Abstract
This paper presents a classification-driven biomedical image retrieval framework based on image filtering and similarity fusion by employing supervised learning techniques. In this framework, the probabilistic outputs of a multiclass support vector machine (SVM) classifier as category prediction of query and database images are exploited at first to filter out irrelevant images, thereby reducing the search space for similarity matching. Images are classified at a global level according to their modalities based on different low-level, concept, and keypoint-based features. It is difficult to find a unique feature to compare images effectively for all types of queries. Hence, a query-specific adaptive linear combination of similarity matching approach is proposed by relying on the image classification and feedback information from users. Based on the prediction of a query image category, individual precomputed weights of different features are adjusted online. The prediction of the classifier may be inaccurate in some cases and a user might have a different semantic interpretation about retrieved images. Hence, the weights are finally determined by considering both precision and rank order information of each individual feature representation by considering top retrieved relevant images as judged by the users. As a result, the system can adapt itself to individual searches to produce query-specific results. Experiment is performed in a diverse collection of 5 000 biomedical images of different modalities, body parts, and orientations. It demonstrates the efficiency (about half computation time compared to search on entire collection) and effectiveness (about 10%–15% improvement in precision at each recall level) of the retrieval approach.
Index Terms—: Classification, classifier combination, content-based image retrieval (CBIR), medical imaging, relevance feedback (RF), similarity fusion
I. Introduction
With the advent of imaging, clinical care could be significantly impacted with improved image handling. In recent years, rapid advances of software and hardware technology have easied the problem of maintaining large medical image collections. These images constitute an important source of anatomical and functional information for the diagnosis of diseases, medical research, and education. Effectively and efficiently searching in these large image collections poses significant technical challenges as the characteristics of the medical images differ significantly from other general purpose images.
The image modality reveals anatomical and/or functional information of different body parts and pathologies. Each imaging modality presents challenges for acquisition, storage [1], and retrieval. Currently, the images are retrieved primarily using text-based searches [1], [2]. Search results in medical collections might be improved by combining text attribute-based search capability with low-level visual features computed directly on the image content, commonly known as the content-based image retrieval (CBIR) [2], [3].
During the last decade, several medical CBIR prototypes have been proposed [2], [4]–[8]. Majority of these are developed around a specific imaging modality and retrieval methods in these systems are task specific. There are a few systems that have a goal of creating CBIR systems for heterogeneous image collections [7]–[9]. To enable effective search in a large medical image collection of diverse modalities, it might be advantageous for a retrieval system to be able to recognize the image class prior to any kind of postprocessing or similarity matching [10]. A successful categorization of images would greatly enhance the performance of a CBIR system by filtering out irrelevant images and, thereby, reducing the search space. For example, to search posteroanterior (PA) chest X-rays with an enlarged heart in a radiographic collection, database images at first can be prefiltered with automatic categorization according to modality (e.g., X-ray), body part (e.g., chest), and orientation (e.g., PA) at different levels. The later similarity matching can be performed between query and images in the filtered set to find the enlarged heart as a distinct visual property. CBIR search under a specific modality and body part based on the automatic categorization of images would likely to perform better. In addition, the automatic classification of images could be utilized to adjust the feature weights in similarity matching at query time. For example, a color feature should have more weight for the images under the category of microscopic pathology or dermatology, whereas edge features are more important for the radiographs and texture-related features are important for CT or MRI images.
Some approaches have been explored in recent years to automatically classify medical image collections into multiple semantic categories for effective retrieval [10]–[12]. For example, in [11], the automatic categorization of 6 231 radiological images into 81 categories is examined by utilizing a combination of low-level global texture features with low-resolution scaled images and a K-nearest-neighbor (KNN) classifier. Although these approaches demonstrate promising results for medical image classification at a global level, they do not directly relate classification to retrieval. Another effective approach for improving retrieval is to include user feedback, known as relevance feedback, (RF) [13], [14].
Due to the limitations in low-level feature representations and motivated by advances in machine learning, we present a learning-based retrieval framework that uses novel image filtering and similarity matching approaches. In this framework, several different image features are extracted to train multiclass support vector machines (SVMs) and perform similarity matching. Probabilities output from the SVM are considered as category-specific information for query and database images, and are used to first filter out irrelevant images before applying a linear combination of similarity matching functions. The features are finally unified by a dynamically weighted linear combination of similarity matching functions to overcome machine learning or user classification errors. The feature weights are calculated by considering both the precision and the rank order information of the top set of retrieved relevant images. This paper is an extended version of our previous work [15]. Here, we report advances in feature representation, similarity matching, and evaluation. The “bag of concepts” and different low-level features are augmented with a “bag of keypoints”-based feature, which is invariant to affine transformations. An image filtering approach is also proposed to filter out irrelevant images and, thereby, reducing the search space for similarity matching. The similarity matching function is now dynamically updated (fused) based on the user’s feedback information. In addition, we performed exhaustive experimental evaluation and result analysis by considering different parameters and retrieval scenarios.
The block diagram of the proposed image retrieval framework is shown in Fig. 1. As can be seen from the top portion of Fig. 1, various image features are extracted offline (A) in a feature extraction subsystem (C) and stored in a feature index (E) for the database images. In addition, image features are combined and classified by the SVM to generate a category index file where for each images the class confidence or probability scores are stored for later filtering purpose. For a query image, similar feature extraction is performed (D) as database images as shown in the bottom portion of Fig. 1. However, instead of performing the similarity matching, the category of a query image is determined as probabilistic outputs or class confidence scores based on the classification subsystem. Next, this output is sent to the filtering subsystem to select candidate images for further similarity matching. In addition, based on the online category prediction (B), the predefined category-specific feature weights are utilized in a linear combination of similarity matching function as shown in the middle portion (F). After obtaining a ranked-based retrieval result (as shown in the left side of the Fig. 1), users next provide the feedback about relevant images and that information is utilized to update the final feature weights for the next retrieval iterations.
Fig. 1.
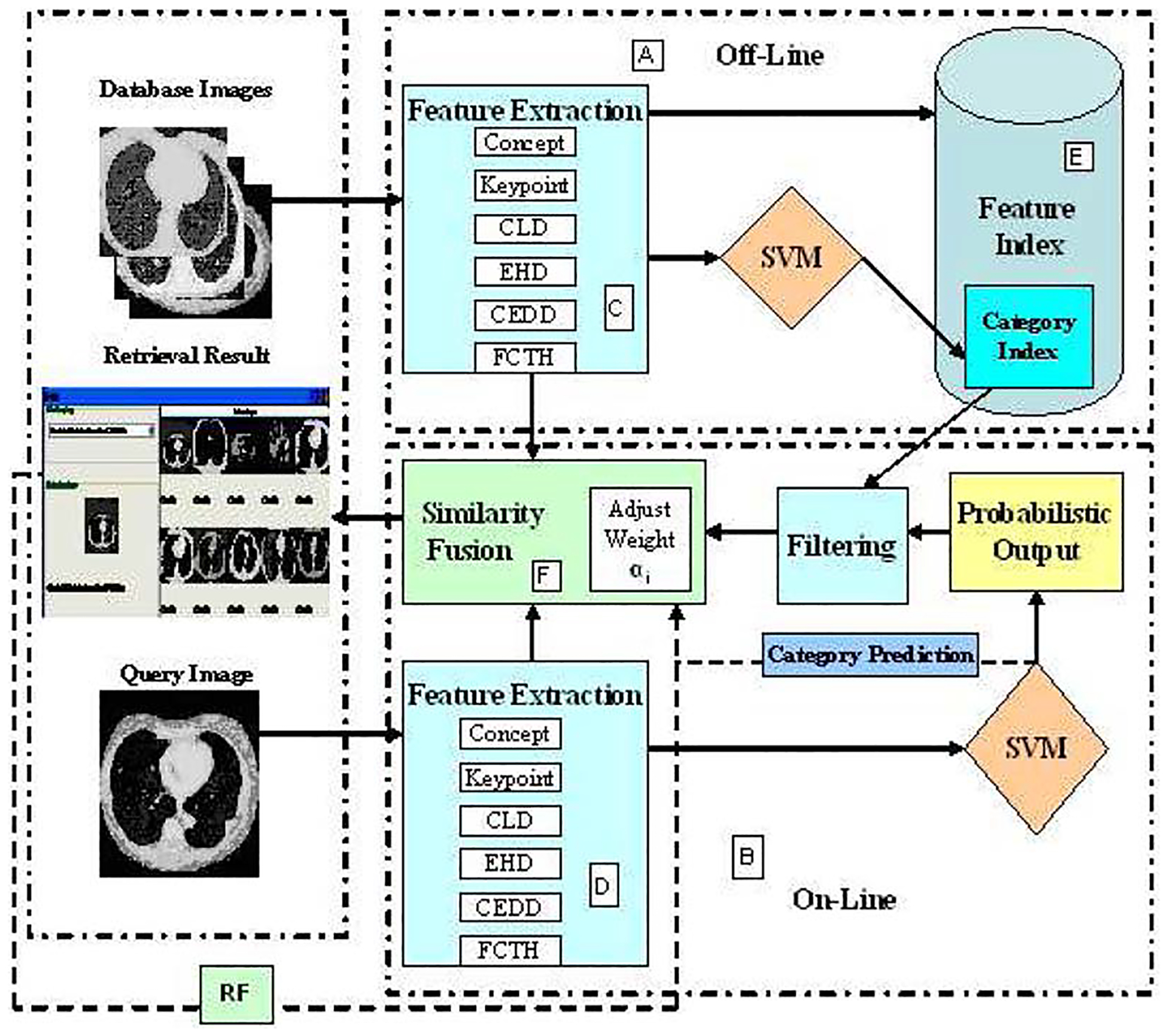
Block diagram of the classification-driven image retrieval framework.
The rest of the paper is organized as follows. Section II presents the different feature representation schemes at the concepts, keypoints, and low-level feature spaces. Section III describes the image categorization and filtering approaches at a global level by utilizing the multiclass SVM. The similarity fusion approach based on image classification and RF is presented in Section IV. The experiments and the analysis of the results are presented in Sections V and VI, respectively, and finally Section VII provides the conclusion.
II. Image Feature Representation
The performance of a classification and/or retrieval system depends on the underlying image representation, usually in the form of a feature vector. In a heterogeneous medical image collection, it is possible to identify specific local patches in images that are perceptually and/or semantically distinguishable, such as homogeneous texture patterns in gray level radiological images, differential color, and texture structures in microscopic pathology and dermoscopic images. The variation in the local patches can be effectively modeled as local concepts [16] analogous to the keywords in text documents by using any supervised learning-based classification techniques, such as the SVM [16].
For the SVM training, the initial input to the system is the feature vector set of the patches along with their manually assigned corresponding concept labels. Images in the dataset are annotated with the concept labels by fixed partitioning each image into regions as , where each is a combined color and texture feature vector. For each , the concept probabilities are determined by the prediction of the multiclass SVMs as [17]
| (1) |
Finally, the category label of is determined as , which is the label of the category with the maximum probability score. Based on this encoding scheme, an image is represented as a vector of weighted concepts as
| (2) |
where each denotes the weight of a concept in image , depending on its information content. The popular “tf–idf” term-weighting scheme [18] is used in this paper, where the element is expressed as the product of local and global weights.
In a heterogeneous medical collection with multiple modalities, images are often captured with different views, imaging and lighting conditions, similar to the real world photographic images. Ideally, the representation of such images must be flexible enough to cope with a large variety of visually different instances under the same category or modality, yet keeping the discriminative power between images of different modalities. In this paper, we extract such robust and invariant features from images as “bag of keypoints” [19].
In addition to the previous features, the MPEG-7 [20]-based color layout descriptor (CLD) and edge histogram descriptor (EHD) and descriptors from the lucene image retrieval library [21], such as fuzzy color texture histogram (FCTH) and color edge direction descriptor (CEDD) are extracted to represent images from different perspectives.
III. Multiclass SVM-Based Categorization
The variation of the medical image categories at a global level (e.g., modalities, body parts, and orientations) can be effectively modeled by using any supervised learning techniques. The SVM [22] is used to classify images to multiple categories. A number of methods have been proposed for its extension to multiclass problems to separate mutually exclusive classes essentially by solving many two-class problems and combining their predictions in various ways [23]. In this research, we utilize a multiclass classification method by combining all pairwise comparisons of binary SVM classifiers, known as one-against-one or pairwise coupling (PWC) [17]. PWC constructs binary SVM’s between all possible pairs of classes. Hence, this method uses binary classifiers, each of which provides a partial decision for classifying a data point. During the testing of a feature , each of the classifier votes for one class. The winning class is the one with the largest number of accumulated votes. In [24], it was shown that the one-against-one method is more suitable for practical use then the other method, such as one-against-all.
For the SVM training, the input is a feature vector set of training images in which each image is manually annotated with a single category label selected out of categories. So, a set of labels are defined as , where each characterizes the representative global image category. In this context, given a feature vector , the multiclass estimates the probability or confidence scores of each category as
| (3) |
The final category of a feature is determined based on the maximum probability score.
A. Classifier Combination
The feature descriptors as described in Section II are complementary in nature and represent image data that can lead to a better or robust classification result. There are two approaches to using different features: 1) concatenate features to a long composite vector and 2) use multiple classifiers on individual feature vectors—here the order and selection of classifiers of importance [25].
Four popular classifier combination techniques derived from Bayes’s theory, such as the product, sum, max, and mean rules [25] are considered for the expert combination strategies. Since the outputs of the classifiers are to be used in combination, the a posterior probabilities in the range of [0, 1] for each category will serve this purpose [15]. A multiclass SVM classifier on a combined feature space (e.g., early fusion) or several classifiers on individual feature spaces are combined or fused by the previous rules (e.g., late fusion) and finally classify the unknown query images to the category with the highest obtained probability value.
Algorithm 1.
Classification-Based Image Filtering
| (Off-line): Select a set training images of categories for SVM learning. |
| (Off-line): Based on the classifier prediction, store the category vectors (4) of database images as a category index. |
| (On-line): For a query image , determine the category vector as based on (4). |
| for to do |
| Consider the top ranked category labels for and after sorting the elements in the category vectors. |
| Construct the category label sets as and for the top ranked categories of and respectively. Here, and . |
| if then |
| Consider for further similarity matching. |
| end if |
| end for |
B. Image Filtering
We utilize the information about category prediction of query and database images for image filtering to reduce the search space. The output of the previous classification approach form a -dimensional category vector of an image as follows:
| (4) |
Here, , denotes the probability or class confidence score that an image belongs to the category in terms of the feature vectors based on applying the previous early or late fusion strategies for classification.
During the offline indexing process, this outputs as category vector of the database images are stored as category index along with the feature indices in a logical database. When the system is searched based on an unknown query image, similar feature extraction and category prediction stages are performed online. The category vector of a query image based on (4) and the similar vectors of the database images (stored as a category index) are considered to find out whether a target image is a good candidate for similarity matching, thereby filtering out irrelevant images for further consideration.
In this approach, instead of only considering the image categories based on the highest obtained probability values, nearest classes of the query and target images are considered based on their sorted outputs of the classification. Next it is verified whether there is any overlap of classes between the query and target images. Generally, the value of is kept much smaller compared to the total number of classes in order to ignore the distant classes and perform the filtering effectively. A target image is only selected for further matching if at least one common category is found out between the top categories of the query image and itself. By performing this step, we decrease the risk of searching images entirely in a wrong place due to the missclassification error.
The steps of the filtering algorithm are presented in Algorithm 1.
IV. Similarity Fusion
It is challenging to find a unique feature representation to compare images accurately for all types of queries. Feature descriptors at different levels of image representation are in diverse forms and may be complementary in nature. In information retrieval (IR), more specifically in text retrieval, data fusion or multiple-evidence combination describes a range of techniques where multiple pieces of information are combined to achieve improvements in retrieval effectiveness [26], [27]. Many researchers have argued that better retrieval effectiveness may be gained by exploiting multiple query representations, retrieval algorithms, or feedback techniques and combining the results of a varied set of techniques or representations [27].
CBIR also adopts some of the ideas of the data fusion research in document retrieval. The most commonly used approach here is the linear combination of similarity matching of different features with predetermined weights. In this framework, the similarity between a query image and target image is described as
| (5) |
where and are the similarity matching function (generally Euclidean) in individual feature spaces and are weights (generally decided by users or hard coded in the systems) within the different image representation schemes within our framework. However, there is a problem with the aforementioned hard-coded or fixed weight-based similarity matching approach. In this approach, for example, a color feature will have the same weight for the search of the microscopic pathology or X-ray images. Although, the importance of the color feature is negligible for many modalities, such as X-ray, CT, or MRI. The following sections presents a fusion-based linear combination scheme based on the online category prediction of a query image.
A. Category-Specific Similarity Fusion
In this approach, for a query image, its category at a global level is determined by employing the SVM learning. Based on the online category prediction of a query image, precomputed category-specific feature weights (e.g., ) are utilized in the linear combination of the similarity matching function. Based on this scheme, for example, a color feature will have more weight for microscopic pathology and dermatology images, whereas edge and texture related features will have more weights for the radiographs. The steps involved in this process are depicted in Algorithm 2.
From the previous algorithm, we can see that instead of using the predetermined hard-coded or fixed weight-based approach, the precomputed category-specific feature weights (e.g., ) based on the online category prediction are utilized in the linear combination similarity matching function for each query.
B. RF-Based Dynamic Similarity Fusion
A user might have a different interpretation of the semantic description in his/her mind or the prediction of the classifier might go wrong. Hence, it may be advantageous to have the option to interact with the system to refine the search process, such as RF. This section presents a RF-based similarity fusion technique where feature weights are updated at each iteration by considering both the precision and the rank order information of relevant images in the individual result lists based on the feedback from the users. As a result, the final rank-based retrieval is obtained through an adaptive and linear weighted combination of overall similarity fusing individual level similarities.
Algorithm 2.
Category-Specific Similarity Fusion Approach
| (Off-line): Store manually-defined category specific feature-weights for similarity matching. |
| (On-line): For a query image , calculate individual feature vectors , where . |
| For each feature, get a category prediction based on the probabilistic output of (3) by applying SVM. |
| Combine the outputs by applying any of the combination rules (e.g., sum, max, prod, min). |
| Get the final category label as of the query image. |
| Consider the individual features weights for the query image category . |
| Finally, combine the similarity scores with the weights based on similarity fusion in (5). |
| Finally return the images based on the similarity matching values in descending order to obtain a final ranked list of images. |
In this approach, to update the feature weights (e.g., ), we at first perform similarity matching based on equal weighting of each feature. After the initial retrieval result, a user needs to provide a feedback about the relevant images from the top returned images. For each ranked list based on individual similarity matching, we also consider top images and measure the effectiveness as
| (6) |
where if image in the rank position is not relevant based on user’s feedback and for the relevant images. Hence, the function monotonically decreasing from 1 (if the image at rank position 1 is relevant) down to zero (e.g., for a relevant image at rank position ). On the other hand, is the precision at top , where be the number of relevant images in the top retrieved result. Equation (6) is basically the product of two factors, rank order and precision. The rank order factor takes into account the position in the retrieval set of the relevant images, whereas the precision is a measure of the retrieval accuracy, regardless of the position. Generally, the rank order factor is heavily biased for the position in the ranked list over the total number of relevant images and the precision value totally ignores the rank order of the images. To balance both the criteria, we use a performance measure that is the product of the rank order factor and precision. If there is more overlap between the relevant images of a particular retrieval set and the one from which a user provides the feedback, then the performance score will be higher. Both terms on the right side of (6) will be 1, if all the top returned images are considered as relevant. The raw performance scores obtained by the previous procedure are then normalized by the total score as to yield numbers in [0, 1] where . For the next iteration of retrieval, these normalized scores are utilized as the weights for the respective features in the linear combination of similarity measures as
| (7) |
where . The steps involved in the weight updating and similarity matching processes are described in Algorithm 3.
Algorithm 3.
RF-based Similarity Fusion Approach
| 1: Initially, consider the top images by applying similarity fusion (5) based on an equal feature weighting. |
| 2: Obtain the user’s feedback about relevant images from the top images. |
| 3: Calculate the new query vector as the mean vector of the relevant images. |
| 4: For each ranked list based on individual similarity matching, also consider top images and measure the effectiveness as by utilizing equation (6). |
| 5: Normalize the effectiveness or weight score to be in the range [0,1]. |
| 6: Utilize the normalized scores as updated weights in the similarity function of equation (7) for the final retrieval. |
| 7: Continue, Steps 2 to 6 until no changes are noticed. |
V. Experiments
To evaluate the effectiveness of the proposed retrieval approach, exhaustive experiments were performed in a medical image collection. The collection comprises of 5 000 biomedical images of 30 manually assigned disjoint global categories, which is a subset of a larger collection of six different datasets used for retrieval evaluation campaign in ImageCLEF1 under the medical image retrieval track in 2007 [28]. In this collection, images are classified into three hierarchical levels. In the first level, images are categorized according to the imaging modalities (e.g., X-ray, CT, MRI, etc.). Next level is the image bodypart, and the final level is the orientation. The categories are selected based on analyzing the visual and some mixed-mode query topics during the last three years (2005, 2006, and 2007) of ImageCLEF campaign under the medical retrieval task. Around 80% of the images are gray level (e.g., X-ray, CT, MRI) and 20% are color images (e.g., microscopic pathology, histology, dermatology) with varying resolutions.
A. Training
A training set of about 2 400 images is used for SVM learning to categorize images at a global level. The images are classified into one of eight modalities (viz., computerized tomography (CT), graphics (GX), magnetic resonance imaging (MR), X-ray (XR), positron emission tomography (PET), optical imaging (PX), and ultrasound (US)) as defined in the the modality detection task in ImageCLEF 2010 [29]. There can be considerable intraclass heterogeneity in this classification, e.g., the PX class contains both microscopic images as well as photographs. For the SVM training, the RBF kernel is used with a tenfold cross validation (CV) to find the best values of tunable parameters and . The kernel parameter controls the shape of the kernel and regularization parameter controls the tradeoffs between margin maximization and error minimization.
The best values of and for different feature representations are computed and used to train the SVM and generate model files. We use the LIBSVM software package [30] for the implementation of the SVM classifiers. For the local concept model generation based on the SVM learning, 30 local concept categories are manually defined, e.g., CT-Tissue-Brain, X-ray-Chest-Bone, etc. The training set consists of only 5% images of the entire dataset of 5 000 images (i.e., 250 images). To generate the local patches, each image in the training set is at first partitioned into an 8 × 8 grid generating 64 nonoverlapping regions. Only the regions that conform to at least 80% of a particular concept category are selected and labeled with the corresponding category label. For the SVM training, we again use the RBF kernel with a tenfold CV to find the best values of tunable parameters and . After finding the best values of the parameters and of the RBF kernel with a CV accuracy of 81.01%, they are utilized for the final training to generate the local concepts model. We utilized the LIBSVM software package [30] for implementing the multiclass SVM classifiers for both global and local concept classification.
To construct the codebook of keypoints based on the Self-Organizing Map (SOM)-based clustering, the similar training set of images as used for local concept learning are utilized. To find the optimal codebook that can provide the best retrieval accuracy in this particular image collection, the SOM is trained at first to generate 2-D codebook of four different sizes as 256 (16 × 16), 400 (20 × 20), 625 (25 × 25), and 1600 (40 × 40) units. After the codebook construction process, all the images in the collection are encoded and represented as “bag of keypoints” as described in Section II. For training of the SOM, we set the initial learning rate as due to its better performance. By comparing the retrieval performances based on precision recall, we finally choose a codebook of size 400 for the generation of the keypoints-based feature representation and the consequent classification and retrieval evaluation.
VI. Results
To measure classification performance, we use a test set of 2620 images provided by the ImageCLEFmed’10 organizers [29]. Individual classification accuracy is as follows: Concept: 71%, Keypoint: 63%, EHD, and CLD: 52%, respectively, FCTH: 63%, CEDD: 69%, and Combined: 79%. The best accuracy is achieved when classification is performed in the combined feature space, but at the computational expense of a much larger feature vector. Different combination method accuracies are as follows: 80%, sum: 80%, max: 76%, and mean: 79%., which is in line with the observation in [25]. As expected, combining classifiers on uncorrelated and complementary features benefits the performance.
For a quantitative evaluation of the retrieval results, we selected all the images in the collection as query images and used query-by-example as the search method. A retrieved image is considered a match if it belongs to the same category as the query image out of the 32 disjoint categories at the global level. Precision (percentage of retrieved images that are also relevant) and recall (percentage of relevant images that are retrieved) are used as the basic evaluation measure of retrieval performances [18]. The average precision and recall are calculated over all the queries to generate the precision–recall (PR) curves in various settings. Fig. 2 presents the PR curves of the individual feature spaces when the similarity matching is performed based on applying the Euclidean distance measure. The performances are compared to a similarity fusion approach by providing equal weight to each feature in a linear combination. By analyzing Fig. 2, we can observe that the best performance is obtained in terms of precision at each recall level when the similarity scores are combined for individual features. More specifically, it is noticeable that the performance is improved in terms of early recall (e.g., 0.0–0.4) when compared to the best performed single concept-based feature. The performance of the proposed similarity fusion approaches is shown in Fig. 3. For the category-specific fusion, the weights of individual features are assigned as follows: 1) for medical imaging modalities (CT, MR, US, XR, NM, PET, and US), Concept, CLD, and EHD features are weighted equally (in general) and twice as much as the remaining features and 2) for photographs (PX), keypoint features are most significant. For RF-based fusion, we consider top 20 (e.g., ) retrieved images from the previous retrieval iteration as the positive or relevant images to be considered for the fusion algorithm in next iteration. In the IR, community retrieval precision at the top 5, 10, and 20 retrieved items has been accepted as an adequate measure of performance. It is clear from Fig. 3 that the performance of the category-specific similarity fusion [e.g., Fusion (Category)] is improved when it is compared with the fusion based on equal weighting [e.g., Fusion (Equal Weight)] approach. In addition, the precision is further improved when similarity fusion is performed by considering one [e.g., Fusion (RF-1)] and two [e.g., Fusion (RF-2)] iterations of feedback information on top of the retrieval result obtained from the category specific fusion. In general, we achieved around 10%–15% performance improvement in precision at most recall levels (0.1–0.9) when our proposed similarity fusion approaches are compared to the equal weight-based similarity fusion. Overall, from the PR curves in Fig. 3, we can conjecture that the supervised learning, either in the form of classification or RF, helps to improve the retrieval results in terms of precision.
Fig. 2.

PR graphs in different feature spaces.
Fig. 3.

PR graphs for different similarity fusion approaches.
To test the retrieval effectiveness of the RF-based similarity fusion in terms of considering the number of top retrieved images as relevant one, experiment is performed with as shown is Fig. 4. The performances are compared by considering only one iteration of feedback information. From Fig. 4, it is observed that the best performance is achieved when we consider only top ten images (e.g., ). There is decrease in performances when the number of is increases as shown by the PR curves in Fig. 4. One possible reason is that due to the automatic feedback for experimental purpose, there can be many irrelevant images being considered as relevant when the number of increases and this affects the retrieval performance. On the other hand, this is actually a benefit for an interactive system like this as the users need to judge less images to provide feedback information to achieve an optimal precision.
Fig. 4.

Effect of the values of .
To check the consistency in RF-based similarity fusion in terms of number of iterations, we also consider 5 iterations of feedback and compared the performances by calculating average precision within the top 20 [e.g., P(20)] retrieved images. The performances are compared by considering only one iteration of feedback and two different setting for and . As the system converged, It was observed that the retrieval performance is consistent across number of iterations and for different values of
Finally, the retrieval experiment was performed with and without filtering on different fusion-based approaches to test the effectiveness and efficiency of the proposed method. Top three ranked category labels were used for the experiment and applying the similarity fusion approaches in the filtered image set, we achieved same PR curves as depicted in Fig. 3. This suggests that search performed on a relevant subset of image collection does not cause any degradation in retrieval accuracy due to the fact that the filtering algorithm only discard those images. Further, an important benefit of searching on a filtered image set is gain in computation time. We tested the efficiency of the fusion-based search schemes by comparing the average retrieval time with and without applying the filtering scheme. The experiment was performed in an Intel Pentium Dual-Core CPU at 3.40 GHz with 3.5 GB of RAM running Microsoft Windows XP SP2 Professional operating system. The linear search time without filtering was twice as much as search on the filtered image set, suggesting that the proposed method is both effective and efficient.
VII. Conclusion
In this paper, a novel learning-based and classification-driven image retrieval framework is proposed for diverse medical image collections of different modalities. In our approach, we directly link classification to retrieval. In this framework, the image category information is utilized directly to filter out irrelevant images and adjust the feature weights in a linear combination of similarity matching. We use the RF-based technique to update the feature weights based on positive user feedback. Retrieval performance is promising and clearly shows the advantage of searching images based on similarity fusion and filtering in terms of effectiveness and efficiency. Overall, this retrieval framework is useful as a front end for large medical databases where a search can be performed in diverse images for teaching, training and research purposes.
Acknowledgment
The authors would like to thank the CLEF [28], [29] organizers and the Radiological Society of North America (RSNA), for making the database available.
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM), and Lister Hill National Center for Biomedical Communications (LHNCBC).
Footnotes
References
- [1].Wong TC, Medical Image Databases. New York, LLC: Springer–Verlag, 1998. [Google Scholar]
- [2].Muller H, Michoux N, Bandon D, and Geissbuhler, “A review of content-basedimage retrieval applications—Clinical benefits and future directions,” Int. J. Med. Informat, vol. 73, no. 1, pp. 1–23, 2004. [DOI] [PubMed] [Google Scholar]
- [3].Smeulder A, Worring M, Santini S, Gupta A, and Jain R, “Content-based image retrieval at the end of the early years,” IEEE Trans. Pattern Anal. Machine Intell, vol. 22, no. 12, pp. 1349–1380, Dec. 2000. [Google Scholar]
- [4].Shyu CR, Brodley CE, Kak AC, Kosaka A, Aisen AM, and Broderick LS, “ASSERT: A physician-in-the-loop content-based image retrieval system for HRCT image databases,” Comput. Vis. Image Understand, vol. 75, pp. 111–132, 1999. [Google Scholar]
- [5].Hsu W, Antani S, Long LR, Neve L, and Thoma GR, “SPIRS: A web-based image retrieval system for large biomedical databases,” Int. J. Med. Informat, vol. 78, pp. 13–24, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rahman MM, Bhattacharya P, and Desai BC, “A framework for medical image retrieval using machine learning and statistical similarity matching techniques with relevance feedback,” IEEE Trans. Inf. Tech. Biomed, vol. 11, no. 1, pp. 59–69, Jan. 2007. [DOI] [PubMed] [Google Scholar]
- [7].Lehmann TM, Wein BB, Dahmen J, Bredno J, Vogelsang F, and Kohnen M, “Content-based image retrieval in medical applications—A novel multi-step approach,” Proc. SPIE, vol. 3972, pp. 312–320, 2000. [Google Scholar]
- [8].Müller H, Rosset A, Vallee J, and Geissbuhler A, “Integrating content-based visual access methods into a medical case database,” in Proc. Med Inf. Eur., St Malo, France, pp. 480–485. [PubMed] [Google Scholar]
- [9].Müller H, Kalpathy-Cramer J, Eggel I, Bedrick S, Radhouani S, Bakke B, Kahn CE Jr., and Hersh W, “Overview of the CLEF 2009 Medical Image Retrieval Track,” 10th Workshop of the Cross-Language Evaluation Forum, CLEF 2009, Corfu, Greece, Sep. 30–Oct. 2, 2009, Proceedings of LNCS, vol. 6242, pp. 72–84, 2010. [Google Scholar]
- [10].Mojsilovis A and Gomes J, “Semantic based image categorization, browsing and retrieval in medical image databases,” in Proc. IEEE Int. Conf. Image Process, 2002, vol. 3, pp. 145–148. [Google Scholar]
- [11].Lehmann TM, Güld MO, Deselaers T, Keysers D, Schubert H, Spitzer K, Ney H, and Wein BB, “Automatic categorization of medical images for content-based retrieval and data mining,” Comput. Med. Imag. Graph, vol. 29, pp. 143–155, 2005. [DOI] [PubMed] [Google Scholar]
- [12].Florea F, Müller H, Rogozan A, Geissbuhler A, and Darmoni S, “Medical image categorization with MedIC and MedGIFT,” in Proc. Med. Inf. Eur., Maastricht, Netherlands, pp. 3–11. [Google Scholar]
- [13].Zhou XS and Huang TS, “Relevance feedback for image retrieval: A comprehensive review,” Multimedia Syst, vol. 8, no. 6, pp. 536–544, 2003. [Google Scholar]
- [14].Rui Y and Huang TS, “Relevance feedback: A power tool for interactive content-based image retrieval,” IEEE Circuits Syst. Video Technol, vol. 8, no. 5, pp. 644–655, Sep. 1999. [Google Scholar]
- [15].Rahman MM, Antani SK, and Thoma GR, “A classification-driven similarity matching framework for retrieval of biomedical images,” presented at the 11th ACM Int. Conf. Multimedia Inf. Retrieval, National Constitution Center, Philadelphia, Pennsylvania, Mar. 29–31. [Google Scholar]
- [16].Rahman MM, Antani SK, and Thoma GR, “A medical image retrieval framework in correlation enhanced visual concept feature space,” in Proc. 22nd IEEE Int. Symp. Comput.-Based Med. Syst., Albuquerque, NM, Aug. 3–4, 2009, pp. 1–4. [Google Scholar]
- [17].Wu TF, Lin CJ, and Weng RC, “Probability estimates for multi-class classification by pairwise coupling,” J. Mach. Learning Res, vol. 5, pp. 975–1005, 2004. [Google Scholar]
- [18].Baeza-Yates R and Ribeiro-Neto B, Modern Information Retrieval, Reading, MA: Addison-Wesley, Jun. 1999. [Google Scholar]
- [19].Rahman MM, Antani SK, and Thoma GR, “Biomedical image retrieval in a fuzzy feature space with affine region detection and vector quantization of a scale-invariant descriptor,” presented at the 6th Int. Symp. on Visual Computing, Las Vegas, NV, Nov. 29–Dec. 1. [Google Scholar]
- [20].Chang SF, Sikora T, and Puri A, “Overview of the MPEG-7 standard,” IEEE Trans. Circ. Syst. Video Technol, vol. 11, no. 6, pp. 688–695, Jun. 2001. [Google Scholar]
- [21].Grubinger M, Clough P, Hanbury A, and Müller H, “Lire: Lucene image retrieval: An extensible java CBIR library,” in Proc. 16th ACM Int. Conf. Multimedia, Vancouver, British Columbia, Canada, 2008, pp. 1085–1088. [Google Scholar]
- [22].Vapnik V, Statistical Learning Theory. New York: Wiley, 1998. [Google Scholar]
- [23].Duan K and Keerthi SS, “Which is the best multiclass SVM method? An empirical study,” presented at the 6th Int. Workshop on Multiple Classifier Syst., LNCS 3541, Seaside, CA, Jun. 13–15, 2005. [Google Scholar]
- [24].Hsu CW and Lin CJ, “A comparison of methods for multi-class support vector machines,” IEEE Trans. Neural Netw, vol. 13, no. 2, pp. 415–425, Mar. 2002. [DOI] [PubMed] [Google Scholar]
- [25].Kittler J, Hatef M, Duin RPW, and Matas J, “On combining classifiers,” IEEE Trans. Pattern Anal. Mach. Intell, vol. 20, no. 3, pp. 226–239, Mar. 1998. [Google Scholar]
- [26].Fox EA and Shaw JA, “Combination of multiple searches,” in Proc. 2nd Text Retrieval Conf, 1994, pp. 243–252 (NIST Special Publication; ). [Google Scholar]
- [27].Lee JH, “Combining multiple evidence from different properties of weighting schemes,” in Proc. 18th Annu. ACM-SIGIR, 1995, pp. 180–188. [Google Scholar]
- [28].Müller H, Deselaers T, Kim E, Kalpathy C, Jayashree D, Thomas M, Clough P, and Hersh W, “Overview of the ImageCLEFmed 2007 medical retrieval and annotation tasks,” in Proc. LNCS 8th Workshop Cross-Lang. Eval. Forum, 2008, vol. 5152, pp. 472–491. [Google Scholar]
- [29].Müller H, Kalpathy-Cramer J, Eggel I, Bedrick S, Kahn CE Jr., and Hersh W, “Overview of the CLEF 2010 medical image retrieval track,” presented at the Working Notes for the CLEF Workshop, Padova, Italy, 2010. [Google Scholar]
- [30].Chang CC and Lin CJ. (2001). “LIBSVM: A library for support vector machines,” [Online]. Software available at http://www.csie.ntu.edu.tw/cjlin/libsvm
- [31].Tang LH, Hanka R, Ip HHS, and Lam R, “Extraction of semantic features of histological images for content-based retrieval of images,” in Proc. IEEE Symp. Comput.-Based Med. Syst, 2000, p. 193. [Google Scholar]
- [32].Mikolajczyk K, Tuytelaars T, Schmid C, Zisserman A, Matas J, Schaffalitzky F, Kadir T, and Van Gool L, “A comparison of affine region detectors,” Int. J. Comput. Vis, vol. 65, pp. 43–72, 2005. [Google Scholar]
- [33].Xu L, Krzyzak A, and Suen CY, “Methods of combining multiple classifiers and their applications to handwriting recognition,” IEEE Trans. Syst. Man. Cybern, vol. 23, no. 3, pp. 418–435, May/Jun. 1992. [Google Scholar]


