight-chain fatty acids in higher plants. Such a hypothesis is consistent with the observations that the peroxisomal short-chain acyl-CoA oxidase is able to oxidize isobutyryl-CoA (Hayashi et al., 1999), and the demonstration that several other enzymes involved in Ile and Val catabolism are localized in peroxisomes in Arabidopsis (Lange et al., 2004; Taylor et al., 2004). The mitochondrial isovaleryl-CoA dehydrogenase does utilize 2-methyl-butyryl-CoA as a substrate in vitro; however, the Km value of the enzyme for the isobutyryl-CoA is ∼10 times larger than that for isovaleryl-CoA (Daschner et al., 2001). Since the relative concentration of Val is only half that of Leu, the large difference in Km values suggests that isovaleryl-CoA dehydrogenase is not involved in Val oxidation in vivo. The recent finding that the Arabidopsis mitochondrial branched-chain aminotransferase (BCAT-1) is able to initiate degradation of Leu, Ile, and Val (Schuster and Binder, 2005) along with previous proteomic results (Taylor et al., 2004) and those of this study provide an important foundation for understanding the degradative pathways of branched-chain amino acids. Intriguingly, the etfqo mutants also displayed elevated accumulation of aromatic amino acids, hinting that the ETF/ETFQO system may also be involved in the metabolism of this family of amino acids.
The Involvement of ETFQO in the Phytol Catabolic Pathway in Plants
Our observations show that there was a significant accumulation of phytanoyl-CoA in etfqo mutants during extended dark treatment (Figures 5 and 6), which parallels a decrease in chlorophyll (Figure 4B). It is well known in humans as a breakdown intermediate of phytol, which is derived from chlorophyll in the diet. In humans, phytol is oxidized to phytanic acid, subsequently converted into phytanoyl-CoA by a long-chain fatty acyl-CoA synthetase, and further degraded in peroxisomes by the α-oxidation system followed by β-oxidation in mitochondria (Wierzbicki et al., 2002; Wanders et al., 2003). Disorders in peroxisomal α-oxidation result in a syndrome called Refsum's disease, which results in a dramatic accumulation of phytanic acid in plasma- and lipid-containing tissues (Wierzbicki et al., 2002). In plants, phytol is derived from the first step of chlorophyll degradation, which is catalyzed by chlorophyllase (EC 3.1.1.14) (Tsuchiya et al., 1999); however, the latter enzymatic steps of phytol degradation are poorly understood, especially in comparison with the pathway in humans. Previous studies have suggested that phytol degradation during leaf senescence occurs via photooxidative conversion into various isoterpenoid compounds (Rontani et al., 1996; Matile et al., 1999; Rontani and Aubert, 2005). This obviously cannot be the case for chlorophyll breakdown during the extended dark treatments in this study. Therefore, increasing accumulation of phytanoyl-CoA in response to the length of dark treatment in etfqo mutants suggests the existence of an enzymatic pathway of phytol degradation in Arabidopsis mitochondria. In the extended dark condition, chlorophyll breakdown was greater in the etfqo mutant than in the wild type (Figure 4B), which could explain the dramatically increased concentration of phytanoyl-CoA in etfqo mutants (Figure 4B). However, it is also possible that ETFQO is directly involved in the phytanoyl-CoA degradation pathway, and phytanoyl-CoA degradation is blocked in etfqo mutants. In the analysis of leaf lipid derived fatty acid content of etfqo mutants during the extended dark treatment, no accumulation of phytanic acid, which is an upstream intermediate of phytanoyl-CoA, was observed (data not shown), suggesting rapid conversion from phytol to phytanoyl-CoA in Arabidopsis, at least under the conditions studied.
The Function of ETFQO in Mammals and Plants
In mammals, it has been shown that one of the main roles of ETF/ETFQO is the reoxidation of four straight-chain acyl-CoA dehydrogenases that are essential for the first step of fatty acid β-oxidation (Bartlett and Eaton, 2004). β-Oxidation of fatty acids primarily occurs in the mitochondria in mammals, while β-oxidation in higher plants occurs exclusively in peroxisomes (Graham and Eastmond, 2002). Our observations are consistent with a peroxisomal localization for β-oxidation in higher plants in that straight-chain acyl-CoAs do not accumulate in the etfqo mutants during the dark treatment (Figure 5). Furthermore, measurement of the fatty acid content of etfqo mutants and the wild type during germination indicated that fatty acid breakdown was the same in etfqo mutants and the wild type (data not shown). This result suggests that the ETFQO is not involved in β-oxidation of straight-chain fatty acids in Arabidopsis, supporting the postulate that β-oxidation of straight-chain fatty acids is localized exclusively in peroxisomes in Arabidopsis.
In mammals, ETF and ETFQO are also involved in the catabolism of several amino acids via dehydrogenation of isovaleryl-CoA dehydrogenase (Leu) catabolism, 2-methyl branched-chain acyl-CoA dehydrogenase (Ile and Val), and glutaryl-CoA dehydrogenase (Lys, Hyl, and Trp) (Frerman and Goodman, 2001). While the accumulation of isovaleryl-CoA in the etfqo mutants showed the involvement of ETFQO in Leu catabolism via isovaleryl-CoA dehydrogenase as in mammals, the lack of accumulation of 2-methyl-butyryl-CoA and isobutyryl-CoA does not allow us to formally conclude a similar role for ETFQO in Ile and Val catabolism. However, it should be noted that Ile and Val accumulate dramatically in the etfqo mutants, and it is possible that ETFQO is involved in catabolism of Ile and Val upstream of the intermediates 2-methyl-butyryl-CoA and isobutyryl-CoA since neither of these accumulates. The increase of Trp in GC-MS profiles of the etfqo mutants suggests involvement of ETFQO in Trp catabolism as found in mammals. The accumulation of phytanoyl-CoA and also amino acids in the etfqo mutants, as revealed by GC-MS profiling, implies the involvement of ETFQO in the unique metabolic processes that are not found in animals. As we discussed above, given the importance of the structure of the mammalian ETFQO to its basic functionality, the high amino acid sequence conservation (70%) between mammalian and Arabidopsis ETFQO suggests that the basic function of ETFQO as the electron donor between ETF and ubiquinone is conserved. However, the specific cellular role will be dependent on the interacting partners of the ETF. Thus, further understanding of these roles in plants will require identification of the ETF interacting partners.
Conclusion
This study has revealed that ETFQO is essential for plants to survive in sucrose-depleted conditions induced by extended growth in the dark and is involved in the Leu catabolic pathway. The study also shows that phytanoyl-CoA accumulates when ETFQO is disrupted, thus implicating the mitochondrial ETF/ETFQO in the degradation of the phytol tail of chlorophyll. The phytol tail represents ∼50% of the carbon in chlorophylls, the other 50% belongs to the porphyrin ring. On a global scale, the annual cycle of synthesis and breakdown of chlorophyll is massive, and more work is now needed to fully understand the role of the mitochondrial ETFQO in this important process. The identification of the ETFQO protein in plants clearly demonstrates the existence and importance of a novel mitochondrial electron transport complex in higher plants. In mammals, ETF is associated with ETFQO and several flavoprotein dehydrogenases (Parker and Engel, 2000). A similar relationship is likely to function in higher plants with at least some interacting partners conferring plant-specific roles, and it is on these that future studies need to focus.
METHODS
Plant Material and Dark Treatment
Arabidopsis thaliana seeds were surface-sterilized and imbibed for 4 d at 4°C in the dark on 0.8% (w/v) agar plates containing half-strength Murashige and Skoog (MS) media (Sigma-Aldrich; pH 5.7). Seeds were subsequently germinated and grown at 22°C under long-day conditions (16 h light/8 h dark) with 150 μmol m−2 s−1 white light. For examination of phenotype in the long-day treatment, seedlings were transferred to soil 7 to 10 d after germination and placed in a growth chamber at 22°C under long-day conditions (16 h light/8 h dark). For dark treatments, 7- to 10-d-old seedlings were transferred to soil and then grown at 22°C under short-day conditions (8 h light/16 h dark) for 4 weeks. Following bolting, plants were grown at 22°C in the dark in the same growth cabinet. The fully expanded, 7th to 12th, rosette leaves were harvested at intervals of 0, 3, 7, and 10 d from control and dark-grown plants for the subsequent experiments.
RNA Gel Blot Analysis
Total RNA was isolated from light-grown control, from dark-treated leaves, and from cell suspension culture using the TRIzol reagent (Invitrogen). cDNA fragments for ETFQO, IVD, the E1α subunit of BCKDH, chlorophyll a/b binding protein (Cab4), and nitrate reductase (NR2), were amplified from Arabidopsis cDNA by PCR. RNA was fractionated by electrophoresis on 1% (w/v) agarose gels, blotted onto positively charged nylon membrane, and hybridized at 65°C with the appropriate cDNA probes labeled with [α-32P]dCTP using the MegaPrime kit (Amarsham-Pharmacia). The hybridized membrane was washed for 1 h in a solution containing 2× SSC and 0.1% (w/v) SDS at 25°C followed by two washes with 0.1× SSC and 0.5% SDS at 55°C for 1 h.
Isolation of T-DNA Insertion Mutants and Genotype Characterization
The three mutant lines, SALK T-DNA line SALK_007870 (etfqo-1, background ecotype Col), Syngenta Arabidopsis insertion line SAIL_91_E03 (etfqo-2, background ecotype Col), and John Innes Centre Gene Trap line GT_3_4705 (etfqo-3, background ecotype Ler) were obtained from the Nottingham Arabidopsis Stock Centre (University of Nottingham). The mutants were isolated according to published procedures: SIGnAL (Alonso et al., 2003), Syngenta SAIL line (Sessions et al., 2002), and John Innes Centre Gene Trap line (Sundaresan et al., 1995). Genotypes of the different knockout mutant lines were analyzed by PCR using primers specific for the ETFQO ORF (for etfqo-1, L1 5′-AAGGTGGTACCGTGCTTCAG-3′ and R1 5′-CACCACCTTCAGGCACTGTC-3′; for etfqo-2 and etfqo-3, L2 5′-AAAGCTATCTTCTTCTTCTTCACCA-3′ and R2 5′-GGTTGCAATCCCAACAACTT-3′) and primers specific for the insertion elements (for etfqo-1, LBa1 5′-TGGTTCACGTAGTGGGCCATCG-3′; for etfqo-2, TMR1_LB1 5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′; for etfqo-3, Ds3-1 5′-ACCCGACCGGATCGTATCGGT-3′ and Ds5-1 5′-GAAACGGTCGGGAAACTAGCTCTAC-3′).
Complementation of etfqo-1 with the CaMV 35S Promoter–Driven ETFQO ORF
The full-length ORF including stop codon of ETFQO was amplified by RT-PCR from wild-type Arabidopsis (ecotype Col-0) and subcloned into the pH2GW7 vector (www.psb.ugent.be/gateway/) (Karimi et al., 2002) using the Gateway recombination system (Invitrogen). Heterozygous etfqo-1 mutant plants were subsequently transformed by Agrobacterium tumefaciens–mediated gene transfer utilizing the floral dip method (Clough and Bent, 1998) because of the partial sterility exhibited by the homozygous etfqo-1 plant. Positive transformants were selected on half-strength MS medium containing 0.8% (w/v) agar and 30 μg/mL hygromycin. Homozygous etfqo-1 lines containing the CaMV 35S promoter–driven ETFQO ORF were characterized by genomic PCR using the primer set L1 and R1 described above in order to check the transformed ETFQO ORF and the primer set L1 and RI1 (5′-TTGTGCAGTGGATGGTTTTG-3′) in order to confirm that there was no amplification from a disrupted copy of ETFQO. The primer RI1 was designed on the basis of the intron sequence of ETFQO in order to eliminate amplification from the transformed ETFQO ORF.
Analysis of ETFQO mRNA Expression by RT-PCR
Total RNA was isolated from rosette leaves of 3 d dark-treated plants using TRIzol reagent. First-strand cDNA was synthesized from 10 μg of total RNA with Superscript II Rnase H− reverse transcriptase (Invitrogen) and oligo(dT) primer. PCR amplification of ETFQO cDNA-specific sequence was performed using primers specific for the ORF, L1 and R1 and L2 and R2, described above. PCR amplification of the cDNA encoding the β-tubulin of Arabidopsis (TUB9) with a forward primer (5′-GATATCTGTTTCCGTACCTTGAAGC-3′) and a reverse primer (5′-ACCGACTGTAGCATCTTGATATTGC-3′) served as a control.
Subcellular Localization Experiments
In order to generate CaMV 35S promoter–driven ETFQO:eGFP fusion protein constructs, the 1899-bp coding region of ETFQO was amplified by RT-PCR from wild-type Arabidopsis (ecotype Col-0) with primers flanked with attB1 and attB2 sites. The resultant sequence was then cloned into the entry vector pDONR201 by Gateway recombination (Invitrogen). This cloned sequence was then transferred from the pDONR201 vector to the pK7WFG2 vector (www.psb.ugent.be/gateway/) (Karimi et al., 2002) by Gateway recombination (Invitrogen). Wild-type Arabidopsis Col-0 was subsequently transformed by Agrobacterium-mediated gene transfer utilizing the floral dip method (Clough and Bent, 1998). Positive transformants were selected on half-strength MS medium containing 0.8% (w/v) agar and 50 μg/mL kanamycin. Following selection, transformants were transferred to soil and seeds collected. The T2 generation was used for the observation of GFP fluorescence. Five-day-old seedlings were harvested into a freshly made staining solution of 200 nM CMTMRos (MitoTracker Orange; Molecular Probes) in half-strength MS medium and allowed to stain for 15 min at room temperature. After staining, the seedlings were submerged in a half-strength MS solution for 10 min. The T2 seedlings were visualized by confocal microscopy with a LSM510 confocal microscope (Zeiss) set to measure an emission band of 475 to 525 nm for GFP (excitation 488, emission 522) and an emission band of 565 to 615 nm for MitoTracker Orange CMTMRos (excitation 551, emission 576). Excitation for GFP was provided by the 458-nm line of a 25-mW argon laser and for MitoTracker by a 543-nm HeNe laser. The software LSM examiner (Zeiss) was used for postacquisition image processing. Several T2 generation lines for each construct were investigated, and all showed the same fluorescence pattern.
Measurement of Senescence Parameters
For chlorophyll content, 20 mg of leaf tissue were frozen and ground in liquid nitrogen and extracted by heating in 1 mL of methanol at 65°C for 20 min. After clarification by centrifugation, the chlorophyll concentration per fresh weight of leaf tissue was calculated as described previously (Porra et al., 1989). Photochemical efficiency was examined at the given time points for the dark treatment, using attached leaves of the wild type (Col-0) and etfqo mutants. The photochemical efficiency of PSII was deduced from the characteristics of chlorophyll fluorescence using a portable plant efficiency analyzer (Hansatech Instruments) (Oh et al., 1996). The ratio of Fv to Fm, which corresponds to the potential quantum yield of the photochemical reactions of PSII, was used as the measure of the photochemical efficiency of PSII (Oh et al., 1996).
Acyl-CoA and Fatty Acid Profiling
Ten-milligram portions of leaf material were frozen in liquid nitrogen and extracted for subsequent quantitative analysis of fluorescent acyl-etheno-CoA derivatives by HPLC with a LUNA 150 × 2 mm C18(2) column (Phenomenex) using the gradient conditions described previously (Larson and Graham, 2001; Larson et al., 2002). The lipid portion of the acyl-CoA extracts was transmethylated for fatty acid analysis by GC with flame ionization detection (Larson and Graham, 2001).
Commercially available phytanic acid (3,7,11,15-tetramethylhexadecanoic acid) (Sigma-Aldrich) was used to synthesize phytanic acid methyl ester by acidic transmethylation (Larson and Graham, 2001). Phytanoyl-CoA, for use as an HPLC retention time standard, was enzymatically synthesized from phytanic acid using Pseudomonas acyl-CoA synthetase from Sigma-Aldrich (Taylor et al., 1990). Putative phytanoyl-CoA peaks from HPLC runs of synthetic and leaf extracts were further characterized by LC-MS to determine molecular weight and verify acyl-CoA specific fragmentation patterns (Larson and Graham, 2001). Additional confirmation of phytanoyl-CoA identity was obtained by GC-MS analysis of collected HPLC fractions transmethylated to their fatty acid methyl esters. Putative phytanoyl-CoA fractions from 10-fold concentrated and derivatized extracts run on the HPLC were collected, dried under vacuum, and transmethylated with 500 μL 0.1 M sodium methoxide (Sigma-Aldrich) for 2 h at 85°C. The formed methyl ester was partitioned against 0.25 mL 0.9% (w/v) KCl and removed in 200 μL hexane. The hexane phase was dried under vacuum and reconstituted in 20 μL fresh hexane, and a 2 μL aliquot was analyzed by GC-MS as described previously (Tonon et al., 2005), with the exception that a ZB5 30 m × 0.25 mm i.d. × 0.25 μm film thickness capillary column (Phenomenex) was used.
Short branched-chain acyl-CoA esters were commercially unavailable, and Pseudomonas acyl CoA synthetase was unable to catalyze the formation of short branched-chain acyl-CoA esters from branched-chain 5-carbon acids (Sigma-Aldrich). Instead, these were synthesized by the carbonyldiimidazole method (Kawaguchi et al., 1981). The acyl-CoA products were derivatized to their etheno derivatives and characterized by LC-MS. In order to increase the resolution between the C5 isomers, a Hypercarb 100 × 3 mm porous graphitic carbon column (Thermo Hypersil) was used in place of the LUNA 150 × 2 mm C18(2) column, using the same gradient program.
Extraction, Derivatization, and Analysis of Arabidopsis Leaf Metabolites Using GC-MS
Metabolite analysis by GC-MS was performed by a method modified from that described by Rossener-Tunai et al. (2003). Arabidopsis leaf tissue (∼180 mg) was homogenized using a mortar and pestle precooled with liquid nitrogen and extracted in 1400 μL of methanol, and 60 μL of internal standard (0.2 mg ribitol mL−1 water) was subsequently added as a quantification standard. The extraction, derivatization, standard addition, and sample injection were exactly as described previously (Roessner-Tunali et al., 2003). The GC-MS system and settings were exactly as described by Roessner et al. (2001). Both chromatograms and mass spectra were evaluated using the MASSLAB program (ThermoQuest), and the resulting data were prepared and presented as described by Roessner et al. (2001).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: for Arabidopsis ETFQO, At2g43400; IVD, At3g45300; BCKDH, At1g21400; Cab4, At3g47470; NR2, At1g37130; TUB9, At4g20890; for rice OsETFQO, AAK39567; for human HsETFQO, Q16134; for rice MmETFQO, Q921G7; for fly DmETFQO, NP_610536; for worm CeETFQO, D88483; for yeast ScETFQO, Q08822; for protobacteria LpETFQO, YP_095306.
Supplementary Material
Acknowledgments
We thank Ian Moore and Ooi-kock Teh for assistance with the confocal microscopy and Nicholas J. Kruger for help with the measure of the photochemical efficiency of PSII. We also thank Lee J. Sweetlove for invaluable suggestions. This work was supported in part by a grant from the Biotechnology and Biological Science Research Council (to C.J.L.), by a research fellowship of the Japan Society for the Promotion of Science for Young Scientists (to K.I.), and by a grant from the German-Israeli Cooperation Project, Deutsch-Israelisches-Projekt (to N.S. and A.R.F.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Christpher J. Leaver (chris.leaver@plants.ox.ac.uk).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035162.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Anderson, M.D., Che, P., Song, J., Nikolau, B.J., and Wurtele, E.S. (1998). 3-Methylcrotonyl-coenzyme A carboxylase is a component of the mitochondrial leucine catabolic pathway in plants. Plant Physiol. 118, 1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Aubert, S., Gout, E., Bligny, R., Marty-Mazars, D., Barrieu, F., Alabouvette, J., Marty, F., and Douce, R. (1996). Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: Control by the supply of mitochondria with respiratory substrates. J. Cell Biol. 133, 1251–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett, K., and Eaton, S. (2004). Mitochondrial beta-oxidation. Eur. J. Biochem. 271, 462–469. [DOI] [PubMed] [Google Scholar]
- Beckmann, J.D., and Frerman, F.E. (1985). Electron-transfer flavoprotein-ubiquinone oxidoreductase from pig liver: Purification and molecular, redox, and catalytic properties. Biochemistry 24, 3913–3921. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston, V., Page, T., Harrison, E., Breeze, E., Lim, P.O., Nam, H.G., Lin, J.F., Wu, S.H., Swidzinski, J., Ishizaki, K., and Leaver, C.J. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42, 567–585. [DOI] [PubMed] [Google Scholar]
- Christensen, C.A., Gorsich, S.W., Brown, R.H., Jones, L.G., Brown, J., Shaw, J.M., and Drews, G.N. (2002). Mitochondrial GFA2 is required for synergid cell death in Arabidopsis. Plant Cell 14, 2215–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros, M.G., and Vincens, P. (1996). Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Contento, A.L., Kim, S.J., and Bassham, D.C. (2004). Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol. 135, 2330–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daschner, K., Couee, I., and Binder, S. (2001). The mitochondrial isovaleryl-coenzyme a dehydrogenase of Arabidopsis oxidizes intermediates of leucine and valine catabolism. Plant Physiol. 126, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden, A., and Benvenisty, N. (1999). Involvement of branched-chain amino acid aminotransferase (Bcat1/Eca39) in apoptosis. FEBS Lett. 457, 255–261. [DOI] [PubMed] [Google Scholar]
- Frerman, F.E. (1987). Reaction of electron-transfer flavoprotein ubiquinone oxidoreductase with the mitochondrial respiratory chain. Biochim. Biophys. Acta 893, 161–169. [DOI] [PubMed] [Google Scholar]
- Frerman, F.E. (1988). Acyl-CoA dehydrogenases, electron transfer flavoprotein and electron transfer flavoprotein dehydrogenase. Biochem. Soc. Trans. 16, 416–418. [DOI] [PubMed] [Google Scholar]
- Frerman, F.E., and Goodman, S.I. (2001). Defects of electron transfer flavoprotein and electron transfer flavoprotein-ubiquinone oxidoreductase: Glutaric acidemia type II. In The Metabolic and Molecular Bases of Inherited Disease, C.R. Scriver, W.S. Sly, B. Childs, A.L. Beaudet, and D. Valle, eds (New York: McGraw-Hill), pp. 2357–2365.
- Fujiki, Y., Ito, M., Nishida, I., and Watanabe, A. (2001). Leucine and its keto acid enhance the coordinated expression of genes for branched-chain amino acid catabolism in Arabidopsis under sugar starvation. FEBS Lett. 499, 161–165. [DOI] [PubMed] [Google Scholar]
- Goodman, S.I., Axtell, K.M., Bindoff, L.A., Beard, S.E., Gill, R.E., and Frerman, F.E. (1994). Molecular cloning and expression of a cDNA encoding human electron transfer flavoprotein-ubiquinone oxidoreductase. Eur. J. Biochem. 219, 277–286. [DOI] [PubMed] [Google Scholar]
- Graham, I.A., and Eastmond, P.J. (2002). Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog. Lipid Res. 41, 156–181. [DOI] [PubMed] [Google Scholar]
- Graham, I.A., Leaver, C.J., and Smith, S.M. (1992). Induction of malate synthase gene expression in senescent and detached organs of cucumber. Plant Cell 4, 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, M.R., and Bentolila, S. (2004). Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16 (suppl.), S154–S169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, H., De Bellis, L., Ciurli, A., Kondo, M., Hayashi, M., and Nishimura, M. (1999). A novel acyl-CoA oxidase that can oxidize short-chain acyl-CoA in plant peroxisomes. J. Biol. Chem. 274, 12715–12721. [DOI] [PubMed] [Google Scholar]
- Heazlewood, J.L., Tonti-Filippini, J.S., Gout, A.M., Day, D.A., Whelan, J., and Millar, A.H. (2004). Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16, 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortensteiner, S. (2004). The loss of green color during chlorophyll degradation—A prerequisite to prevent cell death? Planta 219, 191–194. [DOI] [PubMed] [Google Scholar]
- Ikeda, Y., Dabrowski, C., and Tanaka, K. (1983). Separation and properties of five distinct acyl-CoA dehydrogenases from rat liver mitochondria. Identification of a new 2-methyl branched chain acyl-CoA dehydrogenase. J. Biol. Chem. 258, 1066–1076. [PubMed] [Google Scholar]
- Ikeda, Y., and Tanaka, K. (1983). Purification and characterization of isovaleryl coenzyme A dehydrogenase from rat liver mitochondria. J. Biol. Chem. 258, 1077–1085. [PubMed] [Google Scholar]
- Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kawaguchi, A., Yoshimura, T., and Okuda, S. (1981). A new method for the preparation of acyl-CoA thioesters. J. Biochem. (Tokyo) 89, 337–339. [DOI] [PubMed] [Google Scholar]
- Lange, P.R., Eastmond, P.J., Madagan, K., and Graham, I.A. (2004). An Arabidopsis mutant disrupted in valine catabolism is also compromised in peroxisomal fatty acid beta-oxidation. FEBS Lett. 571, 147–153. [DOI] [PubMed] [Google Scholar]
- Larson, T.R., Edgell, T., Byrne, J., Dehesh, K., and Graham, I.A. (2002). Acyl CoA profiles of transgenic plants that accumulate medium-chain fatty acids indicate inefficient storage lipid synthesis in developing oilseeds. Plant J. 32, 519–527. [DOI] [PubMed] [Google Scholar]
- Larson, T.R., and Graham, I.A. (2001). Technical advance: A novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant J. 25, 115–125. [DOI] [PubMed] [Google Scholar]
- Lin, J.F., and Wu, S.H. (2004). Molecular events in senescing Arabidopsis leaves. Plant J. 39, 612–628. [DOI] [PubMed] [Google Scholar]
- Matile, P., Hortensteiner, S., and Thomas, H. (1999). Chlorophyll degradation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 67–95. [DOI] [PubMed] [Google Scholar]
- Nakai, K., and Horton, P. (1999). PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–36. [DOI] [PubMed] [Google Scholar]
- Ogier de Baulny, H., and Saudubray, J.M. (2002). Branched-chain organic acidurias. Semin. Neonatol. 7, 65–74. [DOI] [PubMed] [Google Scholar]
- Oh, S.A., Lee, S.Y., Chung, I.K., Lee, C.H., and Nam, H.G. (1996). A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol. Biol. 30, 739–754. [DOI] [PubMed] [Google Scholar]
- Parker, A., and Engel, P.C. (2000). Preliminary evidence for the existence of specific functional assemblies between enzymes of the beta-oxidation pathway and the respiratory chain. Biochem. J. 345, 429–435. [PMC free article] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394. [Google Scholar]
- Roessner, U., Luedemann, A., Brust, D., Fiehn, O., Linke, T., Willmitzer, L., and Fernie, A. (2001). Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13, 11–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali, U., Hegemann, B., Lytovchenko, A., Carrari, F., Bruedigam, C., Granot, D., and Fernie, A.R. (2003). Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 133, 84–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rontani, J.F., and Aubert, C. (2005). Characterization of isomeric allylic diols resulting from chlorophyll phytyl side-chain photo- and autoxidation by electron ionization gas chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 19, 637–646. [DOI] [PubMed] [Google Scholar]
- Rontani, J.F., Cuny, P., and Grossi, V. (1996). Photodegradation of chlorophyll phytyl chain in senescent leaves of higher plants. Phytochemistry 42, 347–351. [Google Scholar]
- Ruzicka, F.J., and Beinert, H. (1977). A new iron-sulfur flavoprotein of the respiratory chain. A component of the fatty acid beta oxidation pathway. J. Biol. Chem. 252, 8440–8445. [PubMed] [Google Scholar]
- Schuster, J., and Binder, S. (2005). The mitochondrial branched-chain aminotransferase (AtBCAT-1) is capable to initiate degradation of leucine, isoleucine and valine in almost all tissues in Arabidopsis thaliana. Plant Mol. Biol. 57, 241–254. [DOI] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner, D.J., Baker, S.C., Meister, R.J., Broadhvest, J., Schneitz, K., and Gasser, C.S. (2001). The Arabidopsis HUELLENLOS gene, which is essential for normal ovule development, encodes a mitochondrial ribosomal protein. Plant Cell 13, 2719–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Taylor, D.C., Weber, N., Hogge, L.R., and Underhill, E.W. (1990). A simple enzymatic method for the preparation of radiolabeled erucoyl-CoA and other long-chain fatty acyl-CoAs and their characterization by mass spectrometry. Anal. Biochem. 184, 311–316. [DOI] [PubMed] [Google Scholar]
- Taylor, N.L., Heazlewood, J.L., Day, D.A., and Millar, A.H. (2004). Lipoic acid-dependent oxidative catabolism of alpha-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol. 134, 838–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., Selbig, J., Muller, L.A., Rhee, S.Y., and Stitt, M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Tonon, T., Qing, R., Harvey, D., Li, Y., Larson, T.R., and Graham, I.A. (2005). Identification of a long-chain polyunsaturated fatty acid acyl-coenzyme A synthetase from the diatom Thalassiosira pseudonana. Plant Physiol. 138, 402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, T., Ohta, H., Okawa, K., Iwamatsu, A., Shimada, H., Masuda, T., and Takamiya, K. (1999). Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: Finding of a lipase motif and the induction by methyl jasmonate. Proc. Natl. Acad. Sci. USA 96, 15362–15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders, R.J., Jansen, G.A., and Lloyd, M.D. (2003). Phytanic acid alpha-oxidation, new insights into an old problem: A review. Biochim. Biophys. Acta 1631, 119–135. [DOI] [PubMed] [Google Scholar]
- Wierzbicki, A.S., Lloyd, M.D., Schofield, C.J., Feher, M.D., and Gibberd, F.B. (2002). Refsum's disease: A peroxisomal disorder affecting phytanic acid alpha-oxidation. J. Neurochem. 80, 727–735. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.