Abstract
In the mouse, random mutagenesis with N-ethyl-N-nitrosourea (ENU) has been used since the 1970s in forward mutagenesis screens. However, only in the last decade has ENU mutagenesis been harnessed to generate a myriad of new mouse mutations in large-scale genetic screens and focused, smaller efforts. The development of additional genetic tools, such as balancer chromosomes, refinements in genetic mapping strategies, and evolution of specialized assays, has allowed these screens to achieve new levels of sophistication. The impressive productivity of these screens has led to a deluge of mouse mutants that wait to be harnessed. Here the basic large- and small-scale strategies are described, as are the basics of screen design. Finally, and importantly, this review describes the mechanisms by which such mutants may be accessed now and in the future. Thus, this review should serve both as an overview of the power of forward mutagenesis in the mouse and as a resource for those interested in developing their own screens, adding onto existing efforts, or obtaining specific mouse mutants that have already been generated.
INTRODUCTION
Scientists have come to use the mouse for many, diverse purposes: to study cancer; to model human diseases; to understand development, neurobiology, and behavior; and to examine specific cellular and molecular mechanisms. Mouse mutants have been appreciated for millenia, and the development of technologies, such as gene targeting, has allowed functional analysis of specific genes. However, the sequencing of the mouse genome and the explosion in mouse forward genetic screens have made systematic genetic interrogation of mammalian biologic processes truly feasible. In a remarkable global effort, the mouse community has devoted itself to generating genetic resources that will help us understand all biologic processes better and has set the stage for an unprecedented productive time in mouse and mammalian biology. This review presents an overview of forward genetic approaches in use by established centers and smaller groups and, it is hoped, will act as a resource for those interested in designing their own screens or wishing to access the archives of mutants generated in ongoing genetic screens. It should also highlight the continued need for support of large- and small-scale screens and mouse archives as invaluable scientific community resources.
A Brief History of ENU Mutagenesis in the Mouse
Because spontaneous mutations occur at a low frequency (∼5 × 10−6 per locus), mouse geneticists have searched for mutagens to generate new mutations efficiently. Towards this end, X-ray mutagenesis was explored. Whereas the frequency of X-ray mutagenesis (13 × 10−5 to 50 × 10−5 per locus) is 20 to 100 times that of spontaneous mutants, X rays cause a wide variety of chromosomal rearrangements, such as inversion, deletions, and translocations, which usually affect multiple genes. Similarly, the chemical chlorambucil also causes a range of chromosomal rearrangements, but with greater frequency (127 × 10−5 per locus) (49). In the 1970s, investigators at Oak Ridge National laboratory under the guidance of Bill Russell began to systematically explore the use of chemical mutagenesis with N-ethyl-N-nitrosourea (ENU) to produce new mouse mutations. These researchers found that chemical mutagenesis with ENU introduced primarily point mutations into spermatogonial stem cells at a frequency of ∼150 × 10−5 per locus (50). In addition, ENU is easy to administer, and ENU-treated males can be used to generate mutant progeny for many months (25). Next, a few mouse geneticists, including Monica Justice, Vernon Bode, Bill Dove, and Jean-Louis Guenet, embraced ENU mutagenesis to dissect the enigmatic T complex (23, 52). Mutagenesis of animals heterozygous for an 11-centimorgan (cM) deletion spanning the albino (c) and pink-eyed dilute (p) loci demonstrated that such a strategy could be used to efficiently recover novel recessive mutations in a chromosomal region-specific manner (41, 45, 47). In the early 1990s, the directed use of ENU led to identification of a new model for intestinal neoplasia and a novel behavioral mutant (the circadian rhythm Clock mutant) and aided in the positional cloning of novel genes (10, 29, 30, 60). Steve Brown at the Medical Research Council Mammalian Genetics Unit at Harwell, United Kingdom, and Rudi Balling at the Gesellschaft fuer Strahlenforschung (GSF) Research Center for Environment and Health in Munich, Germany, independently initiated two bold, large-scale mutagenesis programs and thereby ushered in a new era in mouse forward mutagenesis (21, 35). These programs were founded on the strong belief that the systematic production of mutations in nearly every mouse gene was feasible and would be invaluable, not just to mouse geneticists but to the scientific and clinical communities as a whole. As these initial efforts proved successful, additional centers have arisen around the world (Table 1) (4). Each center or research group engaged in ENU mutagenesis is contributing unique mutations, genetic screens, and expertise. In many ways, this global effort has brought out the best in the mouse genetics community. All centers have been striving to standardize phenotypes; develop and share assays; set up archiving, database, and genotyping resources; interface with gene trapping efforts; and facilitate the distribution of the resulting mutants to mouse geneticists and any other interested parties.
TABLE 1.
Centers performing ENU mutagenesis-based genetic screensa
| ENU mutagenesis center | Contact or website | Genetic approach | Mouse region(s)b | Summary of some major screens |
|---|---|---|---|---|
| ENU Mutagenesis Programme | www.mgu.har.mrc.ac.uk/research/mutagenesis | Dominant Recessive | Genome-wide Chr 13 36H | Basic neurobiology, hearing, vision, development, clinical chemistry |
| German ENU Mouse Mutagenesis Screen Project | www.gsf.de/ieg/groups/enu-mouse.html | Dominant Recessive | Genome-wide Genome-wide | Dysmorphology, immunology, clinical chemistry |
| Australian Phenomics Facility | www.apf.edu.au/ | Recessive | Genome-wide | Immunology |
| Baylor College of Medicine Mouse Genome Project | www.mouse-genome.bcm.tmc.edu/ENU/ENUHome.asp | Recessive | Chr 11 Chr 4 | Development |
| Centre for Modeling Human Disease | www.cmhd.ca/ | Dominant | Genome-wide | Development, bone, cardiac and kidney function, learning and memory, hematopoiesis |
| Genomics Institute of the Novartis Research Foundation | web.gnf.org/ | Recessive | Genome-wide | Neurobiology, vision, hearing, immunology, metabolic disorders, and cancer |
| Jackson Laboratory Neuroscience Mutagenesis Facility | www.jax.org/nmf/ | Dominant Recessive Recessive | Genome-wide Genome-wide Chr 5 | Neurobiology |
| Jackson Laboratory Mouse Heart, Lung, Blood and Sleep Disorders Center | www.jax.org/hlbs/documents/about_hlbs.html | Dominant Recessive | Genome-wide Genome-wide | Heart, lung, blood, and sleep disorders |
| Molecular Neurobiology at Northwestern University | http://www.northwestern.edu/neurobiology/faculty/takahashi.html | Dominant Recessive | Genome-wide Genome-wide | Circadian rhythm, general behavior, learning and memory, stress and psychostimulant response, vision |
| Mutagenesis Project at MRI | www.montana.edu/wwwmri/enump.html | Recessive | Genome-wide | Behavior, neurodegeneration, prion disease, hearing, ear and kidney development, peripheral myelination |
| Oak Ridge National Laboratory | http://lsd.ornl.gov/mgd/mutagenesis/ | Recessive Recessive Recessive Recessive | Chr 7 Chr 10 Chr 15 Chr X | Aging, auditory, drug and ethanol abuse, epilepsy, eye, general behavioral, neurohistology, social behavior |
| RIKEN Mutagenesis Center | www.gsc.riken.go.jp/Mouse/ | Dominant Recessive | Genome-wide Genome-wide | Behavior, hematological, urinalysis, clinical biochemical analysis, X-ray imaging; late onset: vision, blood pressure, hearing, tumorigenesis |
| Tennessee Mouse Genome Consortium | tnmouse.org/ in collaboration with Oak Ridge | Recessive | Chr 7 Chr 10 Chr 15 Chr X | Aging, auditory, drug and ethanol abuse, epilepsy, eye, neurohistology, behavior |
| University of Pennsylvania, Philadephia | bucan@upenn.edu, in collaboration with Jackson Laboratory | Recessive | Chr 5 | Circadian and general behavior |
Adapted from reference 4 with permission of the publisher.
Chr, chromosome.
Efficient Mutagenesis of Mice with ENU
ENU causes random single-base-pair mutations by direct alkylation of nucleic acids (25). During DNA replication, these ethylated base pairs cause mistaken identity and introduction of point mutations. Occasionally small deletions can occur (53). In the mouse testis, ENU acts most effectively on spermatogonial stem cells. In mice, the most commonly reported mutations are AT-to-TA transversions or AT-to-GC transitions. In a survey of ENU-induced mutations, Noveroske et al. determined that 63% of mutations were missense mutations, 26% caused abnormal splicing, 10% resulted in nonsense mutations, and ∼1% caused “make-sense” mutations, in which a stop codon was converted to an amino-acid-coding codon (36). Results from subsequent sequence-based analyses are in line with these original observations (8, 40).
After male mice have been treated with ENU, a single mutagenized male can produce 100 to 150 progeny (“first generation” [G1]), each one of which represents one mutagenized gamete. Because ENU mutagenesis targets spermatogonial stem cells, of which a male mouse has roughly 150 to 200, all G1 animals are nonmosaic. ENU treatment reduces the number of spermatogonia in seminiferous tubules and thus results in temporary sterility (50). To avoid repeat identification of the same mutation in separate G1 progeny from the same ENU-treated G0 male, no more than 30 to 50 gametes or G1 animals are sampled per G0 male. The mutation rate depends directly on the ENU dosage and inbred strain chosen. In general, most groups choose to administer 250 to 300 mg ENU per kg of mouse body weight in three fractionated weekly doses or, less commonly, a single dose of 150 to 200 mg/kg. Optimized protocols for a variety of inbred strains have been established (24). So far, many strain backgrounds can be used in ENU mutagenesis experiments, although some strains survive ENU treatment significantly better than others.
On average, the high efficiency of ENU mutagenesis results in identification of a new mutation in any single locus in one out of 500 to 1,500 G1 animals (25). Since the mouse is estimated to have ∼22,000 to 25,000 genes, each G1 animal is expected to carry ∼25 mutations with functional consequences. These mutations result most commonly in hypomorphic alleles. Roughly one out of 10 mutations in a given gene is expected to cause a null allele. In addition, point mutations occasionally result in alleles with hypermorphic (increased), neomorphic (novel), or antimorphic (dominant negative) function (36). The mutation rate of individual loci can vary between genes. Because ENU preferentially alters AT base pairs, genes with higher GC contents may be affected less frequently than ones with lower GC contents. Furthermore, the sizes of the gene and its functional domains and sensitivity to improper folding all contribute to, but do not act as reliable predictors of, its mutability.
FORWARD GENETIC APPROACHES IN THE MOUSE
While initially only the most straightforward genetic approaches were used in the mouse, now genetic screens ranging from simple dominant screens to designer region-specific screens have been successful. An overview of these strategies and some of their successes is provided below.
High-Throughput Dominant Screens
At present roughly 70% of the 38,000 mutations identified in ∼1,500 genes responsible for human disorders are caused by point mutations, and many of these act dominantly or semidominantly (http://archive.uwcm.ac.uk//uwcm/mg/docs/hahaha.html). These observations have made a compelling case for dominant genetic screens to identify novel mutations that model human diseases or conditions. Thus, many large centers are engaged in using dominant screens that focus on physiological, developmental, immunological, and neurobiological phenotypes relevant to human diseases (Table 1). The design of such dominant genetic screens is straightforward. ENU-treated (G0) males are bred with normal females, and subsequently the resulting G1 progeny are assayed for phenotypes of interest (Fig. 1). A G1 male is considered to carry a mutation if multiple pups in a litter and in several subsequent litters show the same phenotype (21, 35). In a dominant screen ∼0.1 to 2% of the animals are expected to exhibit scorable deficits in a given pathway, and indeed the results from current efforts are consistent with this expectation (Table 2). The major centers engaged in dominant screens are listed in Table 1.
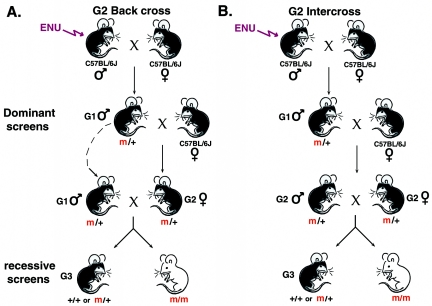
FIG. 1.
Schematic diagram of basic dominant and recessive schemes in the mouse. After a male G0 mouse of a chosen inbred (such as C57BL/6J) or hybrid strain is treated with ENU, he is bred with normal females. The resulting G1 mice are screened for dominant phenotypes of interest. To perform recessive screens, G3 animals may be generated in several ways. A. In the G2 backcross scheme, G1 males are bred with normal females to produce G2 animals. The original G1 male is crossed with three to six of his G2 daughters, and the resulting G3 animals are examined for the phenotype of interest. B. In the G2 intercross scheme, G2 animals are intercrossed with each other to produce G3 mice for recessive screens.
TABLE 2.
Mouse requirements for screening 1,000 G1 mice (ca. one genome equivalent)a
| Type of screen | G2 mice | G3 mice | Total mice screened to identify mutant G1 | Expected % of mutations affecting processb |
|---|---|---|---|---|
| Dominant | ∼40 G2/G1 animal with phenotype | None | 1,000 G1; ∼400-1,000 G2 | ∼0.1-2 (1-20) |
| Sensitized | ∼40 G2/G1 animal with phenotype | None | 1,000 G1 on sensitized background; ∼400-1,000 G2 | ∼0.1-5 (1-50) |
| Recessive (backcross) | 5-7 G2/G1 male; 5,000-7,000 G2 | ∼40 mice/G1; 40,000 G3 | 40,000 G3 | ∼10-20 (100-200) |
| Recessive (intercross) | 6-16 G2/G1 mouse; 6,000-12,000 G2 | ∼40-80 mice/G1; 40,000-80,000 G3 | 40,000-80,000 G3 | ∼10-20 (100-200) |
Estimates of mouse numbers that must be produced and screened to examine mice in dominant and recessive screens are shown. The expected numbers of mutations recovered are derived from current published screens and depend on the specificity, reliability, and sensitivity of the assay and screen design. For example, in dominant screens using highly variable assays, more G2 animals must be scored in order to identify a reliable, heritable mutant G1.
Shown in parentheses are the numbers of mutations expected upon screening 1,000 G1 mice or their progeny.
Accessing Specific Genetic Pathways with Sensitized Screens
While dominant mutations can unequivocally cause some human diseases, often mutations in multiple genes interact and contribute to disease progression. For example, while mutations in the presenilins cause Alzheimer's disease in all known carriers, mutations in apoE predispose to Alzheimer's disease and as-yet-unidentified genetic and environmental events determine whether patients actually develop Alzheimer's disease (9). Mutations in such “predisposing” genes, which otherwise might have effects too subtle to detect in standard dominant screens, can be uncovered if “sensitized” mouse strains and specialized assays are used. “Sensitization” can occur by genetic background or environmental or pharmaceutical challenge, such as salt challenge to detect susceptibility to hypertension. This strategy has been exploited extensively to identify modifiers in lower organisms and can be used in the mouse as well. For example, recently a large-scale suppressor screen identified mutations that ameliorate thrombocytopenia, the reduction in or lack of blood platelets. In this suppressor screen, mice lacking c-Mpl, the receptor for the cytokine thrombopoietin, showed severe thrombocytopenia and a reduction in megakaryocytes, megakaryocyte progenitor cells, and stem cells. Screening of 1,575 Mpl−/− mice carrying random ENU-induced mutations led to the recovery of two independent partial-loss-of-function alleles of c-Myb that rescued the thrombocytopenia. Thus, c-Myb−/+ Mpl−/− mice showed an increase in platelet number. Previously, embryonic lethality of mice homozygous for a c-Myb null allele had precluded any further analyses of later phenotypes. In the homozygous state, these hypomorphic c-Myb mutations lead to expansion of megakaryocyte and platelet production in the absence of thrombopoietin signaling (5). Similarly, many other extant mutations could serve as sensitized backgrounds in other modifier screens. An indicator that an existing mutation might be well suited as a sensitized strain is if it exhibits dramatically different phenotypes on various inbred backgrounds. Investigators have only begun to exploit this approach in the mouse, and doubtlessly these endeavors will be as fruitful as they have been in other organisms.
While genetic sensitization has been used extensively to identify modifiers in lower organisms, the mouse is ideally suited for recovering genes by environmental sensitization or pharmaceutical challenge. For example, small-scale genetic screens have recently identified mice with altered responsiveness to serotonin or dopamine (56, 63). Such pharmaceutically oriented strategies may be of particular value in uncovering the often mystifying molecular and physiologic mechanisms of drug action and thereby may facilitate development of individually tailored treatments for human patients.
High Output of Recessive Screens
Around the world, many centers are engaging in high-throughput dominant screens but fewer are engaging in recessive screens, because managing large-scale genome-wide recessive screens is a more complex logistical task and requires more mouse holding space, personnel, and money. Nonetheless, a few centers are actively and productively performing recessive screens (Table 1). For instance, the GSF in Munich and the ENU Mutagenesis Programme at Harwell are performing genome-wide, phenotype-wide recessive screens; the Australian ENU Mutagenesis Center is focusing primarily on recessive immunologic phenotypes; and the Baylor College of Medicine Mouse Genome Project is performing a largely chromosome region-specific recessive screen (21, 22, 28, 32). In many ways recessive screens are ideally suited for focused screens, both large and small scale, because of their extreme productivity (Table 2) (19, 26, 65, 68). Thus, for example, in a small-scale recessive screen for neurodevelopmental mutations, our group recovered eight neurodevelopmental mutants upon screening 40 pedigrees with an immunohistological assay (L. Mar, E. Rivkin, D. Kim, J. Yu, and S. P. Cordes, unpublished data). In such cases, the specialized interests and expertises of investigators rapidly concentrate resources towards the genetic and phenotypic analyses of only mutations affecting the desired process, thus reducing costs greatly.
To screen for recessive mutations, two breeding schemes are most commonly used (Fig. 1). In the first of these each G1 male is used to create a three-generation pedigree, in which he is mated with three to six of his daughters (G2 females) (Fig. 1A). Six progeny from each G2 female must be analyzed to ensure 80% efficiency of “scanning of the genome.” The advantage of this approach is that the genotype of the G1 male remains “fixed” during the initial screen. Here, in a given litter one-fourth will exhibit the phenotype of interest, while litters from noncarrier females will not exhibit the deficit at all. Although this procedure appears daunting, its feasibility is shown by the recovery of a series of mutations that affect the sonic hedgehog signaling pathway, telencephalic development, and neonatal lethal mutations (2, 13, 15, 19, 26, 70).
An alternative strategy involves the intercrossing of G2 progeny (Fig. 1B). In this case, only 1/16 of G3 progeny will show the phenotype of interest, and larger numbers of mating cages must be maintained (Table 2). In the first of these schemes, a three-eighths genomic contribution from the original mutagenized strain will be present in the G3 progeny, while in the latter case this contribution has been reduced to one-fourth. Phenotypic contributions from multiple unlinked loci are theoretically reduced in the progeny from G2 intercrosses relative to those from G2 backcrosses. Thus, it would appear that the G2 intercross scheme may be advantageous in situations where strain-specific or newly induced quantitative trait loci may affect the phenotype significantly. Both schemes have been successful. The G2 backcross scheme has been used extensively by smaller groups, while the G2 intercross scheme has been used in an immunological screen by a larger consortium and in the chromosome-specific screens described in more detail below (Table 2) (2, 13, 15, 19, 22, 26, 28, 32, 70).
Chromosome Engineering and Region-Specific Screens
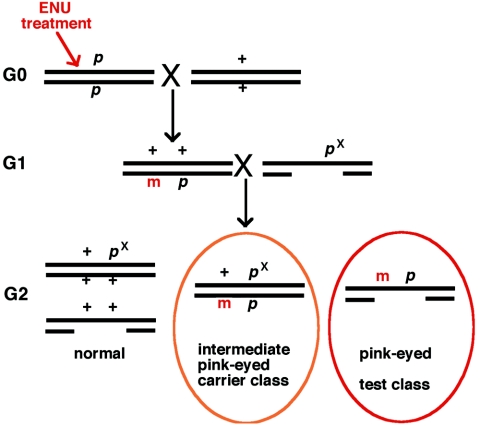
Gene Rinchik and his colleagues first showed that reasonably sized, visibly marked deletions could be used to detect recessive mutations in two cross hemizygosity screens, as shown in Fig. 2 (41, 45, 47). In this scheme, coat color alleles of various severity for albino (c) or pink-eyed dilute (p) were used to distinguish between chromosomes contributed by different parents and, thus, to allow identification of progeny carrying new mutations in the interval of interest (43, 46). In this manner, embryonic lethal, developmental mutations, such as eed and fit-1, were first recovered (39, 44). In the example shown in Fig. 2, after treatment with ENU, male mice that were homozygous for the pink-eyed dilute mutation, which causes pink eyes and severe dilution of the eumelanin (black) in the mouse's coat color, were crossed with normal females. The resulting G1 males were crossed with female mice that were compound heterozygotes for a deletion spanning the p locus and an intermediate allele p, known as pX. G2 animals were identified by their coat colors: normal agouti-colored mice did not carry the new potentially mutated chromosome 7 region, and animals with the intermediate color potentially carried new mutations in this chromosomal interval. Finally, pink-eyed dilute mice uncovered the effects of any new ENU-induced mutations in this region of chromosome 7. The absence of pink-eyed dilute weanlings from a given G1 male indicated that a new recessive lethal mutation had been generated within the deleted region of chromosome 7. Furthermore, mating of mutant mice to mice carrying smaller deletions within this interval expedited localization of the affected loci. Many extant “classical” chromosomal deletions and inversions have been actively maintained or cryopreserved and are listed in the International Mouse Strain Resources database of the Jackson Laboratory and the Medical Research Council Mammalian Genetics Unit (www.jax.org/pub-cg/imrlist) or in the Mutant Mouse database of the Oak Ridge National Laboratory (www.bio.org.gov/htmouse). Currently, other research groups and mutagenesis centers are using some existing deletions to identify new region-specific mutations (57). The regions chosen in these efforts are often gene-rich regions implicated in specific human disorders.
FIG. 2.
Screening for recessive mutations by using region-specific deletions. In the example shown, which is adapted from reference 42 with permission of the publisher, male mice homozygous for the pink-eyed dilute mutation are treated with ENU and subsequently mated with normal females. The resulting G1 mice are heterozygous for any new mutation, designated by “m,” and the p mutation and are bred with mice compound heterozygous for a deletion spanning the p locus and the pX allele, which gives distinct intermediate eye and coat colors. In the resulting G2 progeny, all normal agouti-colored progeny do not carry a new ENU-induced mutation on this region of chromosome 7. Animals that show the intermediate pink-eyed phenotype are carriers for any new mutations introduced on this chromosome 7 region. All pink-eyed mice will uncover any newly induced, recessive mutations present on the deleted interval.
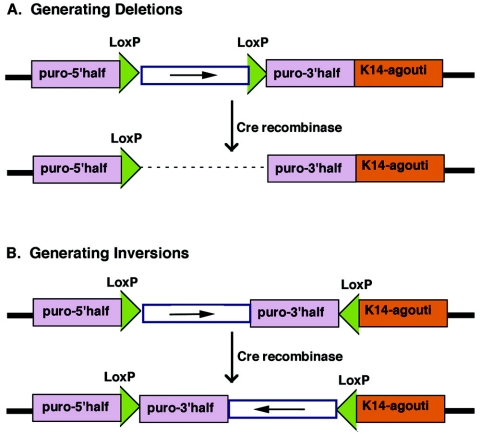
Advances in embryonic stem (ES) cell technology have made it possible to generate “designer” chromosomes containing region-specific rearrangements. Two of these strategies have been paired subsequently with ENU mutagenesis screens. In the first of these, the selectable thymidine kinase (tk) gene is introduced into a locus of interest by standard homologous recombination in ES cells (64). Subsequently, these ES cells are irradiated to induce random deletions. Clones in which the targeted region has been deleted are selected by loss of the tk marker, and the sizes of deletions are defined by using available flanking markers, such as simple sequence length polymorphisms (SSLPs). In this manner, a range of deletions of different sizes can be readily generated for a given interval. These deletions can then be used as reagents in region-specific screens as described above or in genetic mapping experiments. The second scheme makes use of Cre recombinase-mediated excision to generate desired chromosome-specific deletions, duplications, and inversions (66, 69) (Fig. 3). To generate chromosomes with defined region-specific rearrangements, a construct containing a single LoxP site and the 5′ half of the puromycin drug resistance gene is introduced into a specific site in the ES cell genome. Subsequently, another construct containing a LoxP site, the other half of the puromycin gene, and the K14-agouti minigene, which causes yellowish coat color and acts to mark the rearranged chromosome, is introduced in either a site-specific or random manner. Upon addition of Cre recombinase, DNA flanked by LoxP sites that are in the same orientation is deleted, while a DNA region flanked by LoxP sites that are oriented in opposing directions is inverted. To expedite generation of other region-specific chromosomal rearrangements, two ES cell libraries, in which either the 5′ or 3′ constructs have been randomly inserted, have been generated and are available for screening. These libraries can be used to isolate the 5′ or 3′ constructs flanked by region-specific genomic DNA, which can subsequently be used to generate site-specific integration into the partner ES cell line. Thus, researchers can customize mice with desired chromosome rearrangements for genetic screens or mapping.
FIG. 3.
Basic strategy for engineering chromosome-specific deletions and inversions. To generate inversions or deletions, a construct containing a LoxP site and the 5′ half of the puromycin drug resistance gene is inserted either randomly or in a targeted manner into the chromosome in ES cells. A. To create deletions, a construct containing a second LoxP site, the 3′ half of the puromycin gene, and the K14-agouti coat color marker, which causes a yellowish coat color, are introduced either in a targeted manner into a predetermined locus or randomly on the same chromosome. Electroporation of a Cre recombinase-expressing construct results in excision of the sequences flanked by the LoxP site and activation of the puromycin gene. B. To generate inversions, the same construct as was used for generation of deletions is inserted into the chromosome, except this time in the opposite orientation. This time in the presence of Cre recombinase, the puromycin gene is activated but the sequences flanked by LoxP sites are inverted (66).
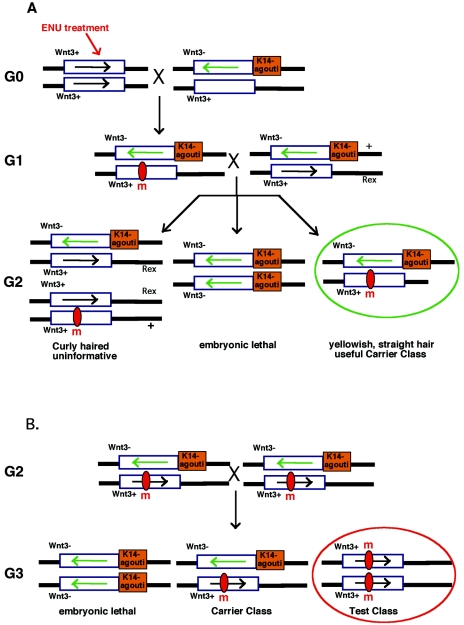
A genetic strategy particularly popular for use in Drosophila melanogaster has involved the use of defined chromosomal inversions as “balancers.” Recombination is suppressed in the inverted region, and thus tracking of the mutagenized noninverted chromosomal region is simplified. A major advantage of this approach is that animals carrying large chromosomal inversions are in general viable and indistinguishable from normal animals. By contrast, many chromosomal deletions, especially larger ones, have deleterious consequences. In the mouse, the power of this approach was demonstrated by Monica Justice's group (28) (Fig. 4). Here a balancer, which contained a 24-cM (34-Mb) inversion of mouse chromosome 11, was used in a recessive screen. In this screen, ENU-treated C57BL/6J males were mated with females carrying the balancer chromosome, which was marked by the K14-agouti coat color marker and by a mutation in the Wnt3 gene, which is embryonic lethal when homozygous. The resulting G1 animals were crossed with mice carrying one copy of the balancer chromosome and one copy of chromosome 11 that carried the Rex mutation, which caused a dominant curly-hair phenotype and marked the nonmutagenized chromosome in the resulting G2 progeny. Informative G2 progeny were identified by their yellowish, straight hair and were intercrossed. As a consequence of the chromosome engineering strategy used to construct this balancer, mice homozygous for the inversion lacked the Wnt3 gene and died in utero by embryonic day 10.5. Thus, only yellowish mice were examined for possible mutant phenotypes linked to chromosome 11. If no yellowish progeny were detected, then the mutation acted as a lethal mutation. Using this approach, 230 recessive mutations that affected processes including patterning defects, growth and endocrine defects, neurological anomalies, and blood defects were identified. Eighty-eight out of these 230 mutations were located in the inverted chromosome 11 region. Fifty-five (∼65%) of the 88 mutations located on chromosome 11 were recessive lethal mutations (28). Thus, such region-specific approaches are highly effective and can begin to approach saturation of specific chromosomal regions with point mutations.
FIG. 4.
A genetic screen using a “balancer” chromosome in the mouse. A. Normal male mice are treated with ENU and subsequently bred with females heterozygous for a balancer chromosome. In the example shown, the balancer chromosome is marked by the presence of the K14-agouti minigene, which causes a yellowish coat color in mice, and the recessive lethal Wnt3 mutation (Wnt3−). The resulting G1 mice are crossed with animals heterozygous for the balancer and for the dominant Rex mutation, which causes curly hair and helps distinguish the unmutagenized chromosome in the resulting G2 progeny. Curly-haired progeny carry the unmutagenized chromosome and are uninformative. Mice homozygous for the balancer and, thus, also for the Wnt3 mutation die at 10.5 dpc in utero. Mice that have yellowish straight hair are carriers of any mutations that may have been introduced in the inverted region. B. To examine any recessive phenotypes, G2 carrier animals are intercrossed and their progeny analyzed. Once again mice homozygous for the balancer chromosome die at 10.5 dpc; mice with yellowish straight hair are carriers, and agouti (normal)-colored mice represent animals that potentially carry a mutation in the region of interest (28).
Generation of Allelic Series with Gene-Driven Approaches
Recent advances in high-throughput mutation detection methods and sperm cyropreservation have made gene-based mutational screens highly feasible. In the past, noncomplementation screens to generate new alleles of a given gene have proven to be highly efficient. Thus, for example, a new allele of the Kreisler (kr) segmentation gene was recovered upon mating ENU-treated males with females heterozygous for the recessive kr allele and screening 597 G1 progeny (10). Now it is possible to screen archives of DNA from G1 mutant progeny of ENU-treated males via a gene-based approach to generate an allelic series for a given gene. Mutations have been identified by analyzing PCR-amplified products of genomic DNA spanning individual exons and at least 30 to 50 bp of the flanking intronic regions by using a Transgenomic Wave machine that uses denaturing high-performance liquid chromatography (DHPLC) (40). Upon identification of a desired allele, the mouse strain is recovered from frozen sperm by in vitro fertilization. In this manner, investigators at Harwell have screened an archive of genomic DNA and cryopreserved sperm from over 6,000 G1 males and have identified 27 mutations in 15 genes tested. Out of these 27 mutations, 15 are predicted to have functional consequences and represent new alleles for 9 out of the original 15 genes. It should be noted that no new mutations were identified in an additional six genes examined. Such screening is available to academic collaborators as a community resource. Details of how to access this resource are provided by Quwailid et al. (40).
Similarly, the RIKEN mutagenesis center has prepared such an archive from over 7,000 G1 male mice for such a gene-driven approach. Here the temperature gradient capillary electrophoresis method has been chosen to identify mutations of interest. Notably, productive collaborations with academic scientists outside Japan have been established (J. Roder, personal communication) (http://gsc.riken.go.jp/Mouse/main.htm). Other centers are no doubt also moving towards incorporating this approach.
An auxiliary approach is to recover new alleles via a gene-based approach from ENU-mutagenized ES cells (7, 31). Here, once ES cells have been mutagenized with ENU, they are replicated for freezing and DNA preparation. So far, DHPLC-based heteroduplex analysis of the PCR products was performed by the WAVE fragment analysis system and has been used to detect these mutations. Fortunately, ENU-treated ES cells remain germ line competent. Thus, they can be used to generate chimeric mice, which will eventually produce progeny derived entirely from the ES cell of choice. The power of this approach was demonstrated by the recovery of 29 mutations in SMAD2 and SMAD4 upon screening of 2,060 ENU-mutagenized ES cell clones (61). For the detection of these mutations, DHPLC-based heteroduplex analysis of the PCR products was performed by the WAVE fragment analysis system, as well. Clearly, this is an effective way to generate allelic series in the mouse, and the creation of screening and distribution centers would be highly beneficial.
Recent advances in ES cell technology, in particular the development of hybrid ES cell lines, will make this ES cell-based approach even more attractive in the near future (51). Traditional ES cell lines have been derived primarily from 129/SvJ mice and, upon tetraploid aggregation, can yield entirely ES cell-derived embryos that can survive up to 10.5 days postcoitum (dpc). By contrast, upon tetraploid aggregation, 129 × C57 hybrid ES cell lines can produce entirely ES cell-derived, fertile adult mice. Thus, this modification would forego the time-consuming and at times frustrating breeding of chimeric mice to obtain germ line transmission from the ES cell line.
In addition to the expansion of mutagenized mouse and ES cell archives, mutation detection strategies are steadily improving and direct sequencing is becoming an increasingly accessible and reasonably priced alternative. For example, recently splice mutations of cKIT were detected in highly pooled cDNA samples from a library of ∼40,000 mutagenized ES cell clones by using exon-skipping PCR primers (17). Ultimately, if such archiving and screening efforts are performed on a charge-back basis by academic centers or companies, it should be possible for any investigator to order an allelic series for any gene of choice.
ASSAYS FOR MUTANT IDENTIFICATION
The success of any genetic screen relies heavily on the assays chosen to identify mutants. The mouse genetic community has been very diligent and sometimes ingenious in designing assays to screen for mouse mutations that affect physiological, neurobiological, developmental, cellular, and gene regulatory processes. The two most important considerations when designing a genetic screen are the overall genetic strategy and the choice and design of robust assays. Some examples of such assays will be described below. The assay(s) used in a primary screen for mutants should ideally be simple, reasonably rapid, and standardized for the inbred strains and hybrid strains that will be used in the screen. These primary assays can be followed up with more specific and perhaps time-consuming secondary assays to define the phenotype more precisely. Furthermore, to avoid an overabundance of time-consuming false-positive results, assay variability among individual normal inbred or hybrid animals should be as low as possible. As specific traits vary greatly among inbred strains, care should be taken to choose strains for mutagenesis and genetic mapping that perform well in the desired assay(s). Strain choice will be discussed in more detail below. Finally, in the ideal scenario, extant mutants can be used to validate and optimize the assay chosen and test its ability to identify outliers on an individual or population basis.
The Issue of Inbred Strain Choice
The roughly 500 inbred mouse strains available provide a unique and invaluable resource for genetic screens. Because physiologic, behavioral, and developmental traits can vary substantially between inbred strains, strain choice is a critical feature in screen design. A variety of considerations, such as availability of genomic sequence and markers and performance in assays of interest, inform the strain choice. For example, in quite a few instances the C57BL/6J strain has been chosen as either the ENU-mutagenized strain or the backcross strain, because its genomic sequence is available in the public domain, many physiological assays have been performed on this background, and it performs well in many behavioral assays. In addition, mutant mice generated by homologous recombination in embryonic stem cells have been generated primarily on the 129Sv/J inbred background, which breeds poorly and is not ideally suited for behavioral or neurobiologic studies. Often the gene-targeted mutation has been subsequently bred onto the C57BL/6J background for phenotypic analyses. Other strains or strain combinations are chosen for similar reasons, and this choice is tailored to a given center's needs. There is no single strain that performs well in all assays or is ideally suited for recovery of mutations in all genetic pathways. Ultimately, this diversity in strain choice among centers is a strength.
Investigators can choose to perform the primary screen on either a uniform inbred background or a hybrid strain. These two approaches offer distinct advantages and disadvantages. By performing a screen on a uniform genetic background, one can assess phenotypic variability immediately. Thus, even though ultimately the mutation must be crossed onto a different inbred genetic background for genetic mapping, confidence of the initial phenotypic presentation will inform selection of which mutants to pursue. However, genetic mapping is slowed, as a suitable strain must be chosen for the mapping backcrosses and carrier hybrid animals must be identified. The choice of a hybrid strain offers the opposite pros and cons. In this instance the mutation is introduced onto an inbred G0 strain of choice, and the ENU-treated male is subsequently bred with females from a different inbred strain. The resulting G1 animals can be screened for dominant mutations or backcrossed to the mapping strain to produce G2 animals for use in recessive screen pedigrees. In this instance, phenotypes may be more variable in later G2 progeny tested, but if a phenotype is suitably robust, mapping is accelerated.
Physiologic Assays
Because mutagenesis centers are interested in using mouse mutations to model human diseases and conditions, a wide range of physiologic, clinically relevant assays have been developed, standardized, and streamlined. In general, an animal is considered a possible mutant if its assay results are ∼2 to 3 standard deviations outside the norm. However, in some instances subtler differences can be reliably detected. Descriptions of many physiological assays are available and regularly updated on center websites (Table 1). Common clinically relevant phenotypes examined include defects in diabetes, hematology, immunology, bone mineralization, cardiovascular and renal function, and behavior. In many ways the assays are miniaturized versions of those used in human clinics. For example, cardiovascular function can be assessed by measuring blood pressure and heart rate via a tail cuff (rather than the “arm cuff” used for humans) apparatus and obtaining an electrocardiogram (20). By contrast, some assays, such as those using low-energy X rays to determine body composition and bone density, are much easier to perform on a high-throughput basis in the mouse. Overall, an impressive array of physiologic assays has been developed. Ultimately, the recovery of mouse mutants affected in clinically relevant processes has proven the power of these assays and has set new standards for mouse biology as a whole.
Behavioral Assays
At present, the mouse offers the best system in which to study mammalian behavioral genetics. The recovery of the circadian activity Clock mutant demonstrated that robust, simple, specialized assays could allow identification of exciting behavioral mutations and intensified interest in forward behavioral genetic screens in the mouse (3, 60). Some of the most successful and robust behavioral screens include those for hearing and vision, which have identified a wide range of mutants (21, 27, 35, 58). However, many behavioral assays, such as the Morris water maze, which tests learning and memory, are too involved for primary high-throughput assays. At present all centers assess potential mutant animals for simple motor behavioral abnormalities by using the SHIRPA protocol (48). In this protocol, an observer assesses whether a mouse shows obvious neurobiologic abnormalities, such as an unsteady gait, decreased grip strength, or overall jumpiness. Quite a few mutations have been identified using this protocol. While these mutations provide important neurobiologic information, it is questionable how effectively they model common human neurobiologic or affective disorders. In human psychiatric genetics and mouse behavioral genetics, the concept of endophenotypes has become quite useful. Endophenotypes are specific neurobiologic or behavioral correlates of the disorder that act as reliable hallmarks of some but not necessarily all aspects of the disease. Thus, genetic screens have begun to focus on identifying mice the exhibit psychiatric endophenotypes. For example, prepulse inhibition (PPI) is a suppression of the startle reflex that occurs when an intense startling stimulus is preceded by a weaker “prepulse” stimulus. PPI deficits have been implicated in the biological bases of schizophrenia. Researchers are identifying possible mouse models for schizophrenia by screening animals for deficits in PPI. Other behavioral assays have been developed and are being refined for these purposes (11, 16, 34). In addition, there has been some interest in devising screens that uncover subtler deficits in specific neurotransmission systems. Thus, we have devised a screen to assess serotonin responsiveness in the mouse, while in independent efforts others have assessed dopaminergic responsiveness (56, 63). The results from these “pharmacologically” and genetically sensitized screens have been promising.
Developmental Assays
Genetic screens for developmental deficits have been strikingly productive in lower organisms and the mouse. In the past, many classical mouse mutations were identified by morphological abnormalities, which had developmental origins. The first ENU-based recessive screen performed in the mouse relied on screening for morphologically abnormal embryos and provided mutants that have dramatically enhanced our understanding of the sonic hedgehog signaling pathway (26). Subsequently, large-scale dominant screens, a neonatal lethal screen, and the aforementioned balancer screen have all used morphology to identify new and exciting developmental mutations (19, 21, 28, 35). Further sophistication has been achieved by adding a variety of visualization techniques as primary assays. For instance, new mutations that affect cortical development were identified using a transgenic mouse line that expresses the LacZ reporter in specific cortical regions (68). Many transgenic mouse lines with tissue-specific expression of LacZ or green fluorescent protein exist and could easily be used in similar forward genetic screens. Other techniques, which do not rely on the existence of transgenic mice, can be used to visualize specific tissues. For instance, in a recessive screen, we recovered recessive neurodevelopmental mutants by using an immunohistochemical assay to visualize neurons in 10.5-dpc embryos. Yu and colleagues used ultrasound visualization of embryonic cardiac function and structure to identify a series of recessive mutants (65). Such visualization techniques may be particularly well suited for focused recessive screens.
Assays for Mammalian Cellular Processes: Beyond Homology Searches
In yeast, Caenorhabditis elegans, and flies, forward genetic approaches have identified key molecules that govern cellular processes. Subsequent analyses of vertebrate homologues have expedited our understanding of these pathways in mammals. This approach, while productive, does not identify molecules acting specifically in mammalian processes. Forward genetic screens performed directly in the mouse would resolve this problem. For example, John Schimenti's group has performed an elegant screen to recover mutations in DNA double-strand break (DSB) repair (54). The majority of mammalian DSB repair genes have been identified by their homology to yeast genes. However, mammalian cells use homologous recombination and nonhomologous end-joining mechanisms, while yeast uses primarily homologous-recombination-based mechanisms. Furthermore, the observation that yeast lacks homologues for BRCA1, BRCA2, or protein kinase, DNA-activated, catalytic polypeptide points towards the existence of DSB repair genes that are unique to mammals. In the mouse, the efficiency of DSB repair can be assessed by examining micronucleus formation. When acentric or whole chromosomes are not incorporated into nuclei, micronuclei are formed. Micronucleus formation can be stimulated by DNA-damaging agents, such as gamma irradiation, and assessed by flow cytometry of small amounts of blood. A screen using this assay determined that micronucleus formation was increased in mice carrying mutations in known DNA repair genes, such as ataxia telangiectasia (ATM), and in novel genes, such as that responsible for the chaos-1 mutation (54, 55). One significant advantage of this strategy is that the association between cancer and DSB repair can be immediately assessed in these mutant mice. In the future, this assay and similar cellularly based assays will enrich our appreciation of features unique to cellular mechanisms in mammals. Thus, this area should prove to be particularly fertile ground for collaborative studies between cell and molecular biologists and mouse geneticists.
Assays for Regulatory Processes
ENU mutagenesis can be used to uncover genes involved in regulatory processes, such as imprinting or X inactivation, that are unique to mammals. The feasibility of this approach was demonstrated by a genetic screen for mutations that affect X inactivation (37). Three elements within the X-inactivation center (Xic) located on the X chromosome govern X inactivation. In addition, autosomal factors interact with the cis-acting elements within the Xic to determine X chromosome choice. In this genetic screen, the X-inactivation pattern was measured by using an efficient, quantitative, allele-specific reverse transcription-PCR assay to measure transcript levels from the X-linked gene Pctaire-1 (Pctk1) (6, 38). Female progeny inherited a maternal Cast/Ei X chromosome, which was more likely to be activated than the weak X from Mus dosmesticus and Mus musculus strains. Quantification of Pctk1 expression from the Mus castaneus and M. dosmesticus/musculus X chromosomes led to recovery of three loci that affect X inactivation, but not genomic imprinting, upon screening of 336 G1 females. Possible future extensions of this approach include the use of microarray-based technology and robotics to assay a larger set of gene expression patterns. In addition, other creative schemes that take advantage of phenotypes caused by loss of imprinting or X inactivation are possible.
GENE IDENTIFICATION
Advances in Genetic Mapping
The mouse genome sequence and advances in mapping schemes have accelerated genetic mapping of mutations (62). The identification of strain-specific polymorphisms, such as SSLPs and single-nucleotide polymorphisms (SNPs), now allows genetic mapping to occur on most strain combinations. As these can be detected by PCR-based techniques, small labs or larger centers can perform these experiments. Due to mouse genome sequencing efforts, the genome-wide coverage of SNPs exceeds that of validated SSLPs. The following databases can currently be used to identify potentially useful SNPs: http://www.ensembl.org/Mus_musculus/, http://mousesnp.roche.com/cgi-bin/msnp_public.pl, http://www.broad.mit.edu/snp/mouse/, http://www.nervenet.org/MMfiles/MMlist.html, http://snp.gnf.org, and http://www.ncbi.nlm.nih.gov/SNP/MouseSNP.cgi. Genotyping may proceed by assaying each affected individual with a total of ∼150 to 160 markers placed at 10- to 20-cM intervals along the chromosomes. In some cases, it is possible to bin affected and unaffected animals into two separate pools and to use these two pools to determine the basic chromosomal location. Furthermore, interval haplotype analysis, which assumes that meiotic recombination occurs only a few times per chromosome, requires far fewer markers (∼40 markers) and has been shown to be an effective and efficient mapping strategy (19, 33). These refinements simplify and expedite genetic mapping and ultimately make positional cloning of ENU-mutagenized genes highly feasible.
Gene Identification and Confirmation
Positional cloning of mutations often provides key entry points into understanding biological pathways. The completion and molecular annotation of the mouse genome sequence, advances in detection of point mutations, and the expansion of genetic techniques facilitate identification of mutated genes. While standard meiotic recombination mapping remains at the heart of all positional cloning efforts, extant or newly generated region-specific deletions can help refine critical intervals. Once a mutation in a candidate cDNA has been identified, genetic complementation experiments can be performed with existing targeted mutations, ENU-induced mutations, mice derived from gene-trapped ES cell lines, or mice carrying new alleles identified in gene-based screens of ENU-mutagenized mice or ES cells. Alternatively, gene confirmation can be accomplished in rescue experiments using mice transgenic for a bacterial artificial chromosome, cosmid, or minigene construct containing the candidate gene (3). The precise strategy chosen rests on the candidate gene, type of mutation, and availability of appropriate mouse lines. For predicted hypomorphic or null alleles, gene confirmation by noncomplementation experiments or transgenic rescue are straightforward. Things become more complicated if an anti- or neomorphic allele may have been generated or if a novel protein has been altered. Some clues regarding the stability, localization, and possibly function of the affected protein may be garnered from experiments done in tissue culture. In some instances, designing a “mimic” experiment, in which the altered protein is expressed in transgenic mice, or generating knock-down mice via small interfering RNA technology may provide a solution to this dilemma. In other cases, the more labor-intensive but ultimately most rigorous approach of recreating the identical mutation via targeted “knock-in” approaches in ES cells may be the only solution.
ACCESSING THE WEALTH OF ENU-INDUCED MOUSE MUTATIONS
From the beginning, mouse mutagenesis centers were designed with the hope that researchers worldwide would be able to obtain these newly induced mutants or participate directly in the mutant identification process. Often larger centers have been amenable to adding simple screens onto their efforts or to “hoteling,” which involves having visiting scientists screen potentially mutant mice with assays not commonly used by the center. In other cases, whole classes of mice have been distributed to outside researchers. A genetic screen for dark-skinned mice nicely illustrates a productive collaboration between a larger center and an individual research group. Upon screening of ∼30,000 mutants, 10 mutations that cause dark skin (Dsk) were identified by the GSF in Munich (14). These mice were then shipped to and analyzed further by Greg Barsh's group at Stanford. At Stanford, several of the affected genes, including a hyperactive mutation in the epidermal growth factor receptor, a mutation in keratin 2e, and mutations in two Galphaq subunits, were identified (14, 59). None of these genes had previously known roles in pigmentation. So far, such collaborations have been conducted on a case-by-case basis. However, more formalized protocols for obtaining and distributing desired mice are being established.
To facilitate access to mouse mutant resources, each center has set up the database resources to manage mouse husbandry, assay and image data, mouse archiving, and distribution and have made this information accessible to their affiliates and the general scientific community. Thus, for each center, a regularly updated web-based interface records the protocols for and data obtained from assays, lists general classes of mutants identified, and flags potential mutations and, if relevant, their genetic map positions. An interlinked web-based management system records the cryopreservation status of organs, DNAs, and, in some cases, sperm from ENU-induced mutants. In addition, mutant alerts, which record any recently identified mutants, further phenotypic analyses, or genetic map information, are sent via electronic mail to web subscribers. Web subscription to these mutant alert systems is freely available. Finally and very importantly, mouse request forms are available online, so researchers in the mouse community may acquire the mutant through this system. To facilitate access to these bioinformatics pipelines, the websites for each of the larger centers are provided in Table 1.
With the astounding successes of large- and small-scale ENU mutagenesis programs, it has become apparent that larger, centralized archiving and distribution centers are needed. Ideally, a single centralized, searchable database would list the phenotype, the map position, and center of origin for validated mutations and would direct interested individuals to the appropriate web links outlining the exact steps required by the actual distributing center. Already such a database exists for gene-trapped ES cell lines. At a meeting in November 2004, the existing archiving centers pledged to share information through a single website, avoid duplication of lines, and facilitate distribution and archiving of mutants (1). Thus, the International Mouse Mutant Federation, which consists of four repositories so far (the U.S. Mutant Mouse Regional Resource Centers, Canadian Mouse Mutant Repository, European Mouse Mutant Archive, and Japanese RIKEN BioResource Center), was born (Table 3). The massive number of mutants generated makes it logistically necessary and safer to have multiple repositories. Mice are being stored primarily as cyropreserved sperm or embryos. Upon being requested, the desired strain will be “reanimated” at the distribution center. This procedure normally takes 3 to 4 months. Alternatively, the mouse strain could be shipped as cryopreserved sperm or embryos to the researcher's host institution for reanimation. The ability of outside institutions to perform such reanimation themselves would expedite this process greatly. For the moment, interested researchers should search websites of and contact both the mutant archives (Table 3) and mutagenesis centers (Table 1).
TABLE 3.
Centers involved in archiving and distributing mice from ENU screens
| Mouse archiving center | Website | Affiliated centers |
|---|---|---|
| Mutant Mouse Regional Resource Centers | http://www.mmrrc.org/ | Harlan/Missouri Consortium, Harlan Sprague Dawley, Indianapolis, Ind. (www.harlan.com); University of Missouri-RADIL (www.mmrrc.missouri.edu); University of North Carolina, Chapel Hill, MMRRC (www.med.unc.edu/mmrrc/pages); Taconic/University at Albany SUNY (www.taconic.com); UC Davis MMRRC, Davis, Calif. (ccm.ucdavis.edu/mmrrc); The Jackson Laboratory MMRRC Informatics Coordinating Center, Bar Harbor, Maine (www.jax.org); National Center for Research Resources, National Institutes of Health, Bethesda, Md. (www.NCRR.NIH.gov) |
| European Mouse Mutant Archive | http://www.emmanet.org/ | Consiglio Nazionale delle Ricerche, Monterotondo, Italy |
| Centre National de la Recherche Scientifique, Orleans, France | ||
| Medical Research Council, Harwell, United Kingdom | ||
| Karolinska Institutet, Stockholm, Sweden | ||
| Fundação Calouste Gulbenkian Institute, Oeiras, Portugal | ||
| GSF National Research Center for Environment and Health, Neuherberg/Munich, Germany | ||
| European Molecular Biology Laboratory-European Bioinformatics Institute, Hinxton, United Kingdom | ||
| Mouse Functional Genomics Research Group RIKEN Gsc | http://www.gsc.riken.go.jp/Mouse/main.htm | RIKEN BioResource Center, Tsukuba, Japan |
| Canadian Mouse Mutant Repository | http://www.cmmr.ca/index.html | Mount Sinai Hospital, The Hospital for Sick Children, St. Michael's Hospital, and UHN, Toronto, Canada |
| Gene Trap Programs at University of Manituba, University of British Columbia, and University of Toronto | ||
| National Cancer Institute Mouse Models of Human Cancers Consortium | http://web.ncifcrf.gov/researchresources/mmhcc/ | |
| International Mouse Strain Resource, Jackson Laboratory, Bar Harbor, Maine | http://www.informatics.jax.org/imsr/index.jsp | |
| MRC Frozen Sperm and Embryo Archive, Harwell, United Kingdom | http://www.mgu.har.mrc.ac.uk/facilities/fesa/ |
FUTURE DIRECTIONS
In addition to analyses performed in the mouse, synteny between vertebrates and advances in comparative genomics allow us to take advantage of cross-organismal genetic information from human diseases, zebra fish and rat mutations, and an array of sequenced genomes. In zebra fish, large scale ENU-based screens and an insertion mutagenesis-based screen have focused on developmental abnormalities, but a wide array of other efforts, both large and small scale, are examining many other processes (12, 18). Differences in genomic organization may make distinct sets of genes accessible to mutational analyses in different organisms. For example, in the fly the redundancy and tight genetic linkage of netrin 1 and netrin 2 made identification of mutations in these genes in primary mutagenesis screens impossible, and the tetraploidization of the zebra fish genome sometimes results in splitting the function of a single mouse gene between two of its zebra fish homologues. Nonetheless, there are certainly examples of mutations in orthologous genes that have been recovered in zebra fish and mouse genetic screens. Ideally, in the very near future existing zebra fish and mouse mutants will be recorded in interlinked searchable databases to further facilitate cross-organismal studies and interactions. Directed genetic studies with the rat have become feasible with the advent of ENU-induced rat mutations (67). Such mutant animals may be particularly well suited to extended behavioral and physiologic studies, but generating them is most efficient via gene-directed approaches rather than forward phenotype-based screens. Thus, the selection of which rat mutants to study could be heavily and productively informed by initial studies on mouse mutants. As already discussed, many human disorders are the consequence of point mutations. The mouse is the mammalian system most amenable to experimental genetic study, and it is an important intermediate for understanding human biology. Thus, the remarkable explosion in mouse mutant numbers, the availability of zebra fish mutations, the ability to generate rat mutations, more informative comparative genomics, and innovations in human mutation identification are heralding an era of unprecedented advances in our understanding of protein structure-function relationships and biologic and disease mechanisms in vertebrates. Integrating investigators with expertise in other disciplines by providing appropriate mouse mutations, designing additional genetic screens, and fostering cross-disciplinary training will contribute immeasurably to this progress.
The dedication and contributions of many mouse geneticists have led to the remarkable advances in mouse forward genetic screens, and no single review can do all of these individuals and their efforts justice. The examples chosen in this review were chosen for illustrative and instructive purposes and are by no means exhaustive. The reader is encouraged to use this review as merely a starting point for further investigating any specific processes, assays, existing mutant classes, or mutagenesis centers.
REFERENCES
- 1.Abbott, A. 2004. Geneticists prepare for deluge of mutant mice. Nature 432:541. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, K. V. 2000. Finding the genes that direct mammalian development: ENU mutagenesis in the mouse. Trends Genet. 16:99-102. [DOI] [PubMed] [Google Scholar]
- 3.Antoch, M. P., E.-J. Song, A.-M. Chang, M. H. Vitaterna, Y. Zhao, L. D. Wilsbacher, A. M. Sangoram, D. P. King, L. H. Pinto, and J. S. Takahashi. 1997. Functional identification of the mouse circadian clock gene by transgenic BAC rescue. Cell 89:655-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, S. D., and R. Balling. 2001. Systematic approaches to mouse mutagenesis. Curr. Opin. Genet. Dev. 11:268-273. [DOI] [PubMed] [Google Scholar]
- 5.Carpinelli, M. R., D. J. Hilton, D. Metcalf, J. L. Antonchuk, C. D. Hyland, S. L. Mifsud, L. Di Rago, A. A. Hilton, T. A. Willson, A. W. Roberts, R. G. Ramsay, N. A. Nicola, and W. Alexander, S. 2004. Suppressor screen in Mpl−/− mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc. Natl. Acad. Sci. USA 101:6553-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrel, L., P. A. Hunt, and H. F. Willard. 1996. Tissue and lineage-specific variation in inactive X chromosome expression of the murine Smcx gene. Hum. Mol. Genet. 5:1361-1366. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y., D. Yee, K. Dains, A. Chatterjee, J. Cavalcoli, E. Schneider, J. Om, R. P. Woychik, and T. Magnuson. 2000. Genotype-based screen for ENU-induced mutations in mouse embryonic stem cells. Nat. Genet. 24:314-317. [DOI] [PubMed] [Google Scholar]
- 8.Coghill, E. L., A. Hugill, N. Parkinson, C. Davison, P. Glenister, S. Clements, J. Hunter, R. D. Cox, and S. D. Brown. 2002. A gene-driven approach to the identification of ENU mutants in the mouse. Nat. Genet. 30:255-256. [DOI] [PubMed] [Google Scholar]
- 9.Corder, E. H., A. M. Saunders, W. J. Strittmatter, D. E. Schmechel, P. C. Gaskell, G. W. Small, A. D. Roses, J. L. Haines, and M. A. Pericak-Vance. 1993. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261:921-923. [DOI] [PubMed] [Google Scholar]
- 10.Cordes, S. P., and G. S. Barsh. 1994. The mouse segmentation gene kr encondes a novel basic domain-leucine zipper transcription factor. Cell 79:1025-1034. [DOI] [PubMed] [Google Scholar]
- 11.Crawley, J. N. 2000. What's wrong with my mouse? Behavioral phenotyping of transgenic and knock-out mice. Wiley-Liss, New York, N.Y.
- 12.Driever, W., L. Solnica-Krezel, A. F. Schier, S. C. Neuhauss, J. Malicki, D. L. Stemple, D. Y. Stainier, F. Zwartkruis, S. Abdelilah, Z. Rangini, J. Belak, and C. Boggs. 1996. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123:37-46. [DOI] [PubMed] [Google Scholar]
- 13.Eggenschwiler, J. T., E. Espinoza, and K. V. Anderson. 2001. Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature 412:194-198. [DOI] [PubMed] [Google Scholar]
- 14.Fitch, K. R., K. A. McGowan, C. D. van Raamsdonk, H. Fuchs, D. Lee, A. Puech, Y. Herault, D. W. Threadgill, M. Hrabe de Angelis, and G. S. Barsh. 2003. Genetics of dark skin in mice. Genes Dev. 17:214-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Garcia, M. J., and K. V. Anderson. 2003. Essential role of glycosaminoglycans in Fgf signaling during mouse gastrulation. Cell 114:727-737. [DOI] [PubMed] [Google Scholar]
- 16.Goldowitz, D., W. N. Frankel, J. S. Takahashi, M. Holtz-Vitaterna, C. Bult, W. A. Kibbe, J. Snoddy, Y. Li, S. Pretel, J. Yates, and D. J. Swanson. 2004. Large-scale mutagenesis of the mouse to understand the genetic bases of nervous system structure and function. Brain Res. Mol. Brain Res. 132:105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greber, B., H. Lehrach, and H. Himmelbauer. 2005. Mouse splice mutant generation from ENU-treated ES cells—a gene-driven approach. Genomics 85:557-562. [DOI] [PubMed] [Google Scholar]
- 18.Haffter, P., M. Granato, M. Brand, M. C. Mullins, M. Hammerschmidt, D. A. Kane, J. Odenthal, F. J. van Eeden, Y. J. Jiang, C. P. Heisenberg, R. N. Kelsh, M. Furutani-Seiki, E. Vogelsang, D. Beuchle, U. Schach, C. Fabian, and C. Nusslein-Volhard. 1996. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123:1-36. [DOI] [PubMed] [Google Scholar]
- 19.Herron, B. J., W. Lu, C. Rao, S. Liu, H. Peters, R. T. Bronson, M. J. Justice, J. D. McDonald, and D. R. Beier. 2002. Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nat. Genet. 30:185-189. [DOI] [PubMed] [Google Scholar]
- 20.Hoit, B. D., S. Kiatchoosakun, J. Restivo, D. Kirkpatrick, K. Olszens, H. Shao, Y. H. Pao, and J. H. Nadeau. 2002. Naturally occurring variation in cardiovascular traits among inbred mouse strains. Genomics 79:679-685. [DOI] [PubMed] [Google Scholar]
- 21.Hrabe de Angelis, M. H., H. Flaswinkel, H. Fuchs, B. Rathkolb, D. Soewarto, S. Marschall, S. Heffner, W. Pargent, K. Wuensch, M. Jung, A. Reis, T. Richter, F. Alessandrini, T. Jakob, E. Fuchs, H. Kolb, E. Kremmer, K. Schaeble, B. Rollinski, A. Roscher, C. Peters, T. Meitinger, T. Strom, T. Steckler, F. Holsboer, T. Klopstock, F. Gekeler, C. Schindewolf, T. Jung, K. Avraham, H. Behrendt, J. Ring, A. Zimmer, K. Schughart, K. Pfeffer, E. Wolf, and R. Balling. 2000. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat. Genet. 25:444-447. [DOI] [PubMed] [Google Scholar]
- 22.Jun, J. E., L. E. Wilson, C. G. Vinuesa, S. Lesage, M. Blery, L. A. Miosge, M. C. Cook, E. M. Kucharska, H. Hara, J. M. Penninger, H. Domashenz, N. A. Hong, R. J. Glynne, K. A. Nelms, and C. C. Goodnow. 2003. Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity 18:751-762. [DOI] [PubMed] [Google Scholar]
- 23.Justice, M. J., and V. C. Bode. 1988. Genetic analysis of mouse t haplotypes using mutations induced by ethylnitrosourea mutagenesis: the order of T and qk is inverted in t mutants. Genetics 120:533-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Justice, M. J., D. A. Carpenter, J. Favor, A. Neuhauser-Klaus, M. Hrabe de Angelis, D. Soewarto, A. Moser, S. Cordes, D. Miller, V. Chapman, J. S. Weber, E. M. Rinchik, P. R. Hunsicker, W. L. Russell, and V. C. Bode. 2000. Effects of ENU dosage on mouse strains. Mamm. Genome 11:484-488. [DOI] [PubMed] [Google Scholar]
- 25.Justice, M. J., J. K. Noveroske, J. S. Weber, B. Zheng, and A. Bradley. 1999. Mouse ENU mutagenesis. Hum. Mol. Genet. 8:1955-1963. [DOI] [PubMed] [Google Scholar]
- 26.Kasarskis, A., K. Manova, and K. V. Anderson. 1998. A phenotype-based screen for embryonic lethal mutations in the mouse. Proc. Natl. Acad. Sci. USA 95:7485-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiernan, A. E., A. Erven, S. Voegeling, J. Peters, P. Nolan, J. Hunter, Y. Bacon, K. P. Steel, S. D. Brown, and J. L. Guenet. 2002. ENU mutagenesis reveals a highly mutable locus on mouse chromosome 4 that affects ear morphogenesis. Mamm. Genome. 13:142-148. [DOI] [PubMed] [Google Scholar]
- 28.Kile, B. T., K. E. Hentges, A. T. Clark, H. Nakamura, A. P. Salinger, B. Liu, N. Box, D. W. Stockton, R. L. Johnson, R. R. Behringer, A. Bradley, and M. J. Justice. 2003. Functional genetic analysis of mouse chromosome 11. Nature 425:81-86. [DOI] [PubMed] [Google Scholar]
- 29.King, D. P., Y. Zhao, A. M. Sangoram, L. D. Wilsbacher, M. Tanaka, M. P. Antoch, T. D. Steeves, M. H. Vitaterna, J. M. Kornhauser, P. L. Lowrey, F. W. Turek, and J. S. Takahashi. 1997. Positional cloning of the mouse circadian clock gene. Cell 89:641-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moser, A. R., H. C. Pitot, and W. F. Dove. 1990. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 247:322-324. [DOI] [PubMed] [Google Scholar]
- 31.Munroe, R. J., R. A. Bergstrom, Q. Y. Zheng, B. Libby, R. Smith, S. W. John, K. J. Schimenti, V. L. Browning, and J. C. Schimenti. 2000. Mouse mutants from chemically mutagenized embryonic stem cells. Nat. Genet. 24:318-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelms, K. A., and C. C. Goodnow. 2001. Genome-wide ENU mutagenesis to reveal immune regulators. Immunity 15:409-418. [DOI] [PubMed] [Google Scholar]
- 33.Neuhaus, I. M., and D. R. Beier. 1998. Efficient localization of mutations by interval haplotype analysis. Mamm. Genome. 9:150-154. [DOI] [PubMed] [Google Scholar]
- 34.Nolan, P. M., D. Kapfhamer, and M. Bucan. 1997. Random mutagenesis screen for dominant behavioral mutations in mice. Methods Cell Biol. 13:379-395. [DOI] [PubMed] [Google Scholar]
- 35.Nolan, P. M., J. Peters, M. Strivens, D. Rogers, J. Hagan, N. Spurr, I. C. Gray, L. Vizor, D. Brooker, E. Whitehill, R. Washbourne, T. Hough, S. Greenaway, M. Hewitt, X. Liu, S. McCormack, K. Pickford, R. Selley, C. Wells, Z. Tymowska-Lalanne, P. Roby, P. Glenister, C. Thornton, C. Thaung, J. A. Stevenson, R. Arkell, P. Mburu, R. Hardisty, A. Kiernan, A. Erven, K. P. Steel, S. Voegeling, J. L. Guenet, C. Nickols, R. Sadri, M. Nasse, A. Isaacs, K. Davies, M. Browne, E. M. Fisher, J. Martin, S. Rastan, S. D. Brown, and J. Hunter. 2000. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat. Genet. 25:440-443. [DOI] [PubMed] [Google Scholar]
- 36.Noveroske, J. K., J. S. Weber, and M. J. Justice. 2000. The mutagenic action of N-ethyl-N-nitrosourea in the mouse. Mamm. Genome 11:478-483. [DOI] [PubMed] [Google Scholar]
- 37.Percec, I., J. L. Thorvaldsen, R. M. Plenge, C. J. Krapp, J. H. Nadeau, H. F. Willard, and M. S. Bartolomei. 2003. An N-ethyl-N-nitrosourea mutagenesis screen for epigenetic mutations in the mouse. Genetics 164:1481-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plenge, R. M., I. Percec, J. H. Nadeau, and H. F. Willard. 2000. Expression-based assay of an X-linked gene to examine effects of the X-controlling element (Xce) locus. Mamm. Genome 11:405-408. [DOI] [PubMed] [Google Scholar]
- 39.Potter, M. D., M. L. Klebig, D. A. Carpenter, and E. M. Rinchik. 1995. Genetic and physical mapping of the fitness 1 (fit1) locus within the Fes-Hbb region of mouse chromosome 7. Mamm. Genome 6:70-75. [DOI] [PubMed] [Google Scholar]
- 40.Quwailid, M. M., A. Hugill, N. Dear, L. Vizor, S. Wells, E. Horner, S. Fuller, J. Weedon, H. McMath, P. Woodman, D. Edwards, D. Campbell, S. Rodger, J. Carey, A. Roberts, P. Glenister, Z. Lalanne, N. Parkinson, E. L. Coghill, R. McKeone, S. Cox, J. Willan, A. Greenfield, D. Keays, S. Brady, N. Spurr, I. Gray, J. Hunter, S. D. Brown, and R. D. Cox. 2004. A gene-driven ENU-based approach to generating an allelic series in any gene. Mamm. Genome 15:585-591. [DOI] [PubMed] [Google Scholar]
- 41.Rinchik, E. M. 1991. Chemical mutagenesis and fine-structure functional analysis of the mouse genome. Trends Genet. 7:15-21. [DOI] [PubMed] [Google Scholar]
- 42.Rinchik, E. M. 2000. Developing genetic reagents to facilitate recovery, analysis, and maintenance of mouse mutations. Mamm. Genome. 11:489-499. [DOI] [PubMed] [Google Scholar]
- 43.Rinchik, E. M., and D. A. Carpenter. 1999. N-Ethyl-N-nitrosourea mutagenesis of a 6- to 11-cM subregion of the Fah-Hbb interval of mouse chromosome 7: completed testing of 4557 gametes and deletion mapping and complementation analysis of 31 mutations. Genetics 152:373-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rinchik, E. M., and D. A. Carpenter. 1993. N-Ethyl-N-nitrosourea-induced prenatally lethal mutations define at least two complementation groups within the embryonic ectoderm development (eed) locus in mouse chromosome 7. Mamm. Genome 4:349-353. [DOI] [PubMed] [Google Scholar]
- 45.Rinchik, E. M., D. A. Carpenter, and M. A. Handel. 1995. Pleiotropy in microdeletion syndromes: neurologic and spermatogenic abnormalities in mice homozygous for the p6H deletion are likely due to dysfunction of a single gene. Proc. Natl. Acad. Sci. USA 92:6394-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rinchik, E. M., D. A. Carpenter, and D. K. Johnson. 2002. Functional annotation of mammalian genomic DNA sequence by chemical mutagenesis: a fine-structure genetic mutation map of a 1- to 2-cM segment of mouse chromosome 7 corresponding to human chromosome 11p14-p15. Proc. Natl. Acad. Sci. USA 99:844-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rinchik, E. M., D. A. Carpenter, and P. B. Selby. 1990. A strategy for fine-structure functional analysis of a 6- to 11-centimorgan region of mouse chromosome 7 by high-efficiency mutagenesis. Proc. Natl. Acad. Sci. USA 87:896-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogers, D. C., E. M. Fisher, S. D. Brown, J. Peters, A. J. Hunter, and J. E. Martin. 1997. Behavioral and functional analysis of mouse phenotype: SHIRPA, a proposed protocol for comprehensive phenotype assessment. Mamm. Genome 8:711-713. [DOI] [PubMed] [Google Scholar]
- 49.Russell, L. B., W. L. Russell, E. M. Rinchik, and P. R. Hunsicker. 1989. Chlorambucil effectively induces deletion mutations in mouse germ cells. Proc. Natl. Acad. Sci. USA 86:3704-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russell, W. L., P. R. Kelly, P. R. Hunsicker, J. W. Bangham, S. C. Maddux, and E. L. Phipps. 1979. Specific locus test shows ethylnitrosourea to be the most potent mutagen in mouse. Proc. Natl. Acad. Sci. USA 76:5918-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwenk, F., B. Zevnik, J. Bruning, M. Rohl, A. Willuweit, A. Rode, T. Hennek, G. Kauselmann, R. Jaenisch, and R. Kuhn. 2003. Hybrid embryonic stem cell-derived tetraploid mice show apparently normal morphological, physiological, and neurological characteristics. Mol. Cell. Biol. 23:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shedlovsky, A., J. L. Guenet, L. L. Johnson, and W. F. Dove. 1986. Induction of recessive lethal mutations in the T/t-H-2 region of the mouse genome by a point mutagen. Genet. Res. 47:135-142. [DOI] [PubMed] [Google Scholar]
- 53.Shibuya, T., and K. Morimoto. 1993. A review of the genotoxicity of 1-ethyl-1-nitrosourea. Mutat. Res. 297:3-38. [DOI] [PubMed] [Google Scholar]
- 54.Shima, N., S. A. Hartford, T. Duffy, L. A. Wilson, K. J. Schimenti, and J. C. Schimenti. 2003. Phenotype-based identification of mouse chromosome instability mutants. Genetics 163:1031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shima, N., R. J. Munroe, and J. C. Schimenti. 2004. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol. Cell. Biol. 24:10381-10389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Speca, D. J., N. Rabbee, D. Chihara, T. P. Speed, and A. S. Peterson. A genetic screen for behavioral mutations that perturb dopaminergic homeostasis in mice. Genes Brain Behav., in press. [DOI] [PubMed]
- 57.Stephenson, D. A., K. H. Lee, D. L. Nagle, C. H. Yen, A. Morrow, D. Miller, V. M. Chapman, and M. Bucan. 1994. Mouse rump-white mutation associated with an inversion of chromosome 5. Mamm. Genome. 5:342-348. [DOI] [PubMed] [Google Scholar]
- 58.Thaung, C., K. West, B. J. Clark, L. McKie, J. E. Morgan, K. Arnold, P. M. Nolan, J. Peters, A. J. Hunter, S. D. Brown, I. J. Jackson, and S. H. Cross. 2002. Novel ENU-induced eye mutations in the mouse: models for human eye disease. Hum. Mol. Genet. 11:755-767. [DOI] [PubMed] [Google Scholar]
- 59.Van Raamsdonk, C. D., K. R. Fitch, H. Fuchs, M. Hrabe de Angelis, and G. S. Barsh. 2004. Effects of G-protein mutations on skin color. Nat. Genet. 36:961-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vitaterna, M. H., D. P. King, A. M. Chang, J. M. Kornhauser, P. L. Lowrey, J. D. McDonald, W. F. Dove, L. H. Pinto, F. W. Turek, and J. S. Takahashi. 1994. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264:719-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vivian, J. L., Y. Chen, D. Yee, E. Schneider, and T. Magnuson. 2002. An allelic series of mutations in Smad2 and Smad4 identified in a genotype-based screen of N-ethyl-N-nitrosourea-mutagenized mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 99:15542-15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waterston, R. H., K. Lindblad-Toh, and M. G. S. Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 63.Weiss, K. C., D. Y. Kim, C. T. Pawson, and S. P. Cordes. 2003. A genetic screen for mouse mutations with defects in serotonin responsiveness. Brain Res. Mol. Brain Res. 115:162-172. [DOI] [PubMed] [Google Scholar]
- 64.You, Y., R. Bergstrom, M. Klemm, B. Lederman, H. Nelson, C. Ticknor, R. Jaenisch, and J. Schimenti. 1997. Chromosomal deletion complexes in mice by radiation of embryonic stem cells. Nat. Genet. 15:285-288. [DOI] [PubMed] [Google Scholar]
- 65.Yu, Q., Y. Shen, B. Chatterjee, B. H. Siegfried, L. Leatherbury, J. Rosenthal, J. F. Lucas, A. Wessels, C. F. Spurney, Y. J. Wu, M. L. Kirby, K. Svenson, and C. W. Lo. 2004. ENU induced mutations causing congenital cardiovascular anomalies. Development 131:6211-6223. [DOI] [PubMed] [Google Scholar]
- 66.Yu, Y., and A. Bradley. 2001. Engineering chromosomal rearrangements in mice. Nat. Rev. Genet. 2:780-790. [DOI] [PubMed] [Google Scholar]
- 67.Zan, Y., J. D. Haag, K. S. Chen, L. A. Shepel, D. Wigington, Y. R. Wang, R. Hu, C. C. Lopez-Guajardo, H. L. Brose, K. I. Porter, R. A. Leonard, A. A. Hitt, S. L. Schommer, A. F. Elegbede, and M. N. Gould. 2003. Production of knockout rats using ENU mutagenesis and a yeast-based screening assay. Nat. Biotechnol. 21:645-651. [DOI] [PubMed] [Google Scholar]
- 68.Zarbalis, K., S. R. May, Y. Shen, M. Ekker, J. L. Rubenstein, and A. S. Peterson. 2004. A focused and efficient genetic screening strategy in the mouse: identification of mutations that disrupt cortical development. PLoS Biol. 2:E219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng, B., M. Sage, W. W. Cai, D. M. Thompson, B. C. Tavsanli, Y. C. Cheah, and A. Bradley. 1999. Engineering a mouse balancer chromosome. Nat. Genet. 22:375-378. [DOI] [PubMed] [Google Scholar]
- 70.Zoltewicz, J. S., N. W. Plummer, M. I. Lin, and A. S. Peterson. 1999. oto is a homeotic locus with a role in anteroposterior development that is partially redundant with lim1. Development 126:5085-5095. [DOI] [PubMed] [Google Scholar]