Abstract
The arrest of DNA replication in Escherichia coli is triggered by the encounter of a replisome with a Tus protein-Ter DNA complex. A replication fork can pass through a Tus-Ter complex when traveling in one direction but not the other, and the chromosomal Ter sites are oriented so replication forks can enter, but not exit, the terminus region. The Tus-Ter complex acts by blocking the action of the replicative DnaB helicase, but details of the mechanism are uncertain. One proposed mechanism involves a specific interaction between Tus-Ter and the helicase that prevents further DNA unwinding, while another is that the Tus-Ter complex itself is sufficient to block the helicase in a polar manner, without the need for specific protein-protein interactions. This review integrates three decades of experimental information on the action of the Tus-Ter complex with information available from the Tus-TerA crystal structure. We conclude that while it is possible to explain polar fork arrest by a mechanism involving only the Tus-Ter interaction, there are also strong indications of a role for specific Tus-DnaB interactions. The evidence suggests, therefore, that the termination system is more subtle and complex than may have been assumed. We describe some further experiments and insights that may assist in unraveling the details of this fascinating process.
INTRODUCTION
Scope
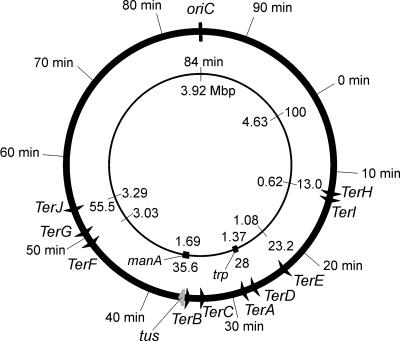
DNA replication in Escherichia coli initiates at oriC, the unique origin of replication, and proceeds bidirectionally (119). This creates two replication forks that invade the duplex DNA on either side of the origin. The forks move around the circular chromosome at a rate of about 1,000 nucleotides per second and so meet about 40 min after initiation in a region opposite oriC. In this region are located a series of sites, called termination or Ter sites, that block replication forks moving in one direction but not the other (Fig. 1). This creates a “replication fork trap” that allows forks to enter but not to leave the terminus region (66, 67).
FIG. 1.
Positions of Ter sites and the tus gene on the E. coli chromosome. All Ter sites are oriented so that the replication forks can travel in the origin-to-terminus direction but not the opposite direction. The tus gene is just downstream of TerB.
Here we give a historical overview of the development of this model for the process of replication termination in E. coli, and then we examine in molecular detail the current hypotheses concerning the mechanism by which interaction of the replication terminator protein (Tus) at Ter sites leads to polar arrest of advancing replication forks. Some new insights are developed.
Several aspects of replication termination (7, 13, 19, 26, 58, 67, 78, 108, 120, 145, 153) and Tus-Ter interaction (85, 170) have been reviewed previously. Although discussion here is limited to the system as it has evolved in E. coli and closely related eubacteria, understanding of termination in E. coli has developed in parallel with work on the mechanistically related system in Bacillus subtilis (26, 169). The B. subtilis termination system is the only other one where the molecular structure of the replication terminator protein (RTP) in complex with a cognate Ter site is known and the only one where structures of both the free (27, 134) and DNA-bound (172) forms of the protein have been determined. Although the Ter sites in B. subtilis were initially thought to be similar to those from E. coli (71), the two terminator proteins are completely unrelated in sequence and in structure and bind their respective Ter sites in quite different ways (85, 172). RTP binds as a dimer of dimers to two symmetric half-sites within a full B. subtilis Ter site (discussed recently in detail in reference 44), while as described below, Tus binds as a monomer to a full (asymmetric) E. coli Ter site.
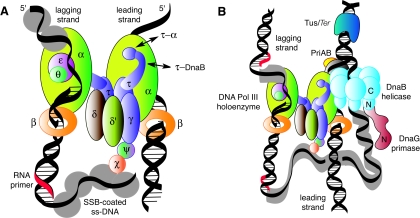
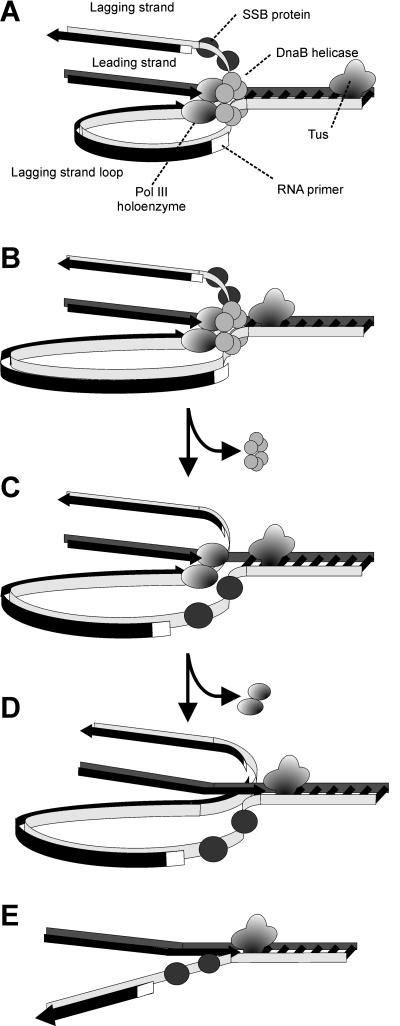
DNA synthesis at replication forks is mediated by a multiprotein assembly called the replisome, which accomplishes concerted DNA synthesis on both the leading and lagging strands (Fig. 2). The roles of the individual protein components of the replisome and the macromolecular interactions that determine its structure and function have been the subject of intensive study over the past 25 years, and this has led to sophisticated models for how the complex works. These have been the subject of recent reviews (8, 15, 34, 35, 118, 153).
FIG. 2.
Protein-protein interactions in the Escherichia coli replisome as it approaches the Tus-Ter termination block. (A) The DNA polymerase III (Pol III) holoenzyme is an asymmetric dimer containing 10 different subunits that include the twin polymerase (α) subunits that simultaneously replicate the two strands of the DNA template. (B) The replisome is a multiprotein complex made up of the DnaB helicase, the DnaG primase, and the Pol III holoenzyme. Each replicated strand commences with a short RNA primer synthesized by DnaG primase recruited from solution by interaction with DnaB. Single-stranded DNA is protected by SSB. Adapted from Fig. 2 of reference 153 with the permission of the authors.
Each replisome (Fig. 2B) is comprised of an asymmetric dimeric DNA polymerase III holoenzyme (118), which is responsible for concerted duplication of both template strands (Fig. 2A), together with a primosome that repeatedly synthesizes short RNA primers on the lagging strand. The primosome moves on the lagging strand in the 5′-3′ direction, powered by the ring-shaped hexameric DnaB helicase, which is also responsible for separation of the template DNA strands. Thus, if we were to propose for the moment that a complex of Tus with a Ter site provides a physical block to progress of a replication fork, we might expect this to be manifested as an inhibition of strand separation by DnaB at the apex of the replication fork (Fig. 2B). We will return later to examine these processes in detail.
Origins of the Concept of Replication Termination
Interest in the process of replication termination was largely sparked by the discovery that replication in E. coli proceeds bidirectionally from oriC, located at 85 min on the 100-min linkage map of the circular chromosome (17, 146). It was clear, therefore, that two replication forks moving in opposite directions would meet at some point approximately halfway around the chromosome from the origin (Fig. 1). Two early reports placed the site of termination at some point close to the trp operon at 28 min (22, 117). Within the error of the mapping by Bird et al. (22), the termination site was observed to be diametrically opposite oriC. Those workers briefly discussed two mechanisms for termination, favoring simple collision of replication forks over termination at a specific site. They noted, however, that there was no strong evidence in favor of either mechanism.
The question of whether replication terminated at a specific site was examined in various experimental systems, and the first indication of the existence of a discrete terminus was found in studies with the conjugative R plasmid R6K and a deletion mutant of it, RSF1040 (33, 112). Electron microscopic examination of RSF1040 replication intermediates showed two origins (α and β) and a single terminus (33). Replication was initiated from the α origin, progressing first towards the “right,” halting at the terminus, and then progressing towards the “left” from the same origin to the same terminus. Replication could also occur from the β origin in the same asymmetric, bidirectional manner to the same terminus. The terminus is thus responsible for converting unidirectional replication into a sequential bidirectional mode (33). These conclusions are unaffected by recent studies that show the initiation of R6K replication to be more complicated, involving looping interactions of the π replication initiator protein bound at a third origin (γ) with the α and β origins (1, 2).
Soon after, in 1977, evidence for a discrete site for termination in the E. coli chromosome was reported. Louarn et al. (110) changed the position of replication initiation by integrating R-plasmid origins at various sites in the chromosome of a temperature-sensitive mutant with a mutation in dnaA, the gene that encodes the replication initiator protein DnaA (119). These strains could not initiate replication from oriC at the nonpermissive temperature, but replication could still initiate at the integrated origins and proceed bidirectionally. It was found to terminate diametrically opposite oriC (between attφ80 at 28 min and attP2H at 45 min) even when the new origin was displaced by 26 min from it. Using a similar system, Kuempel et al. (103, 104) located the terminus between aroD and rac at 38 and 30 min, respectively, and Louarn et al. (111) later reduced this interval to the 6 min between man and rac.
It was still an open question whether the R6K and E. coli termini worked by the same mechanism. Both termini blocked replication at specific sites, and both seemed to work independently of the site of initiation and the type of origin. The E. coli terminus could block bidirectional replication initiated from oriC or (symmetric or asymmetric) replication from various integrated plasmid or phage origins at various locations (103, 104, 110), while the R6K terminus could block replication from the R6K origins in RSF1010 (33) and from a ColE1 origin in two different positions in a plasmid (98). Both termini apparently blocked replication forks arriving at the terminus from both directions. However, as described above, the modes of replication are quite different. In addition, while the R6K terminus region was located to a 216-bp segment of DNA (12), the continuing difficulty in pinpointing the precise location of the chromosomal terminus was beginning to suggest that it was a large region rather than a specific site.
This problem was solved in 1987 with the realization that the E. coli terminus was made up of discrete loci that separately blocked replication forks moving in opposite directions in a polar manner (38, 68, 138). The first two termination sites identified were situated at either end of the terminus region (Fig. 1); one was located close to trp at 28 min and the other near manA at 36 min (68, 138). This polar block to progress of the fork therefore appeared to be different from that at the R6K terminus, which was known to block fork movement from either direction (11, 33, 98).
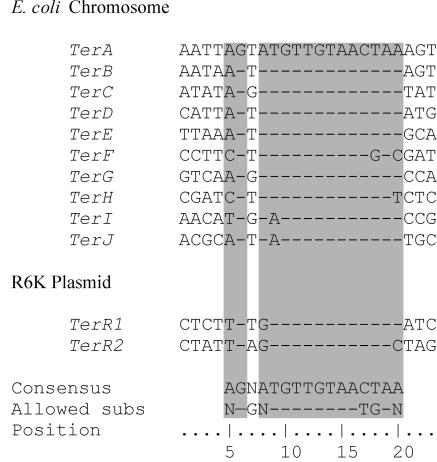
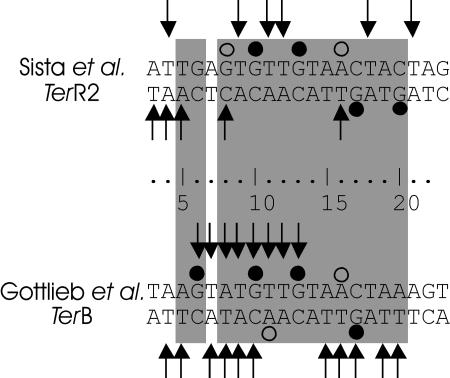
Resolution of the similarity of the two systems had to wait one more year for nucleotide sequences from the E. coli terminus to become available (62, 71). The terminators that would eventually be named TerA and TerB (Fig. 3) had a strong similarity to the two halves of an imperfect inverted repeat in the R6K terminus (62, 71, 75). The two R6K sequences (named TerR1 and TerR2) were identical to TerA and TerB at 15 and 12 positions, respectively (Fig. 3). In both the R6K plasmids and the E. coli chromosome, the Ter sequences were placed so as to form a “replication fork trap” that would allow a replisome to enter the region between the two Ter sites but not to leave. Ter sequences were also found in a variety of other plasmids as well as in other bacteria (30), and the number of Ter sites identified in the E. coli chromosome also increased, first to 4 (48, 62), then to 5 (63), and finally to 10, after the publication of the entire genome sequence (23) and an in-depth study of nucleotide substitutions by Coskun-Ari and Hill (30).
FIG. 3.
Nucleotide sequences of Ter sites from the E. coli chromosome and R6K plasmids. Base pairs that interact with the Tus protein are indicated by the shaded regions. In the orientation shown for these sequences, replication forks approaching from the left are blocked, while those entering from the right are unimpeded.
COMPONENTS OF THE REPLICATION TERMINATION SYSTEM
The Terminator (Ter) Sequences
Sequences of the known 23-bp Ter sites are shown in Fig. 3. The strictly conserved GC6 base pair is followed by a very highly conserved 13-bp core region in which a few substitutions are allowed. The sequence is asymmetric, mirroring the asymmetry of the replication fork block. In termini oriented as in Fig. 3, replication forks arriving from the left are blocked while those from the right pass through unimpeded. The core sequence is usually associated at the fork-blocking side with a preceding AT-rich region (30).
Once small DNA fragments containing TerA and TerB as well as the two TerR sites were available, it was shown that they could block replication forks in ColE1 plasmids in vivo (139, 159), and proof that the minimal Ter sequences were indeed sufficient to block replication forks in a polar manner came after they had been inserted into plasmids as synthetic oligonucleotides (62, 71, 75).
A trans-Acting Factor
Attention was at the same time beginning to be focused on the mechanism of termination. It had been suspected since the early 1980s that a DNA-binding protein might be involved. Bastia et al. (12) had shown that the R6K terminus did not have any significant twofold symmetry, effectively ruling out steric hindrance due to DNA secondary structure as a mechanism for replication fork blockage. Moreover, the plasmid terminus was capable of blocking replication forks in extracts prepared from cells which did not contain an R6K-derived plasmid, indicating that any protein involved is encoded by the host chromosome (53).
The second line of evidence for involvement of a DNA-binding protein arose from deletion studies used to narrow down the locations of TerA and TerB. TerB was quickly located to a 4-kb region, while TerA was more difficult to locate precisely. However, deletion of the TerB region inactivated arrest activity at TerA, implicating a trans-acting factor encoded near the TerB arrest site (69). Kuempel and coworkers named the putative gene tus for “termination utilization substance.”
The first description of the trans-acting factor was by Hill et al. (72), who isolated the gene encoding a DNA-binding protein by screening deletion and insertion mutants with mutations in the TerB region. They reported the gene sequences and the construction of tus strains that were deficient in termination activity. These mutants were complemented by plasmid-borne copies of tus. The gene was predicted to encode a 36-kDa polypeptide, and it directed overproduction of a protein estimated by gel electrophoresis to be this size (72).
Soon after, two other groups isolated a protein that bound to R6K Ter DNA. Sista et al. (159) purified an ∼40-kDa protein that bound the TerR sequence and defined its binding site by using copper-phenanthroline footprinting. A mutated Ter site with changes at six of the protected residues lost both the ability to bind the purified protein and the ability to arrest replication forks in vivo. Kobayashi et al. (96) reported isolation of a fragment of DNA encoding terminus-binding activity, together with insertion mutants that had lost the ability to bind a Ter site, whether on a plasmid or in the chromosome. The activity associated with the gene was sensitive to treatment with proteases and heat but not to treatment with RNase (96). They also determined the sequence of the gene, overproduced and purified the gene product, and demonstrated its binding to both TerR sites by DNase I footprinting (65).
All three activities were soon shown to be those of the same protein, encoded by the tus gene situated just following TerB (Fig. 1 and 4). The Tus protein bound to all known Ter sites and, once bound, could block the progress of a replication fork. A remaining question was how the moving fork was blocked. Did the Tus-Ter complex interact specifically with some component of the moving replisome, or did it merely act as a clamp on the DNA preventing its passage through the Ter site? With the gene, the protein, and hypotheses in hand, several groups tackled the mechanism of replication termination.
FIG. 4.
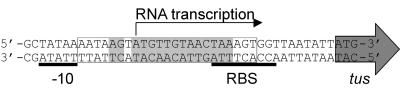
Relationship between TerB and the tus gene. The tus gene and its −10 promoter region and ribosome-binding site (RBS) are shown. The Tus protein regulates tus gene expression by binding to the TerB sequence and blocking the initiation of transcription of tus. The TerB sequence is enclosed in the box, and base pairs that interact with Tus are shaded as in Fig. 3.
The tus Gene and Tus Protein
The tus gene lies 11 base pairs downstream of the TerB site (Fig. 4). Both its ribosome-binding site and the −10 region of its promoter overlap TerB, which suggested transcription of the gene to be regulated by the binding of Tus to its recognition sequence. Two reports confirmed this in 1991. Primer extension studies on templates containing TerB showed that the presence of active Tus reduced transcription of tus and that the addition of more TerB sites on a high-copy-number plasmid increased its transcription (144). Moreover, Natarajan et al. (130) showed that Tus could block its own transcription in vitro and that the protein-DNA complex could prevent RNA polymerase from binding to the promoter. Roecklein and Kuempel (143) later mapped accurately the transcriptional start site in vivo to a site within TerB (Fig. 4) and confirmed that expression of Tus is autoregulated.
The gene coded for a protein of 308 amino acids (after removal of the N-terminal methionine residue) with a mass of 35,652 Da. The protein sequence showed no similarity to any known DNA-binding motif. The purified protein had a pI of 7.5, significantly lower than the value of 10.5 calculated from its amino acid composition. Since there was no indication that the protein was phosphorylated, this suggested that the tertiary structure had a large effect on the ionization state of several basic residues. Gel filtration and sucrose density gradient centrifugation showed that Tus was a monomer in solution with a Stokes radius of 23 Å and an axial ratio of two (31). This would allow it to cover 13 bp of DNA on binding, which was in good agreement with the results of the earlier footprinting studies (159).
Tus was shown by footprinting with copper-phenanthroline (159), DNase I (65, 130), and hydroxyl radicals (54, 158) to bind to several Ter sites. It bound extremely avidly to the TerB site; the Tus-TerB complex had a measured dissociation constant (KD) of 3.4 × 10−13 M and a dissociation half-life in vitro of 550 min at pH 7.5 in a buffer containing 150 mM potassium glutamate (54). Its binding to R6K TerR2 under identical conditions was weaker; the measured value of KD was 30 times higher, primarily due to a higher dissociation rate (54). The protein was shown to bind to TerB as a monomer, which is unusual for a DNA-binding protein but consistent with the asymmetry of the Ter sites and replication fork arrest (31).
PROPOSED MECHANISMS OF REPLICATION FORK ARREST
The basis of the mechanism of fork arrest was soon established. The Tus-TerB complex was shown to block the action of the major replicative DNA helicase, DnaB in vitro in an orientation-dependent manner (91, 106). The orientation of the block was the same as for the arrest of replication fork movement both in vivo and in vitro (61, 70, 106, 113).
In the normal process of replication, DnaB is at the front of the replisome (Fig. 2B). It is a ring-shaped homohexameric enzyme that translocates in the 5′-to-3′ direction on the lagging-strand template to unwind double-stranded DNA in front of the DNA polymerase III holoenzyme, the multisubunit replicase (118, 153) that simultaneously synthesizes both strands (Fig. 2A). One strand (the leading strand) is replicated continuously, while the other (lagging) strand is synthesized discontinuously in a series of (Okazaki) fragments. The replicative RNA-priming enzyme, DnaG primase (49), is recruited by DnaB for the priming of each new fragment on the discontinuous strand (133). The single-stranded sections that result from helicase action are coated with single-stranded DNA-binding protein (SSB). DnaB is physically associated with the replicase through the τ subunit of the holoenzyme (93).
When progress of the replisome was halted by the Tus-Ter complex, both in vitro (70) and in vivo (126), DNA synthesis continued right up to 4 base pairs before the conserved GC6 base pair in the TerB site (Fig. 3). This is a surprising result given the size of the polymerase holoenzyme, let alone the enormity of the entire replisome. Since the leading-strand template is known to be excluded from the central channel of DnaB (80, 87), it is conceivable that the active site of the leading-strand polymerase is very close to the point of strand separation by the helicase (Fig. 2B). However, it appears more likely that dissociation of DnaB from the replisome occurs as part of the arrest process. In the presence of DnaG primase, the distribution of leading-strand stop sites changed, showing a degree of sensitivity of leading-strand synthesis to the protein complement of the lagging strand (70).
Lagging-strand synthesis stopped 50, 66, or 82 bp before the TerB site (70). The 50-bp (17-nm) gap could be envisaged as a loop bound by one or two tetramers of SSB on the lagging strand (Fig. 5). This implies that the loop is either topologically or physically constrained from closing any farther to allow priming by DnaG before dissociation of DnaB. The 16-bp spacing between the lagging-strand priming sites may reflect some aspect of protein organization on the lagging strand that affects the site of priming or subsequent primer extension or may simply be due to the sequence specificity of the DnaG primase (49, 70).
FIG. 5.
Replisome of E. coli and mechanism of replication fork arrest by a Tus-Ter complex. (A) The replisome moving along the DNA template approaches Tus, and the DnaB helicase assists primase to lay down the last lagging-strand primer. (B) DnaB helicase action isblocked by Tus, and DnaB dissociates from the template. (C) DNA polymerase III (Pol III) holoenzyme completes leading-strand synthesis up to the Tus-Ter complex and (D) synthesizes the last Okazaki fragment on the lagging strand, which will eventually be ligated by DNA ligase to the penultimate fragment following removal of its RNA primer by DNA polymerase I (not shown). (E) The holoenzyme then dissociates, leaving a Y-forked structure that is single stranded on the lagging strand near the Tus-Ter complex.
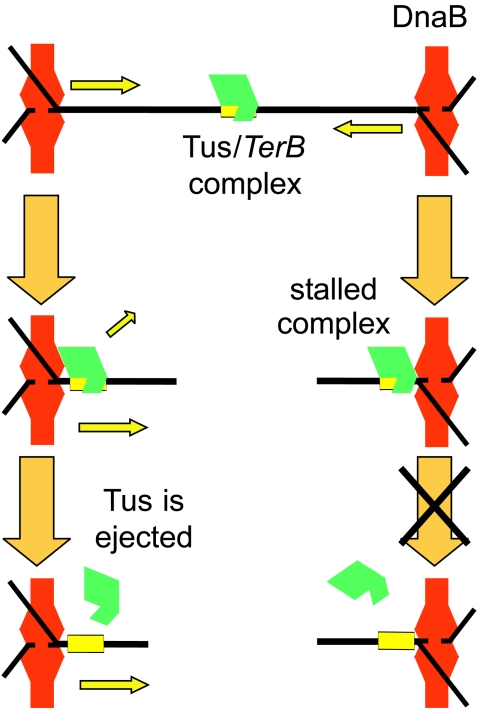
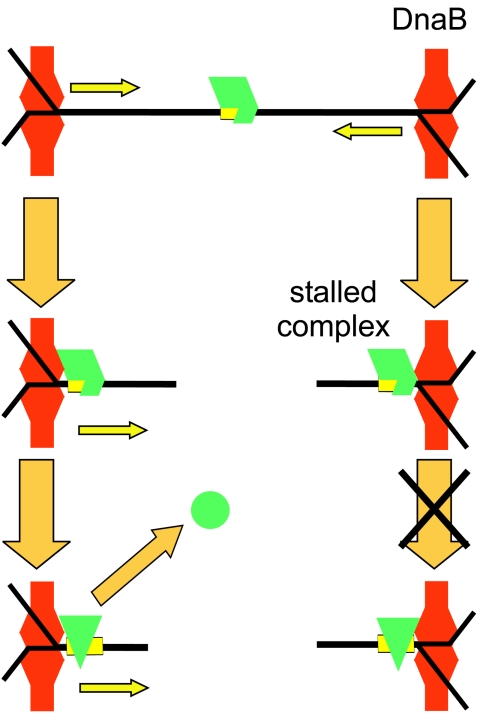
This information allows the development of a quite detailed model of the replication arrest process (Fig. 5). Tus bound to a Ter site faces in one direction towards an oncoming replication fork. The DnaB helicase approaches the Tus-Ter complex and is blocked from proceeding. Before it dissociates, its interaction with primase leads to synthesis of a final lagging-strand primer at a distance that may be dictated by the phase of binding of SSB tetramers to the lagging-strand template. Dissociation of DnaB then leaves a Y-forked structure which is single stranded very close to the Ter site. A further tetramer (or two) of SSB then binds rapidly to the exposed single-stranded DNA to protect it. DNA polymerase III holoenzyme then synthesizes the leading strand of DNA right up to the Ter site and completes synthesis of the last-primed Okazaki fragment on the lagging strand. In vivo the replisome must either reassemble and eventually pass through the block or dissociate, leaving the Y-structure behind. In the latter case, the single-stranded loop might persist (bound by SSB), or the synthesis might be completed by DNA repair mechanisms or by elongation of the leading strand of the other replication fork. The Y-fork structures are known to persist in vivo in plasmids whose replication has been blocked by correctly oriented Ter sites (76). A question that remains to be examined in a satisfactory way is the precise definition of the protein complement of a fork stalled at Tus-TerB and, in particular, at which point the DnaB helicase dissociates.
What occurs when a replication fork approaches from the other (permissive) direction is much less clear. Khatri et al. (91) suggested that the Tus protein remains associated with one strand (the strand shown in Fig. 3) of the unwound DNA after DnaB has passed through the Ter site from the permissive side. However, Gottlieb et al. (54) found that Tus had no affinity for either strand of DNA in the single-stranded form, and Neylon et al. (131) also reported that the affinity of Tus for each separate strand of the TerB site was the same as that for a nonspecific single-stranded DNA under low-salt conditions where binding could be observed. Very little work has been reported on the process by which the helicase passes through the Tus-Ter complex when it approaches from the permissive direction.
Another remaining issue is the nature of the interaction between Tus and DnaB. Does Tus merely act as a clamp on the DNA, or are there specific protein-protein or protein-DNA-protein interactions between Tus and the oncoming helicase (or other component of the replisome)? These two possibilities can be broadly described as the “clamp model” and the “interaction model.” These two simple mechanisms were initially proposed with the expectation that the question would be resolved rapidly. However, it still remains controversial in spite of publication during the ensuing years of a high-resolution crystal structure of the Tus-TerA complex (85). A third potential mechanism that has been recently suggested (131) is one in which Tus interacts with the helicase (or other elements of the replisome) through the DNA. That is, that Tus engineers a structure in the DNA on the nonpermissive side that prevents the further passage of the helicase. A fourth and related alternative, apparently yet to be tested experimentally, is that the helicase generates a structure in the DNA at the permissive face that actively promotes dissociation of Tus and/or a structure at the nonpermissive face that increases the affinity of Tus for the Ter site. In the remainder of this review, we will examine the available evidence for these possible molecular mechanisms of Tus-mediated polar replication fork arrest at Ter sites.
Evidence for Specific Protein-Protein Interactions
A large number of publications on assays of Tus activity appeared soon after the tus gene and Ter sequences became available, and the effects of Tus protein on a range of replication assays, both in vitro and in vivo, were reported. These led rapidly to the description of the first two classes of model described above. The first studies examined the effect of the Tus-Ter complex on the DNA-unwinding activities of a range of helicases in in vitro assay systems. Lee et al. found that the nonpermissive face of Tus-TerB blocked the actions of the four helicases they tested: DnaB, UvrD, Rep, and PriA (105, 106). On the other hand, Bastia and coworkers described a tendency in their results with the Tus-TerR2 complex in a different assay system for the complex to specifically block the subset of replication fork helicases (14, 91, 147). From results of a further study, Hiasa and Marians suggested that while Tus-TerB could block translocation of DnaB, PriA, and the primosome (but not UvrD) in a polar manner, it did not inhibit bone fide DNA helicase activity (60). The controversy over the mechanism of antihelicase activity can therefore be traced to different results obtained from examining the effects of Tus binding to different Ter ligands in different experiments. The difficulties in interpretation of the action of Tus in these in vitro reactions have continued to the present day.
In the experiments of Bastia and coworkers, the Tus-TerR complex was observed to block the replicative helicases DnaB and simian virus 40 (SV40) T antigen, but it failed to block helicases involved in DNA repair or plasmid rolling-circle replication, including Rep, Dda, TraI, and UvrD (14, 91, 147). Even though the block to the action of T antigen (a 3′-5′ helicase) seemed to be at the face permissive for DnaB, they nonetheless favored a mechanism that involves specific protein-protein interactions between Tus and a domain of the replicative helicases. In support of this, they cited the (unpublished) observation of a direct interaction between Tus and DnaB (114). More recently, the same group has described experiments using a yeast two-hybrid system that provide evidence of in vivo interaction between the two proteins (127). They also describe the binding of DnaB to an immobilized glutathione S-transferase-Tus fusion protein and isolation of mutants of Tus that have reduced binding to DnaB and similarly reduced fork-blocking activity but near-normal TerR binding. This is the strongest evidence to date for a specific interaction between Tus-TerR and the oncoming helicase.
In contrast to the results of Bastia and coworkers, in the experiments of Lee et al. and other groups studying the Tus-TerB interaction, the complex impeded the progress of both replicative and repair helicases (60, 105, 106). In addition, it did so in a polar manner. That is, the same face of the Tus-Ter complex blocked DnaB translocating in the 5′-3′ direction but also blocked SV40 T antigen (5, 64), PriA (60, 105), UvrD (106), and TraI (64) translocating on the opposite strand in the 3′-5′ direction. This would suggest that the action of the complex is either as a clamp or directed against some aspect of helicase structure and/or function that is sufficiently general to be exhibited by all those tested. The idea that a clamp might be sufficient is supported by a report that a mutant EcoRI restriction endonuclease that binds to its recognition sequence with a dissociation constant of ∼2.5 × 10−13 M, but does not cleave DNA (95, 173), was capable of blocking the helicase action of DnaB, UvrD, and SV40 T antigen (14). The block was orientation independent, since EcoRI binds to DNA as a symmetric dimer. Later, it was shown that the lac repressor-operator complex can substantially inhibit the action of a range of helicases in vitro, including DnaB (175). The effectiveness of these unrelated protein-DNA complexes in blocking replication forks would appear to indicate that a simple clamp is sufficient to halt helicases in vitro.
Experiments with surrogate systems do not support this view. In an ingenious series of experiments, Andersen et al. (6) compared the effectiveness and polarity of the Tus-Ter complex in vivo in E. coli and B. subtilis. Alongside this, the functionally similar but unrelated replication termination system of B. subtilis was compared in both organisms. While B. subtilis RTP-TerI worked well to terminate replication in both organisms, the E. coli Tus-TerB complex was very much more effective in its natural host. In earlier similar experiments, Kaul et al. (90) had also shown the B. subtilis termination system to be effective in E. coli. These data might indicate a fundamental difference in mechanism between the two systems and support the existence of a specific interaction between Tus-Ter and a replisomal protein(s) in E. coli, at least.
On the other hand, in evolutionary terms, it is not surprising that the systems work somewhat better in their natural context. Natural systems under selection pressure would be expected to take advantage of opportunities to improve their efficiency. Indeed, it would be surprising in the specific case of the Tus-Ter acting against E. coli DnaB if there was not a functional interaction that had developed to improve the efficiency of replication arrest. However, it is not clear how highly specific interactions could develop to play a general role in antihelicase activity. Perhaps the more pertinent question is whether Tus-DnaB interactions are limited to small improvements in a single protein-protein interface or whether they play an important role in the more general case of Tus activity against the full range of helicases.
STRUCTURE OF THE Tus-Ter COMPLEX AND MOLECULAR BASIS OF REPLICATION ARREST
A large amount of data is available on the Tus-Ter interaction, including results of DNA footprinting, kinetic studies, effects of mutations to both Tus and the Ter sequence, and the gene sequences of Tus proteins from related bacteria. In this section, we will analyze the published data on the Tus-Ter interaction, starting with the crystal structure of the complex (85), followed by footprinting and kinetic studies. This will be followed by the data on Ter DNA mutations and mutational studies of the Tus protein itself and then by an analysis of the protein sequences from three related bacterial species, as well as two further proteins with sequence similarity to Tus. There has been no previous analysis of all the available data within the framework of the crystal structure. Finally, we will summarize the results and examine a series of models of protein-DNA and protein-protein interactions at the site of replication arrest.
The Crystal Structure of the Tus-Ter Complex
The first crystal structure of a replication terminator protein to be reported was that of the dimeric B. subtilis RTP in 1995 (27). This was followed quickly by models for the structures of the complex of the RTP dimer and tetramer with half and full Ter sites, derived from consolidation of the structure of the free protein with an extensive series of biochemical data (115, 125, 134, 135). The structure of the half-site complex determined subsequently by a combination of nuclear magnetic resonance and crystallographic studies (172) was largely in accord with these models.
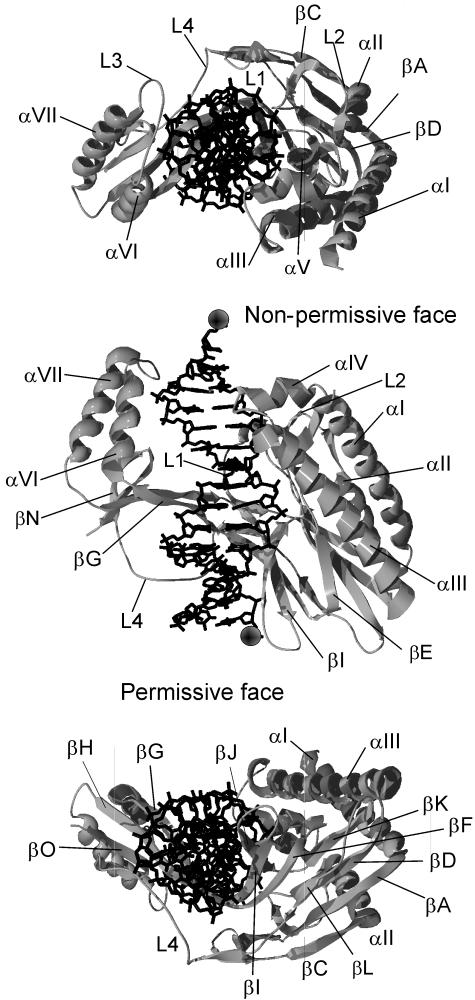
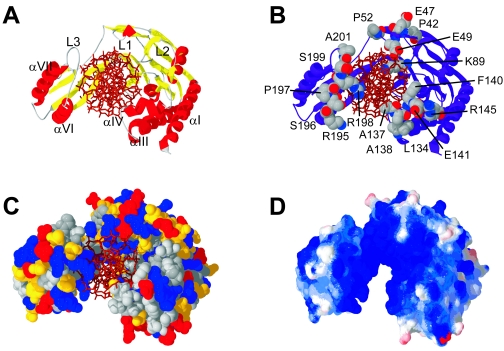
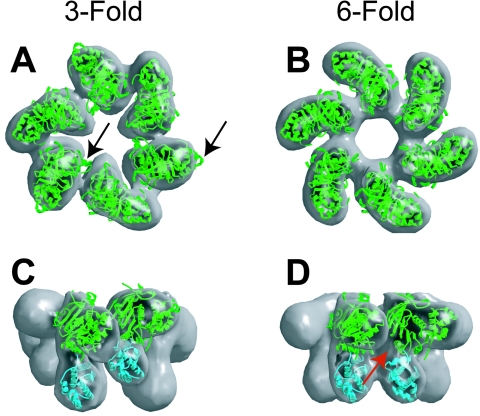
The E. coli Tus-TerA complex was crystallized by Kamada et al., and the X-ray crystal structure was reported in 1996 (85, 86). The structure (Protein Data Bank code 1ECR), shown in Fig. 6, is a unique protein fold consisting of two discontinuous domains that straddle the TerA double helix. The two domains are joined by two antiparallel pairs of β strands that make up the core DNA-binding domain (βIF and βGH) and also by the L4 loop. These two pairs of strands lie in the major groove of TerA. The structure of Tus in the complex is 37% helix, 28% sheet, and 35% loops and turns. The αI, αII, and αIII amphipathic helices form an antiparallel bundle that runs parallel to the DNA but makes no contact with it. The αIV and αV helices along with the L1 and L2 loops lie at the top of the larger (N-terminal) domain. With the αVI-αVII region in the smaller C-terminal domain, they complete the face of Tus that blocks the progressing helicase (the nonpermissive or fork-blocking face). Three of the four main loops (L1 to L3) are at the nonpermissive end of the complex. The remaining loop (L4) lies at the permissive end in the minor groove, making a number of DNA contacts.
FIG. 6.
The crystal structure of the Tus-TerA complex, PDB code 1ECR (85). Three views of the Tus-Ter complex are shown. The top view is looking down the DNA from the nonpermissive face of the complex. The middle view is rotated 90° from the first to show the front of the complex. The bottom view, rotated a further 90°, is along the DNA from the permissive end of the complex. The permissive and nonpermissive faces are indicated in the middle view. The balls indicate the (5′) strands that would pass through the central channel of the DnaB helicase. Images of protein structures in this and succeeding figures were generated in SWISS-PDB VIEWER version 3.7 (http://ca.expasy.org/spdbv/) (56) and rendered using POV-RAY version 3.1g.watcom.win32 (www.povray.org).
There are three main regions of β structure. The βGHON and βJIFL regions have strands in the major groove of the TerA DNA and are involved in base recognition. The other main β sheet (βEKDAC) sits at the bottom of the N domain and is involved in stabilizing the βJIFL region through hydrophobic contacts as well as contributing to the hydrophobic core of the N domain. The hydrophobic cores of both domains are largely made up of residues in the α helices. The core of the N domain consists of residues from helices αI to αIII as well as the βEKDAC sheet, while the core of the smaller C domain is made up mostly of residues from αVI and αVII. Contributions from the βGHON sheet make up the remainder of the hydrophobic core of the C domain.
The double-stranded TerA captured within the complex is significantly deformed from the canonical structure of B-form DNA. The average helical twist is 29.5°, compared to the canonical value of 34.6° (85). The DNA backbone is also deformed between G17 and A14 (Fig. 7) due to it being sandwiched between the βF and βG strands and the L4 loop. The propeller angle of the AT16 base pair is −24.2°. The DNA is consequently underwound, making the major groove deeper and expanding the minor groove, and it is bent overall through about 20° (85). The TerA fragment in the crystal does not extend beyond the protein and therefore provides little information about the DNA structure at the permissive end of the complex; it is thus possible that the DNA would be further deformed by contacts with the protein beyond the extremity of the cocrystallized fragment (Fig. 7).
FIG. 7.
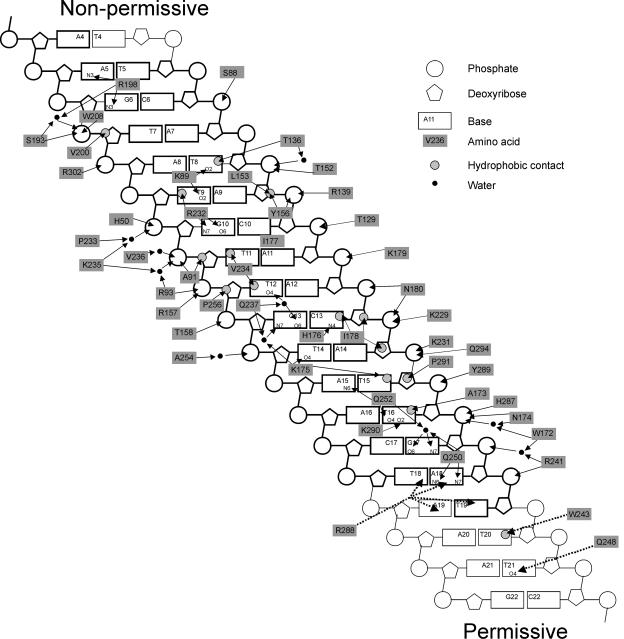
Summary of contacts between Tus and TerA. Adapted from reference 30 with permission of the publisher. Arrows show interactions between amino acid side chains and groups in the base pairs. Residues in the TerA oligonucleotide used for determination of the crystal structure were A4 to T18 on one strand and T19 to T5 on the other and are shown with boldface outlines. Dashed lines indicate possible interactions at the permissive end that were not seen in the crystal structure (see the text for details).
The protein is folded about the DNA ligand, and the complex cannot be disrupted without deforming the protein structure (Fig. 6). Kamada et al. (85) speculated that Tus may be capable of binding a single strand of DNA extending from the permissive face of the complex and proposed a model for Tus removal by a helicase approaching the permissive face that involves association of Tus with the single-stranded DNA product, leading to deformation of the structure and its unfolding from the DNA. Conversely, it is also possible that a single DNA strand extending from the nonpermissive face could be bound by Tus, leading to a tighter interaction that impedes disassembly of the Tus-Ter complex.
Protein-DNA Binding Interactions
The core DNA-binding domain of Tus is the twisted β-sheet structure made up of the βIF and βGH strands. Each of the four strands is seven or 8 residues long (Fig. 8). The gap between βF and βG is one residue in length, and that between βH and βI is two residues. The twist in the DNA ligand is stabilized by a variety of protein interactions, both with the DNA backbone and with the bases (Fig. 7). Within the protein, the twist is facilitated by Pro238, which allows βI to turn through almost 90° and pass underneath βF to the inside of the major groove. Hydrogen bonds between Asn174 (in the βF-βG turn) and Tyr280 (in βM), between Lys175 (in βG) and Gln252 (in βJ), and between Lys235 (in the βH-βI turn) and Asn51 (in L1) further stabilize the twist in the DNA.
FIG. 8.
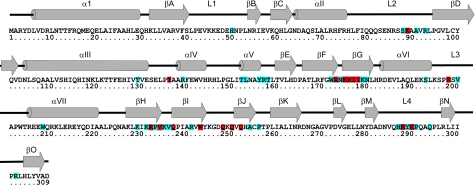
Sequence and secondary structure of the Tus protein (data are from reference 85). The 31 residues that make nonspecific contacts to the DNA backbone are in blue. The 17 residues that make direct or water-mediated specific contacts with the DNA bases are in red.
Between them, βFG and βHIJ contain close to half of the residues making DNA contacts; remaining residues that make contacts are concentrated in other β strands and loops (85). Only eight of the residues that contact DNA are in α helices. Although the DNA contacts are distributed throughout the length of the TerA fragment, they exhibit a striking strand specificity in the sense that they are concentrated near the 5′ end of each strand (Fig. 7).
There are 17 residues that make sequence-specific contacts with TerA DNA (Fig. 8). Nearly half of these are hydrophobic, and the remainder are mainly hydrogen-bonded interactions between charged or polar amino acid side chains and polar donor/acceptor atoms of the bases in the major groove. Several of the latter interactions are mediated by water molecules. Only the hydrophobic contact between Thr136 and T8 involves a residue in an α helix.
In contrast, no fewer than 31 residues make nonspecific contacts with the deoxyribose phosphate backbone of the DNA (Fig. 8). While these residues are still concentrated in the central DNA-binding motif, they are more widely distributed than those that make sequence-specific contacts. The majority of the phosphate interactions involve charged or polar side chains, particularly guanidine, amine, and amide groups, and nearly half are water mediated. Most of these residues lie in β sheets or in loop regions. On the other hand, nearly all the protein-deoxyribose interactions are hydrophobic, usually involving the C4′ and C5′ atoms of the sugar, which protrude into the minor groove of the DNA. The only residue that interacts with the C1′ and C2′ of the deoxyribose in the major groove, Ile178, also makes a sequence-specific hydrophobic contact in the major groove. Arg198 makes the only hydrogen bond contacts with a sugar, from the side chain N(ζ)H2 to the O4′ of A5 and G6. Other residues that may make contacts that are not explicit in the crystal structure are Lys249, His253, and His304, which could make water-mediated contacts, and Gln294, which can be rotated to make a contact with the 5′-phosphate of A14.
Notably, residues that make nonspecific contacts are often positioned such that they flank those that make sequence-specific contacts. It may be that the nonspecific interactions are required to position the backbone interactions correctly for optimal binding or, conversely, that the nonspecific interactions provide a means to allow Tus to slide along DNA searching for its specific binding contacts.
DNA Modification and Protection Studies
The Tus-Ter interaction was examined by DNA footprinting and protection studies soon after both protein and DNA were available. Sista et al. (159) used copper-phenanthroline footprinting to show protection by Tus binding of 14 to 16 nucleotides on both strands of TerR1 and TerR2. The footprint showed no preference for binding to one strand over the other. DNase I footprinting showed protection of a similar, but larger, region due to lesser accessibility of the enzyme compared to the copper-phenanthroline cleavage agent. This assay showed a slight preference for protection of the upper strand shown in Fig. 9 (65). In later studies with both TerB (54) and TerR1/2 (158), more detailed experiments using hydroxyl radical footprinting, methylation protection, and ethylation interference gave broadly consistent results.
FIG. 9.
Summary of the results of footprinting studies by Sista et al. (158) and Gottlieb et al. (54). Arrows indicate protection from hydroxyl radical cleavage. Filled circles indicate protection from methylation by dimethyl sulfate. Open circles show enhanced methylation. The base pairs that interact with Tus are shaded as in Fig. 3.
Both the Hill and Bastia groups (54, 158) reported G10, G13, and G17 to be protected from methylation by Tus binding (Fig. 9), as would be expected from the crystal structure (Fig. 7). The TerR2 site was also protected at the guanosine substituted for T20 of the TerB sequence, while methylation at A16 was enhanced at both Ter sites, consistent with its solvent exposure and distortion from the B form in the crystal structure. TerB also showed enhanced methylation at A11, again a reflection of the solvent exposure and deviation from a B-form structure, while TerR2 DNA showed enhanced methylation at the guanosine substituted at A8, another solvent-exposed residue that may be further distorted as a result of the substitution. Ethylation interference showed that the phosphates between G10 and T14 (on the top strand as shown in Fig. 9) as well as those between A18 and C13 (on the bottom strand) were necessary for Tus binding (54, 158). The phosphates of all these nucleotides interact with Tus in the crystal structure (Fig. 7).
Although most of the protected nucleotides were within the region bound by Tus in the crystal structure, from G6 to A18 on the top strand and from T19 to C6 on the bottom strand (Fig. 3), both groups reported protected sites outside this region. Sista et al. (158) found four such sites, between 1 and 3 base pairs preceding and 1 following the Tus-binding site in the TerR2 sequence (Fig. 9). While Gottlieb et al. (54) described two protected sites preceding TerB, these were only 1 base pair from the binding site and could be explained by occlusion by the overhanging protein. The TerR2 protection sites are more difficult to explain on the basis of the static crystal structure.
The KD of the Tus-TerR2 complex has been estimated to be 30-fold higher than that for Tus-TerB (54). If this is largely the result of the loss of sequence-specific interactions, then the protected sites on TerR2 may reflect greater mobility of the protein on this DNA. Conversely, it may simply be the case that the crystal structure does not accurately represent the mobility in solution of the amino acid side chains in the vicinity.
Another explanation is that Tus engineers structures in the DNA at each end of the complex that are resistant to hydroxyl radical cleavage. At the permissive face of the complex (Fig. 6), this may be the result of strand separation. This is suggested by the run of four AT base pairs, the twisted conformations of the AT16 and TA18 base pairs, and the nucleotide substitution data that will be discussed below. At the nonpermissive face, strand separation may be indicated by the severe twist induced in the AT5 base pair. The high AT content in DNA at the nonpermissive end of most Ter sites (Fig. 3), the nucleotide substitution data (below), and the very close approach of DNA polymerase inferred from the position of the end of leading-strand synthesis (70) may also suggest that strand separation occurs at this point.
Nucleotide Substitution Studies
The effects on Tus binding of substitution of base pairs at various points in the TerB sequence were examined by a variety of approaches. Duggan et al. (42) investigated the effect on the free energy (ΔG) of binding (or an apparent ΔG‡ based on dissociation rate constants) of replacing the base in each of the four conserved deoxyguanosine residues (Fig. 9) with 7-deazaguanine, 2-aminopurine, and inosine. Each of these substitutions removes a specific functional group from the guanine base, and replacement by 2-aminopurine also disturbs base pairing with cytosine. They also replaced GC base pairs with 2-aminopurine · uracil base pairs, which form a more stable hydrogen bonding arrangement. Furthermore, to investigate the role of thymine methyl groups in the binding interaction, six thymine bases were replaced with uracil, as well as with 5-bromo- and 5-iodouracil (41, 42). Bromine and iodine atoms are approximately the same size as a methyl group and could compensate for the loss of this group. Due to the greater electronegativity of iodine, an increase in binding by the substitution of iodo- over bromouridine would also confirm the presence nearby of a polarizable amino acid.
Where a thymine methyl group is involved in a hydrophobic interaction, there was found to be a positive ΔΔG‡ (i.e., more rapid dissociation) for the substitution of halogenated uracil. The two main thymine methyl interactions are at nucleotides T12 and T16, and these are the two thymines with the highest ΔΔG‡ for conversion to uracil and the halogenated analogs. A negative ΔΔG‡ (slower dissociation) for iodo- and bromouracil substitution was observed for modifications of T8, T14, and T19 and indicates the presence of a polarizable group in the minor groove (41). This is confirmed by the crystal structure for T8 (interacts with Lys89) and T14 (interacts with Lys175), and a contact with T19 can be formed by rotating the side chain of Arg288. In most cases the Tus complex with TerB replaced with a 2-aminopurine:uracil base pair was observed to be slightly more stable than a 2-aminopurine · cytosine base pair, indicating that unfavorable base pairing contributes part (GC10) or most (CG17) of the increase in ΔG. However at GC13, where the N4 of cytosine interacts with His176, the substitution of uracil for cytosine opposite 2-aminopurine greatly destabilized the Tus-Ter interaction (42).
Coskun-Ari and Hill (30) chose an alternative approach of replacement of base pairs in TerB with all three natural alternates and produced a near-complete set of all possible substitutions in the region GC6 to AT21. This allowed them to identify three new Ter sites in the E. coli genome sequence, to define in general terms which Ter sites are strong or weak Tus-binding sites, and to specify precisely which residues in the consensus sequence are important for binding as well as for replication fork arrest activity in vivo.
The nucleotide substitution data need to be interpreted carefully. A single substitution could affect DNA stability, the entropic cost of removing water from its hydration shell, and even the internal structure of the Tus protein, as well as directly affecting binding. As expected, the combined substitution data agree broadly with the crystal structure and conservation of residues within the Ter sites. The most important base pairs for Tus binding were found to lie in the most conserved regions (Fig. 3). For example, the TA7 base pair, which is not conserved and does not contact Tus in the crystal structure, was found to be dispensable, and the partially conserved AT8 base pair showed tolerance for the GC substitution found in a number of natural Ter sites (30).
In general, there was a correlation between binding energy and replication arrest activity in vivo. However, at the nonpermissive end, the three substitutions at GC6 all had a much larger effect on replication arrest than expected on the basis of the change in binding energy, indicating that this base pair is important for replication arrest for reasons that are not related primarily to the stability of the Tus-TerB complex (30).
It is also difficult to correlate the crystal structure (85) with the effects of some substitutions at the permissive face (30). Although changes to the conserved AT19 base pair caused a large change in ΔG for binding and abolished replication arrest activity, the crystal structure shows no explicit sequence-specific interaction at this site. The Arg288 side chain can be brought into contact with either O2 and N3 or N3 and O4 of this thymidine, depending on whether its Nɛ is simultaneously positioned to interact with adenine or thymine at TA18 (Fig. 7). The N3-O4 interaction could also be strengthened if the strands were separated, but the quantitative data offer little guidance about which interactions are most likely. Substitution at AT20, which lies beyond the DNA used for crystallization, reduced arrest activity while having only a modest effect on binding (30). This may be due to interactions (not seen in the crystal structure) with Trp243 and Gln248 (Fig. 7) or to structural changes in the DNA required for fork arrest activity. Finally, at the adjacent AT21 site, now well away from the protein, there was also an effect on both binding and arrest activity, again suggesting a role for DNA structure in the binding reaction, the arrest reaction, or both (30). We note that Gln248 can be positioned for potential interactions with T21 (Fig. 7).
The role of the four base pairs GC6 and AT19 to AT21 may well be concerned with engineering of a structure in the DNA that affects helicase passage through the Tus-Ter complex. This structure might include the separation of the DNA strands at one or the other end of the complex, and this is supported by other elements of the crystal structure (85, 170). The results are also consistent with a dynamic complex in which partial unbinding processes play a role in the antihelicase activity. In this case these anomalous base pairs would be involved in binding of intermediates on the binding-unbinding pathway but not in the final steady-state Tus-Ter complex.
Mutants of Tus
Reported mutants of Tus (summarized in Table 1) fall into two main groups, those isolated by screening for defective replication arrest activity or reduced helicase interaction and those generated deliberately to test hypotheses based on structural or biochemical data. As with the nucleotide substitution data, comparison of the effects of these mutations on both DNA binding and replication fork arrest activity has the potential to identify factors involved in fork arrest beyond those that relate simply to binding of Tus to the Ter sequences. It is also tempting to infer the relative contributions of the various contacts revealed by the crystal structure to the specificity of Tus-Ter binding.
TABLE 1.
Effects of amino acid modifications on the activities of Tus
| Tus structure | Mutation(s) | Effect on Tus activity
|
|
|---|---|---|---|
| DNA and DnaB helicase binding | Replication arrest and antihelicase activity | ||
| Wild type | None | t1/2 = 150 mina,b | 100%; no growth, 13% full length rep, 2.4 fmolb |
| L1 | P42S | t1/2 = 9 mina | 37% of wt arrest activitya |
| P42L | KDTeR2 three-fold increase, DnaB binding reducedc | No antihelicase activityc | |
| E43Q | No growth, 14% full length repb | ||
| V44T | No growth, 19% full length repb | ||
| K45A | t1/2 = 48 minb | No growth, 11% full length rep, 2.8 fmolb | |
| K46A | t1/2 = 15 minb | Growth, 54% full length rep, 5.0 fmolb | |
| E47Q | t1/2 = 348 minb | Growth, 56% full length rep, 2.4 fmolb | |
| KDTerR2 fourfold decrease, DnaB binding reducedc | Full anti-helicase and in vitro replication arrest activityc | ||
| D48N | t1/2 = 195 minb | No growth, 10% full length rep, 2.7 fmolb | |
| E49A | t1/2 = 274 minb | Growth, 26% full length rep, 2.5 fmolb | |
| E49K | t1/2 = 175 min, KDTerB unchangeda,b | 38% of wild-type arrest activitya | |
| KDTerR2 twofold increase, DnaB binding reducedc | Defective antihelicase and in vitro replication arrest activityc | ||
| 51%, 7.9 fmolb | |||
| E47Q/E49A | Growth, 39% full length rep, 3.3 fmolb | ||
| E47Q/E49Q | t1/2 = 212 minb | Growth, 30% full length rep, 4.0 fmolb | |
| H50N | t1/2 = 109 minb | No growth, 13% full length rep, 2.7 fmolb | |
| H50Y | t1/2 = 26 min, KDTerB sixfold increasea | 81% of wild-type arrest activitya | |
| N51D | No growth, 17% full length repb | ||
| P52L | KDTerR2 twofold increase, DnaB binding reducedc | Antihelicase activity reducedc | |
| L2 | E84A | No growthb | |
| N85D | No growthb | ||
| K89A | KDnsDNA unaffected, KDTerB 200-fold increased | ||
| kaTerB 10-fold decrease, kdTerB 20-fold increased | |||
| R93H | Partial or complete defectc | Growthc | |
| βD | P95S | Not detectablea | Growtha |
| P95H | Partial or complete defectc | Growthc | |
| P95L | Partial or complete defectc | Growthc | |
| αIV | E141A/R145A | No growthb | |
| αIV-αV | L150Q | Partial or complete defecte | Growth |
| αV | Y156C | Partial or complete defecte | Growth |
| αV-βE | L159P | Not detectablea | Growtha |
| βF | G171D | Partial or complete defecte | Growth |
| A173T | KDTerB 4,000-fold increase, kdTerB 1000-fold increasea | Inactivea | |
| t1/2 = 0.5 minb | Growth, 182% full length rep, 10.9 fmolb | ||
| KDnsDNA unaffected, KDTerB 4,000-fold increased | |||
| kaTerB 40-fold decrease, kdTerB 100-fold increased | |||
| A173V | KDTerB 100-fold increase, kdTerB 100-fold increasea | Activea | |
| βG | K175E | Growthe | |
| L3 | R198A | KDnsDNA 10-fold increase, KDTerB 150-fold increased | Growthb |
| kaTerB 50-fold decrease, kdTerB fourfold increased | |||
| t1/2 = 2 minb | |||
| αVII | R205A/E206A | No growthb | |
| R210A/R214A | No growthb | ||
| βH | R232S | Partial or complete defecte | Growth |
| βI | Q237R | Partial or complete defecte | Growth |
| P238L | Not detectable;a partial or complete defecte | Growtha,e | |
| R241L | Partial or complete defecte | Growth | |
| βJ | Q250A | KDnsDNA unaffected, KDTerB 400-fold increased | |
| kaTerB eightfold decrease, kdTerB 50-fold increased | |||
| Q252R | Partial or complete defecte | Growthe | |
| A254D | 7% DNA binding of wild type | Inactivea | |
| βJ-βK | P256L | 4% DNA binding of wild type | Inactivea |
| βK-βL | D266A | No growthb | |
| D266N | No growthb | ||
Skokatas et al. (160, 161) reported mutant Tus proteins selected from a survival assay where cell growth is associated with defective replication arrest activity. Equilibrium dissociation constants, dissociation and association rate constants, and the half-life (t1/2) of dissociation measured by nitrocellulose filter binding were also reported, together with the percentage of replication arrest activity.
Henderson et al. (59) reported a similar growth assay of replication arrest activity as well as a quantitative assay of in vivo arrest (increasing percentage of full-length plasmid replication from 13% for wild-type Tus to 100% in the absence of Tus shows loss of replication arrest activity) and an in vitro helicase assay (increasing quantity of liberated DNA [to a maximum of 10.7 fmol] shows loss of antihelicase activity), as well as the half-life of dissociation from TerB.
Mulugu et al. (127) reported equilibrium dissociation constants measured by a gel shift assay.
Neylon et al. (131) reported equilibrium and dissociation and association rate constants for binding to TerB and nonspecific DNA obtained from an SPR assay (in 250 and 100 mM KCl, respectively).
Kamada et al. (85) reported mutant Tus proteins selected from a survival assay where cell growth is associated with defective replication arrest activity; all mutant proteins were reported to have a partial or complete defect in Ter binding.
It should be noted, however, that many of the amino acid substitutions that have been studied resulted in a decrease in positive charge in the neighborhood of the changed residue and so might also affect the nonspecific interaction of Tus with DNA sequences that do not resemble Ter sites. Tus binds reasonably avidly to such sites; measured values of KD indicate binding to be 104- to 105-fold weaker than that to TerB (55, 131). Electrostatic interactions clearly make a major contribution to binding of Tus to both specific and nonspecific sites. In a study of the effect of KCl concentrations on the Tus-TerB interaction, using surface plasmon resonance (SPR), Neylon et al. (131) showed that a plot of ln KD versus ln [KCl] had a slope of about −11 and that this very substantial salt dependence is essentially completely due to effects on the association rate constant. Kapur et al. (89) further showed that the dissociation constants of complexes of TerB with various mutant forms of Tus were correlated with the ionic strength dependence of their dissociation, as determined by electrospray ionization mass spectrometry. Thus, in using measurements with Tus variants with charge change substitutions to comment on specificity, it is clearly necessary to separate general electrostatic effects from those due to disruption of sequence-specific contacts. While the work of Neylon et al. (131) indicates that this could be done by comparing binding of the variant proteins to Ter and nonspecific DNA sequences as a function of ionic strength, this has not yet been done for any variant of Tus.
Genetic methods have been developed to select directly for Tus mutants defective in replication arrest (160, 161). In one of these, developed by Skokotas et al. (160) and also used by Kamada et al. (85), a Ter site was placed so as to disrupt replication of the chromosome of a tus recA strain, and tus mutants were introduced into cells on a plasmid. Active Tus binds to the Ter site and prevents chromosome replication, while Tus mutants defective in fork arrest allow replication and cell survival. Mutants of this type could reflect not only the effects of substitutions on specific and/or nonspecific DNA binding but also aspects of fork arrest not related to DNA binding or even the folding of Tus into a stable structure. These effects could of course be separated by further investigation of the properties of the isolated variant proteins, but this has not always been done.
Of the 18 residues (Table 1; Fig. 10) identified in this way as being important for activity, 11 (His50, Arg93, Tyr156, Ala173, Lys175, Arg232, Gln237, Arg241, Gln252, Ala254, and Pro256) are directly involved in DNA binding and 3 others (Glu49, Leu159, and Pro238) are adjacent to residues that make contact with the DNA. The other identified residues which are probably important in maintaining tertiary structure include four prolines (residues 42, 95, 238, and 256); Leu150, which contributes to the hydrophobic core of the helices in the N domain; and Gly171, which provides the flexibility necessary for βF to twist as it bends through ∼90° to pass under βHI to follow the major groove of the bound DNA molecule (Fig. 6). Thus, it is reasonably easy to explain why each residue affects replication arrest activity in terms of effects on Tus stability or Tus-DNA binding. While these studies clearly identify important residues, the absence of mutations at a particular site does not indicate that the residue is unimportant. Only one of the selected mutants (P238L) was obtained in more than one experiment (85, 161), indicating that the sampling processes were not exhaustive.
FIG. 10.
The nonpermissive face of the Tus-Ter complex. Four equivalent views of this face are shown, highlighting the features that might come into contact with the DnaB helicase. (A) Secondary structure elements that could contact DnaB; (B) residues at the nonpermissive face of the complex, including Glu49; (C) space-filling representation colored by residue type (red, acidic; blue, basic; yellow, polar; gray, aliphatic); (D) charge distribution on the Tus surface at the nonpermissive face. Charge was calculated without the TerA DNA in place, using atomic charges and Poisson-Boltzman calculation as implemented in SWISS-PDB VIEWER version 3.7 (56).
Of particular interest is the conversion of Glu49 to Lys. Although this mutation led to an increase in strength of the Tus-TerB interaction, it reduced replication arrest activity in vivo (160). Glu49 lies in the L1 region, near the nonpermissive face of the complex (Fig. 10). It is not well situated for direct interaction with the oncoming helicase, as it is partially occluded by other residues in the L1 loop and the Ter DNA. Perhaps the movement of the helicase into the region of the Ter site leads to structural alterations that reposition Glu49.
The characteristics of this mutant prompted Henderson et al. (59) to use oligonucleotide-directed mutagenesis to examine a larger range of mutants with mutations in the L1 loop (residues 41 to 53). This loop is expected to be a reasonably autonomous folding unit, as it is separated from adjoining secondary structure elements by proline residues at positions 42 and 52. Only His50 interacts directly with Ter DNA, but other mutations in L1 may also destabilize interactions with L2; Lys46 has interactions with Asn85 and Ser88 (which makes a contact to the phosphate of A7), and in solution, Glu47 could interact with Asn85. The E43Q, V44T, K45A, K46A, E47Q, D48N, E49A, E49K, H50N, and N51D mutants were examined in a quantitative assay of arrested plasmid replication intermediates, a growth assay similar to the selection system described above, and for binding to TerB and inhibition of DnaB helicase activity in vitro (Table 1). Of the mutants that had defects in replication arrest (K46A, E47Q, E49K, and E49A), all except for K46A showed more stable rather than weaker TerB binding (59).
However, this in vivo replication arrest defect was not mirrored precisely in in vitro antihelicase assays. Both E47Q and E49A mutants were as effective as wild-type Tus in preventing helicase action, while the E49K and K46A mutants were less effective. These results were confirmed by Mulugu et al. (127), who found that the E47Q mutant protein was an effective block to DnaB helicase action and replication forks in in vitro assays but that the E49K mutant was defective in both. These data indicate a role for some residues (most especially Glu49) in replication arrest beyond simple DNA binding, and this strengthens the case for a role of Tus-DnaB interactions. The differences between results obtained with the in vitro assays and the more complex in vivo systems presumably reflect not only the ability of Tus to block progress of the replication fork but also the efficiency with which replication restart mechanisms operate to reestablish a functional fork following its stalling and dissociation of some of the replisomal components. Replication restart mechanisms are of current interest (32, 120, 148), and these studies have been extended to the specific case of forks stalled by a Tus-Ter block (18-20, 73, 74, 77, 78, 120-122, 140, 145, 155). However, these investigations are beyond the scope of this review and are not discussed further.
Mulugu et al. (127) reported additional mutational evidence for Tus-helicase interactions. An in vivo interaction between Tus and DnaB was detected using a yeast two-hybrid system. A library of randomly mutated tus genes was then screened using a reverse two-hybrid screen for reduced binding to DnaB. Three selected colonies all yielded the same mutation, a conversion of Pro42 to Leu. This mutation resulted in a slightly increased KD for the complex of Tus with a TerR2 oligonucleotide, and the complex dissociated more rapidly. It also had a reduced in vitro affinity for DnaB and was incapable of blocking helicase activity. Pro42 is on the surface of the protein, well away from the helicase-blocking face of the complex. It is not clear from the structure how it could directly affect Tus-helicase interactions. Three other mutations in the L1 region were also examined for effect on Tus-DnaB binding. Like P42L, the E49K mutant had reduced Tus-DnaB binding and was almost completely defective in in vitro antihelicase activity. The P52L mutant had reduced Tus-DnaB binding and somewhat reduced antihelicase activity, while the E47Q mutant had increased binding and normal activity. The reduction in antihelicase activity correlated broadly with the measured strength of binding to DnaB.
In spite of the extensive work with these selected mutant proteins, no single mutation or combination of mutations has been observed to completely eliminate the fork arrest activity of Tus while retaining its strong binding to Ter DNA. In fact, the most defective, the E49K mutant, still showed significant replication arrest activity (59, 160). Taken together, these results suggest that part of the activity of Tus resides in the strength of DNA binding and part resides in interactions with a replisomal component that is probably DnaB.
Another recent study of Tus mutants focused on residues that make specific DNA-binding contacts. Neylon et al. (131) used SPR to measure the effect of converting three of the outlying DNA-binding residues, Lys89, Arg198, and Gln250, to Ala, as well as examining the previously characterized A173T mutant. These measurements were done with buffer conditions different from those used previously, most importantly having significantly higher salt concentrations. The measured KD of the Tus-TerB complex under these conditions (in 250 mM KCl) was about 0.5 nM, while the values for the K89A, R198A, and Q250A mutants were in the range of 90 to 220 nM, and that for the A173T mutant was 2 μM. The large increase in KD for the A173T mutant under these conditions compared well with that reported for the same protein in a very different buffer (161).
The change in the dissociation constant for the complexes of the K89A, Q250A, and A173T mutants with TerB was due mainly to very large increases in the dissociation rate constant (131), suggesting that these residues have an important role in maintaining the complex once formed. The effect of the R198A mutation, however, was due largely to a 50-fold decrease in the association rate. This mutation had only a modest (<4-fold) effect on the dissociation rate. In addition, the R198A mutant had markedly decreased binding to (nonspecific) DNA that did not contain a Ter site. The magnitude of the change in KD for R198A-Tus binding to nonspecific DNA was comparable to the change seen in specific Ter binding, suggesting that a large part of the effect on specific binding was due to a defect in nonspecific binding (e.g., due to the decrease in positive charge). The other mutations had no significant effect on binding to DNA that did not contain a Ter site. The effect of mutations at these residues on antihelicase activity was not reported.
A Stepwise Mechanism for Tus-Ter Binding and Unbinding
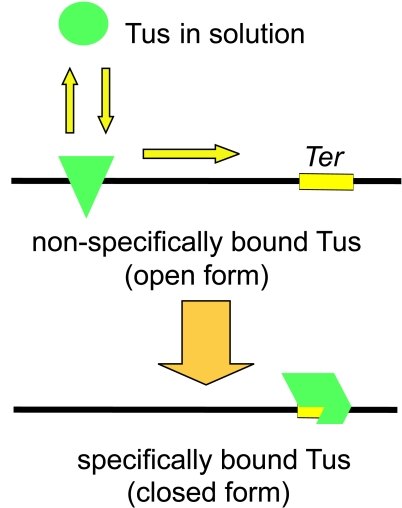
The SPR results of Neylon et al. (131), including also measurement of the salt concentration dependence of rate constants for the Tus-Ter binding equilibrium, were interpreted as supporting a stepwise binding/unbinding mechanism (Fig. 11). The value of KD was highly salt dependent, due almost entirely to a strong effect on the association rate constant, which implies the existence of intermediates after the initial collision step in the binding process (142). Stepwise binding involving one or several intermediate complexes could, in turn, be used to explain the polarity of replication fork arrest and several other outstanding data (131).
FIG. 11.
Tus-DNA and Tus-Ter binding. The solution form of Tus binds nonspecifically to DNA and scans along the double helix searching for a Ter site. On finding a Ter site, a series of conformational changes leads to formation of the closed Tus-Ter complex.
In this model, one crucial step in both binding and removal of Tus from the DNA is the conversion between a nonspecific Tus-DNA complex and the specific Tus-Ter complex. The approach of the helicase from the permissive side of the complex would promote the formation of a lower-affinity nonspecific complex that would then rapidly dissociate. Approach of the helicase from the other, nonpermissive, side would prevent formation of the nonspecific complex, and the Tus protein would be kinetically locked on the Ter DNA. This dynamic equilibrium could be affected by the mode of action and structure of the helicase, the overall strength of Tus binding to the specific Ter site, and the identity of base pairs that do not form explicit bonds in the crystal structure but would have a role in formation of the nonspecific complex. Within this model, the mutations in the L1 loop (59, 127, 160) could be described as having an effect on the internal equilibrium between specific and nonspecific complexes without reducing the overall strength of binding. This could occur if the proteins bind nonspecific DNA more strongly but are destabilized with respect to the specific interaction with Ter DNA. This might be expected, for example, for mutations that increase positive charge (or decrease negative charge) near the bound DNA.
In summary, the mutations isolated by screening procedures (Table 1) identify several residues that are important for in vivo fork arrest activity. Most confirm the importance of particular residues in DNA binding. The effects of the remainder can be explained in terms of disturbing the structure of the protein that provides the scaffolding for DNA-binding residues. The properties of the E49K and some other L1 mutants suggest strongly that there is more to the process of replication arrest than simple DNA binding, and there is evidence from the correlation of Tus-DnaB binding and replication arrest for a role for protein-protein interactions, at least in the specific case of the Tus-Ter complex blocking the DnaB helicase. On the other hand, the differential effect of some residues on specific Tus-Ter binding as opposed to nonspecific Tus-DNA binding suggests a dynamic model of the Tus-Ter complex that can also be used to explain a significant amount of otherwise difficult data.
Comparison of Tus Sequences
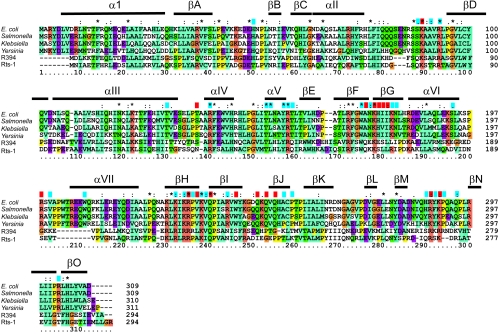
Neither the tus gene sequence nor the protein structure have any significant similarity to the sequence or structure of proteins with other functions in E. coli or any other species for which the chromosomal sequence is known. Furthermore, components of the replication termination system of B. subtilis, while functionally very similar to those of the Tus-Ter system, also have no significant sequence or structural similarity (26, 27, 169, 170, 172). This appears to be a classic demonstration of convergent evolution. The proteins from well-characterized organisms with significant similarity are Tus (or putative Tus) proteins from bacterial species related to E. coli (40, 59, 84, 136) and the products of genes for what appear to be highly diverged Tus proteins carried on plasmids, including R394 of Salmonella enterica serovar Typhimurium (97) and R27 of Salmonella enterica serovar Typhi (156). The Rts-1 plasmid of Proteus vulgaris (128) carries two genes related to tus, one of which encodes a protein identical to the R394 protein. Recent large-scale sequence determination of environmental DNA samples from the Sargasso Sea (167) yielded (only) five complete or near-complete protein sequences with > 25% identity to Tus.
The existence of a Tus-Ter system in S. enterica serovar Typhimurium was reported by Rocklein et al. (144). The sequences of both this tus gene and those of Klebsiella pneumoniae subsp. ozaenae and Yersinia pestis have been reported and analyzed in detail (59). The protein sequences are nearly identical in length and show 78% (S. enterica serovar Typhimurium), 70% (K. pneumoniae), and 53% (Y. pestis) identity to E. coli Tus (Fig. 12). The degree of sequence divergence is consistent with the placement of the host species in phylogenetic trees. BLAST searches (3, 4) identify multiple DNA sequences similar to the core of TerB in the genomes of S. enterica serovar Typhimurium, Yersinia enterocolitica, Clostridium acetobutylicum, Erwinia amylovora, Erwinia chrysanthemi, a Buchnera sp., and a variety of plasmids (C. Neylon, unpublished data), suggesting that the Ter sequences and, by implication, a termination system related to Tus-Ter might be conserved across a wider, but limited, range of bacterial species.
FIG. 12.
Sequence alignment of some Tus and Tus-like proteins. An alignment of the Tus protein sequences from E. coli, Salmonella enterica serovar Typhimurium, Klebsiella pneumoniae subsp. ozaenae, and Yersinia pestis, along with sequences of Tus-like proteins from the R394 plasmid of S. enterica serovar Typhimurium and the Rts-1 plasmid of Proteus vulgaris, was carried out and colored using the default parameters in CLUSTAL_X (64). Essentially, residues are colored where more than a given percentage of residues belong to one class: cyan, aliphatic and hydrophobic residues; orange, basic residues; purple, acidic residues; green, neutral hydrogen bonding residues. All glycines are colored brown, and all prolines are colored yellow. Secondary structure elements from the Tus-Ter crystal structure are shown above the alignment. Residues that make DNA backbone contacts in the crystal structure are shown with a blue block above the alignment. Those residues that make sequence-specific contacts with the Ter DNA are shown with a red block. Tus and Tus-like proteins were identified using PSI-BLAST (4).
Every residue identified as being important by screening for arrest-defective mutants is conserved in the four closely related Tus proteins (i.e., those from E. coli, S. enterica serovar Typhimurium, Y. pestis, and K. pneumoniae), with the single exception of Ala254 in the Yersinia protein (Fig. 12). Both cysteines are conserved in all four species, and apart from Pro295, which is substituted in the Klebsiella protein, and Pro197, which is substituted in the Yersinia protein, every proline is conserved.
Nearly all those residues identified as making DNA contacts in the crystal structure (Fig. 7 and 8) are conserved in all four proteins (Fig. 12). The exceptions are Thr136, Ile177, His253, Ala254, Val285, His287, Arg288, Tyr289, and Gln294. Thr136, Ile177, and Arg288 make (or probably make) sequence-specific contacts, while the others make sugar-phosphate backbone contacts. Ala254, His287, and Tyr289 interact with the DNA through the peptide backbone. The Thr136 interaction is probably unimportant and is conservatively substituted in two of the three cases. Ile177 and Val285 are conservatively substituted by other nonpolar amino acids, and Arg288 is conservatively substituted with lysine. The interactions of His253 and Gln294 with the DNA backbone, if they occur in solution, can be restored in modeled structures by the observed tyrosine and lysine substitutions, respectively.
Interpretation of patterns of conservation in the more highly diverged plasmid-encoded proteins is less straightforward. The R394 and Rts-1 proteins are more closely related to each other than to the chromosomally encoded homologs; sequences have 20 to 30% identity with the other Tus sequences. The R394 gene is associated with one encoding a MucAB lesion bypassing DNA polymerase, which might suggest that it maintains some role in DNA metabolism, and the plasmid contains a number of Ter-like sites, including one that precedes the tus open reading frame but upstream of the promoter (97). In the E. coli gene, TerB lies between the ribosome-binding site and the −10 sequence of the tus promoter (Fig. 4). This position in the R394 gene is occupied by a LexA box, placing the protein under the control of the SOS response (97). The tus gene in the Rts-1 plasmid (128) is not closely associated with an obvious Ter site, although a search of the DNA for elements with similarity to the Ter consensus sequence identifies a number of potential Ter-like sites elsewhere on the plasmid.
By comparison with E. coli Tus, a number of short insertions and deletions occur in the plasmid-encoded proteins, primarily at points corresponding to loops in the Tus structure (Fig. 12). This, along with the fact that conserved residues are often found in interacting pairs in a modeled structure, confirms that the overall topologies of the proteins are similar.
Where known, the sequences of DnaB helicases from these species are more highly conserved than those of Tus (i.e., 92% for S. enterica serovar Typhimurium and 84% for Y. pestis), and the host DnaB is most likely required for replication of the plasmids. Thus, if Tus makes specific contacts with DnaB, it would be expected that elements involved would be conserved and that these would be distinct from residues required for specific Ter DNA binding. Among the three most closely related proteins (those of E. coli, Klebsiella, and Salmonella), there are such conserved regions at the nonpermissive end of the complex (the face that would come into contact with the blocked helicase). In contrast, residues at the permissive face are completely conserved only close to the DNA ligand (59).
When the Yersinia protein is considered, however, this large conserved nonpermissive face is not so apparent. There are some specific regions that are still clear as more highly conserved than their surroundings (Fig. 12). The L3 loop in the C domain (between αVI and αVII) is highly conserved, as are residues in a region of the N domain defined by the N terminus of αI and the region around αIII. The L3 loop may be conserved due to the requirement to position Arg198. The region at the top of the N domain is not intimately involved in DNA binding, except that Arg139 makes a close contact to a backbone phosphate. The completely conserved Glu141 points directly out into the solvent from the middle of αIV. The Arg-Phe-Glu motif (residues 139 to 141) is also conserved in the R394 protein and partially conserved in the Rts-1 protein (Fig. 12).
In summary, the tus gene sequences from S. enterica serovar Typhimurium, K. pneumoniae, and Y. pestis provide some information on residues that are important for the action of Tus in vivo. Overall the proteins are quite similar, and DNA-binding residues and those important for secondary structure are generally conserved. Where apparently important residues have not been conserved, it is usually possible to make a plausible argument to explain how the change could be accommodated. The existence of conserved residues at the fork-blocking end of the molecule that are not involved directly in DNA binding provides further support for the notion that functions of the protein beyond simple DNA binding are important in replication arrest.
STRUCTURAL INSIGHTS INTO THE INTERACTIONS OF Tus WITH HELICASES
Structures of DnaB and Related Hexameric Helicases
Aspects of the structures and functions of E. coli DnaB and related hexameric helicases have recently been reviewed (24, 28, 36, 37, 116, 137). There is no atomic resolution structure of the complete hexameric DnaB molecule available. Each DnaB monomer (471 residues) is made up of two domains linked by a region that may function as a flexible hinge (129). The N-terminal domain, comprising residues 24 to 136 (123), undergoes a monomer-dimer equilibrium in solution (171), is dimeric in the crystalline state (46), and appears to participate in interaction with the DnaG primase (21, 29, 133). The larger C-terminal domain (containing residues from about 170 to 471) is a hexamer and bears the ATPase and DNA-binding sites (21, 129). Two independently determined high-resolution structures of the N-terminal domain are available (46, 171), but since this part of the molecule is believed to face away from the replication fork (80, 82, 83) (Fig. 2), these structures do not provide useful information about the face of DnaB that may come into contact with the Tus-Ter complex.
Low-resolution structures of intact full-length DnaB and its complex with its loading partner DnaC have been obtained by image reconstruction from electron micrographs, using both negatively stained preparations (151, 176, 178) and samples of the native proteins frozen in ice (9, 150). The general structure is a toroid of three- or sixfold rotational symmetry (depending on conditions) with a channel through the center wide enough to accommodate one or both strands of DNA (Fig. 13). The symmetry state varies with pH (39), and these changes are presumed to indicate significant flexibility that may be required for conformational changes that occur during translocation on DNA templates or during loading of the helicase onto DNA at origins of replication or during replication restart at stalled forks. It is clear from work on DnaB (80, 87) and the distantly related phage T7 helicase (45, 57, 177) that the DNA single strand on which the enzymes translocate passes through the central channel, while the other strand is excluded. There is also now clear evidence that under some circumstances DnaB can undergo ATP-dependent translocation on double-stranded DNA, with both strands passing through the central channel (87, 88). The channel is about 30 Å across and 60 to 80 Å deep (150, 176) and could therefore accommodate about 20 bp of double-stranded DNA. It has been estimated independently that the central channel binds a single-stranded DNA fragment 20 ± 3 nucleotides in length (25, 79, 81, 82).
FIG. 13.
Reconstruction of model atomic resolution structures of the DnaB helicase with threefold (A and C) and sixfold (B and D) symmetries. The helicase would approach the Tus-Ter complex with the upper face in C and D. Atomic resolution structures of the T7 gene 4 helicase domain (green) (157) and the N-terminal domain of DnaB (blue) (46) were docked into electron density maps determined by electron microscopy. The arrows in A indicate regions of the helicase structure that penetrate the electron microscopy surface envelope in the compressed helicase domains, suggesting that additional conformational changes in the atomic structure are necessary to fit the electron microscopy map. In both the threefold and sixfold models, there is additional unfilled density between the helicase and N-terminal domain (D, red arrow) that is likely due to 51 residues of the linker region not accounted for in the atomic structures. The figure is reproduced from reference 176 with permission from the Journal of Molecular Biology.
The other hexameric helicase whose progress has been reported to be blocked by Tus is SV40 T antigen. San Martin et al. have reported on the low-resolution structure of this enzyme (149), and a high-resolution structure of a central region of the protein that is hexameric and active as a helicase (residues 251 to 627) has recently appeared (109). The protein is unrelated in sequence to DnaB, but has a similar toroidal structure in spite of the fact that it translocates on single-stranded DNA in the opposite (3′-5′) direction.
Similar low-resolution images have been obtained for other hexameric replicative helicases that are not yet known to interact (in the functional sense) with Tus. These include the coliphage T7 gene 4 helicase-primase (45, 177), the B. subtilis phage SPP1 helicase (10), the papillomavirus E1 helicase (47), and the plasmid RSF1010-encoded RepA protein (154, 174).
The sequence similarities among the hexameric helicases led to a prediction that they all possess hexameric regions that have structures similar to that of the C-terminal domain of DnaB and that this domain has a structure related to that of the DNA recombination factor RecA, which forms a helical structure with a hexameric repeat (28, 162, 163). This prediction has been vindicated by recent determinations of the crystal structures of two hexameric helicases that are distant relatives of DnaB: the RSF1010 RepA protein (132, 174) and the C-terminal helicase domain of T7 gene 4 protein (152, 157). The overall structures of these hexamers are similar. They are ring-shaped structures about 12 nm across with a central channel wide enough to accommodate at least a single strand of DNA. Thus, they resemble their reconstructions from electron-microscope images, as well as those of DnaB and the other hexameric helicases. Very recently, the atomic structure of the complete T7 helicase-primase was reported (165). It crystallized as a ring-shaped particle, surprisingly with seven subunits forming the toroid.
By modeling the X-ray structures of the N-terminal domain of DnaB and the C-terminal domain of the T7 helicase together into the low-resolution electron density maps obtained from electron micrographs, Yang et al. have elaborated a model that locates the N- and C-terminal domains of DnaB in the intact molecule, in both the C3 and the C6 symmetry states (176). In the model, the face that first encounters the Tus-Ter complex is made up of the C-terminal (ATPase/helicase) domains and presents a rather flat surface to the fork-blocking complex (Fig. 13). It is not possible at this stage satisfactorily to model potential direct interactions between the helicase and Tus.
Interaction of Tus with Helicases
There are several other ways by which Tus could interact functionally with an oncoming hexameric helicase to facilitate fork blockage. The face that Tus presents to the helicase is 4 to 5 nm across at its widest. In each of the hexameric helicases, the internal channel is thus smaller than the shortest transverse section of the terminator protein. Tus could therefore act as a plug in the helicase if the association between the two molecules were to become this close. It is also possible that Tus engineers a structure in the DNA that blocks the progress of the helicase. The small fragment of DNA used for the crystallization of Tus does not extend far enough beyond the protein to allow comment on this (170). It is also conceivable that the helicase causes a rearrangement in the Tus structure, either by direct interaction or through the DNA that bridges the two molecules. If this was the case, it would be plausible for Tus to fit partially inside the channel of DnaB, providing a kinetic block to its removal from the DNA.
The fork-blocking face of Tus (Fig. 10) shows no obvious feature that could prevent the passage of the helicase. The concentration of positive charge near the bound TerA DNA (Fig. 10D) is contributed by DNA-binding residues and is neutralized on DNA binding. The solvent-exposed residues that the helicase would contact most closely are predominantly polar residues (Fig. 10C), but there is no apparent bias towards negative or positive polarity, and at least two of the groups are aliphatic. The L1 loop has been reported to be involved in Tus-helicase interactions. In particular, Glu49 when replaced by Lys increases DNA binding affinity but reduces replication arrest activity by 62% (59, 160), suggesting that it may have a role in Tus-helicase interactions. The mutations of Pro42 to Leu, Glu47 to Gln, Glu49 to Lys, and Pro52 to Leu have been reported to reduce Tus-helicase interaction (127). However, neither Pro42 nor Pro52 is exposed near the nonpermissive face in the structure (Fig. 10B), so substantial structural rearrangement would be necessary to enable direct contact with the helicase. Glu49 is situated so that it could contact DnaB, but it is still well below the upper face of the complex (Fig. 10). It is therefore probable that the effects of these mutations are through some indirect mechanism or that they become uncovered by the action of the helicase at the nonpermissive end of the Ter site. On the basis of the crystal structure, exposed residues at the nonpermissive face of the complex (e.g., those in L3 or αIV [Fig. 6]) are better situated to interact with the oncoming helicase. It is of interest that mutations in these residues have not been selected in the screen for those that interact with DnaB.
Other residues that might make contact with the oncoming helicase are in L2, αIV, αV, and the αVI-L3-αVII region (Fig. 6). These regions each make at least one contact with the DNA. In fact, these residues account for nearly all of the DNA contacts outside of the core binding domain. However, it is not possible to determine if secondary structure elements are placed to facilitate the interaction of the binding residues with DNA or if DNA binding is required to position residues within the secondary structure elements in the correct place to block the passage of the helicase. With the exception of residues in L1 and two others (Lys89 in L2 and Arg198 in L3), the effects of mutations at the nonpermissive end of the complex have not been studied in detail (Table 1).
Thus, in contrast to expectations, the structure of the Tus-Ter complex offers no convincing evidence concerning the mechanism of polar replication fork arrest. It may be stated in general terms that the Tus-Ter interaction would have to be disrupted during unwinding of the DNA double helix and that the complex is too large to allow helicases to pass over it. Atomic resolution structural and further mechanistic information about relevant helicases should ultimately allow further comment on the nature of specific Tus-helicase interactions.
MECHANISMS OF POLAR REPLICATION ARREST
In spite of the large volume of information available on Tus and its involvement as an antihelicase in replication fork blockage, there are many mechanistic aspects of the process that are uncertain. Under these circumstances it makes sense to briefly summarize the established data.
(i) The details of antihelicase activity and replication arrest appear to be strongly dependent on the identity of the Ter site, the mode of action of the helicase, and the complement of other proteins in the translocating replisome. The in vitro experiments, while they shed light on the action of the Tus-Ter complex, probably do not fully reflect the details of replication arrest and subsequent replication restart processes in vivo.
(ii) A simple molecular clamp can be an effective antihelicase in vitro. The EcoRI E111Q mutant binds strongly to its DNA recognition sequence and prevents the passage of a variety of helicases (14). This protein binds to its DNA recognition sequence with a KD (95, 173) that is comparable to that of the Tus-TerB complex (54), and other protein-DNA complexes can have a comparable effect (175).
(iii) For a monomeric DNA-binding protein like Tus, a simple thermodynamic clamp cannot account for the polarity of replication fork arrest. A plausible clamp model must include kinetic or structural details to explain polarity.
(iv) There is evidence from both protein mutant and nucleotide substitution studies that the effect of some substitutions on replication arrest cannot be explained in terms of their effect on DNA binding. In particular, substitutions at Pro42, Glu49, GC6, and AT19 have a much greater negative effect on replication or helicase arrest than would be expected from their effect on DNA binding. There is a general but not absolute correlation between the strength of Tus binding to DnaB and in vitro antihelicase activity (Table 1).
(v) Under some circumstances, when bound to TerB, Tus appears to be capable of antihelicase action against a wide variety of helicases, including 5′-3′, 3′-5′, replicative, and nonreplicative. Therefore, if interactions between Tus and these helicases are relevant to general antihelicase activity, the interaction must be with a portion of the helicase that is sufficiently well conserved.
(vi) While the crystal structure of the Tus-TerA complex is in accord with most of the other available data, there are some that are not easily explained. In particular, the structure does not provide an explanation for the protection of DNA from cleavage at base pairs beyond the reach of the protein in the complex, the effect on DNA binding of several amino acid and nucleotide substitutions at positions where no interaction is observed, and the evolutionary conservation of regions of the protein that are not involved in DNA binding. In particular, the fork-blocking face of the protein appears to be more highly conserved than the permissive face.
Having accepted these generalizations, we can make further statements. A simple clamp mechanism is necessary but not sufficient. This leads to the question of what other mechanisms could be used. Two broad classes are possible. The first invokes a role for dynamics of the protein-DNA structure as the helicase competes with Tus for the DNA. The second invokes protein-protein interactions between Tus, the helicase, and potentially other proteins. This could include a role for DNA structural changes engineered by Tus to block the progress of the helicase or engineered by the helicase to affect the affinity of Tus for the Ter DNA. It is likely that aspects of all these mechanisms operate; evolution does not select intelligently among simple and defined mechanisms but is presumed to take advantage of any physical effect that fine tunes the selected function. It should be noted in this context that it is not even clear what is the selective advantage of maintaining the Tus-Ter system (61, 68, 100, 144).
Prospects for the Future
The mechanism of polar replication fork arrest by the Tus-Ter complex is a problem worthy of resolution, because it represents a well-developed model system for an unusual kind of protein-DNA interaction. In what follows, we describe some of the experiments required to further define the process. The most significant question reduces to how to explain the polarity of the process. Fundamentally, and it may seem to be stating the obvious, all that is required to explain polarity is that the pathway for dissociation of Tus from its complex with a Ter site should be different depending on whether the replisome is approaching from the permissive, as opposed to the nonpermissive, face.
We note that protein oligomerization (e.g., monomer to dimer or dimer to tetramer) during DNA binding occurs in many comparable systems, including many repressor-operator interactions, and that it often occurs in a stepwise fashion. A multistep (cooperative) process is capable of solving the problem of achieving high overall binding affinity and specificity while still allowing the dissociation rate to be high enough to allow quick physiological responses (141). In the case of a replication fork approaching Tus-TerB from the permissive direction, a high rate of dissociation of Tus could also result from breaking the process down into a series of steps.
Polarity can also be achieved in this way in the case of multimeric proteins. For example, the B. subtilis replication termination system consists of a series of imperfect inverted repeat Ter sequences and a protein, RTP, that binds sequentially as a homodimer to each of two adjacent half-sites during formation of the fork-arresting complex (44, 101, 169). It is clear that in the case of RTP, polarity of replication termination could be adequately explained by cooperativity in binding of the second dimer, coupled with differential affinity of binding of the protein at each of the half-sites (43, 101, 172). However, even in this case, this is not the whole story (reviewed in reference 44). It may well be that the Tus-Ter and RTP-Ter complexes share aspects of mechanism, even though their structures do not.
Can a clamp mechanism be used to explain polarity of action of a monomeric protein-DNA complex, such as that between Tus and Ter sites? Within the class of clamp mechanisms there are a variety of possibilities. The simplest (Fig. 14) is one for which the passage of a helicase through the Ter site requires the complete dissociation of Tus in a single step. With this mechanism, however, it is not possible to explain polarity. A helicase approaching either face of the complex would have to overcome the same energetic barrier to pass through. Kinetic or other aspects need to be added to explain polarity.
FIG. 14.
A “complete dissociation” model of Tus action. As shown, the permissive face of the Tus-Ter complex is on the left and the nonpermissive face is on the right. DnaB approaching the permissive face for replication comes into contact with the Tus-Ter complex, leading to complete dissociation of Tus. DnaB approaching the nonpermissive (fork-blocking) face is blocked from proceeding farther by the Tus-Ter complex.
More complicated clamp mechanisms involve a concerted stepwise process by which the helicase moves into the Ter site, removing Tus. The simplest form of a stepwise dissociation model, a two-step mechanism, can explain the polarity of the Tus-Ter complex (Fig. 15). Here, the binding residues are divided into two classes, residues at the permissive end of the complex and residues at the nonpermissive (fork-blocking) face. A helicase arriving from the permissive side can successfully compete with the Tus residues binding to this end of the Ter DNA, causing a conformational change in the remaining DNA-binding residues that either removes Tus from the DNA directly or allows the helicase to further compete successfully for the remaining binding sites. When the helicase approaches from the other side, the competition between helicase and Tus is such that Tus cannot so easily be removed. The most complex model of this type would describe the conformational changes and change in binding energy resulting from the removal of each DNA-binding residue as the helicase progresses from either face: a zipper model (131). This could be a good analogy because a zipper is itself inherently polar.
FIG. 15.
A simple two-step model of Tus-Ter and DnaB interactions. (Left) DnaB approaching the permissive face of the Tus-Ter complex promotes the formation of the open, nonspecifically bound form of Tus, which may dissociate directly or slide along the DNA. If DnaB moves into the Ter site before Tus can return to the specifically bound closed form, then helicase activity continues. (Right) DnaB approaching from the nonpermissive face cannot promote the formation of the open form of the complex, and further DNA unwinding is blocked.
A variant of this model involves a progressive change in the affinity of Tus for the DNA as a result of the presence or action of the helicase. Direct helicase-Tus interaction would be one way of accomplishing this. Another would be for Tus to bind with different affinity to an intermediate forked DNA structure engineered by helicase action at either the nonpermissive or the permissive end. As we have noted earlier, there are several experimental data that support the latter possibility. It is also notable that amino acid interactions seen in the crystal structure at the permissive face are almost entirely with the strand that would pass through the central channel of the helicase (85), and there is a cluster of basic residues (i.e., Lys119, His163, Lys245, Lys249, and His253) positioned just out of reach of the duplex DNA in the structure such that they might interact with the displaced strand at the permissive face, progressively driving further destabilization of the duplex DNA (85, 170). An alternate explanation comes from examination of basic residues similarly placed at the nonpermissive face (i.e., Arg145, Lys192, Lys195, and Arg205). Strand separation by the helicase could bring DNA phosphate groups into close proximity with these residues, thereby simply strengthening the Tus-Ter interaction.
The fundamental requirement of this form of model is that the pathway for dissociation of Tus from the Ter DNA is limited. That is, there is an intermediate in the dissociation pathway that is accessible only when the helicase approaches from the permissive (or the nonpermissive) direction. A simple explanation for this behavior could be that the helicase, sitting as a cup over the fork-blocking face of Tus, physically prevents the removal of residue contacts that would ordinarily be disrupted early in the dissociation pathway. However, this would not so simply explain the polarity of action of the Tus-TerB complex against the dimeric helicases, such as Rep (106).
While the dissociation pathway is difficult to probe directly, by examining the association pathway in detail it may be possible to define intermediates that are disfavored when the replisome approaches from the nonpermissive side. Neylon et al. (131) proposed a multistep (zipper) model for the binding of Tus to Ter DNA based on SPR studies of Tus and Tus mutants. Minimally, the protein first binds nonspecifically to the DNA before specific interactions come into play, closing the structure and leading to deformation of the DNA (Fig. 11). This is suggested by the observation that Arg198 plays an important part in nonspecific DNA binding but has a relatively minor role in determining specificity, whereas Lys89, Ala173, and Gln250 appear to be important for specific, but not nonspecific, binding. That is, some residues involved in a general nonspecific association with DNA appear to be separate from those involved in determining the sequence specificity of the interaction. In addition, the strong salt dependence of the association rate suggests that protein conformational changes take place after the initial collision step (142).
The solution structure of free Tus is significantly different from the bound structure. This is suggested by circular dichroism spectroscopic data indicating a smaller proportion of β-sheet structure in the free protein (31) than in the crystal structure (85). Basic residues, including Arg198, are involved in the initial stages of DNA binding, forming an open nonspecific complex presumably capable of scanning DNA in search of Ter sites. On finding a Ter site, residues involved in sequence-specific binding, including but by no means limited to Lys89, Ala173, and Gln250, are in position to bind specifically to their ligand sites. This leads sequentially to the formation of the bound protein structure, closing the complex and deforming the DNA. This process would be expected to be highly salt dependent, resulting in extensive charge neutralization and burial of a large portion of the solvent-exposed protein and DNA surfaces.
This model can be used to explain some of the outstanding data that appear to contradict a clamp model. The presence of protection sites outside the apparent reach of the protein (Fig. 9) is now predicted for a complex that is in equilibrium between a specifically bound form and a nonspecifically bound form. Furthermore, this suggests that such sites could spread farther in cases where the specific binding is weaker, such as with TerR2, as is observed (54, 158). This can also explain an apparent discrepancy between results on the effects of the R198A mutation. Neylon et al. (131) reported only a minor effect of mutagenesis on the rate of dissociation of Tus from TerB, whereas Henderson et al. (59) reported a 75-fold increase. This may be attributed to the difference in DNA fragments used in each case. Henderson et al. used a significantly longer DNA fragment in their measurements, and this should increase the contribution of nonspecific binding to dissociation. Neylon et al. (131) nevertheless reported a large effect of the R198A mutation on nonspecific binding, leading to an immeasurably high dissociation rate constant in their assay (in 0.1 M KCl). The DNA length dependence of kinetic and thermodynamic parameters for site-specific DNA-protein interactions has been used in several instances to comment on the importance of nonspecific interactions (16, 92, 168). These studies have been especially useful in dissecting the stepwise assembly or disassembly of site-specific DNA complexes with protein oligomers, but similar studies with Tus or other monomeric DNA-binding proteins have not been reported.
Amino acid and nucleotide substitution data can also be explained by the zipper model. Those residues that have a greater or different effect on replication arrest than is expected from the change in binding energy play a role in the kinetics of binding or dissociation. A comprehensive study of the effects of mutations in DNA-binding residues will provide more details of how stepwise binding/unbinding takes place. The solution structure of unliganded Tus would also be very helpful.
If an inaccessible intermediate on the dissociation pathway is similar to the complex between Tus and nonspecific DNA sequences, then some of the discrepancies in the results of helicase assays can also be explained. One of the main differences between assays from different research groups is the use of TerR2 as opposed to TerB. Binding to TerR2 is significantly weaker than binding to TerB (54) and may be part way between an “open” nonspecific complex and a “closed” specific Tus-TerB complex. If this intermediate is more accessible, then helicases with different modes of action, such as those involved in replication as opposed to repair, may be expected to displace Tus more or less efficiently.
Since it seems that under some experimental conditions, the Tus-Ter complex is capable of arresting the progress of the replicative hexameric helicases in a polar manner, but not that of the monomeric or dimeric repair (or rolling-circle) helicases (14, 91, 147), it may be illuminating to consider the differences in structure and mechanism between these two classes of enzymes. Structural studies, for example, with the rolling-circle helicases E. coli Rep and Bacillus stearothermophilus PcrA show that the site of DNA strand separation is within a channel in the protein structures (99, 116, 166). In contrast, given that the hexameric enzymes are believed to work by a strand exclusion mechanism, strand separation may occur right at the face of the oncoming helicase. The functional interactions of the two classes of helicases with Tus-Ter might therefore be quite different: only with the hexameric helicases might strand separation influence the Tus-Ter interaction before the progress of the helicase is physically blocked by direct collision of the proteins. Polar replication fork arrest by the hexameric helicases could then be explained by differential effects of helicase-mediated strand separation on the rate of dissociation of the Tus-Ter complex, depending on whether strands are being separated at the permissive or the nonpermissive face. If strand separation was important in determining polarity, then polarity should not be observed in assays that measure translocation of helicases rather than authentic DNA unwinding. Such assays are technically challenging.
It is thus possible in several ways to explain the polarity of replication fork arrest in terms of a mechanism that does not necessarily involve any direct physical interaction between replisomal components and the termination complex. Nevertheless, there are other studies that suggest that such specific protein-protein interactions exist. The primary functional evidence is from Andersen et al. (6), who showed that the Tus-Ter complex is a much more efficient block to the replication fork in E. coli than it is in B. subtilis and that the converse is true, to a lesser extent, of the B. subtilis replication termination system. This suggests that an element of the replication arrest process is specific to the Tus-Ter complex and the E. coli replisome.
Moreover, as described above, there are other recent reports that provide both direct (127) and indirect (59) evidence for protein-protein interactions. The effects of two L1 mutations, E47Q and E49K, on DNA binding, replication arrest, and binding to DnaB are consistent with a role for Tus-helicase interactions, and the preferential evolutionary conservation of residues on the fork-blocking face of Tus is suggestive of interactions between Tus and the replisome. Further studies on the nature and strength of the Tus-DnaB interaction are required. We note that the conserved GC6 base pair of Ter sites, which when mutated affects replication fork arrest more profoundly than DNA binding (30), is positioned at the nonpermissive face of the complex close to residues in the L1 loop of Tus (including Glu49). This may signal the existence of a new kind of interaction of GC6 and L1 at some stage of a process of helicase-promoted dissociation of the Tus-Ter complex.
There are also other replisomal components that could be involved. The τ subunit of DNA polymerase III holoenzyme is the organizational center of the replicase, coordinating and physically linking the actions of the two replication fork polymerases and DnaB (50, 51, 94, 107, 118). In the absence of these interactions, the progress of both the helicase and the replicase is retarded (93). It is tempting, simply on grounds of elegance, to suggest that Tus could disrupt the interaction of τ and DnaB to destabilize the replisome, leading to dissociation of DnaB. The polymerase could then continue to extend the leading strand, halting when it comes into contact with Tus. In fact, Tus might even compete with τ for its binding site on DnaB. Further interactions of Tus with replisomal components, if they were to exist, would also go some way towards explaining the discrepancies both among assays and between in vivo and in vitro results, as well as the species specificity observed by Andersen et al. (6).
The issue of measurements of DNA binding is an important one. Various research groups have measured equilibrium dissociation constants and association and dissociation rate constants in different experimental systems. In most cases only binding to specific Ter DNA fragments has been examined. The lengths and sequences of these fragments also vary among laboratories. If, as suggested, binding to nonspecific and Ter DNA involves different groups of residues, and if these play different roles in the replication arrest process, then the differences among these assays create a significant problem in interpretation of data.
More work clearly needs to be done. The tools to examine and dissect protein-protein interactions or DNA secondary structure are available and should be brought to bear on the problem. Detailed kinetic studies of the competition between Tus and DnaB for the DNA, combined with cross-linking experiments, should give insights into the process of replication arrest. Detailed and comprehensive examination of the effects of mutations on the association and dissociation processes will provide further clues to the events preceding the removal of Tus or the helicase from the DNA. Attention should also be refocused on the approach of DnaB from the permissive end of the complex. The process by which DnaB removes Tus from the DNA has received little, if any attention despite the fact that understanding the polarity of antihelicase action depends critically on understanding how the helicase overcomes the barrier when translocating in this direction.
Structural studies of the free protein and of the open protein-DNA and closed full-size Tus-Ter complexes would provide a helicase-eye view of the complex as it approaches. The dynamics of the Tus-TerB complex versus the Tus-TerR2 complex may provide important clues to the factors that lead to the experimentally observed differences between them. Simulations could provide clues to the molecular dynamics that occur within the various complexes. Another important requirement is a detailed examination of the effects of the protein complement on in vitro replication and helicase assays. It is clear that some of the elements of the in vivo process may be missing from the in vitro assays.
Further molecular dissection of the Tus-Ter complex, the helicase-DNA complex, the replisome, and the interactions among them can be expected ultimately to unravel the details of this fascinating process. Finally, we note in passing that polar binding protein-mediated replication fork arrest is not restricted to prokaryotic replicons. Study of similar processes in yeast and mammalian systems is under way (see, for example, references 52, 102, and 124 and references therein).
Acknowledgments
We are grateful to Mark Mulcair, Iain Duggin, and Patrick Schaeffer for helpful comments on the manuscript and to E. H. Egelman for a high-resolution copy of Fig. 13.
Work at the Australian National University was supported in part by grants from the Australian Research Council.
REFERENCES
- 1.Abhyankar, M. M., J. M. Reddy, R. Sharma, E. Büllesbach, and D. Bastia. 2004. Biochemical investigations of the control of replication initiation of plasmid R6K. J. Biol. Chem. 279:6711-6719. [DOI] [PubMed] [Google Scholar]
- 2.Abhyankar, M. M., S. Zzaman, and D. Bastia. 2003. Reconstitution of R6K DNA replication in vitro using 22 purified proteins. J. Biol. Chem. 278:45476-45484. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amin, A. A., and J. Hurwitz. 1992. Polar arrest of the simian virus 40 tumor antigen-mediated replication fork movement in vitro by the tus protein-terB complex of Escherichia coli. J. Biol. Chem. 267:18612-18622. [PubMed] [Google Scholar]
- 6.Andersen, P. A., A. A. Griffiths, I. G. Duggin, and R. G. Wake. 2000. Functional specificity of the replication fork-arrest complexes of Bacillus subtilis and Escherichia coli: significant specificity of Tus-Ter functioning in E. coli. Mol. Microbiol. 36:1327-1335. [DOI] [PubMed] [Google Scholar]
- 7.Baker, T. A. 1995. Replication arrest. Cell 80:521-524. [DOI] [PubMed] [Google Scholar]
- 8.Baker, T. A., and S. P. Bell. 1998. Polymerases and the replisome: machines within machines. Cell 92:295-305. [DOI] [PubMed] [Google Scholar]
- 9.Bárcena, M., T. Ruiz, L. E. Donate, S. E. Brown, N. E. Dixon, M. Radermacher, and J. M. Carazo. 2001. The DnaB · DnaC complex: a structure based on dimers assembled around an occluded channel. EMBO J. 20:1462-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bárcena, M., C. San Martín, F. Weise, S. Ayora, J. C. Alonso, and J. M. Carazo. 1998. Polymorphic quaternary organization of the Bacillus subtilis bacteriophage SPP1 replicative helicase (G40P). J. Mol. Biol. 283:809-819. [DOI] [PubMed] [Google Scholar]
- 11.Bastia, D., J. Germino, J. H. Crosa, and P. Hale. 1981. Molecular cloning of the replication terminus of the plasmid R6K. Gene 14:81-89. [DOI] [PubMed] [Google Scholar]
- 12.Bastia, D., J. Germino, J. H. Crosa, and J. Ram. 1981. The nucleotide sequence surrounding the replication terminus of R6K. Proc. Natl. Acad. Sci. USA 78:2095-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bastia, D., and B. K. Mohanty. 1996. Mechanisms for completing DNA replication, p. 177-215. In M. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Bedrosian, C. L., and D. Bastia. 1991. Escherichia coli replication terminator protein impedes simian virus 40 (SV40) DNA replication fork movement and SV40 large tumor antigen helicase activity in vitro at a prokaryotic terminus sequence. Proc. Natl. Acad. Sci. USA 88:2618-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benkovic, S. J., A. M. Valentine, and F. Salinas. 2001. Replisome-mediated DNA replication. Annu. Rev. Biochem. 70:181-208. [DOI] [PubMed] [Google Scholar]
- 16.Berg, O. G., R. B. Winter, and P. H. von Hippel. 1981. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry 20:6929-6948. [DOI] [PubMed] [Google Scholar]
- 17.Berlyn, M. K. B. 1998. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol. Mol. Biol. Rev. 62:814-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bidnenko, V., S. D. Ehrlich, and B. Michel. 2002. Replication fork collapse at replication terminator sequences. EMBO J. 21:3898-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bierne, H., and B. Michel. 1994. When replication forks stop. Mol. Microbiol. 13:17-23. [DOI] [PubMed] [Google Scholar]
- 20.Bierne, H., S. D. Ehrlich, and B. Michel. 1997. Deletions at stalled replication forks occur by two different pathways. EMBO J. 16:3332-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bird, L. E., H. Pan, P. Soultanas, and D. B. Wigley. 2000. Mapping protein-protein interactions within a stable complex of DNA primase and DnaB helicase from Bacillus stearothermophilus. Biochemistry 39:171-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bird, R. E., J. Louarn, J. Martuscelli, and L. Caro. 1972. Origin and sequence of chromosome replication in Escherichia coli. J. Mol. Biol. 70:549-566. [DOI] [PubMed] [Google Scholar]
- 23.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 24.Bujalowski, W. 2003. Expanding the physiological role of the hexameric DnaB helicase. Trends Biochem. Sci. 28:116-118. [DOI] [PubMed] [Google Scholar]
- 25.Bujalowski, W., and M. J. Jezewska. 1995. Interactions of Escherichia coli primary replicative helicase DnaB protein with single-stranded DNA. The nucleic acid does not wrap around the protein hexamer. Biochemistry 34:8513-8519. [DOI] [PubMed] [Google Scholar]
- 26.Bussiere, D. E., and D. Bastia. 1999. Termination of DNA replication of bacterial and plasmid chromosomes. Mol. Microbiol. 31:1611-1618. [DOI] [PubMed] [Google Scholar]
- 27.Bussiere, D. E., D. Bastia, and S. W. White. 1995. Crystal structure of the replication terminator protein from B. subtilis at 2.6 Å. Cell 80:651-660. [DOI] [PubMed] [Google Scholar]
- 28.Caruthers, J. M., and D. B. McKay. 2002. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12:123-133. [DOI] [PubMed] [Google Scholar]
- 29.Chang, P., and K. J. Marians. 2000. Identification of a region of Escherichia coli DnaB required for functional interaction with DnaG at the replication fork. J. Biol. Chem. 275:26187-26195. [DOI] [PubMed] [Google Scholar]
- 30.Coskun-Ari, F. F., and T. M. Hill. 1997. Sequence-specific interactions in the Tus-Ter complex and the effect of base pair substitutions on arrest of DNA replication in Escherichia coli. J. Biol. Chem. 272:26448-26456. [DOI] [PubMed] [Google Scholar]
- 31.Coskun-Ari, F. F., A. Skokotas, G. R. Moe, and T. M. Hill. 1994. Biophysical characteristics of Tus, the replication arrest protein of Escherichia coli. J. Biol. Chem. 269:4027-4034. [PubMed] [Google Scholar]
- 32.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 33.Crosa, J. H., L. K. Luttropp, and S. Falkow. 1976. Mode of replication of the conjugative R-plasmid RSF1040 in Escherichia coli. J. Bacteriol. 126:454-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davey, M. J., D. Jeruzalmi, J. Kuriyan, and M. O'Donnell. 2002. Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell Biol. 3:826-835. [DOI] [PubMed] [Google Scholar]
- 35.Davey, M. J., and M. O'Donnell. 2000. Mechanisms of DNA replication. Curr. Opin. Chem. Biol. 4:581-586. [DOI] [PubMed] [Google Scholar]
- 36.Delagoutte, E., and P. H. von Hippel. 2002. Helicase mechanisms and the coupling of helicases within macromolecular machines. I. Structures and properties of isolated helicases. Q. Rev. Biophys. 35:431-478. [DOI] [PubMed] [Google Scholar]
- 37.Delagoutte, E., and P. H. von Hippel. 2003. Helicase mechanisms and the coupling of helicases within macromolecular machines. II. Integration of helicases into cellular processes. Q. Rev. Biophys. 36:1-69. [DOI] [PubMed] [Google Scholar]
- 38.De Massy, B., S. Béjar, J. Louarn, J.-M. Louarn, and J.-P. Bouché. 1987. Inhibition of replication forks exiting the terminus region of the Escherichia coli chromosome occurs at two loci separated by 5 min. Proc. Natl. Acad. Sci. USA 84:1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donate, L. E., O. Llorca, M. Bárcena, S. E. Brown, N. E. Dixon, and J.-M. Carazo. 2000. pH-Controlled quaternary states of hexameric DnaB helicase. J. Mol. Biol. 303:383-393. [DOI] [PubMed] [Google Scholar]
- 40.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, A. Givaudan, S. Taourit, S. Bocs, C. Boursaux-Eude, M. Chandler, J.-F. Charles, E. Dassa, R. Derose, S. Derzelle, G. Freyssinet, S. Gaudriault, C. Médigue, A. Lanois, K. Powell, P. Siguier, R. Vincent, V. Wingate, M. Zouine, P. Glaser, N. Boemare, A. Danchin, and F. Kunst. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21:1307-1313. [DOI] [PubMed] [Google Scholar]
- 41.Duggan, L. J., P. T. Asmann, T. M. Hill, and P. A. Gottlieb. 1996. Identification of a Tus protein segment that photo-cross-links with TerB DNA and elucidation of the role of certain thymine methyl groups in the Tus-TerB complex using halogenated uracil analogues. Biochemistry 35:15391-15396. [DOI] [PubMed] [Google Scholar]
- 42.Duggan, L. J., T. M. Hill, S. Wu, K. Garrison, X. Zhang, and P. A. Gottlieb. 1995. Using modified nucleotides to map the DNA determinants of the Tus-TerB complex, the protein-DNA interaction associated with termination of replication in Escherichia coli. J. Biol. Chem. 270:28049-28054. [DOI] [PubMed] [Google Scholar]
- 43.Duggin, I. G., P. A. Andersen, M. T. Smith, J. A. Wilce, G. F. King, and R. G. Wake. 1999. Site-directed mutants of RTP of Bacillus subtilis and the mechanism of replication fork arrest. J. Mol. Biol. 286:1325-1335. [DOI] [PubMed] [Google Scholar]
- 44.Duggin, I. G., J. M. Matthews, N. E. Dixon, R. G. Wake, and J. P. Mackay. 2005. A complex mechanism determines polarity of DNA replication fork arrest by the replication terminator complex of Bacillus subtilis. J. Biol. Chem. 280:13105-13113. [DOI] [PubMed] [Google Scholar]
- 45.Egelman, E. H., X. Yu, R. Wild, M. M. Hingorani, and S. S. Patel. 1995. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc. Natl. Acad. Sci. USA 92:3869-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fass, D., C. E. Bogden, and J. M. Berger. 1999. Crystal structure of the N-terminal domain of the DnaB hexameric helicase. Structure 7:691-698. [DOI] [PubMed] [Google Scholar]
- 47.Fouts, E. T., X. Yu, E. H. Egelman, and M. R. Botchan. 1999. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J. Biol. Chem. 274:4447-4458. [DOI] [PubMed] [Google Scholar]
- 48.François, V., J. Louarn, and J.-M. Louarn. 1989. The terminus of the Escherichia coli chromosome is flanked by several polar replication pause sites. Mol. Microbiol. 3:995-1002. [DOI] [PubMed] [Google Scholar]
- 49.Frick, D. N., and C. C. Richardson. 2001. DNA primases. Annu. Rev. Biochem. 70:39-80. [DOI] [PubMed] [Google Scholar]
- 50.Gao, D., and C. S. McHenry. 2001. τ binds and organizes Escherichia coli replication proteins through distinct domains. Partial proteolysis of terminally tagged τ to determine candidate domains and to assign domain V as the α binding domain. J. Biol. Chem. 276:4433-4440. [DOI] [PubMed] [Google Scholar]
- 51.Gao, D., and C. S. McHenry. 2001. τ binds and organizes Escherichia coli replication proteins through distinct domains. Domain IV, located within the unique C terminus of τ, binds the replication fork helicase, DnaB. J. Biol. Chem. 276:4441-4446. [DOI] [PubMed] [Google Scholar]
- 52.Gerber, J.-K., E. Gögel, C. Berger, M. Wallisch, F. Müller, I. Grummt, and F. Grummt. 1997. Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell 90:559-567. [DOI] [PubMed] [Google Scholar]
- 53.Germino, J., and D. Bastia. 1981. Termination of DNA replication in vitro at a sequence-specific replication terminus. Cell 23:681-687. [DOI] [PubMed] [Google Scholar]
- 54.Gottlieb, P. A., S. Wu, X. Zhang, M. Tecklenburg, P. Kuempel, and T. M. Hill. 1992. Equilibrium, kinetic, and footprinting studies of the Tus-Ter protein-DNA interaction. J. Biol. Chem. 267:7434-7443. [PubMed] [Google Scholar]
- 55.Guajardo, R., and R. Sousa. 1999. Characterization of the effects of Escherichia coli replication terminator protein (Tus) on transcription reveals dynamic nature of the Tus block to transcription complex progression. Nucleic Acids Res. 27:2814-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 57.Hacker, K. J., and K. A. Johnson. 1997. A hexameric helicase encircles one DNA strand and excludes the other during DNA unwinding. Biochemistry 36:14080-14087. [DOI] [PubMed] [Google Scholar]
- 58.Harry, E. J. 2001. Coordinating DNA replication with cell division: lessons from outgrowing spores. Biochimie 83:75-81. [DOI] [PubMed] [Google Scholar]
- 59.Henderson, T. A., A. F. Nilles, M. Valjavec-Gratian, and T. M. Hill. 2001. Site-directed mutagenesis and phylogenetic comparisons of the Escherichia coli Tus protein: DNA-protein interactions alone can not account for Tus activity. Mol. Genet. Genomics 265:941-953. [DOI] [PubMed] [Google Scholar]
- 60.Hiasa, H., and K. J. Marians. 1992. Differential inhibition of the DNA translocation and DNA unwinding activities of DNA helicases by the Escherichia coli Tus protein. J. Biol. Chem. 267:11379-11385. [PubMed] [Google Scholar]
- 61.Hiasa, H., and K. J. Marians. 1994. Tus prevents overreplication of oriC plasmid DNA. J. Biol. Chem. 269:26959-26968. [PubMed] [Google Scholar]
- 62.Hidaka, M., M. Akiyama, and T. Horiuchi. 1988. A consensus sequence of three DNA replication terminus sites on the E. coli chromosome is highly homologous to the terR sites of the R6K plasmid. Cell 55:467-475. [DOI] [PubMed] [Google Scholar]
- 63.Hidaka, M., T. Kobayashi, and T. Horiuchi. 1991. A newly identified DNA replication terminus site, TerE, on the Escherichia coli chromosome. J. Bacteriol. 173:391-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hidaka, M., T. Kobayashi, Y. Ishimi, M. Seki, T. Enomoto, M. Abdel-Monem, and T. Horiuchi. 1992. Termination complex in Escherichia coli inhibits SV40 DNA replication in vitro by impeding the action of T antigen helicase. J. Biol. Chem. 267:5361-5365. [PubMed] [Google Scholar]
- 65.Hidaka, M., T. Kobayashi, S. Takenaka, H. Takeya, and T. Horiuchi. 1989. Purification of a DNA replication terminus (ter) site-binding protein in Escherichia coli and identification of the structural gene. J. Biol. Chem. 264:21031-21037. [PubMed] [Google Scholar]
- 66.Hill, T. M. 1992. Arrest of bacterial DNA replication. Annu. Rev. Microbiol. 46:603-633. [DOI] [PubMed] [Google Scholar]
- 67.Hill, T. M. 1996. Features of the chromosomal terminus region, p. 1602-1614. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 68.Hill, T. M., J. M. Henson, and P. L. Kuempel. 1987. The terminus region of the Escherichia coli chromosome contains two separate loci that exhibit polar inhibition of replication. Proc. Natl. Acad. Sci. USA 84:1754-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hill, T. M., B. J. Kopp, and P. L. Kuempel. 1988. Termination of DNA replication in Escherichia coli requires a trans-acting factor. J. Bacteriol. 170:662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hill, T. M., and K. J. Marians. 1990. Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc. Natl. Acad. Sci. USA 87:2481-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hill, T. M., A. J. Pelletier, M. L. Tecklenburg, and P. L. Kuempel. 1988. Identification of the DNA sequence from the E. coli terminus region that halts replication forks. Cell 55:459-466. [DOI] [PubMed] [Google Scholar]
- 72.Hill, T. M., M. L. Tecklenburg, A. J. Pelletier, and P. L. Kuempel. 1989. tus, the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA-binding protein. Proc. Natl. Acad. Sci. USA 86:1593-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Horiuchi, T., and Y. Fujimura. 1995. Recombinational rescue of the stalled DNA replication fork: a model based on analysis of an Escherichia coli strain with a chromosome region difficult to replicate. J. Bacteriol. 177:783-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Horiuchi, T., Y. Fujimura, H. Nishitani, T. Kobayashi, and M. Hidaka. 1994. The DNA replication fork blocked at the Ter site may be an entrance for the RecBCD enzyme into duplex DNA. J. Bacteriol. 176:4656-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Horiuchi, T., and M. Hidaka. 1988. Core sequence of two separable terminus sites of the R6K plasmid that exhibit polar inhibition of replication is a 20 bp inverted repeat. Cell 54:515-523. [DOI] [PubMed] [Google Scholar]
- 76.Horiuchi, T., M. Hidaka, M. Akiyama, H. Nishitani, and M. Sekiguchi. 1987. Replication intermediate of a hybrid plasmid carrying the replication terminus (ter) site of R6K as revealed by agarose gel electrophoresis. Mol. Gen. Genet. 210:394-398. [DOI] [PubMed] [Google Scholar]
- 77.Hou, R., and T. M. Hill. 2002. Loss of RecA function affects the ability of Escherichia coli to maintain recombinant plasmids containing a Ter site. Plasmid 47:36-50. [DOI] [PubMed] [Google Scholar]
- 78.Hyrien, O. 2000. Mechanisms and consequences of replication fork arrest. Biochimie 82:5-17. [DOI] [PubMed] [Google Scholar]
- 79.Jezewska, M. J., and W. Bujalowski. 1996. A general method of analysis of ligand binding to competing macromolecules using the spectroscopic signal originating from a reference macromolecule. Application to Escherichia coli replicative helicase DnaB protein-nucleic acid interactions. Biochemistry 35:2117-2128. [DOI] [PubMed] [Google Scholar]
- 80.Jezewska, M. J., S. Rajendran, D. Bujalowska, and W. Bujalowski. 1998. Does single-stranded DNA pass through the inner channel of the protein hexamer in the complex with the Escherichia coli DnaB helicase? Fluorescence energy transfer studies. J. Biol. Chem. 273:10515-10529. [DOI] [PubMed] [Google Scholar]
- 81.Jezewska, M. J., S. Rajendran, and W. Bujalowski. 1997. Strand specificity in the interactions of Escherichia coli primary replicative helicase DnaB protein with a replication fork. Biochemistry 36:10320-10326. [DOI] [PubMed] [Google Scholar]
- 82.Jezewska, M. J., S. Rajendran, and W. Bujalowski. 1998. Functional and structural heterogeneity of the DNA binding site of the Escherichia coli primary replicative helicase DnaB protein. J. Biol. Chem. 273:9058-9069. [DOI] [PubMed] [Google Scholar]
- 83.Jezewska, M. J., S. Rajendran, and W. Bujalowski. 1998. Complex of Escherichia coli primary replicative helicase DnaB protein with a replication fork: recognition and structure. Biochemistry 37:3116-3136. [DOI] [PubMed] [Google Scholar]
- 84.Jin, Q., Z. H. Yuan, J. G. Xu, Y. Wang, Y. Shen, W. C. Lu, J. H. Wang, H. Liu, J. Yang, F. Yang, X. B. Zhang, J. Y. Zhang, G. W. Yang, H. T. Wu, D. Qu, J. Dong, L. L. Sun, Y. Xue, A. L. Zhao, Y. S. Gao, J. P. Zhu, B. Kan, K. Y. Ding, S. X. Chen, H. S. Cheng, Z. J. Yao, B. K. He, R. S. Chen, D. L. Ma, B. Q. Qiang, Y. M. Wen, Y. D. Hou, and J. Yu. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30:4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kamada, K., T. Horiuchi, K. Ohsumi, N. Shimamoto, and K. Morikawa. 1996. Structure of a replication-terminator protein complexed with DNA. Nature 383:598-603. [DOI] [PubMed] [Google Scholar]
- 86.Kamada, K., K. Ohsumi, T. Horiuchi, N. Shimamoto, and K. Morikawa. 1996. Crystallization and preliminary X-ray analysis of the Escherichia coli replication terminator protein complexed with DNA. Proteins 24:402-403. [DOI] [PubMed] [Google Scholar]
- 87.Kaplan, D. L. 2000. The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J. Mol. Biol. 301:285-299. [DOI] [PubMed] [Google Scholar]
- 88.Kaplan, D. L., and M. O'Donnell. 2002. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell 10:647-657. [DOI] [PubMed] [Google Scholar]
- 89.Kapur, A., J. L. Beck, S. E. Brown, N. E. Dixon, and M. M. Sheil. 2002. Use of electrospray ionization mass spectrometry to study binding interactions between a replication terminator protein and DNA. Protein Sci. 11:147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kaul, S., B. K. Mohanty, T. Sahoo, I. Patel, S. A. Khan, and D. Bastia. 1994. The replication terminator protein of the Gram-positive bacterium Bacillus subtilis functions as a polar contrahelicase in Gram-negative Escherichia coli. Proc. Natl. Acad. Sci. USA. 91:11143-11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Khatri, G. S., T. MacAllister, P. R. Sista, and D. Bastia. 1989. The replication terminator protein of E. coli is a DNA sequence-specific contra-helicase. Cell 59:667-674. [DOI] [PubMed] [Google Scholar]
- 92.Khoury, A. M., H. J. Lee, M. Lillis, and P. Lu. 1990. Lac repressor-operator interaction: DNA length dependence. Biochim. Biophys. Acta 1087:55-60. [DOI] [PubMed] [Google Scholar]
- 93.Kim, S., H. G. Dallmann, C. S. McHenry, and K. J. Marians. 1996. Coupling of a replicative polymerase and helicase: a τ-DnaB interaction mediates rapid replication fork movement. Cell 84:643-650. [DOI] [PubMed] [Google Scholar]
- 94.Kim, S., H. G. Dallmann, C. S. McHenry, and K. J. Marians. 1996. τ couples the leading- and lagging-strand polymerases at the Escherichia coli DNA replication fork. J. Biol. Chem. 271:21406-21412. [DOI] [PubMed] [Google Scholar]
- 95.King, K., S. J. Benkovic, and P. Modrich. 1989. Glu-111 is required for activation of the DNA cleavage center of EcoRI endonuclease. J. Biol. Chem. 264:11807-11815. [PubMed] [Google Scholar]
- 96.Kobayashi, T., M. Hidaka, and T. Horiuchi. 1989. Evidence of a ter specific binding protein essential for the termination reaction of DNA replication in Escherichia coli. EMBO J. 8:2435-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Koch, W. H., A. R. F. de Henestrosa, and R. Woodgate. 2000. Identification of mucAB-like homologs on two IncT plasmids, R394 and Rts-1. Mutat. Res. 457:1-13. [DOI] [PubMed] [Google Scholar]
- 98.Kolter, R., and D. R. Helinski. 1978. Activity of the replication terminus of plasmid R6K in hybrid replicons in Escherichia coli. J. Mol. Biol. 124:425-441. [DOI] [PubMed] [Google Scholar]
- 99.Korolev, S., J. Hsieh, G. H. Gauss, T. M. Lohman, and G. Waksman. 1997. Major domain swiveling revealed by the crystal structures of complexes of E-coli Rep helicase bound to single-stranded DNA and ADP. Cell 90:635-647. [DOI] [PubMed] [Google Scholar]
- 100.Krabbe, M., J. Zabielski, R. Bernander, and K. Nordström. 1997. Inactivation of the replication-termination system affects the replication mode and causes unstable maintenance of plasmid R1. Mol. Microbiol. 24:723-735. [DOI] [PubMed] [Google Scholar]
- 101.Kralicek, A. V., P. K. Wilson, G. B. Ralston, R. G. Wake, and G. F. King. 1997. Reorganization of terminator DNA upon binding replication terminator protein: implications for the functional replication fork arrest complex. Nucleic Acids Res. 25:590-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Krings, G., and D. Bastia. 2004. swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 101:14085-14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kuempel, P. L., S. A. Duerr, and P. D. Maglothin. 1978. Chromosome replication in an Escherichia coli dnaA mutant integratively suppressed by prophage P2. J. Bacteriol. 134:902-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kuempel, P. L., S. A. Duerr, and N. R. Seeley. 1977. Terminus region of the chromosome in Escherichia coli inhibits replication forks. Proc. Natl. Acad. Sci. USA 74:3927-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee, E. H., and A. Kornberg. 1992. Features of replication fork blockage by the Escherichia coli terminus-binding protein. J. Biol. Chem. 267:8778-8784. [PubMed] [Google Scholar]
- 106.Lee, E. H., A. Kornberg, M. Hidaka, T. Kobayashi, and T. Horiuchi. 1989. Escherichia coli replication termination protein impedes the action of helicases. Proc. Natl. Acad. Sci. USA 86:9104-9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Leu, F. P., R. Georgescu, and M. O'Donnell. 2003. Mechanism of the E. coli τ processivity switch during lagging-strand synthesis. Mol. Cell 11:315-327. [DOI] [PubMed] [Google Scholar]
- 108.Lewis, P. J. 2001. Bacterial chromosome segregation. Microbiology 147:519-526. [DOI] [PubMed] [Google Scholar]
- 109.Li, D., R. Zhao, W. Lilyestrom, D. Gai, R. Zhang, J. A. DeCaprio, E. Fanning, A. Jochimiak, G. Szakonyi, and X. S. Chen. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423:512-518. [DOI] [PubMed] [Google Scholar]
- 110.Louarn, J., J. Patte, and J.-M. Louarn. 1977. Evidence for a fixed termination site of chromosome replication in Escherichia coli K12. J. Mol. Biol. 115:295-314. [DOI] [PubMed] [Google Scholar]
- 111.Louarn, J., J. Patte, and J.-M. Louarn. 1979. Map position of the replication terminus on the Escherichia coli chromosome. Mol. Gen. Genet. 172:7-11. [DOI] [PubMed] [Google Scholar]
- 112.Lovett, M. A., R. B. Sparks, and D. R. Helinski. 1975. Bidirectional replication of plasmid R6K DNA in Escherichia coli; correspondence between origin of replication and position of single-strand break in relaxed complex. Proc. Natl. Acad. Sci. USA 72:2905-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.MacAllister, T., G. S. Khatri, and D. Bastia. 1990. Sequence-specific and polarized replication termination in vitro: complementation of extracts of tus− Escherichia coli by purified Ter protein and analysis of termination intermediates. Proc. Natl. Acad. Sci. USA 87:2828-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Manna, A. C., K. S. Pai, D. E. Bussiere, C. Davies, S. W. White, and D. Bastia. 1996. Helicase-contrahelicase interaction and the mechanism of termination of DNA replication. Cell 87:881-891. [DOI] [PubMed] [Google Scholar]
- 115.Manna, A. C., K. S. Pai, D. E. Bussiere, S. W. White, and D. Bastia. 1996. The dimer-dimer interaction surface of the replication terminator protein of Bacillus subtilis and termination of DNA replication. Proc. Natl. Acad. Sci. USA 93:3253-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Marians, K. J. 2000. Crawling and wiggling on DNA: structural insights to the mechanism of DNA unwinding by helicases. Structure 8:R227-R235. [DOI] [PubMed] [Google Scholar]
- 117.Masters, M., and P. Broda. 1971. Evidence for the bidirectional replication of the Escherichia coli chromosome. Nat. New Biol. 232:137-140. [DOI] [PubMed] [Google Scholar]
- 118.McHenry, C. S. 2003. Chromosomal replicases as asymmetric dimers: studies of subunit arrangement and functional consequences. Mol. Microbiol. 49:1157-1165. [DOI] [PubMed] [Google Scholar]
- 119.Messer, W. 2002. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 26:355-374. [DOI] [PubMed] [Google Scholar]
- 120.Michel, B. 2000. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 25:173-178. [DOI] [PubMed] [Google Scholar]
- 121.Michel, B., S. D. Ehrlich, and M. Uzest. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Michel, B., M.-J. Flores, E. Viguera, G. Grompone, M. Seigneur, and V. Bidnenko. 2001. Rescue of arrested replication forks by homologous recombination. Proc. Natl. Acad. Sci. USA 98:8181-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Miles, C. S., J. Weigelt, N. P. J. Stamford, N. Dammerova, G. Otting, and N. E. Dixon. 1997. Precise limits of the N-terminal domain of DnaB helicase determined by NMR spectroscopy. Biochem. Biophys. Res. Commun. 231:126-130. [DOI] [PubMed] [Google Scholar]
- 124.Mohanty, B. K., and D. Bastia. 2004. Binding of the replication terminator protein Fob1p to the Ter sites of yeast causes polar fork arrest. J. Biol. Chem. 279:1932-1941. [DOI] [PubMed] [Google Scholar]
- 125.Mohanty, B. K., D. E. Bussiere, T. Sahoo, K. S. Pai, W. J. J. Meijer, S. Bron, and D. Bastia. 2001. Structural and functional analysis of a bipolar replication terminus. Implications for the origin of polarity of fork arrest. J. Biol. Chem. 276:13160-13168. [DOI] [PubMed] [Google Scholar]
- 126.Mohanty, B. K., T. Sahoo, and D. Bastia. 1998. Mechanistic studies on the impact of transcription on sequence-specific termination of DNA replication and vice versa. J. Biol. Chem. 273:3051-3059. [DOI] [PubMed] [Google Scholar]
- 127.Mulugu, S., A. Potnis, Shamsuzzaman, J. Taylor, K. Alexander, and D. Bastia. 2001. Mechanism of termination of DNA replication of Escherichia coli involves helicase-contrahelicase interaction. Proc. Natl. Acad. Sci. USA 98:9569-9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Murata, T., M. Ohnishi, T. Ara, J. Kaneko, C.-G. Han, Y. F. Li, K. Takashima, H. Nojima, K. Nakayama, A. Kaji, Y. Kamio, T. Miki, H. Mori, E. Ohtsubo, Y. Terawaki, and T. Hayashi. 2002. Complete nucleotide sequence of plasmid Rts1: implications for evolution of large plasmid genomes. J. Bacteriol. 184:3194-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nakayama, N., N. Arai, Y. Kaziro, and K. Arai. 1984. Structural and functional studies of the dnaB protein using limited proteolysis. Characterization of domains for DNA-dependent ATP hydrolysis and for protein association in the primosome. J. Biol. Chem. 259:88-96. [PubMed] [Google Scholar]
- 130.Natarajan, S., W. L. Kelley, and D. Bastia. 1991. Replication terminator protein of Escherichia coli is a transcriptional repressor of its own synthesis. Proc. Natl. Acad. Sci. USA 88:3867-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Neylon, C., S. E. Brown, A. V. Kralicek, C. S. Miles, C. A. Love, and N. E. Dixon. 2000. Interaction of the Escherichia coli replication terminator protein (Tus) with DNA: a model derived from DNA-binding studies of mutant proteins by surface plasmon resonance. Biochemistry 39:11989-11999. [DOI] [PubMed] [Google Scholar]
- 132.Niedenzu, T., D. Röleke, G. Bains, E. Scherzinger, and W. Saenger. 2001. Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010. J. Mol. Biol. 306:479-487. [DOI] [PubMed] [Google Scholar]
- 133.Oakley, A. J., K. V. Loscha, P. M. Schaeffer, E. Liepinsh, G. Pintacuda, M. C. J. Wilce, G. Otting, and N. E. Dixon. 2005. Crystal and solution structures of the helicase-binding domain of Escherichia coli primase. J. Biol. Chem. 280:11495-11504. [DOI] [PubMed] [Google Scholar]
- 134.Pai, K. S., D. E. Bussiere, F. G. Wang, C. A. Hutchison III, S. W. White, and D. Bastia. 1996. The structure and function of the replication terminator protein of Bacillus subtilis: identification of the ‘winged helix’ DNA-binding domain. EMBO J. 15:3164-3173. [PMC free article] [PubMed] [Google Scholar]
- 135.Pai, K. S., D. E. Bussiere, F. G. Wang, S. W. White, and D. Bastia. 1996. Structure of the replication terminus-terminator protein complex as probed by affinity cleavage. Proc. Natl. Acad. Sci. USA 93:10647-10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. G. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 137.Patel, S. S., and K. M. Picha. 2000. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69:651-697. [DOI] [PubMed] [Google Scholar]
- 138.Pelletier, A. J., T. M. Hill, and P. L. Kuempel. 1988. Location of sites that inhibit progression of replication forks in the terminus region of Escherichia coli. J. Bacteriol. 170:4293-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pelletier, A. J., T. M. Hill, and P. L. Kuempel. 1989. Termination sites T1 and T2 from the Escherichia coli chromosome inhibit DNA replication in ColE1-derived plasmids. J. Bacteriol. 171:1739-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Peter, B. J., C. Ullsperger, H. Hiasa, K. J. Marians, and N. R. Cozzarelli. 1998. The structure of supercoiled intermediates in DNA replication. Cell 94:819-827. [DOI] [PubMed] [Google Scholar]
- 141.Ptashne, M. 1986. A genetic switch: gene control and phage λ, p. 109-117. Cell Press, Cambridge, Mass.
- 142.Record, M. T., Jr., J.-H. Ha, and M. A. Fisher. 1991. Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability and specificity and mechanism of formation of site-specific complexes between proteins and helical DNA. Methods Enzymol. 208:291-343. [DOI] [PubMed] [Google Scholar]
- 143.Roecklein, B. A., and P. L. Kuempel. 1992. In vivo characterization of tus gene expression in Escherichia coli. Mol. Microbiol. 6:1655-1661. [DOI] [PubMed] [Google Scholar]
- 144.Roecklein, B., A. Pelletier, and P. Kuempel. 1991. The tus gene of Escherichia coli: autoregulation, analysis of flanking sequences and identification of a complementary system in Salmonella typhimurium. Res. Microbiol. 142:169-175. [DOI] [PubMed] [Google Scholar]
- 145.Rothstein, R., B. Michel, and S. Gangloff. 2000. Replication fork pausing and recombination or “gimme a break.” Genes Dev. 14:1-10. [PubMed] [Google Scholar]
- 146.Rudd, K. E. 1998. Linkage map of Escherichia coli K-12, edition 10: the physical map. Microbiol. Mol. Biol. Rev. 62:985-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sahoo, T., B. K. Mohanty, M. Lobert, A. C. Manna, and D. Bastia. 1995. The contrahelicase activities of the replication terminator proteins of Escherichia coli and Bacillus subtilis are helicase-specific and impede both helicase translocation and authentic DNA unwinding. J. Biol. Chem. 270:29138-29144. [DOI] [PubMed] [Google Scholar]
- 148.Sandler, S. J., and K. J. Marians. 2000. Role of PriA in replication fork reactivation in Escherichia coli. J. Bacteriol. 182:9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.San Martín, M. C., C. Gruss, and J. M. Carazo. 1997. Six molecules of SV40 large T antigen assemble in a propeller-shaped particle around a channel. J. Mol. Biol. 268:15-20. [DOI] [PubMed] [Google Scholar]
- 150.San Martin, C., M. Radermacher, B. Wolpensinger, A. Engel, C. S. Miles, N. E. Dixon, and J.-M. Carazo. 1998. Three-dimensional reconstructions from cryoelectron microscopy images reveal an intimate complex between helicase DnaB and its loading partner DnaC. Structure 6:501-509. [DOI] [PubMed] [Google Scholar]
- 151.San Martin, M. C., N. P. J. Stamford, N. Dammerova, N. E. Dixon, and J. M. Carazo. 1995. A structural model for the Escherichia coli DnaB helicase based on electron microscopy data. J. Struct. Biol. 114:167-176. [DOI] [PubMed] [Google Scholar]
- 152.Sawaya, M. R., S. Guo, S. Tabor, C. C. Richardson, and T. Ellenberger. 1999. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell 99:167-177. [DOI] [PubMed] [Google Scholar]
- 153.Schaeffer, P. M., M. J. Headlam, and N. E. Dixon. 2005. Protein-protein interactions in the eubacterial replisome. IUBMB Life 57:5-12. [DOI] [PubMed] [Google Scholar]
- 154.Scherzinger, E., G. Ziegelin, M. Bárcena, J. M. Carazo, R. Lurz, and E. Lanka. 1997. The RepA protein of plasmid RSF1010 is a replicative DNA helicase. J. Biol. Chem. 272:30228-30236. [DOI] [PubMed] [Google Scholar]
- 155.Sharma, B., and T. M. Hill. 1995. Insertion of inverted Ter sites into the terminus region of the Escherichia coli chromosome delays completion of DNA replication and disrupts the cell cycle. Mol. Microbiol. 18:45-61. [DOI] [PubMed] [Google Scholar]
- 156.Sherburne, C. K., T. D. Lawley, M. W. Gilmour, F. R. Blattner, V. Burland, E. Grotbeck, D. J. Rose, and D. E. Taylor. 2000. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28:2177-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Singleton, M. R., M. R. Sawaya, T. Ellenberger, and D. B. Wigley. 2000. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101:589-600. [DOI] [PubMed] [Google Scholar]
- 158.Sista, P. R., C. A. Hutchison, III, and D. Bastia. 1991. DNA-protein interaction at the replication termini of plasmid R6K. Genes Dev. 5:74-82. [DOI] [PubMed] [Google Scholar]
- 159.Sista, P. R., S. Mukherjee, P. Patel, G. S. Khatri, and D. Bastia. 1989. A host-encoded DNA-binding protein promotes termination of plasmid replication at a sequence-specific replication terminus. Proc. Natl. Acad. Sci. USA 86:3026-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Skokotas, A., H. Hiasa, K. J. Marians, L. O'Donnell, and T. M. Hill. 1995. Mutations in the Escherichia coli Tus protein define a domain positioned close to the DNA in the Tus-Ter complex. J. Biol. Chem. 270:30941-30948. [DOI] [PubMed] [Google Scholar]
- 161.Skokotas, A., M. Wrobleski, and T. M. Hill. 1994. Isolation and characterization of mutants of Tus, the replication arrest protein of Escherichia coli. J. Biol. Chem. 269:20446-20455. [PubMed] [Google Scholar]
- 162.Story, R. M., and T. A. Steitz. 1992. Structure of the recA protein-ADP complex. Nature 355:374-376. [DOI] [PubMed] [Google Scholar]
- 163.Story, R. M., I. T. Weber, and T. A. Steitz. 1992. The structure of the E. coli recA protein monomer and polymer. Nature 355:318-325. [DOI] [PubMed] [Google Scholar]
- 164.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Toth, E. A., Y. Li, M. R. Sawaya, Y. Cheng, and T. Ellenberger. 2003. The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol. Cell 12:1113-1123. [DOI] [PubMed] [Google Scholar]
- 166.Velankar, S. S., P. Soultanas, M. S. Dillingham, H. S. Subramanya, and D. B. Wigley. 1999. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97:75-84. [DOI] [PubMed] [Google Scholar]
- 167.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y.-H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 168.von Hippel, P. H., and O. G. Berg. 1989. Facilitated target location in biological systems. J. Biol. Chem. 264:675-678. [PubMed] [Google Scholar]
- 169.Wake, R. G. 1997. Replication fork arrest and termination of chromosome replication in Bacillus subtilis. FEMS Microbiol. Lett. 153:247-254. [DOI] [PubMed] [Google Scholar]
- 170.Wake, R. G., and G. F. King. 1997. A tale of two terminators: crystal structures sharpen the debate on DNA replication fork arrest mechanisms. Structure 5:1-5. [DOI] [PubMed] [Google Scholar]
- 171.Weigelt, J., S. E. Brown, C. S. Miles, N. E. Dixon, and G. Otting. 1999. NMR structure of the N-terminal domain of E. coli DnaB helicase: implications for structure rearrangements in the helicase hexamer. Structure 7:681-690. [DOI] [PubMed] [Google Scholar]
- 172.Wilce, J. A., J. P. Vivian, A. F. Hastings, G. Otting, R. H. A. Folmer, I. G. Duggin, R. G. Wake, and M. C. J. Wilce. 2001. Structure of the RTP-DNA complex and the mechanism of polar replication fork arrest. Nat. Struct. Biol. 8:206-210. [DOI] [PubMed] [Google Scholar]
- 173.Wright, D. J., K. King, and P. Modrich. 1989. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J. Biol. Chem. 264:11816-11821. [PubMed] [Google Scholar]
- 174.Xu, H., N. Sträter, W. Schröder, C. Böttcher, K. Ludwig, and W. Saenger. 2003. Structure of DNA helicase RepA in complex with sulfate at 1.95 Å resolution implicates structural changes to an ‘open’ form. Acta Crystallogr. D 59:815-822. [DOI] [PubMed] [Google Scholar]
- 175.Yancey-Wrona, J. E., and S. W. Matson. 1992. Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res. 20:6713-6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yang, S., X. Yu, M. S. VanLoock, M. J. Jezewska, W. Bujalowski, and E. H. Egelman. 2002. Flexibility of the rings: structural asymmetry in the DnaB hexameric helicase. J. Mol. Biol. 321:839-849. [DOI] [PubMed] [Google Scholar]
- 177.Yu, X., M. M. Hingorani, S. S. Patel, and E. H. Egelman. 1996. DNA is bound within the central hole to one or two of the six subunits of the T7 DNA helicase. Nat. Struct. Biol. 3:740-743. [DOI] [PubMed] [Google Scholar]
- 178.Yu, X., M. J. Jezewska, W. Bujalowski, and E. H. Egelman. 1996. The hexameric E. coli DnaB helicase can exist in different quarternary states. J. Mol. Biol. 259:7-14. [DOI] [PubMed] [Google Scholar]