Abstract
To explore the value of applying the MRI-based radiomic nomogram for predicting lymph node metastasis (LNM) in rectal cancer (RC). This retrospective analysis used data from 430 patients with RC from two medical centers. The patients were categorized into the LNM negative (LNM−) and LNM positive (LNM+) according to their surgical pathology results. We developed a physician model by selecting clinical independent predictors through physician assessments. Additionally, we developed deep learning radscore (DLRS) models by extracting deep features from multiparametric MRI (mpMRI) images. A nomogram model was constructed by combining the physician model and DLRS models. Among the patients, 192 (44.65%, 192/430) experienced LNM+. Six prediction models were developed, namely the physician model, three sequence models, the DLRS, and the nomogram. The physician model achieved AUC of the receiver operating characteristic (ROC) values of 0.78, 0.79, and 0.7, whereas the sequence models, DLRS model, and nomogram model achieved AUC values ranging from 0.83 to 0.99. The predictive performance of the DLRS and nomogram models was superior to that of the physician model. DLRS and nomogram models based on mpMRI provided higher accuracy in predicting LNM status in patients with RC than the other models.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-025-96618-y.
Keywords: Rectal cancer, Deep learning, Magnetic resonance imaging, Lymph node metastasis
Subject terms: Predictive markers, Colorectal cancer, Cancer imaging, Tumour heterogeneity
Introduction
The incidence of colorectal cancer, constituting 9.6% of all cancer cases, has shown a yearly increase. Among cancers, it ranks as the third most frequent both in men and women. Furthermore, it is the second leading cause of cancer-related mortality1,2. Approximately 20% patients with colorectal cancer present with metastases at the time of diagnosis, and 50% experience metastasis from initially localized tumors. The lymphatic system and blood vessels are the primary pathways for the spread of colorectal cancer, with lymph nodes being the most common sites for metastasis3. Patients with positive lymph nodes have higher rates of local recurrence following surgery, underscoring the importance of accurately identifying lymph node metastasis (LNM) in patients before initiating treatment. This information is pivotal in shaping individualized clinical decisions and treatment plans4. Therefore, conducting research and developing related auxiliary diagnostic technologies for assessing LNM in rectal cancer (RC) are essential.
MRI plays a critical role in RC staging. It provides valuable insights into rectal anatomy and the prediction of tumor stages, making it a recommended imaging modality in clinical guidelines5,6. Typically, MRI assesses lymph node characteristics by using morphological criteria such as size, shape, boundaries, and signals. However, these criteria, although straightforward, are subjective and can lead to overly high or low cancer staging in clinical practice. In fact, cancer staging results can vary considerably across different studies and may not consistently provide high accuracy7,8. Therefore, accurately determining LNM status in patients with RC remains a challenge for staging and is crucial in formulating treatment decisions for these patients.
Radiomics demonstrates potential in clinical practice9,10 and demonstrates high efficacy in predicting colorectal cancer outcomes. In recent years, deep learning (DL) models, specifically those using deep convolutional neural networks, have been gaining considerable attention11,12. DL models offer numerous advantages, such as high accuracy, reduced processing time, robust data processing capabilities, the requirement of fewer parameters, and end-to-end data processing capabilities13.
In studies related to noninvasive assessment of LNM in RC, technologies such as MRI-based intravoxel incoherent motion, Diffusion Kurtosis Imaging and dynamic contrast-enhanced MRI have been demonstrated to be able to identify lymph nodes properties8,14,15. However, these methods exhibit some overlap in the LNM-positive and LNM-negative information. Moreover, their complex software operations can make assessment results irreproducible15,16. In recent years, researchers have developed prediction models based on CT or MRI-based radiomics methods in predicting RC LNM status17,18. However, most studies involve manual delineation of tumor boundaries for feature extraction. This process places a significant burden on physicians, is time-consuming, and is relatively subjective. Different physicians may produce different delineations, affecting feature extraction consistency. Therefore, the purpose of this study was to develop user-friendly deep learning (DL) models based on clinical factors and multiparametric MRI (mpMRI) for predicting LNM status, validate these models using internal and external data, and explore their clinical application in rectal cancer.
Methods
Participants
This retrospective study was conducted at Tongde Hospital of Zhejiang Province (Center 1) and Putuo People’s Hospital, School of Medicine, Tongji University (Center 2). All experimental protocols were approved by the ethics committees of at Tongde Hospital of Zhejiang Province (Center 1) and Putuo People’s Hospital, School of Medicine, Tongji University (Center 2), and were performed in accordance with the Declaration of Helsinki. The requirement for informed consent was waived by the ethics committees of at Tongde Hospital of Zhejiang Province (Center 1) and Putuo People’s Hospital, School of Medicine, Tongji University (Center 2) due to the retrospective study design.
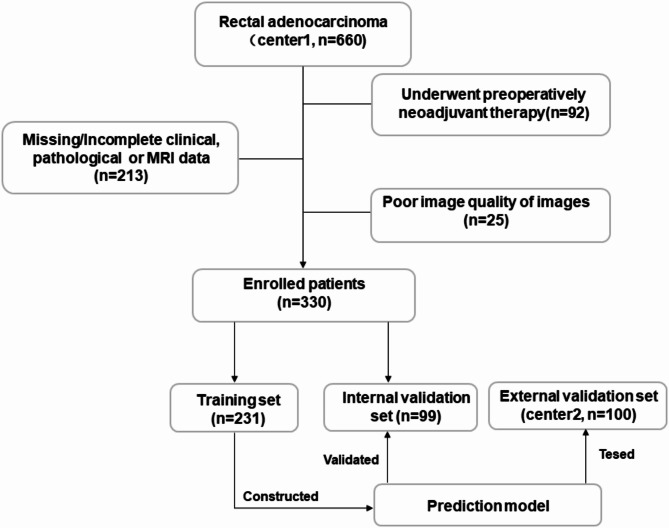
A total of 430 patients with RC from January 2016 to September 2021 were collected. For Center 1, a computer-generated randomization method was used to divide the 330 cases into a training cohort and an internal validation (In-Vad) set at a 7:3 ratio. The 100 cases from Center 2 were designated as the external validation (Ex-Vad) set. The data from the training cohort were used to construct a prediction model, which was subsequently tested using both the In-Vad and Ex-Vad data.
Inclusion criteria: (1) Patients confirmed to have RC through surgical pathology; (2) those who underwent preoperative MRI scans with contrast enhancement; (3) those with complete imaging, clinical, and pathological information available. Exclusion criteria: (1) Patients who underwent preoperative neoadjuvant therapy; (2) patients with incomplete clinical, imaging, or pathological data; (3) poor-quality images that compromised observations. The patient enrollment flowchart is presented in Fig. 1.
Fig. 1.
Enrollment of patients in this study.
MRI scanning protocol
Siemens Verio 3.0T and Siemens Avanto 1.5T MRI scanners were used for scanning, using three sequences (DWI, T2WI, and T1C) for deep learning. The sequence parameters are shown in supplementary Table S1.
Clinical analyses
All patients underwent radical surgical resection, tissue samples resected through radical surgery were pathologically assessed in accordance with the eighth edition of the American Joint Committee on Cancer RC staging system19, and every resected lymph node was studied pathologically in serial sections in order to define the lymph node metastasis status. Patients were categorized into two groups based on lymph node status: LNM-positive for those with lymph node metastasis and LNM-negative for those without. The pathological results were determined by a pathologist who 14 years of abdominal diagnosis experience, and confirmed by a pathologist who has 20 years of diagnosis experience. Discrepancies between the readers were resolved by discussion and consensus after joint re-evaluation of the pathological images.
A total of 14 indicators, including patients’ demographic information, MRI findings, and postoperative pathological information: Sex, Age, Carcinoembryonic antigen (CEA) level, Location, LD (largest tumor diameter), ID (depth of tumor invasion), apparent diffusion coefficient (ADC) value, circumferential resection margin at the primary staging MRI (mrCRM), MRI-detected extramural vascular invasion (mrEMVI), T stages of MRI T stage (mrT), N stages of MRI (mrN), T sages of pathology (pT), differentiation, Ki67 status, were recorded. An abdominal imaging diagnostician with 15 years of experience conducted the initial assessments, which were subsequently corroborated by a radiologist with 17 years of experience. Any disagreement was resolved through discussion and consensus.
In this study, the location of the rectal tumor was categorized on the basis of the distance between the lower end of the tumor and the anal verge, dividing it into upper, middle, and lower segments. Two key measurements were used: LD and ID were measured on sagittal and axial T2WI. For ADC value measurement, the largest tumor slice was identified on the PACS system. A circular region of interest (ROI) was manually delineated to cover the tumor as much as possible while avoiding areas of intestinal gas and necrosis.The mrT staging for RC was defined as follows: T1 stage for tumor invasion of the submucosa, T2 for invasion into the lamina propria, T3 stage for penetration through the lamina propria and invasion of the subserosa (serous-covering section) or mesorectum (no serous-covering section), and T4 stage for invasion into the visceral peritoneum or adjacent organs or structures. mrN positive had the following characteristics: (a) a short diameter measuring ≥ 9 mm; (b) a short diameter ranging from 5 to 8 mm, accompanied by at least two suspicious morphological features (SMFs); (c) a short diameter of less than 5 mm with at least three SMFs; or (d) the presence of SMFs in lymph node, such as irregular borders, round shapes, and heterogeneous signals. mrCRM was defined as the distance from the outermost edge of the tumor to the CRM edge, with a measurement of ≤ 1 mm considered positive. Additionally, mrEMVI was noted when tumor-related signals were observed within dilated blood vessels near the extrarectal wall penetrated by the tumor20.
Deep learning process
The DL process is explained as follows: First, for image preprocessing, data preprocessing was performed using the Pycharm Community Edition (https://pytorch.org/) and Python version 3.9 (https://www.python.org/) software. The largest MR image of the RC lesion, obtained through the T2WI, DWI, and T1C sequence models, was selected, and a ROI in the form of a rectangular area covering the lesion was selected. All images were resized to 256 × 256 pixels and converted to jpg format. To avoid data heterogeneity, a series of operations, including resizing, CenterCrop, ToTensor and normalization, were applied to the images by using the transform.Compose() function. The normalize function was utilized to standardize the images, preparing them for use by the model. Second, for feature extraction, the resnet101 deep transfer neural network learning model was employed. The fully connected layer in the final stage of the model was redefined. Rectal MRI images were input into the model, undergoing a series of operations, including convolution, pooling, and residual connections. The last fully connected layer was removed, and the final global average pooling layer produced a 2048-dimensional feature vector. Third, for model building, minimum redundancy maximum relevance (mRMR) and least absolute shrinkage and selection operator (LASSO) regression were used to methods for filtering and dimension reduction. This step aimed to reduce feature redundancy and prevent model overfitting. Finally, deep learning features were identified, and the coefficients of each depth feature were selected to calculate the deep learning radscores (DLRS).
Development of prediction models and nomogram
All 14 clinical indicators were subjected to univariate and multivariate regression analyses to identify independent predictors in the training set, which were subsequently used to construct a physician model. A backward stepwise selection stopping rule based on the red pool information criterion were used to filter out independent predictors to construct the model. Ultimately, three LNM prediction models were established: A physician model based on independent predictors, Deep learning models based the DLRS, and a combined model (physician + DLRS model) based on both clinical independent predictors and DLRS. The area under the receiver operating characteristic (ROC) curve (AUC), accuracy, sensitivity, specificity, positive predictive value, and negative predictive value of each model were used to evaluate the predictive performance of the model on the training set and internal and external validation sets. Finally, the Delong test was used to compare the prediction performance of the models. Calibration curves, decision curves, and radar charts were used to evaluate the model’s applicability in clinical settings. The experimental flowchart is shown in Fig. 2.
Fig. 2.
Flow chart of this study. First, the largest MR image of the RC lesion, obtained through the T2WI, DWI, and T1C sequence models, was selected. A rectangular region of interest (ROI) covering the lesion was chosen. Second, the ResNet-101 network was employed for feature extraction. A series of operations, including convolution, pooling, and residual connection, were performed to obtain the DL features of the fully connected layers. mRMR and Lasso regression were used to screen for highly collinear features. Third, the coefficients of each deep feature were calculated using logistic regression, completing the construction of the DL model. Subsequently, the physician model was integrated with the DLRS model to create a nomogram model. Forth, clinical application of models.
Statistical analysis
The R software (version 3.6.1, https://www.r-project.org/) and SPSS 23.0 was used for statistical analysis. Continuous variables that followed a normal distribution are presented as means ± standard deviation. Between-group comparisons for these variables were performed using t tests. Variables that did not exhibit a normal distribution are presented as medians (interquartile ranges). Between-group comparisons for these variables were conducted using nonparametric Mann–Whiney U tests. Count data are presented as frequencies, and between-group comparisons for these data were conducted using chi-squared tests. Multiple regression analyses were performed to identify independent predictors, which were used to construct a physician model. The predictive performance of the model was assessed using the region of convergence curve. Statistical significance was set at p < 0.05, and all results were two-tailed.
Results
General data
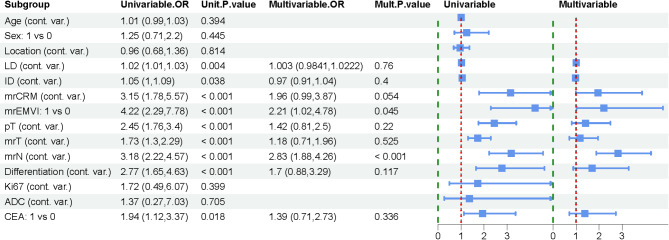
The training, In-Vad, and Ex-Vad cohorts had LNM rates of 38.5% (89/231), 53.5% (53/99), and 47.0% (47/100), respectively. The differences in CEA level, LD, mrCRM, mrEMVI, mrT, mrN, differentiation, and pT between the two patient groups in the training cohort were statistically significant (p < 0.05). Clinical and MRI characteristics of patients in training set, internal and external validation set was showed in Table 1. All variables were initially included in a univariate logistic regression analysis. The results showed that LD (OR = 1.02, p = 0.004), ID (OR = 1.05, p = 0.038), mrCRM (OR = 3.15, p < 0.001), mrEMVI (OR = 4.22, p < 0.001), mrT (OR = 1.73, p < 0.001), mrN (OR = 4.22, p < 0.001), and CEA (OR = 1.94, p = 0.018) were significantly associated with lymph node metastasis. After multivariable analysis, mrEMVI (OR = 2.21, p = 0.045) and mrN (OR = 2.83, p < 0.001) remained as independent predictors and were used to construct the physician model. (Fig. 3).
Table 1.
Clinical characteristics and Radscore of patients in training set, internal and external validation set.
| Characteristic | Training set (n = 231) | In-vad (n = 99) | Ex-vad (n = 100) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| LNM- | LNM+ | P | LNM- | LNM+ | P | LNM- | LNM+ | P | |
| (n = 142) | (n = 89) | (n = 46) | (n = 53) | (n = 53) | (n = 47) | ||||
| Sex | 0.268 | 0.058 | 0.660 | ||||||
| Male | 92 | 62 | 29 | 24 | 39 | 32 | |||
| Female | 50 | 27 | 17 | 29 | 14 | 15 | |||
| Age(y) | 64.6 ± 11.0 | 65.9 ± 11.9 | 0.396 | 61.8 ± 10.5 | 63.8 ± 9.2 | 0.303 | 63.4 ± 8.9 | 63.7 ± 12.0 | 0.846 |
| CEA | 0.017 | 0.106 | 0.042 | ||||||
| Negative | 100 | 49 | 29 | 24 | 37 | 23 | |||
| Positive | 42 | 40 | 17 | 29 | 16 | 24 | |||
| Location | 0.973 | 0.679 | 0.178 | ||||||
| Upper | 35 | 23 | 9 | 11 | 15 | 7 | |||
| Middle | 59 | 37 | 30 | 37 | 34 | 38 | |||
| Low | 48 | 29 | 7 | 5 | 4 | 2 | |||
| LD (mm) | 39.7(25.0) | 34.8(25.6) | 0.001 | 36.0(10.8) | 39.0(17.0) | 0.060 | 39.0(18.5) | 40.0(20.1) | 0.320 |
| ID (mm) | 14.4(8.6) | 14.8(6.7) | 0.066 | 12.3(6.9) | 11.7(7.8) | 0.847 | 12.6(7.1) | 11.5(4.7) | 0.668 |
| ADC | 0.78(0.2) | 0.8(0.2) | 0.477 | 0.82(0.1) | 0.86(0.1) | 0.064 | 0.85(0.2) | 0.83(0.2) | 0.738 |
| mrCRM | < 0.001 | 0.017 | 0.394 | ||||||
| Negative | 110 | 46 | 37 | 30 | 38 | 29 | |||
| Positive | 32 | 43 | 9 | 23 | 15 | 18 | |||
| mrEMVI | < 0.001 | < 0.001 | 0.014 | ||||||
| Negative | 119 | 49 | 35 | 20 | 39 | 23 | |||
| Positive | 23 | 40 | 11 | 33 | 14 | 24 | |||
| mrT | 0.001 | 0.036 | 0.351 | ||||||
| T1 | 25 | 6 | 2 | 0 | 2 | 0 | |||
| T2 | 57 | 23 | 20 | 16 | 12 | 8 | |||
| T3 | 32 | 27 | 19 | 20 | 18 | 23 | |||
| T4 | 28 | 33 | 5 | 17 | 21 | 16 | |||
| mrN | < 0.001 | < 0.001 | 0.021 | ||||||
| N0 | 101 | 24 | 21 | 4 | 19 | 8 | |||
| N1 | 24 | 27 | 19 | 18 | 22 | 17 | |||
| N2 | 17 | 38 | 6 | 31 | 12 | 22 | |||
| pT | < 0.001 | 0.613 | 0.517 | ||||||
| T1 | 23 | 4 | 1 | 0 | 1 | 1 | |||
| T2 | 48 | 14 | 16 | 16 | 7 | 5 | |||
| T3 | 50 | 28 | 20 | 23 | 25 | 29 | |||
| T4 | 21 | 43 | 9 | 14 | 20 | 12 | |||
| Differentiation | < 0.001 | 0.029 | 0.026 | ||||||
| Well | 31 | 7 | 0 | 1 | 4 | 1 | |||
| Moderate | 97 | 58 | 38 | 31 | 43 | 31 | |||
| Low | 14 | 24 | 8 | 21 | 6 | 15 | |||
| Ki67 | 0.6 (0.3) | 0.7(0.25) | 0.470 | 0.8(0.2) | 0.8(0.2) | 0.403 | 0.8(0.2) | 0.8(0.1) | 0.849 |
| DLRSa | -1.49(1.15) | -0.00(1.54) | < 0.001 | -0.72(1.20) | 0.28(0.66) | < 0.001 | -2.23(0.81) | -1.05(1.87) | < 0.001 |
ADC, apparent diffusion coefficient (×10− 3mm2/s); CEA, carcinoembryonic antigen; DLRS, deep learning radscore; Ex-Vad, external validation; ID, depth of tumor invasion; In-Vad, internal validation; LD, largest tumor diameter lymph; LNM, node metastasis; mrEMVI, MRI-reported extramural vascular invasion; mrCRM, MRI-reported circumferential resection margin; mrT, T stages of MRI; mrN, N stages of MRI; pT, T sages of pathology.
aContinuous variables are presented as the median and interquartile range (IQR).
Fig. 3.
Univariate and multivariate LNM analyses conducted by physicians. mrEMVI (OR = 2.21, p = 0.045) and mrN (OR = 2.83, p < 0.001) were identified as independent predictors that can be used to develop the physician model.
Construction of DL and nomogram models
Using the ResNet-101 network, 2048 features were extracted from each image. For each patient, 6144 deep features were extracted from MRI images (2048 each from DWI, T2WI, and T1C, respectively). Subsequently, the most significant 30 DL features from each sequence were selected using the mRMR and LASSO methods. The DWI_RS, T2WI_RS, and T1C_RS were calculated using a linear combination based on the corresponding weighted coefficients. The DLRS was constructed from DWI_RS, T2WI_RS, and T1C_RS by logistic regression analysis. The DLRS values between the LNM-positive and LNM-negative groups were significantly different, with the LNM-positive group having a significantly higher radscore than the LNM-negative group (p all < 0.001 both in training, In-Vad, and Ex-Vad cohorts) (Table 1).
The distribution of the DLRS model in the training, In-Vad, and Ex-Vad cohorts is presented in Supplementary Fig. S1. Based on these findings, the DWI_RS, T2WI_RS, T1C_RS, and DLRS models were developed. Subsequently, the physician model was integrated with the DLRS model to create a nomogram. The nomogram includes variables including mrEMVI, mrN, and DLRS. Each variable is projected onto the points axis to assign a score. The total score is summed and converted via a linear predictor scale to obtain the lymph node metastasis probability (Fig. 4). The formulas for DWI_RS, T2WI_RS, T1C_RS, and DLRS are listed in the supplementary Table S2.
Fig. 4.
The nomogram was constructed based on mrEMVI, mrN, and DLRS. Each variable corresponds to a specific point value, which is determined by projecting it onto the points axis. The total score is obtained by summing the individual point values of all variables. The linear predictor scale, ranging from − 14 to 10, is then used to convert the total score into a linear predictor value. Finally, by mapping this value onto the LNM probability axis, we obtain the corresponding probability of lymph node metastasis.
Models evaluation
The AUC values for the training, In-Vad, and Ex-Vad cohorts for the physician model were 0.75, 0.77, and 0.67, respectively. By contrast, the AUC values for the DWI_RS, T2WI_RS, and T1C_RS models ranged from 0.8 to 0.89; the AUC values for the DLRS model were 0.96, 0.97, and 0.93, respectively; and those for the nomogram model were 0.97, 0.99, and 0.94, respectively (Fig. 5A-C). The DLRS and nomogram model demonstrated highly favorable prediction results. The accuracy, AUC, sensitivity, specificity, NPV, and PPV of all the models are presented in Table 2. DeLong’s tests conducted on the learning models revealed that the DLRS and nomogram models outperformed the physician model (p < 0.001) in the training, In-Vad, and Ex-Vad cohorts. However, the DLRS and nomogram models themselves did not exhibit significant differences (p > 0.05) (Table 3). The decision curve analysis (DCA) revealed that, compared with the physician model, both the nomogram and DLRS models yielded greater net benefits (Fig. 6). The clinical impact curves confirmed the favorable Clinical applicability of the nomogram model (Supplementary Fig. S2). Furthermore, the radar chart indicated that among the six models, the nomogram and DLRS models exhibited the most comprehensive prediction performance, followed by the three sequence models and finally the physician model (Supplementary Fig. S3).
Fig. 5.
The receiver operating characteristic (ROC) curve for different prediction models in training (a), internal (b) and external validation set (c).
Table 2.
Comparison of predictive performance among six models.
| Cohort | Model | Accuracy | AUC | Sensitivity | Specificity | NPV | PPV |
|---|---|---|---|---|---|---|---|
| Training | Physician | 0.75 | 0.78 | 0.74 | 0.78 | 0.84 | 0.65 |
| DWI_RS | 0.86 | 0.89 | 0.86 | 0.85 | 0.9 | 0.79 | |
| T2WI_RS | 0.83 | 0.85 | 0.82 | 0.83 | 0.89 | 0.75 | |
| T1C_RS | 0.81 | 0.83 | 0.8 | 0.81 | 0.87 | 0.72 | |
| DLRS | 0.96 | 0.97 | 0.96 | 0.96 | 0.97 | 0.94 | |
| Nomogram | 0.97 | 0.98 | 0.97 | 0.97 | 0.98 | 0.96 | |
| In-vad | Physician | 0.77 | 0.79 | 0.78 | 0.75 | 0.73 | 0.8 |
| DWI_RS | 0.88 | 0.91 | 0.91 | 0.85 | 0.84 | 0.92 | |
| T2WI_RS | 0.84 | 0.85 | 0.87 | 0.81 | 0.8 | 0.88 | |
| T1C_RS | 0.86 | 0.89 | 0.89 | 0.83 | 0.82 | 0.9 | |
| DLRS | 0.97 | 0.99 | 0.98 | 0.96 | 0.96 | 0.98 | |
| Nomogram | 0.99 | 0.99 | 1 | 0.98 | 0.98 | 1 | |
| Ex-vad | Physician | 0.67 | 0.7 | 0.66 | 0.68 | 0.7 | 0.64 |
| DWI_RS | 0.81 | 0.83 | 0.85 | 0.77 | 0.8 | 0.82 | |
| T2WI_RS | 0.84 | 0.86 | 0.87 | 0.81 | 0.84 | 0.84 | |
| T1C_RS | 0.83 | 0.84 | 0.87 | 0.79 | 0.82 | 0.84 | |
| DLRS | 0.93 | 0.95 | 0.92 | 0.94 | 0.94 | 0.92 | |
| Nomogram | 0.94 | 0.95 | 0.94 | 0.94 | 0.94 | 0.94 |
AUC, area under the receiver operating characteristic curve; NPV, negative predictive value; PPV, positive predictive value.
Table 3.
Delong test in different models.
| Model comparison | Z1 | p1 | Z2 | p2 | Z2 | p3 |
|---|---|---|---|---|---|---|
| Physician vs. DLRS | − 6.495 | < 0.001 | − 4.485 | < 0.001 | − 4.688 | < 0.001 |
| Physician vs. nomogram | − 6.821 | < 0.001 | − 4.589 | < 0.001 | − 4.742 | < 0.001 |
| DLRS vs. nomogram | − 1.105 | 0.269 | − 0.351 | 0.725 | − 4.742 | < 0.001 |
P1, Training; p2, Invad; p3, Exvad.
Fig. 6.

A DCA indicating that the net benefits of the nomogram and DLRS models are higher than those of the physician model.
Discussion
In this study, we investigated the effectiveness of a physician and five DL models based on clinical factors and mpMRI in predicting LNM status in patients with RC. Compared with the other four models, both the DLRS and nomogram models demonstrated superior accuracy in predicting LNM status, verifying their stability and reliability. The models were validated using both internal and external data, providing a noninvasive and user-friendly method with significant clinical importance for individualized patient management and decision-making.
Our results showed that, mrEMVI and mrN were used as the independent predictors of RC LNMs. Research has consistently reported significant correlations between RC LNMs and then presence of mrEMVI/EMVI.21,22 mrEMVI is closely related to RC prognoses and has demonstrated superior predictive power for determining treatment and follow-up strategies23. Li et al.24 developed a nomogram model by combining T2WI radiomics and clinical information to predict RC LNM status, and their findings highlighted the significance of mrEMVI as one of the independent predictors. The predicted AUCs of the physician model for the training group and internal and external validation groups (0.67, 0.64, and 0.76, respectively) were significantly lower than those of the nomogram model (0.84, 0.78, and 0.72, respectively), indicating a poorer prediction performance. Another study25 collected MRI data from 91 patients with RC to construct a model for predicting LNM status. The results revealed that, compared with the radiomics and merged models, the sensitivity, specificity, and accuracy (72.09%, 73.81% and 78.12%, respectively) of the subjective radiologist analysis model was even lower, supporting the results of the present study. In the present study, the AUC values of the physician model in the training, In-Vad, and Ex-Vad cohorts were 0.78, 0.79, and 0.7, respectively, with low model accuracy and sensitivity, Specificity, NPV and PPV. In clinical practice, diagnostic and assessment accuracy is dependent on physicians’ experience and expertise and therefore varies considerably among different physicians. Moreover, some LNM images may exhibit atypical features, which can result in false negative or false positive results8. Routine imaging assessments of RC lymph node statuses often fail to meet the precision medicine requirements.
This study employed ResNet101, a 101-layer deep residual network, due to its high classification accuracy. This architecture effectively addresses the problem of network degradation, thereby optimizing model performance13. In this study, the Resnet101 transfer learning neural network was applied to perform unsupervised MRI image learning. Although the DWI_RS, T2WI_RS, and T1C_RS models learned from a single image in each sequence, their AUC and accuracy ranged from 0.81 to 0.88, higher than that of the physician model. However, compared with the accuracy, sensitivity and specificity of individual sequence models, the DLRS and nomogram models achieved even higher values, and these differences were statistically significant. Images obtained from different sequence models capture different characteristics, reflecting the diversity and heterogeneity of tumors. When these different sequence models are combined, they complement each other morphologically. The aggregation of deep features obtained from various sequence models offers a more accurate representation of intratumor heterogeneity.
Shin et al.26 used T2WI and ADC sequences to assess the pathological complete response of patients with locally advanced RC following neoadjuvant chemoradiotherapy. Their results indicated that the AUC of the radiologist analysis, T2WI, ADC and merged models (T2WI + ADC) was 0.74, 0.82, 0.79, and 0.82, respectively. Notably, the T2WI and merged models outperformed both the radiologist and ADC models in terms of classification performance. Li et al.27 collected data from 301 patients with RC and used T2WI and DWI sequence models to construct multiple radiomics models for predicting metachronous liver metastases. In their test set, the AUC of the fusion (T2WI + DWI + clinical factors), radiomics (T2WI + DWI), T2WI, and DWI models was 0.916, 0.986, 0.861, and 0.771, respectively, with the fusion model achieving the most accurate prediction results. The results of DeLong’s test conducted in this study indicated that the prediction performance of both the DLRS and nomogram models was superior to that of the physician model. However, no statistically significant differences were noted between the DLRS and nomogram models, which is in contrast with the findings of Li20. Interestingly, in this study, the prediction performance of the DLRS model was remarkable even without physician assessment information, corroborating that deep features can fully reflect tumor heterogeneity.
Currently, various methods are used for constructing radiomics models, the most common method involves manually delineating the ROI layer by layer and subsequently extracting features28–31. In a study30, ITK-SNAP software was used to manually outline ROIs in multiple MRI sequences layer by layer for feature extraction, leading to the creation of a radiomics model that predicted peripheral fat invasion in patients with RC. The results revealed that, in the training cohort, the AUC values for the clinical, radscore, and combined models were 0.85, 0.82, and 0.89, respectively. Jiang et al.31 used preoperative T2WI images as input data and drew ROIs by using ITK-SNAP software and subsequently extracted features to develop a vision transformer–based DL model for predicting RC prognoses. The best model achieved a C-index hazard ratio of 0.82 for overall survival in the validation set, 3.0 in the high-risk group in the internal test set, and 2.3 in the external test set. However, most of these studies involved manually segmenting or drawing lesions, which has the disadvantages of a heavy workload for physicians, being time-consuming, variation in drawing results among different physicians, and significant differences in the radiomics features selected and prediction results obtained. Furthermore, the manual feature extraction approach may overlook crucial data and fail to adequately capture tumor heterogeneity. Therefore, conventional machine learning methods face significant challenges.
In terms of DL methods, Deng et al.13 utilized multiple convolutional neural network (CNN) models to predict Ki-67 expression in prostate cancer. Their results demonstrated that ResNet-101, Inception-v3, and DenseNet-121 all achieved satisfactory predictive performance. Williams et al.32 employed a ResNet-50 architecture to analyze endoscopic images for assessing non-complete response to total neoadjuvant therapy (TNT) and local regrowth during surveillance. While the CNN model achieved high accuracy, matching the performance of colorectal surgeons in identifying non-complete responses to TNT, its accuracy in detecting local regrowth remained suboptimal. Similarly, Lo et al.33 applied a Vision Transformer to develop a microsatellite instability prediction model for colorectal cancer using colonoscopy images. Their model achieved an accuracy of 84% and an AUC of 0.86, outperforming DenseNet-201.These studies highlight the indispensable role of DL architectures in medical image analysis and recognition. The DL model employed in our study eliminates the need for manual tumor delineation, thereby minimizing operator-dependent variability. Through deep learning, intrinsic data features are directly extracted from imaging datasets and subsequently categorized or clustered based on shared characteristics. This approach facilitates data-driven decision-making, allowing clinicians to derive meaningful insights from the inherent properties of the imaging data.
In the Ex-Vad cohort, despite differences in the MRI machines used and scanning parameters for each sequence, the AUC and accuracy of the five DL models still ranged from 0.83 to 0.95 and from 0.81 to 0.94, respectively, indicating favorable model prediction results and applicability. The clinical application of the DL models was verified in this study by employing various methods. The nomogram model converted complex regression equations into simple visual graphical representations, facilitating the interpretation of prediction results and patient assessments. In clinical practice, this model can be used in conjunction with other clinical indicators to aid in the diagnosis or prediction of disease risks or prognoses. The DCA revealed that the DLRS and nomogram models surpassed the physician model in terms of net benefits. The radar chart provided a multidimensional comparison of performance among the various models. According to the chart, compared with the other models, the DLRS and nomogram models demonstrate superior clinical practicability.
This study has several limitations that should be acknowledged. First, it relies on retrospective data analysis, which may introduce selection bias. Future research should prioritize prospective data collection and external validation to enhance the model’s robustness and reliability. Second, the study focuses solely on primary RC lesions, utilizing ROIs from the largest tumor MRI slice to predict LNM status, without incorporating deep learning analyses of lymph nodes. Future studies should explore whether integrating analyses of both primary tumors and lymph nodes could improve the generalizability of this approach. Third, due to an insufficient sample size, this study did not perform a multiclass analysis of patients with different LNM stages (N0, N1, N2). Expanding the study to include a multi-class classification framework could provide more comprehensive insights into LNM prediction.
In conclusion, several MRI-based DL models were developed to predict LNM in patients with RC, and their performance was evaluated. Among these, the DLRS and nomogram models demonstrated high accuracy in predicting LNM status in patients with RC, offering a reliable and user-friendly prediction tool that can assist physicians in clinical decision-making.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- AUC
Area under the curve
- CNN
convolutional neural network
- DCA
Decision curve analysis
- DL
Deep learning
- DLRS
Deep learning radscore
- Ex-Vad
External validation
- LNMs
Lymph node metastases
- RC
Rectal cancer
- ID
Depth of tumor invasion
- In-Vad
Internal validation
- LD
Largest tumor diameter
- mrEMVI
MRI-detected extramural vascular invasion
- mpMRI
Multiparametric MR
- mrN
N stages of MRI
- mrT
T stages of MRI
- mrCRM
Circumferential resection margin at the primary staging MRI
- NPV
Negative predictive value
- PPV
Positive predictive value
- T1C
Contrast enhanced T1WI
Author contributions
S.D. and W.A. designed the study. W.A. took charge of the writing of this paper. S.D. were responsible for the software and statistics. G.M., J.W. and N.W. to data collection. J.H., S.W. and X.H. for data analysis. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Medical Science and Technology Project of Zhejiang Province (2022KY122).
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.74, 229–263 (2024). [DOI] [PubMed] [Google Scholar]
- 2.Ao, W. et al. Value of apparent diffusion coefficient for assessing preoperative T staging of low rectal cancer and whether this is correlated with Ki-67 expression. Can. Assoc. Radiol. J.71, 5–11 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Ciardiello, F. et al. Clinical management of metastatic colorectal cancer in the era of precision medicine. CA Cancer J. Clin.72, 372–401 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Liu, Y. et al. A magnetic resonance imaging (MRI)-based nomogram for predicting lymph node metastasis in rectal cancer: a node-for-node comparative study of MRI and histopathology. Quant. Imaging Med. Surg.11, 2586–2597 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beets-Tan, R. G. H. et al. Magnetic resonance imaging for clinical management of rectal cancer: updated recommendations from the 2016 European society of gastrointestinal and abdominal radiology (ESGAR) consensus meeting. Eur. Radiol.28, 1465–1475 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gollub, M. J. et al. Use of magnetic resonance imaging in rectal cancer patients: society of abdominal radiology (SAR) rectal cancer disease-focused panel (DFP) recommendations 2017. Abdom. Radiol. (N Y)43, 2893–2902 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Peng, W., Qiao, H., Mo, L. & Guo, Y. Progress in the diagnosis of lymph node metastasis in rectal cancer: A review. Front. Oncol.13, 1167289 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borgheresi, A. et al. Lymph nodes evaluation in rectal cancer: Where do we stand and future perspective. J. Clin. Med.11, 2599 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin, H., Wang, Y., Li, X., Yang, Y. & Qi, R. Radiomics nomogram for predicting chemo-immunotherapy efficiency in advanced non-small cell lung cancer. Sci. Rep.14(1), 20788 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xing, X. et al. A combinatorial MRI sequence-based radiomics model for preoperative prediction of microsatellite instability status in rectal cancer. Sci. Rep.14(1), 11760 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, P. T. et al. Pancreatic cancer detection on CT scans with deep learning: a nationwide population-based study. Radiology306, 172–182 (2023). [DOI] [PubMed] [Google Scholar]
- 12.Shah, A. A., Malik, H. A. M., Muhammad, A. & Alourani, A. Butt. Z.A. Deep learning ensemble 2D CNN approach towards the detection of lung cancer. Sci. Rep.13, 2987 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng, S. et al. Deep learning-based radiomic nomograms for predicting Ki67 expression in prostate cancer. BMC Cancer23, 638 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grøvik, E. et al. Dynamic multi-echo DCE- and DSC-MRI in rectal cancer: Low primary tumor K and ∆R2* peak are significantly associated with lymph node metastasis. J. Magn. Reson. Imaging46, 194–206 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Yang, X. et al. Role of quantitative dynamic contrast-enhanced MRI in evaluating regional lymph nodes with a short-axis diameter of less than 5 mm in rectal cancer. AJR Am. J. Roentgenol.212, 77–83 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Ao, W. et al. Preoperative prediction of extramural venous invasion in rectal cancer by dynamic contrast-enhanced and diffusion-weighted MRI: A preliminary study. BMC Med. Imaging22, 78 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang, Y. S. et al. High-resolution MRI-based radiomics analysis to predict lymph node metastasis and tumor deposits respectively in rectal cancer. Abdom. Radiol. (N Y). 46, 873–884 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Li, J. et al. Development and validation of a radiomics model based on 3-dimensional endoanal rectal ultrasound of rectal cancer for predicting lymph node metastasis. Turk. J. Gastroenterol.34, 542–551 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amin, M. B., Greene, F. L. & Edge, S. AJCC cancer Staging Manual 8th edn (Springer, 2017).
- 20.Horvat, N., Rocha, C. T., Clemente Oliveira, C., Petkovska, B., Gollub, M. J. & I. & MRI of rectal cancer: tumor staging, imaging techniques, and management. Radiographics39, 367–387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abe, T. et al. Combination of extramural venous invasion and lateral lymph node size detected with magnetic resonance imaging is a reliable biomarker for lateral lymph node metastasis in patients with rectal cancer. World J. Surg. Oncol.20, 5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sumii, A. et al. Establishment and validation of a nomogram for predicting potential lateral pelvic lymph node metastasis in low rectal cancer. Int. J. Clin. Oncol.27, 1173–1179 (2022). [DOI] [PubMed] [Google Scholar]
- 23.Lord, A. C. et al. MRI-diagnosed tumor deposits and EMVI status have superior prognostic accuracy to current clinical TNM staging in rectal cancer. Ann. Surg.276, 334–344 (2022). [DOI] [PubMed] [Google Scholar]
- 24.Li, H. et al. MRI-based multiregional radiomics for predicting lymph nodes status and prognosis in patients with resectable rectal cancer. Front. Oncol.12, 1087882 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, J. et al. An MRI-based multi-objective radiomics model predicts lymph node status in patients with rectal cancer. Abdom. Radiol. (N Y)46, 1816–1824 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Shin, J. et al. MRI radiomics model predicts pathologic complete response of rectal cancer following chemoradiotherapy. Radiology303, 351–358 (2022). [DOI] [PubMed] [Google Scholar]
- 27.Li, Z. F. et al. Radiomics based on preoperative rectal cancer MRI to predict the metachronous liver metastasis. Abdom. Radiol. (N Y)48, 833–843 (2023). [DOI] [PubMed] [Google Scholar]
- 28.Ao, W. et al. A novel CT-based radiomic nomogram for predicting the recurrence and metastasis of gastric stromal tumors. Am. J. Cancer Res.11, 3123–3134 (2021). [PMC free article] [PubMed] [Google Scholar]
- 29.Hou, M., Zhou, L. & Sun, J. Deep-learning-based 3D super-resolution MRI radiomics model: Superior predictive performance in preoperative T-staging of rectal cancer. Eur. Radiol.33, 1–10 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, H. et al. Novel multiparametric MRI-based radiomics in preoperative prediction of perirectal fat invasion in rectal cancer. Abdom. Radiol. (N Y). 48, 471–485 (2023). [DOI] [PubMed] [Google Scholar]
- 31.Jiang, X. et al. An MRI deep learning model predicts outcome in rectal cancer. Radiology307, e222223 (2023). [DOI] [PubMed] [Google Scholar]
- 32.Williams, H. et al. Assessing endoscopic response in locally advanced rectal cancer treated with total neoadjuvant therapy: Development and validation of a highly accurate convolutional neural network. Ann. Surg. Oncol.31, 6443–6451 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo, C. M., Jiang, J. K. & Lin, C. C. Detecting microsatellite instability in colorectal cancer using transformer-based colonoscopy image classification and retrieval. PLoS One19, e0292277 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.