Abstract
Background
The spadetail (spt) gene of zebrafish is expressed in presomitic mesoderm and in neural cells previously suggested to be Rohon-Beard neurons. The mechanism(s) generating the apparently irregular rostrocaudal distribution of spt-expressing cells in the developing CNS is unknown.
Results
spt-expressing neural cells co-express huC, a marker of neurons. These cells also co-express the genes islet-1, -2 and -3 but not valentino. The islet-1 gene expression, irregular distribution and dorsolateral position of spt-expressing cells in the developing CNS are characteristic of dorsal longitudinal ascending (DoLA) interneurons. Shortly after their birth, these neurons extend processes rostrally into which spt mRNA is transported. At 24 hours post fertilisation(hpf), spt-expressing neurons occur most frequently at rostral levels caudal of the 5th-formed somite pair. There is no apparent bias in the number of spt-expressing cells on the left or right sides of embryos. Extended staining for spt-transcription reveals expression in the dorsocaudal cells of somites at the same dorsoventral level as the spt-expressing neurons. There is frequent juxtaposition of spt-expression in newly formed somites and in neurons. This suggests that both types of spt-expressing cell respond to a common positional cue or that neurons expressing spt are patterned irregularly by flanking somitic mesoderm.
Conclusions
spt-expressing cells in the developing CNS appear to be DoLA interneurons. The irregular distribution of these cells along the rostrocaudal axis of the spinal cord may be due to "inefficient" patterning of neural spt expression by a signal(s) from flanking, regularly distributed somites also expressing spt.
Background
The spinal cord of vertebrates shows no apparent morphological metamerism. However, the pattern of motor and sensory axonal projection from the spinal cord shows a metameric distribution that is patterned by the flanking somites [1,2].
In developing zebrafish, both metameric and non-metameric patterns of neuron distribution can be observed. When primary motoneurons first arise in the developing ventral spinal cord, three such cells are present per hemisegment [3,4]. Mutation of the gene spadetail (spt) causes changes in somite formation that affect this pattern of motoneuron formation. This shows that motoneuron patterning is controlled by signals from the somites [5-7]. In contrast, the Rohon-Beard sensory neurons in the dorsal central nervous system (CNS) show no segmental distribution and are not affected by mutations affecting somite formation [5]. However, mutations such as bmp2b/swirl, bmp7/snailhouse affecting signalling by members of the bone morphogenic protein (BMP) family, [8]) and changes in Notch signalling [9][10][11] can affect the number/differentiation of these cells.
Rohon-Beard neurons, when they arise, are sufficiently numerous to be found adjacent to every somite (i.e. in each "hemisegment"). However, a third type of neural cell distribution exists with less than one cell per hemisegment. For example, dorsal longitudinal ascending (DoLA) interneurons are found at a frequency of 0.06 per hemisegment for the 5th- to 8th-formed flanking somite pairs in embryos at 18 hpf [12]. The mechanisms that control these irregular distributions are unknown.
The spt mutation was originally described by Kimmel et al. in 1989 [13] as a γ ray-induced mutation affecting trunk development including somite formation. Closer analysis of the effect of this mutation on development has shown that spt controls convergence movements and the differentiation fate of mesodermal precursors of the trunk [13-17].
The locus for spt mutations was identified by Griffin et al. in 1998 [18]. They showed that the spt gene encodes a T-box protein similar to those encoded by the Xenopus gene Xombi (also known as Antipodean, BraT or VegT) and the chick gene Tbx6L. spt is transcribed in caudal paraxial mesoderm before its differentiation to somitic mesoderm. spt is also expressed in irregularly distributed neural cells that have been suggested, on the basis of their position and distribution, to be Rohon-Beard neurons [19].
In the work described in this paper, we show that the neural cells expressing spt have the characteristics of DoLA interneurons. We then examine the distribution of spt-expressing neurons on the rostrocaudal axis and on the left and right sides of embryos. Intriguingly, we have discovered low-level expression of spt in the dorsocaudal extremities of newly formed somites that corresponds in dorsoventral level and, frequently, rostrocaudal position, to newly formed neurons expressing spt. This distribution of spt expression suggests the possible existence of an "inefficient" mechanism producing an irregular pattern of neuron distribution based on a regularly patterned flanking structure (somitic mesoderm).
Results
Neural spt-expressing cells have the characteristics of DoLA neurons
Cells expressing spt in the developing central nervous system have previously been suggested to be Rohon-Beard neurons [18,19]. To confirm their neuronal nature, we double-stained embryos for spt expression and the neuronal marker gene huC[20]. We observed coexpression of spt with huC confirming that these cells are neurons (Figure 1D).
Figure 1.

Whole mount in situ transcript hybridisation analysis of the expression of spt and other genes in the tail and trunk of zebrafish embryos at approximately 22 hpf. In all images, dorsal is up and rostral is to the left. An apparently irregular rostrocaudal distribution of spt-expressing cells is seen in the developing CNS rostral to the domain of expression in the presomitic mesoderm of the extending tail (A). Boxed areas in A indicate parts of the image magnified in B and C. Shortly after their birth, these cells extend a process rostrally (indicated by a black asterisk in B) into which spt transcript is transported. spt is expressed in newly formed somites in a restricted region, the "somitic trail" (bracketed in C), at the same dorsoventral level as spt-expressing cells in the developing CNS (black arrowheads in any panel). The spt-expressing cells in the developing CNS (red stain) co-express huC, a marker of neurons (blue stain in D). A probe that identifies cells transcribing val (blue stain) shows that the spt-expressing neurons (red stain) are not identical with these (E). Transcription of the isl-1 gene (see F) is seen dorsally in Rohon-Beard neurons (black arrows in any panel), and ventrally in motoneurons (white arrow). Intermediate between these two levels are DoLA neurons that also express isl-1 (black arrowhead). Double staining with isl-1 (blue) and spt (red) shows that these intermediate-level neurons express spt (G). Costaining of spt (red) with isl-2 (blue in H) and isl-3 (blue in I) shows that the DoLA neurons also apparently express these genes, although the onset of expression occurs more rostrally than for isl-1. Scale bars equal 100 μm in A, B, C and F and 20 μm in D, E, G, H, I.
To test the idea that spt-expressing neurons are Rohon-Beard neurons we double-stained embryos for expression of spt and the islet (isl)-1, -2 or -3 genes [6,7,21] or valentino (val, [22]) that have been stated to be expressed in these cells. Interestingly, the spt-expressing neurons also express all three known isl genes but not val (Figures 1E,1G,1H,1I). In embryos at 22 hours post fertilisation (hpf, at 28.5°C), spt-expressing cells express isl-1 from the moment of their first detection at the caudal end of the developing CNS. isl-2 and isl-3 coexpression with spt is more easily visible at more rostral levels. In every case, the cells co-expressing the spt and isl genes are located just ventral to dorsally located cells expressing isl genes alone, i.e. Rohon-Beard neurons. The isl-1 expression, rostrocaudal distribution and dorsolateral position of these cells are characteristic of DoLA interneurons [6,21]. We cannot state with certainty that all DoLA neurons express spt, only that all DoLA neurons expressing isl-1 also appear to express this gene. Contrary to an earlier report [7], we observed expression of isl-2 and isl-3 in these interneurons. This might be explained by difficulty in distinguishing DoLA neurons from Rohon-Beard neurons at the rostral levels where isl-2 and -3 expression is more easily observed.
spt mRNA is transported into neurite-like structures
Soon after their differentiation in the central nervous system, a rostrally-projecting process of the spt-expressing neurons can be observed to contain spt mRNA (Figure 1B). This process may, in fact, become the future ascending axon of the DoLA neurons. The transport of spt mRNA into this process presumably is an active rather than passive process since other mRNAs, such as those of huC and the islet genes, are not similarly localised (data not shown).
We attempted to observe the pattern of axonal projection from spt-expressing neurons at later times after their differentiation. We stained embryos at 22 hpf to reveal both spt-transcription and the presence of acetylated tubulin (that labels axons). Confocal imaging of spt-expressing neurons in the region of the spinal cord dorsal to the yolk extension showed that these cells lie alongside the dorsal longitudinal fasciculus (DLF, Figure 2). Their proximity to the DLF obscured the pattern of axonal projection from these cells. We did not observe the presence of spt transcript in axons near these cells.
Figure 2.

Close association of neurons expressing spt with the dorsal longitudinal fasciculus. Images shown are projections of serial 0.5 μm optical sections through a 22 hpf embryo stained to reveal spt transcripts (red) and acetylated tubulin (green) that marks axons. The cell shown lies in that part of the developing spinal cord midway along the yolk extension. Rostral is to the left in both images. A shows a lateral projection with dorsal to the top. B shows a dorsal projection with medial to the bottom and lateral to the top. The size bar in A indicates 10 μm. B has an identical rostrocaudal dimension but the mediolateral dimension is compressed. The size bar in B indicates 10 μm in the mediolateral dimension.
Dorsoventral and rostrocaudal correspondence of caudal spt expression in the somitic mesoderm and developing CNS
Extended staining for spt expression allowed us to observe spt mRNA in recently-formed somites at 24 hpf just rostral to the previously observed, high-level expression of spt in the presomitic mesoderm. This expression is not present throughout the somites but, rather, only at the same dorsoventral level as spt-expressing cells in the developing spinal cord. From a lateral perspective, this gives the impression of a "trail" of spt-expressing cells in the somitic mesoderm left behind by the extending tail tip (Figures 1C, 3A,3B,3D).
Figure 3.
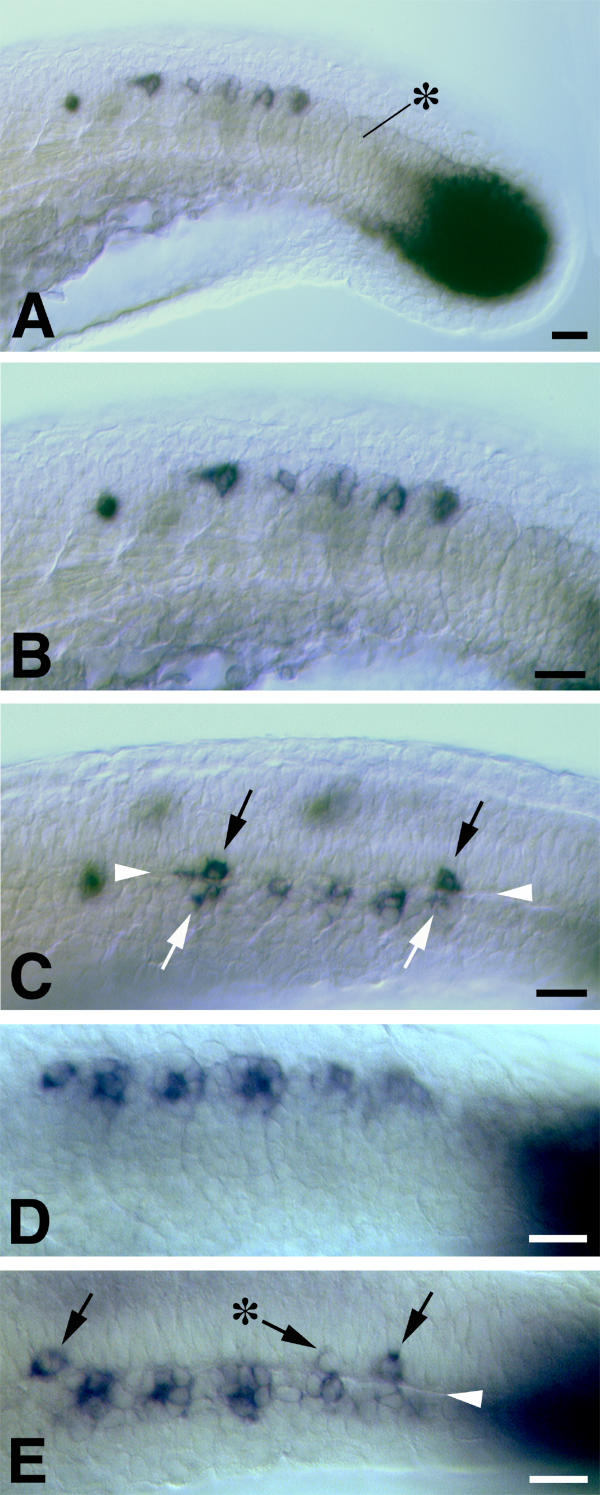
The juxtaposition of spt expression in newly formed somites and the developing CNS at approximately 22 hpf. In all images dorsal is uppermost and rostral is to the left. A, B and C are views from one embryo. A and B show the appearance from a lateral view of the tail in the region of the "somitic trail" of spt expression. A black asterisk indicates the most recently formed somite. spt expression is concentrated to the dorsocaudal extremity of somites. In an optical (DIC) section through the same embryo viewed from a dorsolateral perspective (C), the basal lamina separating the developing CNS and the somitic mesoderm can be seen clearly (arrowheads). Cells expressing spt in the developing CNS (black arrows) are juxtaposed to somitic cells expressing spt (white arrows). The "somitic trail" region of a second embryo is shown in D (lateral view) and E (dorsolateral view). The black asterisk in E indicates a neural cell expressing a lower level of spt. Scale bars equal 20 μm.
The somitic expression of spt is strongest in the dorsocaudal cells of these structures (Figure 3). Observation of this region from a dorsolateral perspective shows that cells expressing spt in the developing spinal cord most commonly form so that they are in direct juxtaposition with these cells across the basal lamina (Figures 3C,3E; at least 76% of observed cases, n = 25). However, they do not form adjacent to every somite. This distribution suggests that: 1) the spt-expressing neurons are either generated in response to signals from the dorsocaudal cells of each somite or, 2) that neural and somitic cells express spt in response to a common patterning signal(s). In either case, an "inefficient" stimulation of neural cells to transcribe spt would result in the observed distribution of spt-expressing neurons.
Occasionally, neural cells transcribing lower levels of spt can be observed adjacent to the most posterior somites (see asterisk in Figure 3E). We have not observed such cells at more rostral levels so these might represent cells in the process of activating spt transcription. Alternatively, neural cells transcribing spt at lower levels might be lost or might repress spt transcription later in spinal cord development.
The earliest formation of spt-expressing neurons
The somitic expression of spt at 24 hpf is only seen in the most recently formed somites. We wished to observe whether newly born spt-expressing neurons are always flanked by spt expression in somitic mesoderm, and to determine the earliest time at which spt-expressing neurons could be observed.
To gain an indication of the time at which spt-expressing neurons might first arise, we observed the somitic juxtaposition of the most rostral spt-expressing neuron in 11 embryos at approximately 24 hpf. The majority of the embryos (n = 10) possessed at least 6 somite pairs rostral to the most rostral spt-expressing neural cell (Figures 4A,4B). Only one embryo showed a lower number (at least 5 rostral somite pairs). Thus, spt-expression in the spinal cord is flanked by the region of somitic mesoderm that shows slower somite formation (occurring after the initial rapid formation of the first six somite pairs, [23]). Since the 5th somite pair forms at approximately 12 hpf (at 28.5°C), we examined embryos between 12 hpf and 16 hpf for spt staining in the CNS. The CNS primordium is relatively flattened at this time and the basal laminae separating CNS, mesoderm and individual somites are difficult to observe in fixed embryos. Nevertheless, the earliest time at which we could observe spt expression confidently in the developing CNS was 15.5 hpf (13 somite pairs). At 16 hpf (14 somite pairs), five of six embryos examined for which spt-expressing neurons could be seen had at least 9 somite pairs rostral to the most rostral spt-expressing neuron (see Figure 5). This observation implies that the spt-expressing neurons at more rostral positions (i.e. adjacent to the 6th to 9th somite pairs) differentiate at later times or that spt-expressing cells migrate rostralwards after their birth (see later). At 16 hpf, the spt-expressing neural cells are also flanked by low level spt expression in somites (white arrowheads in Figure 5). Thus, low level somitic expression of spt occurs during most of somitogenesis. Low level spt expression is observable at 14.5 hpf in laterocaudal cells. However, we could not determine whether these cells were neural or mesodermal (data not shown).
Figure 4.

Lateral views of two embryos (A and B) at approximately 24 hpf stained to reveal spt transcription. Dorsal is up and rostral is to the left. DIC microscopy was used to reveal somite boundaries. Consequently, spt-expressing cells in the developing CNS are not seen clearly because they lie in a different focal plane. However, the most rostral cell in each embryo is indicated by a white arrrowhead. The most rostral visible discernible somite is indicated by a white arrow. In both cases there are 6 somites rostral to the most rostral spt-expressing neuron. Scale bars equal 100 μm.
Figure 5.

Early spt expression in the developing CNS and somites. A and C show lateral views of two embryos at 16 hpf. Rostral is up and dorsal is to the right. B shows a dorsal view of the embryo in A. Rostral is up. White arrowheads indicate the most rostral somitic domain of spt transcription visible. Black arrowheads indicate the most rostral neural cell expressing spt. For B, the light source was concentrated behind the yolk to give greater visibility of staining. All images are composites of smaller images. Scale bars equal 100 μm.
Analysis of left-right bias in spt-expressing neuron number
The irregular distribution of spt-expressing neurons may conceal a left or right bias in the number of these neurons. To investigate this we examined the numbers of neurons on the left and right sides of 48 embryos at 24 hpf. The mean number of cells on the left sides of embryos was found to be 10.5 with a standard deviation of 2.1. The mean number of cells on the right sides of embryos was found to be 10.7 with a standard deviation of 1.9. The differences in the mean number of spt-expressing neurons on the left and right sides of the embryos is considerably smaller than the standard deviations of left and right. This argues against any left-right bias.
The analysis above might not reveal a left or right bias when the variability in the number of spt-expressing neurons in each embryo is high. Thus, we also examined the difference in the numbers of spt-expressing neurons between the left and right sides of individual embryos. For each of the 48 embryos, the number of spt-expressing cells on the left of the embryo was subtracted from the number on the right. The mean difference was +0.2 with a standard deviation of 2.0. Since the standard deviation is far larger that the mean difference, this also argues against any left or right bias in spt-expressing neuron number. Finally, we tested whether there is simply a tendency for an absolute difference in the numbers of spt-expressing cells to exist between the two sides of the embryo, regardless of any left-right bias. The mean absolute bilateral difference for the 48 embryos was 1.5 cells. The standard deviation for this value was 1.4. Thus, there is no significant difference in the numbers of spt-expressing cells between the two sides of embryos.
Preferred positions of spt-expressing neurons on the rostrocaudal axis
While the distribution of spt-expressing neurons along the rostrocaudal axis of the spinal cord appears to be irregular, preferred positions may, nevertheless, exist. To analyse this, 20 embryos were fixed at 24 hpf and stained to reveal expression of spt. The left and right sides of the trunk and tail of the embryos were then photographed under differential interference contrast (DIC) optics to show simultaneously the spt-expressing neurons and the boundaries between the flanking somitic tissue. We then counted the neurons occurring adjacent to each particular somite on the left and right sides of the embryo. Since we have shown that there is no left-right bias in the number of spt-expressing neurons, we combined the data from the two sides. The number of somite pairs present in embryos at 24 hpf can vary [23], as can the visibility in fixed embryos of the most anterior somite boundaries and the most recently formed somite boundaries. Therefore, to make the results from each embryo comparable, we identified the somite pair directly dorsal to the most caudal extent of the yolk extension as somite level 0. We then numbered the other somite pairs according to this reference point (Figure 6). Somite pairs rostral to somite level 0 were given a "+" designation while caudal somite pairs were given a "-" designation. The mean number of cells present at each somite level was then calculated (Table 1).
Figure 6.
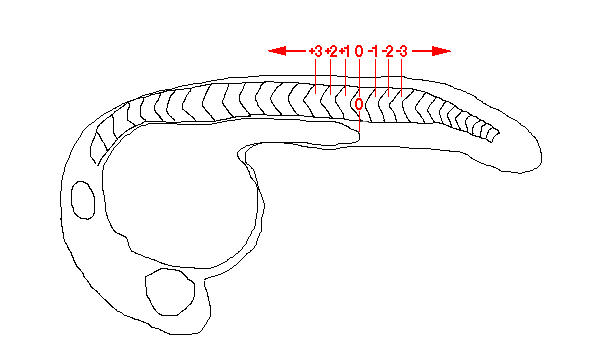
Diagram of somite level designations relative to the caudal tip of the yolk extension in a 24 hpf embryo.
Table 1.
Numbers of spt-expressing neurons per somite level (pair of hemisegments) at 24 hpf
| Somite level | Somite number | Number of embryos | Mean cell number | Standard deviation |
| -14 | 32 | 3 | 0 | 0 |
| -13 | 31 | 7 | 0 | 0 |
| -12 | 30 | 12 | 0 | 0 |
| -11 | 29 | 15 | 0.07 | 0.26 |
| -10 | 28 | 19 | 0.16 | 0.50 |
| -9 | 27 | 19 | 0.47 | 0.61 |
| -8 | 26 | 19 | 0.47 | 0.61 |
| -7 | 25 | 19 | 0.63 | 0.76 |
| -6 | 24 | 20 | 0.55 | 0.69 |
| -5 | 23 | 20 | 0.50 | 0.61 |
| -4 | 22 | 20 | 0.70 | 0.92 |
| -3 | 21 | 20 | 0.55 | 0.69 |
| -2 | 20 | 20 | 0.55 | 0.60 |
| -1 | 19 | 20 | 0.95 | 0.83 |
| 0 | 18 | 20 | 0.95 | 1.05 |
| +1 | 17 | 20 | 0.70 | 0.66 |
| +2 | 16 | 20 | 0.70 | 0.73 |
| +3 | 15 | 20 | 0.95 | 1.00 |
| +4 | 14 | 20 | 0.75 | 0.79 |
| +5 | 13 | 20 | 1.00 | 0.92 |
| +6 | 12 | 20 | 0.75 | 0.72 |
| +7 | 11 | 20 | 1.40 | 0.68 |
| +8 | 10 | 20 | 1.45 | 1.19 |
| +9 | 9 | 19 | 1.11 | 0.88 |
| +10 | 8 | 17 | 1.47 | 1.01 |
| +11 | 7 | 13 | 1.92 | 1.38 |
| +12 | 6 | 7 | 1.14 | 0.69 |
| +13 | 5 | 4 | 0.75 | 0.50 |
| +14 | 4 | 2 | 0 | 0 |
Somite level 0 represents the somite pair immediately dorsal to the most posterior extremity of the yolk extension. Negative values are more caudal to somite level 0 and positive values are more rostral. The common identity of each somite pair (i.e. disregarding variability between embryos) in terms of its order of formation is given as the somite number
A tendency to higher numbers of cells at rostral somite levels is evident. The highest mean number observed was at somite level +11 (1.9 cells per embryo for left and right sides combined). At 24 hpf, somite level +11 commonly corresponds to the 7th somite pair formed. Lower numbers of spt-expressing neurons are observed at somite levels caudal to somite level 0 (commonly the 18th somite pair formed). However, there is great variability between embryos in the number of cells at any somite level (as indicated by the large standard deviation values in Table 1). The increase in cell number at rostral levels is not explained by the increase in the rostrocaudal dimension of somites as they mature since the segmental pattern of neuron distribution in the spinal cord expands correspondingly [2]. The higher number of spt-expressing neurons found rostral to somite level 0 could be due to: 1) continuing birth of these neurons at rostral positions as the CNS develops, 2) programmed cell death of neurons at caudal positions, or 3) rostralwards migration of neurons after their birth. Two observations support the last possibility. First, the mean number of spt-expressing neurons along the entire rostrocaudal axis per embryo was determined for 76 embryos at 24 hpf (21.4 neurons, standard deviation 3.4) and 45 embryos at 30 hpf (22.7 neurons, standard deviation 2.9). Somitogenesis ends at approximately 24 hpf but differentiation along the rostrocaudal axis continues in a rostral to caudal manner. Thus, any later, rostral generation of spt-expressing neurons or programmed cell death of caudal neurons as spinal cord development continues after 24 hpf might be expected to alter the average number of neurons by a greater number than that observed. Second, ipsilateral juxtaposition of spt-expressing neurons (which we defined as instances in which the cell bodies of the neurons appear to contact each other) occurred for 5.5% of cells in the region of somite levels -11 to +4, but for 11.3% of these cells in the region of somite levels +5 to +12. These data, together with the observation of greater neuron numbers at rostral levels, suggest that these neurons accumulate at rostral levels due to rostralwards migration after their birth.
No spt-expressing cells were observed rostral of somite level +13, commonly corresponding to the 5th somite pair formed. This could be an artefact of the low number of embryos for which these somite levels could be distinguished during observation. However, this result is consistent with our earlier failure to observe spt-expressing neurons more rostral than the 5th most rostral somite pair (see Figure 4 and above).
At first glance, the numbers of spt-expressing neurons we observe at each somite level (i.e. per two hemisegments) at 24 hpf does not appear to be comparable to the previous observations of Bernhardt et al. in 1990 [12] of 0.06 DoLA interneurons per hemisegment (0.12 per somite level) flanked by the 5th- to 8th-formed somite pairs in embryos at 18 hpf. However, the fact that we rarely observe spt-expressing neurons anterior to the 6th-formed somite pair at 24 hpf combined with the possibility that these neurons migrate rostrally after birth (see above) suggests that fewer DoLA neurons may be found in the region flanked by the 5th- to 8th-formed somite pairs at 18 hpf compared to 24 hpf. Also, these authors identified DoLA neurons by their pattern of arborisation whereas we have identified these cells by spt expression. At 18 hpf many spt-expressing cells may not yet have developed characteristic DoLA arborisation patterns. In contrast, in a study of GABAergic DoLA neurons in embryos at 27 hpf by Bernhardt et al. in 1992 [24], a mean of 3.89 cells (standard deviation 1.17) were observed in the region of hemisegments 6 to 10. At 24 hpf, we observed a mean of 3.64 cells (standard deviation 1.08) in the same region. The close correspondence of these figures supports that spt-expressing neural cells are DoLA neurons.
Discussion
The identity of spt-expressing neural cells
The spt-expressing cells in the developing spinal cord show coexpression of a number of neural markers such as huC and the islet genes. This, together with the position of these cells just ventral to the Rohon-Beard neurons and their rostrocaudal distribution establishes that these cells are likely to be the DoLA neurons originally described by Bernhardt et al. [12]. Indeed, we are able to observe a rostrally projecting process of these cells similar to the ascending axon of DoLA neurons due to the active transport of spt mRNA along this process.
That DoLA neurons express spt conflicts with observations of the expression in Xenopus embryos of the spt orthologous gene, Xombi. Xombi is transcribed in a very similar pattern to spt in the developing spinal cord. In 1996, Stennard et al. [25] and Zhang and King [26] suggested that this gene (they named it Antipodean and VegT respectively) might be expressed in Rohon-Beard neurons based on the dorsal/dorsolateral position of expressing cells in the spinal cord. However, in a simultaneous publication, Lustig et al. [27] suggested that Xombi expression was in the dorsolateral area of interneuron formation. We expect that closer examination will show that Xombi is expressed in Xenopus DoLA-equivalent cells, probably dorsolateral interneurons (see review by Roberts [28]).
The observation of spt mRNA in an anterior growth process/axon suggests a number of possibilities. First, the mRNA may not be translated but may perform some other (or no) role in the process. Second, Spt protein may be required in this process for a function other than gene regulation. Third, spt mRNA may be required in the process for production of protein that is used to signal back to the nucleus. There is some precedence for the expression of transcription factors in neurites since these are known to be found in dendrites where it is thought that they may be involved in activities such as long term potentiation [29]. Finally, and most intriguing, is the possibility that spt mRNA might be involved in signalling to cells with which the process makes contact. It has been demonstrated that the transcription factor Engrailed and the homeodomains of other proteins can be transported between cells [30-33]. Testing of these possibilities will require observation of the distribution of Spt protein.
spt-expressing DoLA neurons possibly migrate rostrally
Higher numbers of spt-expressing neurons are observed rostrally compared to caudally in the spinal cord. It may be that spt-expressing neurons continue to be born as the developing CNS matures in a rostral to caudal progression or that caudal neurons undergo programmed cell death. However, the marginal change in the number of these neurons between 24 and 30 hpf argues against this. Also, ipsilateral juxtaposition of these neurons is more common at rostral compared to caudal sites. The increased juxtaposition rostrally could be caused by rostral migration of spt-expressing neurons when an anterior limit exists for the migration. spt-expressing neurons are rarely seen anterior of the 6th-formed somite pair suggesting that this position on the rostrocaudal axis may represent such a limit.
spt is expressed in somitic mesoderm
Extended staining for spt mRNA revealed that this gene is transcribed at low levels in the dorsocaudal cells of recently formed somites. It has previously been assumed that spt expression marks only presomitic mesoderm. The function of spt expression in these somitic cells is unknown. Discovery of other genes expressed in a similar pattern in newly formed somites may reveal more of the function or fate of these cells.
The irregular pattern of spt-expressing neurons may be based on an underlying regularity
The question of how irregular patterns of cell distribution or gene expression are controlled is not commonly addressed in studies of developmental biology. Nevertheless, these patterns are common in the central nervous systems of most animals and occur in many other tissues. In the spinal cord of the developing embryo, Rohon-Beard neurons occur at a frequency of more than one per hemisegment [12]. Their positions are not highly ordered and do not depend upon signals from mesoderm [5]. Instead, short-range intercellular interactions controlled by Notch signalling appear to play a role in their differentiation from a field of progenitor cells [10,11].
The ascending commissural neurons that are located just ventral to Rohon-Beard neurons are also found at a frequency of more than one per hemisegment. However, subclasses of these neurons exist with lower frequency. For example, anti-CON1 antibody labels a subclass of ascending commissural neurons in the embryo that probably become commissural primary ascending (CoPA) interneurons in the larva. These are present in an irregular pattern on the rostrocaudal axis at a frequency of 0.87 per hemisegment flanking the 6th- to 11th-formed somite pairs at 28 hpf [12]. Ascending commissural neurons are located at a similar dorsoventral level to the DoLA neurons. We have shown that val expression in the spinal cord occurs just ventral to spt-expressing neurons. Thus, it is possible that val labels a subclass of ascending commissural neurons.
The neuromasts of the posterior lateral line – while part of the peripheral nervous system – are, nevertheless, an example of a neural cell type distributed at a frequency of less than one per hemisegment. These neurons are deposited by the migrating lateral line primordia along the myoseptum at the boundary between somites at four or five positions along each side of the embryo. While their rostrocaudal distribution is not completely irregular, there is considerable variation in the actual position of any one neuromast. The position at which a neuromast is deposited appears to depend more strongly on the distance from the previously deposited neuromast rather than the precise position on the rostrocaudal axis [34]. Interestingly, the recessive, homozygous viable mutation hypersensitive (hps) results in neuromast deposition at nearly every somite boundary [35]. The fact that this (presumably) loss-of-function mutation can increase the regularity of a pattern indicates that the distribution of neuromasts probably results from the combined effect of at least two patterning mechanisms – one controlling inter-neuromast distance and one controlling neuromast localisation to intersomitic boundaries. This raises the question as to whether mutations might exist that increase the frequency of generation of spt-expressing neurons, for example, by increasing the strength of a patterning signal from the mesoderm to the developing CNS.
The dorsoventral and rostrocaudal correspondence of spt expression in newly formed somites and the CNS suggests a functional connection between the spt expression in these two tissues. The somitic and neural cells may be responding to a common patterning signal. Alternatively, the somitic spt expression may mark the source of a signal from the somite to neural tissue. A precedent for the latter alternative exists in the influence of flanking mesoderm on primary motoneuron formation [5-7]. However, the formation of most primary motoneurons occurs with complete regularity (one neuron per hemisegment). An interesting exception to this is the Variable Primary (VaP) motoneuron that occurs at a frequency of less than 0.5 per hemisegment. VaPs arise adjacent to Caudal Primary (CaP) motoneurons midway between hemisegment boundaries [36]. VaPs normally extend an axon to the horizontal myoseptum in the myotomes after which the VaP dies. In contrast, the CaP axon continues from the myoseptum into ventral muscle. These two neurons actually represent an equivalence pair since ablation of a CaP causes the neighbouring VaP to develop a CaP-like arborisation pattern [37]. Thus, rather than VaP formation occurring with less than complete regularity, we can regard this situation as CaP formation at greater than one cell per hemisegment followed by regulation to one cell per hemisegment.
We suggest that the spt-expressing DoLA interneurons might be "inefficiently" patterned by flanking somitic mesoderm. Thus, the initial distribution of these neurons would represent an incomplete pattern based on a regular template. Migration and tissue growth might then scramble this pattern. We are currently testing this hypothesis by examining the role of spt expression and mesodermal signals in DoLA neuron differentiation and distribution.
Conclusions
spt-expressing cells in the developing central nervous system appear to be DoLA interneurons. The irregular distribution of these cells along the rostrocaudal axis of the spinal cord may be due to "inefficient" patterning of neural spt expression by flanking, regularly distributed somites also expressing spt. Rostral migration of spt-expressing neurons might then scramble any residual regularity in their distribution. The idea that irregular patterns of neuron distribution may arise in partial correspondence to regular templates is a parsimonious explanation for the evolution of such patterns.
Materials and Methods
Double whole mount in situ transcript hybridisation
(Cloning of probe sources)
A cDNA clone, (26 M), corresponding to transcription from spt was isolated in a whole mount in situ transcript hybridisation screen of zebrafish embryos [38]. cDNAs corresponding to parts of transcripts from the genes huC, isl-2 and valentino were amplified by RT-PCR from embryos at 24 hpf using the oligonucleotide primers described in Table 2. All cDNA fragments were cloned into the pGEMT vector (Promega Corporation, Madison, WI, USA). The inserts of these clones were amplified by PCR using M13 primers and then transcribed with T3 or SP6 RNA polymerase to produce digoxigenin- or fluorescein-labelled antisense RNA probes (see [38]). The clones for production of probes against isl-1 and isl-3 transcripts were obtained from Hitoshi Okamoto [6,7].
Table 2.
Oligonucleotides used for cDNA fragment isolation for probe synthesis
| Gene transcripts detected | oligo name | PCR oligonucleotide sequence |
| huC | #277 | 5' CAG ATG ACA GCA AAA CTA ACC 3' |
| #278 | 5' AGA GCA ATA GTG ACT AGG CC 3' | |
| isl2 | #351 | 5' GAC GGC AAG ACT TAT TGC 3' |
| #352 | 5' CAT CTT CGG AGA TCA TGC 3' | |
| val | #322 | 5' GGT CCC CCT GTC GCC TC 3' |
| #323 | 5' CCA CGA GCG ACA ACC CG 3' |
Double whole mount in situ transcript hybridisation was performed essentially as described in [39] but the first staining reaction was with BCIP/NBT, inactivation of the first alkaline phosphatase staining reaction was by heating to 65°C for 45 min in PBS and the second staining reaction used the Alkaline Phosphatase Substrate Kit I (Vector Laboratories Inc., Burlingame, CA, USA).
Combined whole mount in situ transcript hybridisation and immunohistochemistry
Staining for the presence of spt transcript and acetylated tubulin was performed essentially as described above for double whole mount in situ transcript hybridisation except that spt staining using the Alkaline Phosphatase Substrate Kit I ("Vector Red", Vector Laboratories Inc.) was performed first followed by washing for 10 min in 100 mM Tris HCl pH 8.5 then 10 min in PBS + 0.1% Tween 20 (Sigma, St. Louis, MO, USA) (PBT) before fixation in 4% formaldehyde in PBT. Embryos were then washed 4 × 5 min in PBT, then 3 h in PBT + 0.3% IPEGAL (Sigma) (PBTI) + 2% BSA (Fraction V, Sigma), then 1 h in PBTI + 2% BSA at 4°C before incubation overnight at 4°C in a 1:2500 dilution of anti-Acetylated Tubulin antibody (Sigma Cat. No. T6793) in PBTI + 0.2% BSA. Embryos were then washed 6 × 1 h in PBTI then 2 × 30 min in PBTI + 2% BSA before incubation overnight at 4°C in a 1:200 dilution of anti-mouse IgG labelled with Alexa Fluor 488 (Molecular Probes Inc., Eugene, OR, USA) in PBTI + 0.2% BSA. Finally, embryos were washed 7 × 1 h in PBTI before equilibration with 80% glycerol in PBT before imaging. Note that all wash series were preceded by three rinses in the wash solution and were at room temperature unless otherwise indicated.
Observation and statistical analysis of cell distribution
Embryos were dechorionated at 15–18 hpf, 22 hpf, 24 hpf or 30 hpf and fixed in 4% formaldehyde in PBS at 4°C before in situ transcript hybridisation with a probe for spt. To ensure observation of all cells expressing spt including any expressing spt at low levels, the staining reaction was allowed to proceed overnight at 4°C before the embryos were fixed in 4% formaldehyde in PBS and then equilibrated with 80% glycerol.
Light field observation of the embryos was conducted under a Zeiss Axiophot™ microscope (Carl Zeiss Jena GmbH, Jena, Germany) at 200× magnification using DIC optics. For examination of cell positions, the trunk-tail region of an embryo was removed from the rest of the body and then laid flat on a slide. Photographs were taken such that the intersomitic boundaries and the spt-expressing neural cells were simultaneously visible. Confocal imaging of embryos was conducted on a Bio-Rad MRC-1000 UV Confocal Laser Scanning Microscope System (Bio-Rad Laboratories Inc., Hercules, CA, USA) using a Nikon Diaphot 300 inverted microscope (Nikon Instech Co., Ltd., Kawasaki, Kanagawa, Japan). Fluorescence was observed using a krypton/argon laser with excitation at 488/10 nm and emission at 522/35 nm excitation for Alexa 488 and with excitation at 568/10 nm and emission at 605/32 nm for Vector Red. Images were processed with Adobe Photoshop version 5.0 (Adobe Systems Inc. San Jose, California, USA) and Confocal Assistant version 4.02 (Todd Clark Brelje).
List of abbreviations used
BCIP, 5-Bromo-4-chloro-3-indolyl-phosphate.p-toluidine-salt
BSA, bovine serum albumin
CaP, Caudal Primary
CNS, central nervous system
CoPA, Commissural Primary Ascending
DIC, differential interference contrast
DLF, dorsal longitudinal fasciculus
DoLA, dorsal longitudinal ascending
isl, islet
hpf, hours post fertilisation
NBT, Nitroblue tetrazolium chloride
PBS, phosphate buffered saline
PBT, PBS + 0.1% Tween 20
PBTI, PBT + 0.3% IPEGAL
RT-PCR, reverse transcription polymerase chain reaction
spt, spadetail
val, valentino
VaP, Variable Primary
Authors' contributions
RT carried out the majority of the in situ transcript hybridisation, cell counting, and statistical analyses and some photography.
SW performed the in situ transcript hybridisation analyses on 12 – 16 hpf embryos and staining for acetylated tubulin.
JGC contributed to the statistical analysis
ML directed the research, performed observation of staining patterns, some cell counting and statistical analysis, some photography and drafted the manuscript.
Acknowledgments
Acknowledgements
The authors wish to thank Dan Kortschak and Judith Eisen for valuable discussion and Simon Koblar for critical reading of the manuscript. However, the conclusions drawn by the authors are theirs alone. We thank Meredith Wallwork for assistance with confocal microscopy. Clones for production of probes against the isl-1 and isl-3 genes were the kind gift of Hitoshi Okamoto. This work was supported by an Australian Research Council small grant and by funds from the Special Research Centre for the Molecular Genetics of Development. RT was supported by an International Postgraduate Research Scholarship from The University of Adelaide.
Contributor Information
Richard Tamme, Email: richard.tamme@adelaide.edu.au.
Simon Wells, Email: simon.wells@adelaide.edu.au.
John G Conran, Email: john.conran@adelaide.edu.au.
Michael Lardelli, Email: michael.lardelli@adelaide.edu.au.
References
- Keynes R, Stern C. Segmentation in the vertebrate nervous system. Nature. 1984;310:786–9. doi: 10.1038/310786a0. [DOI] [PubMed] [Google Scholar]
- Hanneman E, Trevarrow B, Metcalfe WK, Kimmel CB, Westerfield M. Segmental pattern of development of the hindbrain and spinal cord of the zebrafish embryo. Development. 1988;103:49–58. doi: 10.1242/dev.103.1.49. [DOI] [PubMed] [Google Scholar]
- Myers P. Spinal motoneurons of the larval zebrafish. J Comp Neurol. 1985;236:555–61. doi: 10.1002/cne.902360411. [DOI] [PubMed] [Google Scholar]
- Myers P, Eisen J, Westerfield M. Development and axonal outgrowth of identified motoneurons in the zebrafish. J Neurosci. 1986;6:2278–89. doi: 10.1523/JNEUROSCI.06-08-02278.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JS, Pike SH. The spt-1 mutation alters segmental arrangement and axonal development of identified neurons in the spinal cord of the embryonic zebrafish. Neuron. 1991;6:767–776. doi: 10.1016/0896-6273(91)90173-w. [DOI] [PubMed] [Google Scholar]
- Inoue A, Takahashi M, Hatta K, Hotta Y, Okamoto H. Developmental regulation of islet-1 mRNA expression during neuronal differentiation in embryonic zebrafish. Dev Dyn. 1994;199:1–11. doi: 10.1002/aja.1001990102. [DOI] [PubMed] [Google Scholar]
- Tokumoto M, Gong Z, Tsubokawa T, Hew C, Uyemura K, Hotta Y, Okamoto H. Molecular heterogeneity among primary motoneurons and within myotomes revealed by the differential mRNA expression of novel islet-1 homologs in embryonic zebrafish. Dev Biol. 1995;171:578–89. doi: 10.1006/dbio.1995.1306. [DOI] [PubMed] [Google Scholar]
- Nguyen V, Trout J, Connors S, Andermann P, Weinberg E, Mullins M. Dorsal and intermediate neuronal cell types of the spinal cord are established by a BMP signaling pathway. Development. 2000;127:1209–20. doi: 10.1242/dev.127.6.1209. [DOI] [PubMed] [Google Scholar]
- Dornseifer P, Takke C, Campos-Ortega J. Overexpression of a zebrafish homologue of the Drosophila neurogenic gene Delta perturbs differentiation of primary neurons and somite development. Mech Dev. 1997;63:159–71. doi: 10.1016/S0925-4773(97)00037-3. [DOI] [PubMed] [Google Scholar]
- Cornell R, Eisen J. Delta signaling mediates segregation of neural crest and spinal sensory neurons from zebrafish lateral neural plate. Development. 2000;127:2873–82. doi: 10.1242/dev.127.13.2873. [DOI] [PubMed] [Google Scholar]
- Gray M, Moens CB, Amacher SL, Eisen JS, Beattie CE. Zebrafish deadly seven functions in neurogenesis. Dev Biol. 2001;237:306–23. doi: 10.1006/dbio.2001.0381. [DOI] [PubMed] [Google Scholar]
- Bernhardt RR, Chitnis AB, Lindamer L, Kuwada JY. Identification of spinal neurons in the embryonic and larval zebrafish. J Comp Neurol. 1990;302:603–616. doi: 10.1002/cne.903020315. [DOI] [PubMed] [Google Scholar]
- Kimmel C, Kane D, Walker C, Warga R, Rothman M. A mutation that changes cell movement and cell fate in the zebrafish embryo. Nature. 1989;337:358–62. doi: 10.1038/337358a0. [DOI] [PubMed] [Google Scholar]
- Ho R, Kane D. Cell-autonomous action of zebrafish spt-1 mutation in specific mesodermal precursors. Nature. 1990;348:728–30. doi: 10.1038/348728a0. [DOI] [PubMed] [Google Scholar]
- Weinberg E, Allende M, Kelly C, Abdelhamid A, Murakami T, Andermann P, Doerre O, Grunwald D, Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–80. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Amacher S, Kimmel C. Promoting notochord fate and repressing muscle development in zebrafish axial mesoderm. Development. 1998;125:1397–406. doi: 10.1242/dev.125.8.1397. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Amacher S, Kim S, Geissert D, Kimmel C, De Robertis EM. Zebrafish paraxial protocadherin is a downstream target of spadetail involved in morphogenesis of gastrula mesoderm. Development. 1998;125:3389–97. doi: 10.1242/dev.125.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin K, Amacher S, Kimmel C, Kimelman D. Molecular identification of spadetail: regulation of zebrafish trunk and tail mesoderm formation by T-box genes. Development. 1998;125:3379–88. doi: 10.1242/dev.125.17.3379. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Silver LM, Ho RK. Characterization of the zebrafish tbx16 gene and evolution of the vertebrate T-box family. Dev Genes Evol. 1998;208:94–99. doi: 10.1007/s004270050158. [DOI] [PubMed] [Google Scholar]
- Kim CH, Ueshima E, Muraoka O, Tanaka H, Yeo SY, Huh TL, Miki N. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci Lett. 1996;216:109–112. doi: 10.1016/S0304-3940(96)13021-4. [DOI] [PubMed] [Google Scholar]
- Korzh V, Edlund T, Thor S. Zebrafish primary neurons initiate expression of the LIM homeodomain protein Isl-1 at the end of gastrulation. Development. 1993;118:417–25. doi: 10.1242/dev.118.2.417. [DOI] [PubMed] [Google Scholar]
- Moens CB, Yan YL, Appel B, Force AG, Kimmel CB. Valentino: A zebrafish gene required for normal hindbrain segmentation. Development. 1996;122:3981–3990. doi: 10.1242/dev.122.12.3981. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the Zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Bernhardt RR, Patel CK, Wilson SW, Kuwada JY. Axonal trajectories and distribution of GABAergic spinal neurons in wildtype and mutant zebrafish lacking floor plate cells. J Comp Neurol. 1992;326:263–72. doi: 10.1002/cne.903260208. [DOI] [PubMed] [Google Scholar]
- Stennard F, Carnac G, Gurdon J. The Xenopus T-box gene, Antipodean, encodes a vegetally localised maternal mRNA and can trigger mesoderm formation. Development. 1996;122:4179–88. doi: 10.1242/dev.122.12.4179. [DOI] [PubMed] [Google Scholar]
- Zhang J, King M. Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development. 1996;122:4119–29. doi: 10.1242/dev.122.12.4119. [DOI] [PubMed] [Google Scholar]
- Lustig K, Kroll K, Sun E, Kirschner M. Expression cloning of a Xenopus T-related gene (Xombi) involved in mesodermal patterning and blastopore lip formation. Development. 1996;122:4001–12. doi: 10.1242/dev.122.12.4001. [DOI] [PubMed] [Google Scholar]
- Roberts A. Early functional organization of spinal neurons in developing lower vertebrates. Brain Res Bull. 2000;53:585–593. doi: 10.1016/S0361-9230(00)00392-0. [DOI] [PubMed] [Google Scholar]
- Eberwine J, Job C, Kacharmina J-E, Miyashiro K, Therianos S. Transcription factors in dendrites: dendritic imprinting of the cellular nucleus. Results Probl Cell Differ. 2001;34:57–68. doi: 10.1007/978-3-540-40025-7_4. [DOI] [PubMed] [Google Scholar]
- Le RI, Joliot AH, Bloch GE, Prochiantz A, Volovitch M. Neurotrophic activity of the Antennapedia homeodomain depends on its specific DNA-binding properties. Proc Natl Acad Sci U S A. 1993;90:9120–9124. doi: 10.1073/pnas.90.19.9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatelin L, Volovitch M, Joliot AH, Perez F, Prochiantz A. Transcription factor Hoxa-5 is taken up by cells in culture and conveyed to their nuclei. Mech Dev. 1996;55:111–117. doi: 10.1016/0925-4773(95)00478-5. [DOI] [PubMed] [Google Scholar]
- Joliot A, Maizel A, Rosenberg D, Trembleau A, Dupas S, Volovitch M, Prochiantz A. Identification of a signal sequence necessary for the unconventional secretion of Engrailed homeoprotein. Curr Biol. 1998;8:856–863. doi: 10.1016/s0960-9822(07)00346-6. [DOI] [PubMed] [Google Scholar]
- Maizel A, Bensaude O, Prochiantz A, Joliot A. A short region of its homeodomain is necessary for Engrailed nuclear export and secretion. Development. 1999;126:3183–3190. doi: 10.1242/dev.126.14.3183. [DOI] [PubMed] [Google Scholar]
- Gompel N, Cubedo N, Thisse C, Thisse B, Dambly-Chaudiere C, Ghysen A. Pattern formation in the lateral line of zebrafish. Mech Dev. 2001;105:69–77. doi: 10.1016/S0925-4773(01)00382-3. [DOI] [PubMed] [Google Scholar]
- Whitfield TT, Granato M, van Eeden FJM, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg C-P, Jiang Y-J, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Nusslein-Volhard C. Mutations affecting development of the zebrafish inner ear and lateral line. Development. 1996;123:241–54. doi: 10.1242/dev.123.1.241. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Pike SH, Romancier B. An identified motoneuron with variable fates in embryonic zebrafish. J Neurosci. 1990;10:34–43. doi: 10.1523/JNEUROSCI.10-01-00034.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JS. The role of interactions in determining cell fate of two identified motoneurons in the embryonic zebrafish. Neuron. 1992;8:231–40. doi: 10.1016/0896-6273(92)90290-t. [DOI] [PubMed] [Google Scholar]
- Tamme R, Mills K, Rainbird B, Nornes S, Lardelli M. Simple, directional cDNA cloning for in situ transcript hybridisation screens. BioTechniques. 2001;31:938–46. doi: 10.2144/01314rr05. [DOI] [PubMed] [Google Scholar]
- Jowett T. Tissue in situ hybridization. New York: John Wiley & Sons; 1997.


