Abstract
Lactobacillus rhamnosus GR-1 and L. fermentum RC-14 are well-characterized probiotic strains with efficacy in the prevention and treatment of urogenital infections in women. The aim of the present study was to apply a molecular biology-based methodology for the detection of these strains and L. rhamnosus GG (a commercially available intestinal probiotic) in the human vagina in order to assess probiotic persistence at this site. Ten healthy women inserted vaginally a capsule containing either a combination of strains GR-1 and RC-14 or the GG strain for 3 consecutive nights. Vaginal swabs taken before and at various time points after probiotic insertion were analyzed, and the Lactobacillus flora was assessed by randomly amplified polymorphic DNA (RAPD) analysis. This method generated discrete DNA fingerprints for GR-1, RC-14, and GG and enabled successful detection of these strains in the vagina. Strain GR-1 and/or strain RC-14 was found to persist in the vaginal tract for up to 19 days after vaginal instillation, while L. rhamnosus GG was detectable for up to 5 days postadministration. In conclusion, the fates of probiotic L. rhamnosus and L. fermentum strains were successfully monitored in the human vagina by RAPD analysis. This technique provides molecular biology-based evidence that RC-14 and GR-1, strains selected as urogenital probiotics, persist in the human vagina and may be more suited to vaginal colonization than L. rhamnosus GG. This highlights the importance of proper selection of strains for urogenital probiotic applications.
The vaginal ecosystem is a complex environment in which lactobacilli are the most predominant bacterial species in healthy women (30). A recent study showed that this microflora changes constantly, with only 22% of women having a normal Lactobacillus-dominated flora at any given time (27). As depletion of vaginal lactobacilli can have serious consequences, including increasing the risk of preterm labor and susceptibility to infections such as Neisseria gonorrhoeae, Trichomonas vaginalis, human immunodeficiency virus, and urinary tract infections, bacterial vaginosis, and yeast vaginitis (2, 28, 30), restoration and maintenance of a normal microflora are important.
While antibiotics can be effective for eradication of urinary tract infections, they are often unable to cure bacterial vaginosis or prevent complications that arise from it (6). Furthermore, antibiotics have many side effects, multidrug-resistant microorganisms are common, and such chemotherapeutic agents ideally should not be used for prophylaxis or health maintenance.
The use of probiotics, defined as “microbial cell preparations or components of microbial cells that have a beneficial effect on health and well being of the host” (25), offers a potential alternative approach to health restoration and maintenance in the vaginal tract (2, 16, 29). The question becomes which probiotic strains will persist in and colonize the vagina, given that the vaginal microflora fluctuates daily in terms of both the types and the numbers of microorganisms present (27, 30).
Of the many probiotic microorganisms available commercially, the majority of which are used for the intestine (11), only Lactobacillus acidophilus NCFM has been tested for its potential to colonize the vagina, and it was found in a series of in vitro experiments not to have the optimal characteristics required of a urogenital probiotic (17). L. rhamnosus GG is one of the most studied intestinal probiotic microorganisms and has been used successfully to treat and prevent diarrhea (13). It adheres to intestinal epithelial cells and inhibits growth of enteropathogens (26), and one could expect that it would colonize the vagina and reduce the risk of infection; however, vaginal persistence has not been tested, until now.
On the other hand, L. rhamnosus GR-1 and L. fermentum RC-14 are extensively characterized urogenital isolates which possess a number of properties considered important for urogenital probiotics; both strains adhere to uroepithelial cells and inhibit the growth and adhesion of uropathogens, while GR-1 is resistant to the spermicide nonoxynol 9 and RC-14 produces hydrogen peroxide (19). Furthermore, studies with humans have shown that these strains are efficacious in the prevention and treatment of urogenital infections in women (4, 5, 21, 24).
In order to detect probiotic lactobacilli in the vagina, a specific diagnostic methodology must be used to differentiate exogenous from indigenous isolates, as the vaginal microflora of an individual can harbor five or more different strains of lactobacilli at any given time. Molecular biology-based techniques such as pulsed-field gel electrophoresis (PFGE), ribotyping, denaturing gradient gel electrophoresis, analysis with DNA probes, and randomly amplified polymorphic DNA (RAPD) analysis provide the means to do this (10, 15, 20, 33).
The objective of the present study was to use RAPD analysis to determine the ability of two products, one containing a combination of L. fermentum RC-14 and L. rhamnosus GR-1 and another containing L. rhamnosus GG, to colonize the vaginas of healthy women.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The probiotic strains used in this study, L. fermentum RC-14, L. rhamnosus GR-1, and L. rhamnosus GG (ATCC 53103), have been well characterized and were selected as a result of extensive in vitro experimentation and human studies (see references 13 and 18 for reviews). Lactobacillus strains obtained from culture collections (L. fermentum ATCC 9338, L. rhamnosus ATCC 7469, L. reuteri ATCC 23272, L. plantarum ATCC 14917, L. casei ATCC 393, L. vaginalis NCFB 2810, L. acidophilus ATCC 33199, and L. jensenii ATCC 25258) and some Lactobacillus strains previously isolated from the human urogenital tract (23) were included in the study for comparative purposes. All lactobacilli were routinely cultured at 37°C in MRS broth (Merck, Darmstadt, Germany) (9) under anaerobic conditions (anaerobic jars with BBL gas packs; Becton Dickinson and Co., Sparks, Md.).
Genetic fingerprinting by RAPD PCR analysis.
RAPD PCR analysis (32) was performed with each Lactobacillus strain and with vaginal isolates recovered from participating subjects. Genomic DNA was first isolated from 1.5 ml of overnight MRS broth cultures by the method outlined by Coakley et al. (7), which uses shearing with glass beads to lyse the bacterial cells. The extracted DNA was then used as a template in subsequent PCR amplifications, which were performed in a total volume of 50 μl in a DNA thermal cycler (Eppendorf Scientific Inc., Westbury, N.Y.). PCR mixtures contained 1 μl of template DNA, 1×Taq polymerase buffer (Gibco BRL Life Technologies, Burlington, Ontario, Canada), 4 mM MgCl2, each deoxynucleoside triphosphate (Amersham Pharmacia Biotech, Baie d’Urfe, Quebec, Canada) at a concentration of 200 μM, 2.5 U of platinum Taq DNA polymerase (Gibco BRL), and each primer at a concentration of 1 μM. Two primers with arbitrary nucleotide sequences (5′ ACGAGGCAC3′ and 5′ACGCGCCCT3′) (31) were used, and these were synthesized by Gibco BRL. DNA was amplified for 40 cycles by using the following temperature profile: denaturation at 94°C for 30 s, annealing at 36°C for 30 s, and polymerization at 72°C for 2 min. The initial denaturation was performed at 94°C for 5 min, and a final extension step of 72°C for 10 min was also used. The PCR products (10 μl of each reaction mixture) were analyzed on a 1.5% (wt/vol) agarose (Fisher Biotech, Fair Lawn, N.J.) gel with 1× TAE buffer (40 mM Tris acetate, 1 mM EDTA [pH 8]) and ethidium bromide staining. A 100-bp ladder (Gibco BRL) was used as a molecular size standard. Gels were run for approximately 2 h at 100 V, and the DNA was visualized by UV transillumination.
Human study.
Ten women between the ages of 21 and 51 (mean age, 35.5 years; standard deviation, 11 years) participated in the study, which was approved by the Review Board for Health Sciences Research Involving Human Subjects at the University of Western Ontario, London, Ontario, Canada. Although not specifically screened for urogenital disease, the women were considered healthy on entry into the study, as they did not present with any signs or symptoms of such diseases, and all women harbored vaginal lactobacilli at levels between 105 and 107 CFU/swab. Each subject signed an informed consent. Individuals were not included in the study if they were pregnant or breast-feeding, had a history of diabetes, or were on immunosuppressive or antibiotic therapy. The women were divided randomly into two groups, each of which contained five subjects. Immediately following menses, each member of one group inserted into the vagina a capsule containing a combination of L. fermentum RC-14 and L. rhamnosus GR-1 in freeze-dried form (a total of >109 viable bacteria) at night for 3 consecutive days (days 0 to 2). Each member of the second group followed the same regimen with commercially available capsules containing >1010 CFU of L. rhamnosus GG (Con Agra Foods, Omaha, Nebr.). Vaginal swabs were obtained prior to capsule insertion (day 0); immediately after treatment (day 3); and 5 days (day 7), 12 days (day 14), and 19 days (day 21) following treatment.
Probiotic detection in Lactobacillus capsules and vaginal swabs.
Each vaginal swab was placed in a tube containing modified Amies Clear transport medium, and the tubes were held at 4°C until analysis. Suspensions were made from each swab in 1 ml of 0.9% saline solution and were diluted 10-fold in the same medium. Portions (100 μl) of appropriate dilutions were spread plated onto MRS agar, and the plates were incubated anaerobically for 48 h at 37°C. The total number of colonies was counted in order to enumerate the total lactobacilli in the vaginal swabs. Then, on the basis of colony morphology, 4 to 20 representative Lactobacillus colonies from each swab (particularly those suspected of being GR-1, RC-14, or GG) were selected from the MRS agar plates for RAPD PCR analysis, performed as described above. The RAPD fingerprints of the probiotic strains were compared with those of the vaginal isolates and lactobacilli recovered from the probiotic capsules used for vaginal insertion. In addition, PFGE analysis was performed with selected vaginal isolates to confirm their identity as GR-1. PFGE was performed by the method of Brennan et al. (3), with the following modifications: genomic DNA was isolated from 1 ml of stationary-phase cultures, incubation with proteinase K was performed at 37°C, restriction digests were performed overnight at 25°C with ApaI, and gels were run for 18 h with the pulse time ramped from 1 to 15 s.
RESULTS AND DISCUSSION
DNA fingerprinting of probiotic strains by RAPD analysis.
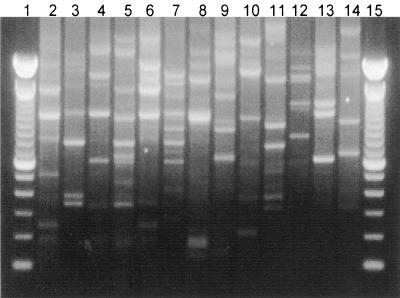
RAPD analysis, a molecular biology-based method which uses PCR with arbitrary primers, was used to generate DNA fingerprints for cultures of L. rhamnosus GR-1, L. fermentum RC-14, and L. rhamnosus GG. The RC-14 and GG strains generated unique, reproducible RAPD profiles (Fig. 1) which were substantially different from the banding patterns of Lactobacillus species representative of those found in the human vagina (Fig. 1). While L. rhamnosus GR-1 could also be differentiated from most of the representative lactobacilli by RAPD analysis, it was difficult to distinguish GR-1 from some other L. rhamnosus strains (e.g., ATCC 7469) (Fig. 1). Therefore, PFGE was investigated and enabled differentiation of the GR-1 strain from these closely related L. rhamnosus strains (data not shown). However, RAPD analysis was chosen for subsequent tracking of probiotic lactobacilli in the human vagina, as it is faster and less laborious than the PFGE technique.
FIG. 1.
RAPD fingerprints of probiotic Lactobacillus strains (L. rhamnosus GR-1 [lane 2], L. fermentum RC-14 [lane 3], and L. rhamnosus GG [lane 4]), culture collection Lactobacillus strains (L. fermentum ATCC 9338 [lane 5], L. rhamnosus ATCC 7469 [lane 6], L. reuteri ATCC 23272 [lane 7], L. plantarum ATCC 14917 [lane 8], L. casei ATCC 393 [lane 9], L. vaginalis NCFB 2810 [lane 10], L. acidophilus ATCC 33199 [lane 11], and L. jensenii ATCC 25258 [lane 12]), and vaginal isolates (L. casei 70 [lane 13] and L. paracasei 55 [lane 14]). Lanes 1 and 15, 100-bp ladders.
Human study.
While previous studies in our laboratory have shown that vaginal insertion of GR-1 and B-54 (a strain almost identical to RC-14) leads to colonization of the vaginal tract and prevention of infection in women with acute or recurrent urinary tract infections (4, 5, 21, 24), probiotic identification was based on the Gram staining result and colony morphology only. Molecular biology-based methods have also been used for identification of these strains, with ribotyping used to identify GR-1 in the vagina of one patient following vaginal instillation (22) and to detect RC-14 and GR-1 vaginally following oral administration (20). The RAPD method has also been used to monitor the survival of RC-14 and GR-1 in the human intestine (12) and to determine the survival of other probiotic lactobacilli in dairy products (10). The present data indicate that RAPD analysis can be used to identify constituents of the vaginal microflora.
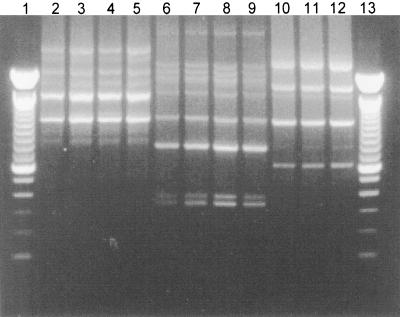
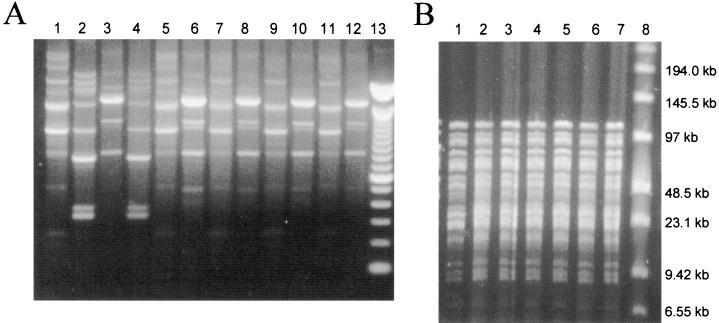
As shown in Fig. 2, the RAPD fingerprints of Lactobacillus isolates from capsules containing the GR-1-RC-14 combination (lanes 3 to 5 and 7 to 9) or GG alone (lanes 11 to 12) were identical to those of the relevant probiotic strains isolated from the vaginal swabs (lanes 2, 6, and 10, respectively). The usefulness of RAPD analysis for the detection of GR-1, GG, and RC-14 in human vaginal swabs was further verified (Fig. 3A and 4). On examination of the vaginal microflora prior to probiotic treatment (day 0), it was found that all of the women were colonized vaginally by lactobacilli at levels of 105 to 107 CFU/swab and so were considered healthy, although no specific screening for urogenital disease was performed. Molecular biology-based analysis showed that no individual harbored strains with RAPD fingerprints corresponding to those for GR-1, RC-14, or GG (Fig. 3A, lane 3; Fig. 4, lane 2; Tables 1 and 2). Immediately following vaginal insertion (day 3) the Lactobacillus strains that had been applied were detected in the vaginas of all of the women except subject 209, in whom only one of the probiotic strains (L. rhamnosus GR-1) from the RC-14-GR-1 combination administered was detected (Tables 1 and 2). Thereafter, strain RC-14 or strain GR-1 or both strains were detectable as part of the dominant Lactobacillus microflora by RAPD analysis in the vaginas of four of the five women who inserted these strains 12 days following the cessation of treatment, and they were still present at 18 to 19 days posttreatment in three of these women (Fig. 3A; Table 1). Interestingly, L. rhamnosus GR-1 was recovered more frequently than L. fermentum RC-14 and so appears to have had a superior survival capability in all except one of the women (subject 206). Bearing in mind that GR-1 was difficult to distinguish from some other L. rhamnosus strains by RAPD analysis (Fig. 1), PFGE was also used in some cases to confirm the identity of GR-1 (Fig. 3B).
FIG. 2.
RAPD fingerprints of L. rhamnosus GR-1 (lane 2), L. rhamnosus GR-1 isolates from a capsule containing the L. rhamnosus GR-1-L. fermentum RC-14 combination (lanes 3 to 5), L. fermentum RC-14 (lane 6), L. fermentum RC-14 isolates from the same L. rhamnosus GR-1-L. fermentum RC-14 capsule (lanes 7 to 9), L. rhamnosus GG (lane 10), and Lactobacillus isolates from a capsule containing the L. rhamnosus GG strain (lanes 11 and 12). Lanes 1 and 13, 100-bp ladders.
FIG. 3.
(A) RAPD fingerprints of L. rhamnosus GR-1 (lane 1), L. fermentum RC-14 (lane 2), and representative lactobacilli isolated from vaginal swabs of subject 210 before (lane 3) and 1 (lanes 4 to 6), 5 (lanes 7 and 8), 12 (lanes 9 and 10), and 18 (lanes 11 and 12) days after vaginal insertion of an L. rhamnosus GR-1-L. fermentum RC-14 capsule for 3 consecutive nights. Lane 13, a 100-bp ladder. (B) PFGE analysis of L. rhamnosus GR-1 (lane 1) and selected vaginal Lactobacillus isolates from subject 207 at 1 day (lanes 2 to 4) and 5 days (lanes 5 to 7) after vaginal insertion of an L. rhamnosus GR-1-L. fermentum RC-14 capsule for 3 consecutive nights. Lane 8, low-range PFGE molecular size markers.
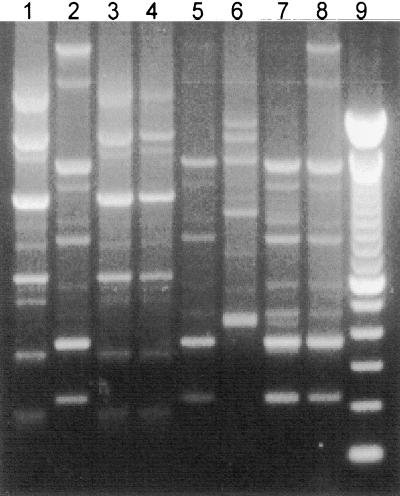
FIG. 4.
RAPD fingerprints of L. rhamnosus GG (lane 1) and representative lactobacilli isolated from vaginal swabs of subject 222 before (lane 2) and 3 (lane 3), 7 (lanes 4 to 6), 14 (lane 7), and 21 (lane 8) days after vaginal insertion of an L. rhamnosus GG capsule for 3 consecutive nights. Lane 9, a 100-bp ladder.
TABLE 1.
RAPD PCR detection of L. rhamnosus GR-1 and L. fermentum RC-14 in the vaginal tracts of women following vaginal instillation of a combination of these strains
| Subject no. | Strain(s) detected on:
|
||||
|---|---|---|---|---|---|
| Day 0 | Day 3 | Day 7 | Day 14 | Day 21 | |
| 210 | −a | RC-14, GR-1 | GR-1 | GR-1 | GR-1b |
| 207 | − | RC-14, GR-1 | GR-1 | GR-1 | − |
| 206 | − | RC-14, GR-1 | RC-14, GR-1 | RC-14 | RC-14 |
| 209 | − | GR-1 | − | − | − |
| 230 | − | RC-14, GR-1 | RC-14 GR-1 | GR-1 | GR-1 |
−, GR-1 or RC-14 was not detected
Swab taken at day 20.
TABLE 2.
RAPD PCR detection of L. rhamnosus GG in the vaginal tract of women following vaginal instillation of this strain
| Subject no. | Detection of GG ona:
|
||||
|---|---|---|---|---|---|
| Day 0 | Day 3 | Day 7 | Day 14 | Day 21 | |
| 223 | −a | + | − | − | − |
| 221 | − | + | − | −b | − |
| 222 | − | + | + | − | − |
| 224 | − | + | − | − | − |
| 225 | − | + | + | − | − |
−, GG not detected; +, GG detected.
Swab taken at day 17.
L. rhamnosus GG was detected in two of five women 5 days after the treatment ceased, and it was undetectable in the vaginas at all test points thereafter (Fig. 4; Table 2). A previous study had suggested that vaginal insertion of L. rhamnosus GG could improve symptoms of yeast vaginitis (14), but no data were reported on the recovery of GG from the vagina. The numbers of subjects evaluated in the present study are too small to conclude that GG is not effective in the vagina, but in vitro studies predicted this outcome (19) and oral administration of this strain did not prevent urinary tract infections (T. Kontiokari, M. Nuutinen, K. Sundqvist, T. Pokka, M. Koskela, and M. Uhari, Abstr. 38th Meet. Infect. Dis. Soc. Am., abstr. 179, 2000). Therefore, it does appear that GG is more suitable for intestinal applications (13, 26) than as a urogenital probiotic.
While a complete analysis of the vaginal microflora was not the aim of the present study, some interesting findings arose regarding indigenous lactobacilli in the vagina. For example, a Lactobacillus strain present in the vagina prior to probiotic treatment in subject 210 (Fig. 3A, lane 3), who subsequently became colonized by GR-1, was still detectable immediately after treatment (Fig. 3A, lane 6) and remained for up to 18 days after discontinuation of treatment (Fig. 3A, lane 12). Similar results were obtained for subjects receiving the L. rhamnosus GG treatment (Fig. 4). This shows that endogenous lactobacilli can also be monitored by RAPD analysis, an advantage of using this method (i.e., PCR with random primers) over a PCR with strain-specific primers. Interestingly, in another subject (i.e., subject 230), all of the Lactobacillus colonies selected were identified as being of the exogenous strain (data not shown), suggesting that it could dominate the vaginal microflora of this individual. Previously, the predominant Lactobacillus species present in the vagina have been identified by molecular biology-based methods (1), and changes in the vaginal microflora over time have been examined by use of vaginal smears (27). Data from the present study suggest that further longitudinal molecular biology-based analysis of vaginal lactobacilli at the strain level would be worthy of pursuit.
In conclusion, to our knowledge, this is the first report of the application of RAPD analysis to the detection of probiotic strains in the vagina. Potentially clinically important probiotic strains L. rhamnosus GR-1 and GG and L. fermentum RC-14 as well as indigenous Lactobacillus microflora were differentiated by this method, which uses arbitrarily chosen PCR primers to amplify randomly sized DNA fragments, separation of which results in a characteristic DNA fingerprint (32). Recently, there has been a more rational approach to choosing probiotic strains both for urogenital and for intestinal use (8, 11, 18), and as a result, strains which have the best chance of survival in the host are selected. In this respect, RC-14 and GR-1 are well-characterized urogenital isolates selected on the basis of their probiotic properties at this site, while GG possesses characteristics considered more important for an intestinal probiotic. The present study provides a good example of how a molecular biology-based methodology can track properly selected probiotic lactobacilli as they colonize the vagina.
Acknowledgments
We thank Warren McDonald, Wendy Kennette, and Andrea Domonkos for technical assistance and Paul Simpson, Teagasc, Dairy Products Research Center, Moorepark, Fermoy, County Cork, Ireland, for assistance with PFGE.
REFERENCES
- 1.Antonio, M. A. D., S. E. Hawes, and S. L. Hillier. 1999. The identification of vaginal Lactobacillus species by the demographic and microbiologic characteristics of women colonized by these species. J. Infect. Dis. 180:1950–1956. [DOI] [PubMed] [Google Scholar]
- 2.Barbes, C., and S. Boris. 1999. Potential role of lactobacilli as prophylactic agents against genital pathogens. AIDS Patient Care STDS 13:747–751. [DOI] [PubMed] [Google Scholar]
- 3.Brennan, N. M., R. Brown, M. Goodfellow, A. C. Ward, T. P. Beresford, P. J. Simpson, P. F. Fox, and T. M. Cogan. 2001. Corynebacterium mooreparkense sp. nov. and Corynebacterium casei sp. nov., isolated from the surface of a smear-ripened cheese. Int. J. Syst. E vol. Microbiol. 51:843–852. [DOI] [PubMed] [Google Scholar]
- 4.Bruce, A. W., and G. Reid. 1988. Intravaginal instillation of lactobacilli for prevention of recurrent urinary tract infections. Can. J. Microbiol. 34:339–343. [DOI] [PubMed] [Google Scholar]
- 5.Bruce, A. W., G. Reid, J. A. McGroarty, M. Taylor, and C. Preston. 1992. Preliminary study on the prevention of recurrent urinary tract infections in ten adult women using intravaginal lactobacilli. Int. Urogynecol. J. 3:22–25. [Google Scholar]
- 6.Carey, J. C., M. A. Klebanoff, J. C. Hauth, S. L. Hillier, E. A. Thom, J. M. Ernest, R. P. Heine, R. P. Nugent, M. L. Fischer, K. J. Leveno, R. Wapner, and M. Varner. 2000. Metronidazole to prevent preterm delivery in pregnant women with asymptomatic bacterial vaginosis. N. Engl. J. Med. 342:534–540. [DOI] [PubMed] [Google Scholar]
- 7.Coakley, M., R. P. Ross, and D. Donnelly. 1996. Application of the polymerase chain reaction to the rapid analysis of brewery yeast strains. J. Inst. Brew. 102:349–354. [Google Scholar]
- 8.Collins, J. K., G. Thornton, and G. O’Sullivan. 1998. Selection of probiotic strains for human applications. Int. Dairy J. 8:487–490. [Google Scholar]
- 9.de Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130–135. [Google Scholar]
- 10.Gardiner, G., R. P. Ross, J. K. Collins, G. Fitzgerald, and C. Stanton. 1998. Development of a probiotic cheddar cheese containing human-derived Lactobacillus paracasei strains. Appl. Environ. Microbiol. 64:2192–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardiner, G., R. P. Ross, P. M. Kelly, J. K. Collins, G. Fitzgerald, and C. Stanton. Microbiology of therapeutic milks. In R. K. Robinson (ed.), Dairy microbiology, vol. 2, 3rd ed., in press. John Wiley & Sons, Inc., New York, N.Y.
- 12.Gardiner, G. E., C. Heinemann, M. L. Baroja, A. W. Bruce, D. Beuerman, J. Madrenas, and G. Reid. Oral administration of the probiotic combination Lactobacillus rhamnosus GR-1 and L. fermentum RC-14 for human intestinal applications. Int. Dairy J., in press.
- 13.Gorbach, S. L. 2000. Probiotics and gastrointestinal health. Am. J. Gastroenterol. 95(Suppl.):S2–S4. [DOI] [PubMed] [Google Scholar]
- 14.Hilton, E., P. Rindos, and H. D. Isenberg. 1995. Lactobacillus GG vaginal suppositories and vaginitis. J. Clin. Microbiol. 33:1433.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Sullivan, D. J. 1999. Methods for analysis of the intestinal microflora, p.23–44. In G. W. Tannock (ed.), Probiotics: a critical review. Horizon Scientific Press, Wymondham, United Kingdom.
- 16.Reid, G. 2000. Probiotic therapy and functional foods for prevention of urinary tract infections: state of the art and science. Curr. Infect. Dis. Rep. 2:518–522. [DOI] [PubMed] [Google Scholar]
- 17.Reid, G. 2000. In vitro testing of Lactobacillus acidophilus NCFM™ as a possible probiotic for the urogenital tract. Int. Dairy J. 10:415–419. [Google Scholar]
- 18.Reid, G. 1999. The scientific basis for probiotic strains of Lactobacillus. Appl. Environ. Microbiol. 65:3763–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid, G., and A. W. Bruce. 2001. Selection of Lactobacillus for urogenital probiotic applications. J. Infect. Dis. 183(Suppl.):S77–S80. [DOI] [PubMed] [Google Scholar]
- 20.Reid, G., A. W. Bruce, N. Fraser, C. Heinemann, J. Owen, and B. Henning. 2001. Oral probiotics can resolve urogenital infections. FEMS Immunol. Med. Microbiol. 30:49–52. [DOI] [PubMed] [Google Scholar]
- 21.Reid, G., A. W. Bruce, and M. Taylor. 1995. Instillation of Lactobacillus and stimulation of indigenous organisms to prevent recurrence of urinary tract infections. Microecol. Ther. 23:32–45. [Google Scholar]
- 22.Reid, G., K. Millsap, and A. W. Bruce. 1994. Implantation of Lactobacillus casei var rhamnosus into the vagina. Lancet 344:1229.. [DOI] [PubMed] [Google Scholar]
- 23.Reid, G., P. L. Cuperus, A. W. Bruce, H. C. van der Mei, L. Tomeczek, A. H. Khoury, and H. J. Busscher. 1992. Comparison of contact angles and adhesion to hexadecane of urogenital, dairy, and poultry lactobacilli: effect of serial culture passages. Appl. Environ. Microbiol. 58:1549–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reid, G., A. W. Bruce, and M. Taylor. 1992. Influence of three-day antimicrobial therapy and Lactobacillus vaginal suppositories on recurrence of urinary tract infections. Clin. Ther. 14:11–16. [PubMed] [Google Scholar]
- 25.Salminen, S., A. G. Ouwehand, Y. Benno, and Y. K. Lee. 1999. Probiotics: how should they be defined? Trends Food Sci. Technol. 10:107–110. [Google Scholar]
- 26.Saxelin, M. 1997. Lactobacillus GG—a human probiotic strain with thorough clinical documentation. Food Rev. Int. 13:293–313. [Google Scholar]
- 27.Schwebke, J. R., C. M. Richey, and H. L. Weiss. 1999. Correlation of behaviors with microbiological changes in vaginal flora. J. Infect. Dis. 180:1632–1636. [DOI] [PubMed] [Google Scholar]
- 28.Sewankambo, N., R. H. Gray, M. J. Wawer, L. Paxton, D. McNaim, F. Wabwire-Mangen, D. Serwadda, C. Li, N. Kiwanuka, S. L. Hillier, L. Rabe, C. A. Gaydos, T. C. Quinn, and J. Konde-Lule. 1997. HIV-1 infection associated with abnormal vaginal flora morphology and bacterial vaginosis. Lancet 350:530–531. [DOI] [PubMed] [Google Scholar]
- 29.Sieber, R., and U. T. Dietz. 1998.Lactobacillus acidophilus and yogurt in the prevention and therapy of bacterial vaginosis. Int. Dairy J. 8:599–607. [Google Scholar]
- 30.Sobel, J. D. 1999. Is there a protective role for vaginal flora? Curr. Infect. Dis. Rep. 1:379–383. [DOI] [PubMed] [Google Scholar]
- 31.Tilsala-Timisjarvi, A., and T. Alatossava. 1998. Strain-specific identification of probiotic Lactobacillus rhamnosus with randomly amplified polymorphic DNA-derived PCR primers. Appl. Environ. Microbiol. 64:4816–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welsh, J., and M. McClelland. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 18:7213–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong, W., K. Millsap, H. Bialkowska-Hobrzanska, and G. Reid. 1998. Differentiation of Lactobacillus species by molecular typing. Appl. Environ. Microbiol. 64:2418–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]