Abstract
Postweaning multisystemic wasting syndrome of swine associated with porcine circovirus (PCV) is a recently reported and economically important disease. Simple and reliable diagnostic methods are needed for detecting antibodies to PCV type 2 (PCV2) for monitoring of PCV infection. Here, we report the development of two modified indirect enzyme-linked immunosorbent assays (ELISAs): a PCV2 ELISA based on cell-culture-propagated PCV2 and an ORF2 ELISA based on recombinant major capsid protein. PCV2 and ORF2 ELISA detected antibodies to PCV2 and the capsid protein, respectively, in sera from pigs experimentally infected with PCV2 as early as 14 and 21 days postinoculation (dpi). The kinetics of the antibody response to PCV2 and the major capsid protein were similar. Repeatability tests revealed that the coefficients of variation of positive sera within and between runs for both assays were less than 30%. To validate the assays, PCV2 and ORF2 ELISAs were performed with 783 serum samples of young and adult pigs collected from different herds in the Midwestern United States and compared with an indirect immunofluorescent assay (IIF). Six out of 60 samples collected from nursery and growing pigs in 1987 were positive by both ELISA and IIF. Compared with IIF, the diagnostic sensitivity, specificity, and accuracy of PCV2 and ORF2 ELISAs were similar (>90%). The tests showed no cross-reactivity with antibodies to porcine parvovirus and porcine reproductive and respiratory syndrome virus. There was good agreement between the two ELISAs and between the ELISAs and IIF. The availability of the two ELISAs should accelerate our understanding of the host immune response to PCV2 and facilitate the development of prevention and control strategies by elucidating the ecology of PCV2 within swine populations.
Porcine circovirus (PCV) was first identified as a PK-15 cell contaminant (46) and was subsequently classified in the family Circoviridae (28). The PCV virions are spherical, have a diameter of 17 nm, and contain single-stranded closed circular genomic DNA 1.7 kb in size (46). In 1991, a wasting disease of weaned pigs emerged in a herd in Saskatchewan, Canada. In 1995, the disease appeared again with higher incidence and at that time was named postweaning multisystemic wasting syndrome (PMWS) (8, 16).
The PK-15 cell contaminant PCV has been shown to be nonpathogenic in experimental studies (1, 47). However in recent studies, PCV was consistently detected in PMWS cases (2, 10, 36). Further genomic analysis revealed that there are two distinct genotypes of PCV (15, 32, 33, 36); PCV type 1 (PCV1) and PCV2 were designated for the nonpathogenic PCV and PMWS-associated PCV, respectively. Differential reactivity with monoclonal antibodies to either PCV1 or PCV2 revealed that they were antigenically different (2, 4). However, a low level of cross-reactivity exists between the two types since antibodies to PCV1 reacted to a low degree with PCV2 (2).
PMWS is a disease of growing pigs with low morbidity but high case mortality. The most common clinical signs are unthriftiness, dyspnea, and jaundice (16). Histological lesions include lymphohistiocytic to granulomatous inflammation in lung, liver, kidney, and lymphoid tissues as well as lymphoid depletion (8). Experiments showed that dual infection of pigs with PCV2 and porcine parvovirus (PPV) caused severe clinical signs and lesions of PMWS (12), while inoculation with PCV2 alone produced only mild to moderate histological lesions (3, 22, 24). Inoculation with PCV1, PPV, or a combination of both PCV1 and PPV failed to produce clinical signs or lesions resembling PMWS (24). In a more recent study, injection of an immunostimulant into PCV2-infected pigs enhanced replication of PCV2 and induced clinical signs and lesions typical of PMWS (25), confirming the role of PCV2 as an essential cause of PMWS. The occurrence of PMWS has been reported worldwide (7, 9, 10, 20, 23, 36, 38, 39, 42, 43, 49).
The genomic organizations of PCV1 and PCV2 are similar. Both contain two major open reading frames (ORFs): ORF1 and ORF2 (15, 32, 33, 36). ORF1 of PCV1 encodes a putative protein involved in viral replication (31). The predicted proteins from ORF2 of PCV1 and PCV2 are similar in size (15, 33, 36). Recently, we reported that ORF2 of PCV2 encodes a major structural protein of approximately 30 kDa and recombinant ORF2 expressed in insect cells self-assembles to form virus-like particles (37). The recombinant ORF2 protein reacted strongly with serum from PCV2-infected swine, suggesting its possible use in diagnostic assays.
The most common diagnostic methods for detecting PCV2 antibodies include indirect immunofluorescent assay (IIF) and immunoperoxidase monolayer assay (IPMA) (2, 4, 10). These techniques require experienced technicians, not only for the preparation of infected cells but also for accurate interpretation of the staining reactions. Additionally, examination of IIF and IPMA plates is tedious and time-consuming. Enzyme-linked immunosorbent assay (ELISA) can be automated, hence decreasing the potential bias that may occur with the interpretation of IIF or IPMA results. Recently, an indirect competitive ELISA (cELISA) with high sensitivity and specificity utilizing monoclonal antibodies to PCV2 was reported (48). Here, we describe the development of two modified ELISA systems: a PCV2-infected cell antigen-based ELISA and a recombinant ORF2-based ELISA. The diagnostic sensitivity (DSN) and diagnostic specificity (DSP) of both assays in comparison with IIF are reported.
MATERIALS AND METHODS
Cells and viruses.
The working stock of PCV2 strain ISU31 (36) was prepared as previously described (37). To prepare antigen for ELISA, 106 50% tissue culture infective doses (TCID50) of PCV2 were mixed with 5 × 105 cells of freshly trypsinized PK-15 cells (kindly provided by K. M. Lager, National Animal Disease Center, Ames, Iowa). After 1 h of absorption, the infected cells were maintained in culture medium MEM (Gibco BRL, Grand Island, N.Y.) with 10% fetal bovine serum (Gibco BRL) and 1% antibiotic-antimycotic (100 U of penicillin G per ml, 100 μg of streptomycin sulfate per ml, and 0.025 μg of amphotericin B per ml; Gibco BRL). Mock-infected PK-15 cells were also prepared for control antigen. At 12 h postinoculation (hpi), mock- and PCV2-infected cells were treated with 300 mM d-glucosamine (Gibco BRL) in Hanks’ balanced salt solution (Gibco BRL) for 30 min at 37°C (47). Cells were harvested at 60 hpi and used for antigen preparation for ELISA.
To prepare plates for IIF, freshly trypsinized PK-15 cells were inoculated with PCV2 at a multiplicity of infection (MOI) of 0.1. One hundred microliters of freshly trypsinized PCV2-infected PK-15 cell suspension at a concentration of 5 × 104 cells/ml was transferred into rows 1, 3, 5, and 7 of a 96-well plate. Mock-infected PK-15 cells were prepared similarly to PCV2-infected cells and seeded in alternate rows. After 1 h of absorption, 100μl of MEM with 20% fetal bovine serum and 2% antibiotic-antimycotic was added into each well. Cells were treated with 300 mM d-glucosamine at 37°C for 20 min at 12 hpi, fixed with absolute methanol at 60 hpi, dried, and stored at −20°C.
Wild type baculovirus (AcMNPV.wt) and recombinant baculovirus carrying the ORF2 gene of PCV2 (AcMNPV.ORF2) were prepared as previously described (37). Both AcMNPV.wt and AcMNPV.ORF2 were inoculated onto 60% monolayers of Hi-Five insect cells (Invitrogen, Carlsbad, Calif.) at an MOI of 5. The virus was allowed to adsorb onto the cells for 1 h at room temperature. Subsequently, the cells were cultured in an insect cell culture medium, Excel 405 (JRH Biosciences, Lenexa, Kans.), with 1% antibiotic-antimycotic (Gibco BRL) at 27°C for 72 h.
Serum samples.
Sera were acquired from cesarean-derived and colostrum-deprived (CD-CD) piglets experimentally infected with PCV2 strain 35358 (18). Three-week-old CD-CD pigs were inoculated with PCV2 or sham inoculated with culture medium. Nine CD-CD pigs were inoculated with culture media, and 21 pigs were infected with PCV2. One to two pigs in the sham group and two to four pigs in the PCV2-infected group were sacrificed at 7, 14, 21, 28, 35, 42, and 49 days postinoculation (dpi). Sera were collected before inoculation and at each day postinoculation.
Seven hundred and eighty-three swine serum samples were obtained from Midwestern U.S. herds and represented three sets of samples. First, 568 total samples were collected from 36 herds in Iowa, 19 herds in Oklahoma, and a herd in Illinois during 2000; 118 samples were from nursery pigs, 243 samples were from grow-finish pigs, and 207 samples were from sows and gilts. Second, 60 serum samples were randomly selected from pooled sera of grow-finish pigs in Illinois, Michigan, and Iowa collected from 1989 to 1999. Finally, 155 samples were collected from a Wisconsin herd defined as clinically negative for PMWS in 1987, 1988, and 1989.
Sixty-seven serum samples containing antibodies against other swine viruses were also included in this study to determine the specificity of assays. Thirty-one samples positive for porcine reproductive and respiratory syndrome virus (PRRSV) antibodies were obtained from CD-CD pigs experimentally inoculated with PRRSV (18). Thirty-six samples were positive for antibodies to PPV as determined by hemagglutination inhibition assay, with titers ranging from 1:32 to 1:16,384.
IIF.
IIF was used as the reference method to detect the presence of antibodies to PCV2. Ninety-six-well plates containing PCV2- and mock-infected cells fixed in methanol and stored at −20°C were thawed and washed with phosphate-buffered saline (PBS). Serum samples (50 μl) diluted either 1 in 10 or 1 in 100 in PBS and 0.1% Tween 20 (Sigma, St. Louis, Mo.) were added to PCV2-infected or noninfected PK-15 cells and incubated at 37°C for 1 h. Unbound antibodies were washed three times with 5-min incubations with PBS containing 0.1% Tween 20. An optimum dilution of fluorescein-conjugated anti-swine immunoglobulin G (Sigma) was added and incubated for 30 min at 37°C. After washing, positive signal was observed under UV light. Serum samples that gave a positive signal at a serum dilution of 1:10 or higher were called positive.
Antigen preparation.
For the PCV2 ELISA, positive and negative antigens were prepared from PCV2-infected and noninfected PK-15 cells, respectively. At 60 hpi, cells in 25-cm2 flasks were washed three times with cold PBS, pH 7.4, and lysed by incubating on ice for 30 min with 0.5 ml of a lysis buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 1% (vol/vol) NP-40, 0.5% (vol/vol) sodium deoxycholate in HEPES, and 0.1% sodium dodecyl sulfate (SDS) (17). Cells were then scraped, transferred into a microcentrifuge tube and incubated on ice for 1 h with periodic mixing on a vortex mixer. The cell suspension was clarified by centrifugation at 10,000 × g for 10 min at 4°C. The supernatant containing either positive antigen (PCV2 Ag) or negative antigen (PK-15 Ag) was dispensed in aliquots and kept at −70°C. The total protein concentration of both antigens was 0.8 mg/ml as determined by Bio-Rad (Hercules, Calif.) protein assay.
Negative and positive antigens for ORF2 ELISA were partially purified proteins from insect cells infected with wild-type or recombinant baculovirus, respectively. At 72 hpi, wild-type baculovirus (AcMNPV.wt)- or recombinant baculovirus (AcMNPV.ORF2)-infected Hi-Five cells were transferred to a 50-ml tube and centrifuged at 800 × g for 10 min. The cell pellet was washed three times with cold PBS, pH 7.4, resuspended with PBS, and frozen at −70°C. The infected cells were frozen and thawed three times to release viral proteins before clarification by centrifugation at 800 × g for 10 min. The supernatant was diluted in PBS, laid over 6 ml of 40% sucrose in PBS, and centrifuged at 270,000 × g for 6 h using a Ti 41 SW rotor (Beckman, Palo Alto, Calif.). The pellet was washed three times with cold PBS before incubation with the lysis buffer (150 μl/original cells from a 75-cm2 flask) for 30 min on ice. Subsequently, the viral antigen was clarified by centrifugation at 10,000 × g at 4°C for 10 min. Supernatants containing positive antigen (ORF2 Ag) and negative antigen (WT Ag) were stored in aliquots at −70°C. Both ORF2 Ag and WT Ag had a protein concentration of 1.4 mg/ml as measured by the Bio-Rad protein assay.
Modified indirect ELISA.
To coat plates for ELISA, the optimum concentration of antigen was determined by checkerboard titration. Two modified ELISA methods, utilizing antigen prepared from PCV2-infected PK-15 cells or recombinant ORF2 protein, were performed as described previously (6). For the conventional PCV2 ELISA, both positive and negative antigens were diluted 1:200 in a carbonate-bicarbonate coating buffer, pH 9.5 (5). Immulon 2HB polystyrene microtiter plates (Dynex Technologies Inc., Chantilly, Va.) were coated with positive and negative antigen by adding 100 μl of diluted PCV2 or PK-15 Ag into wells of alternate columns. The coated plate was incubated at 4°C for 36 to 40 h and then stored at−20°C. For the ORF2 ELISA, ORF2 and WT Ag were diluted 1:800 in the carbonate-bicarbonate coating buffer, pH 9.5 (5). The Immulon 2HB polystyrene microtiter plates were coated with 100 μl of diluted ORF2 or WT Ag as described for the PCV2 ELISA.
To perform the PCV2 ELISA on experimentally infected pig sera, the plate was thawed, equilibrated at room temperature, and washed three times with 30-s incubation using PBST washing buffer (0.1 M PBS [pH 7.2] and 0.1% Tween 20). Sera were diluted 1 to 40 in 5% milk diluent (Kirkegaard & Perry Laboratories Inc., Gaithersburg, Md.). One hundred microliters of each diluted serum was incubated with positive and negative antigen at 37°C for 30 min. Excess antibodies were removed by washing five times with PBST buffer using a microplate washer (BioTEK Instruments). Plates were blotted dry between each wash and gently but firmly tapped and blotted dry after the last wash. An optimum dilution of peroxidase-labeled anti-pig immunoglobulin G (100μl; Sigma) in 5% milk diluent was added into each well. After incubation at 37°C for 30 min, plates were washed five times. Then, 100 μl of freshly prepared and warmed (37°C) 2,2′-azino-di-(3-ethylbenzthiazoline-6-sulfonate) (ABST) substrate (Kirkegaard & Perry Laboratories Inc.) was added and incubated for 30 min at 37°C. The enzyme-substrate reaction was stopped by adding equal amount of 1% SDS into each well.
The ORF2 ELISA was performed as described for the PCV2 ELISA except that substrate was allowed to react with peroxidase enzyme for 15 min instead of 30 min before the reaction was stopped by adding an equal volume of 1% SDS.
To determine the optimum dilution to test field sera, 22 field sera representing negative (IIF titer less than 10), weak positive (IIF titer between 10 and 100), and strong positive (IIF titer more than 100) serum samples were diluted twofold from 1:50 to 1:200 in 5% milk diluent. Each dilution of serum was tested for PCV-specific antibodies with both PCV2 and ORF2 ELISA. A dilution of 1 to 100 of field sera was selected as an optimum dilution for both assays. This dilution provided a high corrected optical density value (COD) while effectively differentiating between negative, weak positive, and strong positive sera. The PCV2 and ORF2 ELISA procedures for testing the field sera were similar to those described for the experimental sera except for the higher (1:100) dilution of tested serum.
The optimum dilution of conjugates for each assay was determined by checkerboard titration. The dilution that gave the highest COD was selected as the optimum dilution. All serum samples were run in quadruplicate, two on positive antigen and two on negative antigen wells. Positive and negative control sera were included in every plate.
The optical density (OD) was measured at 405 nm using a microtiter plate reader (Molecular Devices, Sunnyvale, Calif.). The COD of each sample and control sera was calculated by subtraction of the mean OD of the well containing negative antigen from that of the parallel well containing positive antigen. To normalize the data, the COD of each serum sample, referred to as sample COD, was divided by a positive serum control COD and reported as the sample/positive (S/P) ratio.
Evaluation of assay repeatability.
Evaluation of assay repeatability within and between runs was performed as proposed by Jacobson (21). Twenty-one field serum samples representing negative (IIF titer of less than 10), weak positive (IIF titer between 10 and 100), and strong positive (IIF titer more than 100) serum samples were selected for the repeatability test. For intra-assay (within-plate) repeatability, three replicates of each serum sample were assigned in the same plate. For interassay (between-run) repeatability, three replicates of each sample were run in different plates on different occasions. PCV2 and ORF2 ELISAs were performed using conditions for field sera as described earlier in this work. Mean S/P ratio, standard deviation (SD), and coefficient of variation (CV) of three replicates of each test were calculated.
Selection of negative-positive cutoff.
To set a negative-positive cutoff value for each assay, the S/P ratios of 783 field sera obtained from either PCV2 or ORF2 ELISAs were compared with IIF results. A negative-positive threshold for each assay was determined using a modified receiver-operator characteristic analysis as previously described (44). A cutoff point for each assay was determined so that DSN and DSP were maximized while the sum of false negative and false positive results were minimized. The data is calculated using the Microsoft Excel spreadsheet.
Evaluation of assay performance.
Diagnostic accuracy was determined by calculating DSN and DSP of the test (21). IIF was used as the reference method to classify samples as positive or negative. The DSN of the ELISA is the proportion of IIF-positive serum samples that give positive results in the assay [DSN = TP/(TP+FN), where TP is true positive and FN is false negative]. DSP is the proportion of IIF-negative serum samples that give negative results by the assay [DSP = TN/(TN+FP), where TN is true negative and FP is false positive]. Positive predictive value, negative predictive value, and accuracy of the test were calculated from the following formulas:
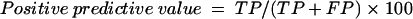 |
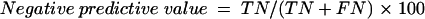 |
 |
Determination of correlation between IIF titers and ELISA S/P ratio.
Thirty-six field serum samples were randomly selected using the SAS program. Serum samples were divided into four groups according to their S/P ratios. Samples were picked proportionally from each group, resulting in serum samples with S/P ratios ranging from 0.097 to 1.307 for PCV2 ELISA and 0.091 to 2.110 for ORF2 ELISA. IIF was performed on twofold serial dilutions of the selected sera from 1:10 to 1:20,480. A correlation between IIF titer and S/P ratio of the corresponding serum was determined using Spearman’s rank correlation method (44), and a regression line was plotted between IIF titer and S/P ratio of the corresponding serum samples using Microsoft Excel. The level of agreement between PCV2 and ORF2 ELISA was determined by the kappa statistic (44), defined as the proportion of potential agreement of two tests beyond chance. We interpreted the agreement of kappa statistic values between the two tests as follows: 0 to 0.2 = slight, 0.2 to 0.4 = fair, 0.4 to 0.6 = moderate, 0.6 to 0.8 = substantial, and 0.8 to 1.0 = excellent.
RESULTS
IIF results.
Results of IIF of field serum samples are summarized in Table 1. Out of 783 field sera tested by IIF, 546 samples were positive and 238 samples were negative for PCV antibodies. PCV-positive pigs were detected as early as 1987 in Wisconsin. This was followed by positive samples from pigs in Michigan in 1989 and Illinois in 1992. Out of 568 samples collected in 2000 from different herds in Iowa and Oklahoma, 471 samples were positive for PCV antibodies.
TABLE 1.
Detection of antibodies to PCV by IIF in swine serum samples obtained from the Midwestern United States
| Yr | PCV antibodies in serum samplesa
|
|||||
|---|---|---|---|---|---|---|
| IA | IL | MI | OK | WI | Total | |
| 1987 | — | — | — | — | 12/60 | 12/60 |
| 1988 | — | — | — | — | 3/70 | 3/70 |
| 1989 | — | — | 6/10 | — | 15/25 | 21/35 |
| 1992 | — | 4/10 | — | — | — | 4/10 |
| 1993 | — | — | 6/10 | — | — | 6/10 |
| 1994 | — | 9/10 | — | — | — | 9/10 |
| 1998 | — | 10/10 | — | — | — | 10/10 |
| 1999 | 10/10 | — | — | — | — | 10/10 |
| 2000 | 291/326 | 17/20 | — | 163/222 | — | 471/568 |
| Total | 301/336 | 40/50 | 12/20 | 163/222 | 30/155 | 546/783 |
Results are presented as number of IIF-positive sera/total sera tested. —, no serum tested. Abbreviations: IA, Iowa; IL, Illinois; MI, Michigan; OK, Oklahoma; WI, Wisconsin.
ELISA results on experimental pigs.
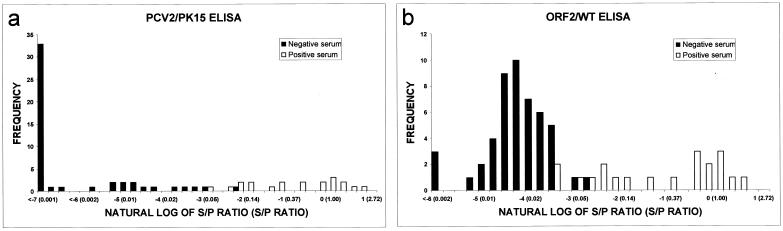
Sixty-nine samples from pigs experimentally infected with PCV2 or culture media were used to evaluate the feasibility of PCV2 and ORF2 ELISA as diagnostic tools. Twenty-five negative sera were obtained from nine pigs in the sham-inoculated group. An additional 24 IIF-negative serum samples were collected from 13 pigs prior to or 1 week postinfection with PCV2. Twenty IIF-positive serum samples were collected at 14 to 49 dpi from seven CD-CD pigs experimentally infected with PCV2. Both PCV2 and ORF2 ELISAs were able to distinguish the sera that were classified by IIF as negative, weakly positive, and strongly positive for PCV2 antibodies. The distribution of IIF-negative and -positive samples according to S/P ratios of pig experimental sera obtained from both ELISAs is shown in Fig. 1. S/P ratios of sera positive by IIF ranged from 0.0497 to 2.0529 for PCV2 ELISA and from 0.0282 to 1.4062 for ORF2 ELISA. S/P ratios of negative sera ranged from −0.0703 to 0.0938 for PCV2 ELISA and −0.0181 to 0.0627 for ORF2 ELISA.
FIG. 1.
Frequency plots of IIF-negative (49 samples from 22 pigs) and IIF-positive (20 samples from 7 pigs) serum samples by natural log of S/P ratios from PCV2 (a) and ORF2 (b) ELISAs show the distribution of pig experimental sera. The numbers in parentheses are S/P ratios.
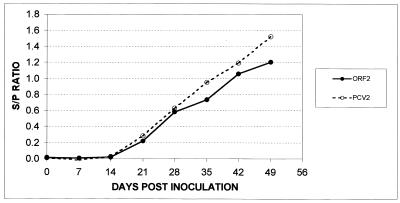
The mean S/P ratios of sera from the pigs surviving the entire duration of the experiment were plotted over time to determine kinetic curves of antibodies to PCV2 and the major capsid protein by PCV2 and ORF2 ELISA, respectively (Fig. 2). Kinetic plots created from both assays were similar. Antibodies to PCV2 and the capsid protein appeared clearly after 14 dpi. The graphs showed gradual increase in antibody levels to PCV2 over the 49 days after infection. S/P ratios of sera from most pigs experimentally infected with PCV2 were more than 0.2 by all assays at 21 dpi.
FIG. 2.
Kinetic curves of antibodies to PCV2 and its major capsid protein ORF2 plotted as means of S/P ratios over time. S/P ratios were calculated for PCV2 ELISA and ORF2 ELISA using sera from three pigs experimentally infected with PCV2 at different days postinfection. Graphs show early detection of antibodies from 14 to 21 days after infection and an increase in concentration of antibodies over time. Note that both assays differentiate among negative, weakly positive, and strongly positive sera as S/P ratios of sera at 49 dpi are higher than those for 21 dpi while those for 0 and 7 dpi were less than the negative cutoff value (0.04).
ELISA results on field sera.
We further validated these assays using field sera. A positive serum that consistently gave a high COD and a negative serum with low COD and background were selected from the experimental pigs and used as positive and negative serum controls, respectively. Usually, this positive control serum had CODs of approximately 0.4 and 0.45 for PCV2 and ORF2 ELISA, respectively.
(i) Evaluation of repeatability.
The repeatability test was done by comparing S/P ratios of each field serum sample tested in triplicate in intra-assay (within plate) and interassay (between run) in PCV2 and ORF2 ELISA. The results showed that both assays were repeatable. The intra-assay CV of 14 IIF positive serum samples in PCV2 ELISA ranged from 0.29 to 25.70%, with a median value of 6.30%, while those of ORF2 ELISA ranged from 2.39 to 13.68%, with a median value of 6.19%. The interassay CV for positive serum samples in PCV2 ELISA was between 1.76 and 14.47%, with a median value of 7.47%, whereas for ORF2 ELISA it was between 0.34 and 8.95%, with a median value of 5.46%. Both ELISAs also showed repeatability for negative field sera as indicated by low variability among each replicate of similar sample (Table 2).
TABLE 2.
Repeatability of PCV2 and ORF2 ELISAs for IIF-negative samples of field pig sera
| Repeatability | Mean S/P ratio ± SD
|
|
|---|---|---|
| PCV2 ELISA | ORF2 ELISA | |
| Intra-assay | 0.0292 ± 0.0041 | 0.0266 ± 0.0101 |
| 0.0393 ± 0.0043 | 0.0397 ± 0.0083 | |
| 0.0362 ± 0.0056 | 0.0718 ± 0.0098 | |
| 0.0298 ± 0.0101 | 0.0791 ± 0.0150 | |
| 0.0074 ± 0.0101 | 0.1097 ± 0.0011 | |
| 0.0504 ± 0.0101 | 0.0549 ± 0.0094 | |
| −0.0016 ± 0.0080 | 0.0333 ± 0.0105 | |
| Interassay | 0.0173 ± 0.0121 | 0.0107 ± 0.0030 |
| 0.0032 ± 0.0047 | 0.0463 ± 0.0047 | |
| 0.0237 ± 0.0051 | 0.0590 ± 0.0018 | |
| −0.0125 ± 0.0116 | 0.0506 ± 0.0059 | |
| −0.0119 ± 0.0184 | 0.0884 ± 0.0146 | |
| 0.0361 ± 0.0108 | 0.0606 ± 0.0030 | |
| −0.0150 ± 0.0077 | 0.0253 ± 0.0270 | |
(ii) PCV2 ELISA.
The results of the PCV2 ELISA using field sera are presented in Table 3. Minimum and maximum values of S/P ratios were −0.1933 and 0.2067 for IIF-negative sera, respectively, and −0.0704 and 2.9405 for IIF-positive sera, respectively. The modified receiver-operator characteristic analysis was used to determine the negative-positive cutoff value. The negative cutoff of 0.125 was selected because it resulted in values of more than 90% for both DSN and DSP. At this cutoff, the sum of false results was lowest, as there were 14 false positives and 49 false negatives. The data from performing the assay at a negative cutoff of 0.125 is summarized in Table 4. When the cutoff was moved to 0.2, the DSP of the assay increased to 99.6% while the DSN decreased to 78.2% compared to IIF. Accordingly, serum samples with S/P ratios equal to 0.125 or less were classified as negative, those with ratios of more than 0.125 to 0.2 were equivocal, and those with ratios of more than 0.2 were positive. A COD of positive serum control of at least 0.4 was also required for the assay to be valid. At the negative and positive cutoff values of 0.125 and 0.2, respectively, 429 samples were positive, 82 samples were equivocal and 272 samples were negative (Table 5).
TABLE 3.
Summary of S/P ratios of IIF-negative and -positive sera in PCV2 or ORF2 ELISAs
| Serum group | Parameter | S/P ratio
|
|
|---|---|---|---|
| PCV2 ELISA | ORF2 ELISA | ||
| IIF negative (n = 237) | Minimum | −0.1993 | −0.0852 |
| Maximum | 0.2067 | 0.2096 | |
| Mean | 0.0476 | 0.0564 | |
| SD | 0.0540 | 0.0411 | |
| Mean + 2SD | 0.1556 | 0.1385 | |
| Mean + 3SD | 0.2097 | 0.1795 | |
| IIF positive (n = 546) | Minimum | −0.0704 | 0.0335 |
| Maximum | 2.9405 | 2.6563 | |
| Mean | 0.5088 | 0.7977 | |
| SD | 0.4196 | 0.6323 | |
TABLE 4.
Performance of PCV2 and ORF2 ELISAs compared with IIFa
| ELISA | Cutoff (S/P ratio) | Diagnostic value
|
Predictive value
|
% Accuracy | ||
|---|---|---|---|---|---|---|
| % Sensitivity | % Specificity | % Positive | % Negative | |||
| PCV2 | 0.125 | 91.0 | 94.1 | 97.3 | 82.0 | 92.0 |
| ORF2 | 0.12 | 90.7 | 93.2 | 96.9 | 81.3 | 91.4 |
Comparison was made at cutoff values of 0.125 for PCV2 ELISA and of 0.12 for ORF2 ELISA.
TABLE 5.
Relative comparison between two ELISAs and IIF for detection of antibodies to PCV in field serum samples
| IIF result | No. of serum samples (%)
|
Total no. of serum samples | |||||
|---|---|---|---|---|---|---|---|
| PCV2 ELISAa
|
ORF2 ELISAb
|
||||||
| Positive | Equivocal | Negative | Positive | Equivocal | Negative | ||
| Positive | 427 (54.5) | 70 (8.9) | 49 (6.3) | 436 (55.7) | 59 (7.5) | 51 (6.5) | 546 |
| Negative | 2 (0.3) | 12 (1.5) | 223 (28.5) | 1 (0.1) | 15 (1.9) | 221 (28.2) | 237 |
| Total | 429 (54.8) | 82 (10.5) | 272 (34.7) | 437 (55.8) | 74 (9.5) | 272 (34.7) | 783 |
S/P ratio cutoffs: positive, >0.20; equivocal, >0.125 and ≤0.20; negative, ≤0.125.
S/P ratio cutoffs: positive, >0.20; equivocal, >0.12 and ≤0.20; negative, ≤0.12.
(iii) ORF2 ELISA.
The results of the ORF2 ELISA on IIF-negative and -positive sera are summarized in Table 3. The S/P ratios of samples varied from a minimum of −0.0852 to a maximum of 0.2096 for negative sera and from a minimum of 0.0335 to a maximum of 2.6563 for positive sera. The negative-positive cutoff value of 0.1 gave the least sum of false results compared to IIF, as out of 783 samples, 30 were false negative and 27 were false positive. This resulted in a DSN of 94.5% and a DSP of 88.6%. By increasing the negative-positive cutoff value to 0.12, the DSP increased to 93.2% while the DSN decreased to 90.7%. Consequently, total false results increased to 67 samples compared with IIF; however, both DSN and DSP were greater than 90%. The performance of the assay at the negative cutoff of 0.12 is shown in Table 4. When the cutoff was increased to 0.2, the DSP of the assay increased to 99.6% but the DSN decreased to 79.9%. Under these conditions, samples with S/P ratios equal to 0.12 or less were considered negative, those with ratios of more than 0.12 to 0.2 were considered equivocal, and those with ratios of more than 0.2 were considered positive. In addition, a COD of 0.45 or more for the positive serum control was also required to achieve the specified negative-positive cutoff. For the ORF2 ELISA, 437, 74, and 272 serum samples were considered positive, equivocal, and negative, respectively (Table 5). When the samples classified as equivocal by PCV2 (82 samples) and ORF2 (74 samples) ELISAs were analyzed, 34 sera were similar. Out of 34 samples, 33 samples were IIF positive and 1 sample was IIF negative.
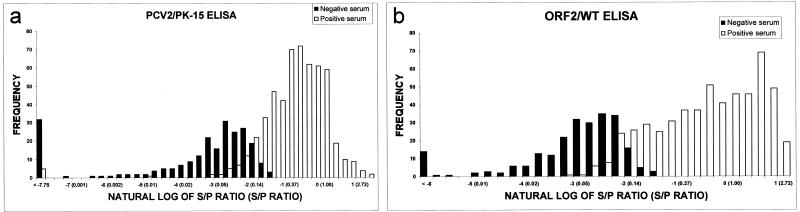
S/P ratios obtained from PCV2 and ORF2 ELISA were also plotted against each other. The results showed that their relationship was linear with high correlation (r2 of 0.7), and the regression equation was S/P ratio (ORF2) = 1.28 S/P ratio (PCV2) + 0.10. Agreement between PCV2 and ORF2 ELISAs was excellent, with a kappa value of 0.81 (P < 0.05). When the frequency of IIF-positive and -negative serum samples was plotted against the natural log of S/P ratios obtained from either PCV2 or ORF2 ELISA, two overlapping populations appeared as depicted in Fig. 3. The overlapping area represented the equivocal zone for each assay.
FIG. 3.
Frequency plots of number of IIF positive (n = 546) and negative (n = 237) samples of field sera and natural log of S/P ratios obtained from either PCV2 (a) or ORF2 (b) ELISA demonstrating two overlapping populations. The number in parentheses is the S/P ratio. The negative and positive cutoffs are 0.125 and 0.2 with a COD of positive serum control of at least 0.4 for the PCV2 ELISA and 0.12 and 0.2 with a COD of positive serum control of at least 0.45 for the ORF2 ELISA. The overlapping area represents an equivocal zone for each assay.
(iv) Specificity of ELISA.
The specificity of both assays was determined by testing the reactivity of PPV- and PRRSV-positive sera. Thirty-six serum samples positive for antibodies to PPV, with reciprocal hemagglutination inhibition titers ranging from 32 to 16,384, had S/P ratios ranging from −0.0063 to 0.0866, with a mean and SD of 0.0298 ± 0.0319 for PCV2 ELISA and from −0.0158 to 0.0971 with a mean and SD of 0.0308 ± 0.0243 for ORF2 ELISA. Thirty-one serum samples containing antibodies to PRRSV from CD-CD pigs experimentally inoculated with PRRSV were also tested for cross-reactivity by both ELISAs. Their S/P ratios were between −0.0629 and 0.0662 with a mean and SD of −0.0037 ± 0.0252 for PCV2 ELISA and between −0.0205 and 0.0250 with a mean and SD of 0.0057 ± 0.0093 for ORF2 ELISA. The results showed that neither ELISA cross-reacted with either PPV- or PRRSV-positive serum.
Correlation between IIF titer and S/P ratio.
The correlation between IIF titer and S/P ratio was determined by plotting endpoint IIF titers of 36 serum samples with different levels of antibodies to PCV2 against S/P ratios of the corresponding serum (Fig. 4). The results indicated that the linear relationships between log10 titer of IIF and S/P ratio obtained from both PCV2 ELISA (Spearman’s rank correlation = 0.62; P < 0.01) and ORF2 ELISA (Spearman’s rank correlation = 0.61; P < 0.01) were similar. There was lower correlation between the sera with low S/P ratios in both ELISA and IIF titers. Conclusively, S/P ratio obtained from either assay could be used to predict IIF titer.
FIG. 4.
Scatter plots of log10 IIF titers of 36 serum samples against S/P ratios of the corresponding serum obtained from PCV2 (•) or ORF2 (+) ELISA. The relationships between IIF and S/P ratios of either PCV2 (dashed line) or ORF2 (solid line) ELISAs are linear, with a Spearman’s rank correlation of 0.6.
DISCUSSION
We have successfully developed and validated two modified indirect ELISAs for the detection of antibodies to PCV2 and its major capsid protein. Both assays could be easily performed and all of the reagents except for the antigens were commercially available. Additionally, extensive purification steps were not necessary for antigen preparation. Every laboratory currently handling PCV2 could easily perform the PCV2 ELISA. However, infection of freshly seeded PK-15 cells with PCV2 at a high MOI (2 or more) was crucial for preparing high quality antigen. Inoculation at a lower MOI of PCV2 decreased the amount of viral protein produced while increasing cellular protein leading to a decrease in COD and sensitivity. Although ORF2 ELISA required the recombinant baculovirus carrying the ORF2 gene of PCV2, there were advantages of ORF2 ELISA over PCV2 ELISA. First, the production of a high-titer baculovirus, either wild type or recombinant, was much easier than the preparation of a high-titer PCV2. Secondly, the recombinant baculovirus produced a much greater amount of PCV2 capsid protein than PCV2 grown in culture. Finally, ORF2 ELISA required a much lower concentration of peroxidase-conjugated anti-swine immunoglobulin G compared with PCV2 ELISA (1:4,000 dilution for ORF2-based ELISA versus 1:1,000 dilution for PCV2-based ELISA), resulting in lower nonspecific background, and the reaction was less dependent on this amplification step.
This is the first published report documenting the existence of PCV2 infection in U.S. swine herds as early as 1987. Six of the 60 serum samples obtained from a herd in Wisconsin in 1987 were positive by all three assays, with IIF titers of more than 100 and S/P ratios ranging from 0.201 to 0.568 for PCV2 ELISA and 0.271 to 0.516 for ORF2 ELISA. This finding was in accordance with several previous reports. PCV2 antigen was detected by immunohistochemistry in pigs necropsied in 1989 in Japan (35). In addition, pooled serum samples from 12 herds in Belgium collected in 1985 (34) and serum samples obtained from sows at a slaughter house in Canada the same year (29) contained antibodies to PCV2.
The results from both ELISAs demonstrated that their DSN and DSP were comparable when IIF was used as the reference method. However, PCV2 and ORF2 ELISAs seemed to be slightly less sensitive than IIF since a considerable number of IIF-positive sera were classified as negative or equivocal by both ELISAs. However, evaluating a newly developed assay by comparison with a widely used assay is not an absolute standard of comparison (21, 44). The relative standard for comparison, in this case the IIF, possesses its own inherent level of sensitivity and specificity (21, 44). In addition, the discrepancy between IIF and ELISA results was possibly due to different epitope recognition. The source of antigen for IIF was fixed cells, while the ELISA antigens were soluble. As expected, both types of antigens contain shared and distinct epitopes which will be recognized by different antibodies.
We were not able to compare our two assays with the previously described cELISA based on monoclonal antibodies (48). Although all three ELISAs were compared against IIF as the reference method, the cutoff dilutions for IIF-positive sera were different. In our study, we defined sera with an IIF titer of 10 or more as positive, while for the cELISA, sera with IIF titers of 50 or more were considered positive.
We felt that it was critical to evaluate the specificity of the ELISAs against PPV and PRRSV as these infections are common in swine and natural coinfections of PCV2 with either PPV or PRRSV have been reported (11, 13, 45). Moreover, experimental coinfection of young pigs with PCV2 and PPV produced clinical signs resembling those observed in field cases of PMWS (3, 12, 22, 24). Our results confirmed that neither PCV2 nor ORF2 ELISA reacted with antibodies to PPV or PRRSV. In addition, we intended to test the specificity of our assays to detect antibodies to PCV1, if PCV1 positive serum was available. We expected that the PCV2 ELISA would detect antibodies to PCV1 and PCV2 since reactivity between PCV1 antibodies and PCV2 antigens has been reported (2, 4). In contrast, we expected that ORF2 ELISA would differentiate between antibodies to PCV1 and PCV2 since antibodies to PCV2 recognized the ORF2 gene product of PCV2 but not that of PCV1 (30). However, at least one shared epitope between ORF2 of PCV1 and PCV2 has been reported (30).
Recently, PCV2 infection in the pig population has been found to be more common than PCV1 infection (13, 26, 27, 29, 40). By PCR, the number of Canadian pigs coinfected with PCV1 and PCV2 was less than the number of those infected with PCV2 alone (26, 27, 40). Thus far, only PCV2 has been detected by in situ hybridization in tissues of pigs with PMWS in the Midwestern United States (S. D. Sorden, unpublished data). Previously, Hines et al. (19) found that sera from 13 swine herds in the United States contained antibodies to the PK-15 contaminant PCV. However, this should be interpreted carefully as antibodies to PCV2 cross-react with PCV1 and titers as high as 5,120 have been detected when PCV1-infected PK-15 cells were used in IPMA (41). Additionally, a strong association between antibody titers to PCV2 and those to PCV1 of identical serum samples suggested that the reactivity to PCV1 was possibly due to cross-reaction with antibodies to PCV2 infection (41). The specificity of assays to detect antibodies to PCV2 versus antibodies to PCV1 may be less important in geographical areas where PCV1 is not present. In contrast, further testing for reactivity with PCV1 is necessary in regions where PCV1 infections are common.
The ELISA procedures reported here are relatively simple and should be applicable worldwide since homology among PCV2 isolates from North America, Europe, and Asia is reported to be 95 to 99% (14). Both assays have been well standardized and validated and thus provide valuable tools for the detection of antibodies to PCV. Due to the semiquantitative nature of ELISA, the application of both assays is not limited to diagnosis of prior infection or the estimation of prevalence in a target population. In addition, both ELISAs could be used for determining the pattern of decaying maternal antibody to PCV2 and the minimum level of antibody for protection against PCV2 infection. Further refinement of both assays to differentiate the immune status between healthy PCV2 antibody positive animals and those with progressive PMWS should be pursued.
Acknowledgments
This study was funded in part with a grant from the Iowa State University Research Foundation and a Thai scholarship for Porntippa Nawagitgul.
We greatly thank Theresa F. Young and Marsha K. Morgan of Veterinary Medical Research Institute at Iowa State University for technical assistance.
REFERENCES
- 1.Allan, G. M., F. McNeilly, J. P. Cassidy, G. A. C. Reilly, B. Adair, W. A. Ellis, and M. S. McNulty. 1995. Pathogenesis of porcine circovirus; experimental infections of colostrum deprived piglets and examination of pig foetal material. Vet. Microbiol. 44:49–64. [DOI] [PubMed] [Google Scholar]
- 2.Allan, G. M., F. McNeilly, S. Kennedy, B. Daft, E. G. Clarke, J. A. Ellis, D. M. Haines, B. M. Meehan, and B. M. Adair. 1998. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J. Vet. Diagn. Investig. 10:3–10. [DOI] [PubMed] [Google Scholar]
- 3.Allan, G. M., S. Kennedy, F. McNeilly, J. C. Foster, J. A. Ellis, S. J. Krakowka, B. M. Meehan, and B. M. Adair. 1999. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J. Comp. Pathol. 121:1–11. [DOI] [PubMed] [Google Scholar]
- 4.Allan, G. M., F. Mc Neilly, B. M. Meehan, S. Kennedy, D. P. Mackie, J. A. Ellis, E. G. Clark, E. Espuna, N. Saubi, P. Riera, A. Botner, and C. E. Charreyre. 1999. Isolation and characterisation of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland. Vet. Microbiol. 66:115–123. [DOI] [PubMed] [Google Scholar]
- 5.Bereiter, M., T. F. Young, H. S. Joo, and R. F. Ross. 1990. Evaluation of the ELISA and comaprison to the complement fixation test and radial immunodiffusion enzyme assay for detection of antibodies against M. hyopneumoniae in swine serum. Vet. Microbiol. 25:117–192. [DOI] [PubMed] [Google Scholar]
- 6.Chiang, S., A. M. Dar, S. M. Goyal, M. A. Sheikh, J. C. Pedersen, B. Panigraphy, D. Senne, D. A. Halvorson, K. V. Nagaraja, and V. Kapur. 2000. A modified enzyme-linked immunosorbent assay for the detection of avian pneumovirus antibodies. J. Vet. Diagn. Investig. 12:381–384. [DOI] [PubMed] [Google Scholar]
- 7.Choi, C., C. Chae, and E. G. Clark. 2000. Porcine postweaning multisystemic wasting syndrome in Korean pig: detection of porcine circovirus 2 infection by immunohistochemistry and polymerase chain reaction. J. Vet. Diagn. Investig. 12:151–153. [DOI] [PubMed] [Google Scholar]
- 8.Clark, E. G. 1997. Post-weaning multisystemic wasting syndrome. Proc. Am. Assoc. Swine Pract., 27th Annu. Meet., p.499–501.
- 9.Daft, B., R. W. Nordhausen, K. S. Latimer, and F. D. Niagro. 1996. Interstitial pneumonia and lymphadenopathy associated with circoviral infection in a six-week-old pig. Proc. Am. Assoc. Vet. Lab. Diagn. 39:32. [Google Scholar]
- 10.Ellis, J., L. Hassard, E. Clark, J. Harding, G. Allan, P. Willson, J. Strokappe, K. Martin, F. McNeilly, B. Meehan, D. Todd, and D. Haines. 1998. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can. Vet. J. 39:44–51. [PMC free article] [PubMed] [Google Scholar]
- 11.Ellis, J., S. Krakowka, G. Allan, E. Clark, and S. Kennedy. 1999. The clinical scope of porcine reproductive and respiratory syndrome virus infection has expanded since 1987: an alternative perspective. Vet. Pathol. 36:262–265. [DOI] [PubMed] [Google Scholar]
- 12.Ellis, J., S. Krakowka, M. Lairmore, D. Haines, A. Bratanich, E. Clark, G. Allan, C. Konoby, L. Hassard, B. Meehan, K. Martin, J. Harding, S. Kennedy, and F. McNeilly. 1999. Reproduction of lesions of postweaning multisystemic wasting syndrome in gnotobiotic piglets. J. Vet. Diagn. Investig. 11:3–14. [DOI] [PubMed] [Google Scholar]
- 13.Ellis, J. A., A. Bratanich, E. G. Clark, G. Allan, B. Meehan, D. M. Haines, J. Harding, K. H. West, S. Krakowka, C. Konoby, L. Hassard, K. Martin, and F. McNeilly. 2000. Coinfection by porcine circoviruses and porcine parvovirus in pigs with naturally acquired postweaning multisystemic wasting syndrome. J. Vet. Diagn. Investig. 12:21–27. [DOI] [PubMed] [Google Scholar]
- 14.Fenaux, M., P. G. Halbur, M. Gill, T. E. Toth, and X. J. Meng. 2000. Genetic characterization of type 2 porcine circovirus (PCV-2) from pigs with postweaning multisystemic wasting syndrome in different geographic regions of North America and development of a differential PCR-restriction fragment length polymorphism assay to detect and differentiate between infections with PCV-1 and PCV-2. J. Clin. Microbiol. 38:2494–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamel, A. L., L. L. Lin, and G. P. S. Nayar. 1998. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J. Virol. 72:5262–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harding, J. C. 1997. Post-weaning multisystemic wasting syndrome (PMWS): preliminary epidemiology and clinical presentation. Proc. Am. Assoc. Swine. Pract., 27th Annu. Meet., p.503.
- 17.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual, p.446–451. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Harms, P. A., S. D. Sorden, P. G. Halbur, S. Bolin, K. Lager, I. Morozov, and P. S. Paul. 2001. Experimental reproduction of severe disease in CD/CD pigs concurrently infected with type 2 porcine circovirus and porcine reproductive and respiratory syndrome virus. Vet. Pathol. 38:528–539. [DOI] [PubMed] [Google Scholar]
- 19.Hines, R. K., and P. D. Lukert. 1995. Porcine circovirus: a serological survey of swine in the United States. Swine Health Prod. 3:71–73. [Google Scholar]
- 20.Illanes, O., A. Lopez, L. Miller, J. McLearon, C. Yason, D. Wadowska, and J. Martinez. 2000. Lesions associated with postweaning multisystemic wasting syndrome in pigs from Prince Edward Island, Canada. J. Vet. Diagn. Investig. 12:146–150. [DOI] [PubMed] [Google Scholar]
- 21.Jacobson, R. H. 1998. Validation of serological assays for diagnosis of infectious disease. Rev. Sci. Tech. Off. Int. Epizoot. 17:469–486. [DOI] [PubMed] [Google Scholar]
- 22.Kennedy, S., D. Moffett, F. McNeilly, B. Meehan, J. Ellis, S. Krakowka, and G. M. Allan. 2000. Reproduction of lesions of postweaning multisystemic wasting syndrome by infection of conventional pigs with porcine circovirus type 2 alone or in combination with porcine parvovirus. J. Comp. Pathol. 122:9–24. [DOI] [PubMed] [Google Scholar]
- 23.Kiupel, M., G. W. Stevenson, S. K. Mittal, E. G. Clark, and D. M. Haines. 1998. Circovirus-like viral associated disease in weaned pigs in Indiana. Vet. Pathol. 35:303–307. [DOI] [PubMed] [Google Scholar]
- 24.Krakowka, S., J. A. Ellis, B. Meehan, S. Kennedy, F. McNeilly, and G. Allan. 2000. Viral wasting syndrome of swine: experimental reproduction of postweaning multisystemic wasting syndrome in gnotobiotic swine by coinfection with porcine circovirus 2 and porcine parvovirus. Vet. Pathol. 37:254–263. [DOI] [PubMed] [Google Scholar]
- 25.Krakowka, S., J. A. Ellis, F. McNeilly, S. Ringler, D. M. Rings, and G. Allan. 2001. Activation of the immune system is the pivotal event in the production of wasting disease in pigs infected with porcine circovirus-2 (PCV-2). Vet. Pathol. 38:31–42. [DOI] [PubMed] [Google Scholar]
- 26.Larochelle, R., M. Antaya, M. Morin, and R. Magar. 1999. Typing of porcine circovirus in clinical specimens by multiplex PCR. J. Virol. Methods 80:69–75. [DOI] [PubMed] [Google Scholar]
- 27.Liu, Q., L. Wang, P. Willson, and L. A. Babiuk. 2000. Quantitative, competitive PCR analysis of porcine circovirus DNA in serum from pigs with postweaning multisystemic wasting syndrome. J. Clin. Microbiol. 38:3474–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukert, P., G. F. de Boer, J. L. Dale, P. Keese, M. S. McNulty, J. W. Randles, and I. Tischer. 1995. The Circoviridae, p.166–168. In F. A. Murphy, C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. P. Martelli, M. A. Mayo, and M. D. Summers (ed.), Virus taxonomy sixth report of the International Committee on Taxonomy of viruses. Springer-Verlag, Vienna, Austria.
- 29.Magar, R., P. Muller, and R. Larochelle. 2000. Retrospective serological survey of antibodies to porcine circovirus type 1 and type 2. Can. J. Vet. Res. 64:184–186. [PMC free article] [PubMed] [Google Scholar]
- 30.Mahe, D., P. Blanchard, C. Truong, C. Arnauld, P. Le Cann, R. Cariolet, F. Madec, E. Albina, and A. Jestin. 2000. Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J. Gen. Virol. 81:1815–1824. [DOI] [PubMed] [Google Scholar]
- 31.Mankertz, A., J. Mankertz, K. Wolf, and H. Buhk. 1998. Identification of protein essential for replication of porcine circovirus. J. Gen. Virol. 79:381–384. [DOI] [PubMed] [Google Scholar]
- 32.Meehan, B. M., J. L. Creelan, M. S. McNulty, and D. Todd. 1997. Sequence of porcine circovirus DNA: affinities with plant circoviruses. J. Gen. Virol. 78:221–227. [DOI] [PubMed] [Google Scholar]
- 33.Meehan, B. M., F. McNeilly, D. Todd, S. Kennedy, V. A. Jewhurst, J. A. Ellis, L. E. Hassard, E. G. Clark, D. M. Haines, and G. M. Allan. 1998. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J. Gen. Virol. 79:2171–2179. [DOI] [PubMed] [Google Scholar]
- 34.Mesu, A. P., G. G. Labarque, H. J. Nauwynck, and M. B. Pensaert. 2000. Seroprevalence of porcine circovirus types 1 and 2 in the Belgian pig population. Vet. Q. 22:234–236. [DOI] [PubMed] [Google Scholar]
- 35.Mori, M., K. Sato, S. Akachi, S. Asahi, S. Taniguchi, and M. Narita. 2000. Retrospective study of porcine circovirus 2 infection in Japan: seven cases in 1989. Vet. Pathol. 37:667–669. [DOI] [PubMed] [Google Scholar]
- 36.Morozov, I., T. Sirinarumitr, S. D. Sorden, P. G. Halbur, M. K. Morgan, K. J. Yoon, and P. S. Paul. 1998. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J. Clin. Microbiol. 36:2535–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nawagitgul, P., I. Morozov, S. R. Bolin, P. A. Harms, S. D. Sorden, and P. S. Paul. 2000. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 81:2281–2287. [DOI] [PubMed] [Google Scholar]
- 38.Nayar, G. P. S., A. Hamel, and L. Lin. 1997. Detection and characterization of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. Can. Vet. J. 38:385–386. [PMC free article] [PubMed] [Google Scholar]
- 39.Onuki, A., K. Abe, K. Togashi, K. Kawashima, A. Taneichi, and H. Tsunemitsu. 1999. Detection of porcine circovirus from lesions of a pig with wasting disease in Japan. J. Vet. Med. Sci. 61:1119–1123. [DOI] [PubMed] [Google Scholar]
- 40.Quardani, M., L. Wilson, R. Jette, C. Montpetit, and S. Dea. 1999. Multiplex PCR for detection and typing of porcine circoviruses. J. Clin. Microbiol. 37:3917–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez-Arrioja, G. M., J. Segales, M. Balasch, C. Rosell, J. Quintana, J. M. Folch, J. Plana-Duran, A. Mankertz, and M. Domingo. 2000. Serum antibodies to porcine circovirus type 1 and type 2 in pigs with and without PMWS. Vet. Rec. 146:762–764. [DOI] [PubMed] [Google Scholar]
- 42.Sato, K., T. Shibahara, Y. Ishikawa, H. Kondo, M. Kubo, and K. Kadota. 2000. Evidence of porcine circovirus infection in pigs with wasting disease syndrome from 1985 to 1999 in Hokkaido, Japan. J. Vet. Med. Sci. 62:627–633. [DOI] [PubMed] [Google Scholar]
- 43.Segales, J., M. Sitjar, M. Domingo, S. Dee, M. Del Pozo, R. Noval, C. Sacristan, A. De las Heras, A. Ferro, and K. S. Latimer. 1997. First report of post-weaning multisystemic wasting syndrome in pigs in Spain. Vet. Rec. 141:600–601. [PubMed] [Google Scholar]
- 44.Smith, R. D. 1995. Veterinary clinical epidemiology: a problem-oriented approach, 2nd ed., p.31–52. CRC Press Inc., Boca Raton, Fla.
- 45.Sorden, S. D. 2000. Update on porcine circovirus and postweaning multisystemic wasting syndrome (PMWS). Swine Health Prod. 8:133–136. [Google Scholar]
- 46.Tischer, I., H. Gelderblom, W. Vettermann, and M. A. Koch. 1982. A very small porcine virus with circular single-stranded DNA. Nature 295:64–66. [DOI] [PubMed] [Google Scholar]
- 47.Tischer, I., W. Meilds, D. Wolff, M. Vagt, and W. Griem. 1986. Studies on epidemiology and pathogenicity of porcine circovirus. Arch. Virol. 91:271–276. [DOI] [PubMed] [Google Scholar]
- 48.Walker, I. W., C. A. Konoby, V. A. Jewhurst, I. McNair, F. McNeilly, B. M. Meehan, T. S. Cottrell, J. A. Ellis, and G. M. Allan. 2000. Development and application of a competitive enzyme-linked immunosorbent assay for the detection of serum antibodies to porcine circovirus type 2. J. Vet. Diagn. Investig. 12:400–405. [DOI] [PubMed] [Google Scholar]
- 49.Wellenberg, G. J., S. Pesch, F. W. Berndsen, P. J. G. M. Steverink, W. Hunneman, T. J. K. Van der Vorst, N. H. M. T. Peperkamp, V. F. Ohlinger, R. Schippers, J. T. Van Oirschot, and M. F. de Jong. 2000. Isolation and characterization of porcine circovirus type 2 from pigs showing signs of post-weaning multisystemic wasting syndrome in the Netherlands. Vet. Q. 22:167–172. [DOI] [PubMed] [Google Scholar]