Abstract
In individuals infected with human immunodeficiency virus type 1 (HIV-1), specific immunity is associated with a more diverse viral repertoire and slower disease progression. Attempts to enhance antiviral immunity with therapeutic vaccination have shown that recombinant glycoprotein (RGP) vaccines are safe, well tolerated, and immunogenic, but the effect of RGP vaccines on the viral repertoire is unknown. We evaluated diversification of the viral envelope in 12 HIV-infected children who received placebo or RGP vaccines. At baseline, 11 of 12 patients had multiple viral variants. On follow-up 6 months later, children who had a strong vaccine-associated lymphoproliferative immune response showed less viral diversification than those in whom the immune response was weak or absent. These results suggest that the immune response elicited by RGP vaccines does not exert a significant selection pressure on the viral quasispecies and therefore may not be helpful in changing the course of the disease.
Infection with human immunodeficiency virus type 1 (HIV-1) leads to eventual immune attrition and the onset of AIDS. The rate of disease progression, however, is variable and appears to be determined, in part, by the nature of the immune response against the virus. Long-term slow progressors, for example, typically exhibit a broad range of antiviral responses, particularly cell-mediated immunity. Such individuals also harbor diverse viral quasispecies, and several recent reports have shown that this high degree of viral diversity is a direct consequence of immune selection pressure. (4, 8, 14)
Attempts to boost specific immunity with HIV-1 recombinant glycoprotein (RGP) vaccines have resulted in some HIV- infected individuals developing augmented humoral and cellular immune responses. (7, 12, 15) Whether these vaccine- associated immune responses correlate with any change in the repertoire of viral quasispecies remains unknown.
The objectives of the current study were to characterize quasispecies diversity at baseline in a group of 12 asymptomatic HIV-1-infected children who received either placebo or RGP vaccines and to determine whether these vaccines generate an added immunologic selection pressure capable of driving viral evolution. The vaccines studied were all recombinant envelope proteins; therefore, we assayed changes in the diversity of the viral envelope—the region of the genome most likely to be affected by a vaccine-associated immune response.
The number of patients evaluated was small; however, our results suggest that RGP vaccination in these children did not drive viral evolution. Indeed, there seemed to be an inverse relationship between vaccine-associated immune response and the degree of viral diversity. Although the study sample was small, these findings are provocative, and we believe that the results merit further study in a larger cohort of patients. These data highlight the importance of measuring viral diversity to evaluate the efficacy of therapeutic HIV vaccination.
MATERIALS AND METHODS
Patients and vaccines.
AIDS Clinical Trials group (ACTG) protocol 218 was a multicenter placebo-controlled phase 1 trial to evaluate the safety and immunogenicity of HIV-1 RGPs in children >1 month old with asymptomatic HIV infection (9). At one of the ACTG sites, Children’s Memorial Hospital in Chicago, 12 children aged 8 to 141 months were enrolled into the study. All were clinically stable, with median CD4 percentages of 34.5% (range, 27 to 38%). Nine children were randomized to receive one of three vaccines: MicroGeneSys HIV-1 IIIB rgp160, Genentech HIV-1 MN rgp120, or Chiron/Biocine HIV-1 SF2 rgp120. Three children received placebo.
Patients were immunized at entry and weeks 4, 8, 12, 16, and 24. Informed consent was obtained according to the guidelines of the Institutional Review Board of Northwestern University Medical School, Chicago, Ill. Following entry into the study, patients were evaluated at monthly intervals, at which times CD4 cell percentages and vaccine-associated lymphoproliferative responses were measured prospectively. Plasma HIV-1 RNA was not evaluated as part of this protocol.
Vaccine-specific lymphoproliferative responses.
To evaluate vaccine immunogenicity, each patient had serial lymphoproliferative studies done with recombinant antigens autologous to those present in the administered vaccines. Each assay was conducted in triplicate as previously described (1), and the result was compared with a control sample to which no antigen had been added to yield the stimulation index (SI), a measure of autologous vaccine antigen-specific lymphoproliferative response. A stimulation index of 3 or greater was considered a significant lymphoproliferative response. The best lymphoproliferative response recorded for the 6-month duration of the study was correlated with the results of the heteroduplex assays.
Heteroduplex assays.
For the analysis of changes in envelope diversity, we examined samples at baseline and at 6 months, just after the final dose of study vaccine. Frozen plasma was not available, and the heteroduplex studies were conducted on stored samples of peripheral blood mononuclear cells (PBMCs). Although the plasma RNA fraction reflects changes in the viral population more rapidly than integrated proviral DNA, Liu et al. have shown that new variants appear in the proviral compartment with a relatively short lag time of 6 weeks, considerably shorter than the 6-month time frame of the study period (10).
Nested HIV-1 DNA PCR was performed on lysate derived from 105 PBMCs, and a 653-bp-long segment of the viral envelope was amplified by PCR as previously described (4) using primers spanning the V3-V5 region (kindly given by David Ho, Aaron Diamond AIDS Research Center, New York). The PCR was calibrated using known titers of pNL4-3 plasmid DNA and found to reliably detect as few as 10 copies of viral template. In order to ensure that enough proviruses were amplified to be representative of the quasispecies repertoire in each sample, proviral load was semiquantified by limiting dilutions. All PBMC samples contained between 50 and 200 viral copies per 105 cells.
The heteroduplex tracking assay (HTA) was modified from Delwart et al. (5). In order to generate a probe for the assays, the downstream inner primer from the second PCR was end-radiolabeled in a phosphorylation reaction using γ-32-labeled ATP and T4 polynucleotide kinase (10 U/μl; Boehringer, Mannheim, Germany) (6). This radioactive primer was then used on a stock of pNL4-3 plasmid in order to amplify a 653-bp-long clonal probe in which only one of the DNA strands was radioactive. Using a single-stranded clonal probe in the HTA ensures that each distinct HTA band can be correlated with a distinct proviral variant.
For the heteroduplex hybridization, probe was mixed with amplified material from each of the specimens in a ratio of 1:120. The mixture was heated to 94°C for 3 min to denature all the DNA present and then cooled to 22°C for 7 min to permit reannealing and the formation of hybrids between the radioactive probe strand and complementary specimen DNA strands. The hybridized material was placed on ice before being loaded into a 5% polyacrylamide gel (acrylamide:bisacrylamide ratio = 30:0.8). An electrophoretic gradient of 230 V was applied for 3 h, after which time the gel was fixed, dried, and imaged using a PhosporImager (Amersham Pharmacia Biotech Inc., Piscataway, N.J.).
The degree of genetic relatedness between probe and specimen is proportional to the electrophoretic mobility of the probe-driver DNA hybrid (5). If the two are identical, a homoduplex forms, which migrates most rapidly. Unhybridized probe forms a single strand which migrates as the slowest band, and heteroduplexes (nonidentical probe-driver hybrids) migrate at an intermediate rate. HTA has been shown to correlate well with measures of genetic relatedness determined by nucleotide sequencing (2). The technique has also been able to detect sequence changes of as little as 1 to 2% within the DNA fragment under investigation (5). Furthermore, assuming that PCR amplifies different variants equally well, the relative intensity of each heteroduplex band is a function of the relative proportions of each variant present in the unamplified sample.
Data analysis.
Gel images were edited and imaged using the graphics program Adobe Photoshop 3.0 (Adobe Systems Inc., San Jose, Calif.). In order to quantify the degree of diversification between the first and second specimen in each pair, a diversity scoring system based on visual inspection of the gels was devised. Patients scored one point for a new variant, one point for loss of an existing variant, and half a point for a change in band intensity (indicating a shift in predominance from one variant to another.) The higher the total score, the greater the degree of diversification.
Statistical analysis.
Comparison of mean diversity scores in patients with strong and weak lymphoproliferative responses was performed using the paired two-tailed t test.
RESULTS
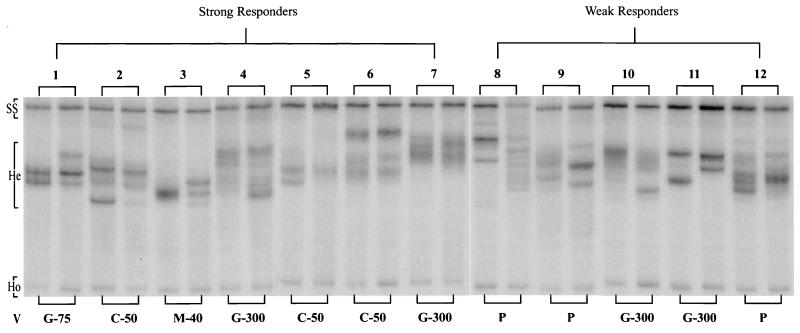
Heteroduplex tracking assays for the 12 patients are shown in Fig. 1. Only the lower half of each gel lane (from the position of the single-stranded probe to the position of the rapidly migrating homoduplex) is shown. Each pair of lanes (baseline and 6 months) represent a single patient. Three patients (numbers 8, 9, and 12) received placebo. The remaining nine patients were immunized with one of the study vaccines as noted (Fig. 1 and Table 1).
FIG. 1.
Paired HTAs are shown for each patient, representing the baseline and 6-month specimens. For convenience, only the lower half of the gel, from the single-stranded probe (SS) to the faint, rapidly migrating homoduplex (Ho), is shown. Heteroduplexes (He) migrate to a position between these two points. The vaccine (V) administered in each case is given underneath each pair of lanes. P, placebo; C-50, Chiron/Biocine HIV-1 SF2 rgp120 at a dose of 50 μg; G-75 and G-300, Genentech HIV-1 MN rgp120 at 75 and 300 μg, respectively; M-40, MicroGeneSys HIV-1 IIIB rgp160 at 40 μg. Patients 1 to 7 had strong lymphoproliferative responses to autologous vaccine antigens (SI > 3), whereas patients 8 to 12 had weak responses (SI < 3).
TABLE 1.
Patient details
| Patient no. | Age (mo) | Vaccine and dose (μg) | Best LPRa obtained/study wk | Diversity score | % CD4 cells
|
|
|---|---|---|---|---|---|---|
| Entry | Wk 24 | |||||
| 1 | 22 | Genentech (75) | 3.3/24 | 1.0 | 34 | 30 |
| 2 | 57 | Chiron (50) | 19.5/16 | 1.5 | 21 | 22 |
| 3 | 8 | MicroGeneSys (40) | 16.8/16 | 2.5 | 41 | 26 |
| 4 | 53 | Genentech (300) | 20.3/16 | 1.5 | 33 | 32 |
| 5 | 31 | Chiron (50) | 11.3/16 | 1.0 | 33 | 27 |
| 6 | 48 | Chiron (50) | 23.6/20 | 0 | 22 | 16 |
| 7 | 9 | Genentech (300) | 5.9/16 | 0 | 55 | 43 |
| 8 | 120 | Placebo | 0.9/16 | 6.5 | 36 | 43 |
| 9 | 16 | Placebo | 1.1/16 | 5.0 | 42 | 38 |
| 10 | 19 | Genentech (300) | 1.5/24 | 2.5 | 31 | 34 |
| 11 | 141 | Genentech (300) | 1.3/16 | 2.5 | 44 | 31 |
| 12 | 37 | Placebo | 1.6/16 | 2.5 | 36 | 40 |
LPR, lymphoproliferative response.
Multiple heteroduplex bands were present at baseline in 11 of the 12 children. The only exception was patient number 3, for whom the baseline specimen had a single broad band. The use of a laboratory strain of HIV-1 as the probe (in lieu of an autologous patient strain) reduces the sensitivity of HTA to detect closely related variants. Therefore, the broad band seen for patient 3 may represent a number of variants which migrate at very similar rates and cannot be appreciated as distinct bands. Most children had a major variant (seen as an intense band), with one or more weaker bands signifying minor variants. Some children (patients 1, 2, 5, and 11) had codominant variants, with two or more bands of equal intensity.
At 6 months, various patterns of change in the viral repertoire were noted. These included loss of an existing variant, appearance of a new variant, and shift in the predominance of one variant to another. The degree of change was quantified for each patient using a diversity scoring system (see above). Diversity scores for each patient are given in the table, which also shows age at enrollment, best lymphoproliferative response, timing of best lymphoproliferative response relative to the start of the study, and CD4 cell percentages at entry and week 24. As expected, patients who received placebo had a negative lymphoproliferative response. However, two of the vaccinees (numbers 10 and 11) also failed to make a significant response.
To further analyze these results, patients were grouped according to the magnitude of the lymphoproliferative response into strong responders (group 1, patients 1 to 7, SI > 3) and weak responders (group 2, patients 8 to 12, SI < 3). The groups were well matched in terms of baseline CD4 cell percentage (34.1% and 37.8%, respectively). However, over the 6-month period of study, CD4 percentages fell by 6.1% in the strong responders and rose by 0.6% in the weak responders. This difference was not statistically significant, but when the two groups were compared in terms of viral diversification, strong responders had a significantly lower mean diversity score than weak responders (1.1 and 3.8, respectively; P = 0.03).
DISCUSSION
Previous studies have documented continuous emergence of HIV-1 variants in individuals with chronic HIV infectio. (3, 6). In keeping with these findings, we noted that most of our asymptomatic HIV-infected children had multiple env variants at baseline. In addition, over the 6 months of follow-up, most patients showed evidence of viral diversification by HTA analysis.
In an attempt to test the hypothesis that HIV vaccines may drive viral evolution in vivo by enhancing immunity and thereby increasing selection pressure, we correlated the degree of envelope diversification with the magnitude of the vaccine-associated lymphoproliferative response. There are several methods of measuring the degree of diversity in HTAs (3, 6). The most widely accepted uses an entropy analysis to determine how ordered the distribution of signal intensity is along the length of a gel lane (3). As the quasispecies become more diverse, the signal becomes more diffusely distributed and entropy approaches maximum. This technique is elegant in principle but has some practical problems. In many cases, entropy analysis does not yield interpretable results until the pattern of signal intensity is enhanced, either by removing an arbitrarily determined amount of “background” or by smoothing the profile along a Gaussian distribution. This is particularly true if the HTA technique yields a dark image.
More recently, authors who have studied viral evolution using HTA have eschewed entropy analysis and opted instead for a technique based on visual inspection of the gels to determine changes in viral diversity (11). In this analysis we also used visual inspection and devised a system to measure the degree of diversification in each patient. Although the RGP vaccines used in this study were able to elicit measurable vaccine-specific cellular immunity in seven of the nine vaccinees, this response did not appear to be associated with greater viral diversification. In fact, when the seven patients who had strong responses were compared with the five patients who had weak responses, there was a significant trend towards less diversification in the responder group. In addition, strong responders had a mean decrease in the CD4 cell percentage of 6.1%, whereas the CD4 percentage increased by 0.6% in the weak responders. Although this difference was not statistically significant, it is noteworthy that the same phenomenon was observed in the entire cohort of ACTG 218 patients (9).
Immunization with both T-cell-dependent and T-cell-independent vaccines is known to cause a transient upregulation of HIV-1 through T-cell activation and tumor necrosis factor alpha (TNF-α) production by activated cells (13). Plasma viral RNA levels were not assayed as part of the ACTG 218 protocol, and stored plasma was not available for retrospective analysis. Therefore, it is not possible to definitively determine whether this was the cause of the reduced diversification or indeed the fall in CD4 count in the strong responder group.
A diverse viral repertoire is associated with potent immunity and good clinical outcome, while a static quasispecies is a marker for rapid disease progression. (3, 14) The unexpected inverse association between viral diversification and lymphoproliferative response raises the possibility that these RGP vaccines may not have been able to generate an added selection pressure and may indeed have had a deleterious impact on antiviral effector mechanisms. This is in keeping with the overall results of ACTG 218, which showed no clinical benefit to vaccine recipients However, it is difficult to extrapolate these findings beyond this small cohort of patients. ACTG 218 was conducted before the advent of highly active antiretroviral therapy (HAART), and it is possible that while RGP vaccines do not augment antiviral responses in untreated patients, they may yet prove effective in patients with low viral loads on HAART (F. Valentine and V. DeGruttola, Abstr. 6th Conf. Retroviruses Opportunistic Infections 1999, p. 46, abstr. 346, 1999).
These preliminary findings underscore the importance of further study to investigate changes in viral repertoire as a measure of efficacy and clinical benefit in studies of therapeutic vaccination for HIV-infected individuals.
Acknowledgments
We acknowledge the assistance of John Lambert and Terry Fenton for providing access to data from the results of ACTG 218 and the support of the General Clinical Research Center at New York University Medical Center.
This work was supported by the General Clinical Research Center, New York University Medical Center (NIH NCRR M01 RR00096), the New York Community Trust, the Shubert Foundation, and the National Institutes of Health (NIH 1 R01 HD3433601). S. M. Essajee is the recipient of a Research Fellowship Award supported by Abbott Laboratories from the Pediatric Infectious Diseases Society.
REFERENCES
- 1.Borkowsky, W., C. Steele, S. Grubman, T. Moore, P. La Russa, and K. Krasinski. 1987. Antibody responses to bacterial toxoids in children infected with human immunodeficiency virus. J. Pediatr 110:563–566. [DOI] [PubMed] [Google Scholar]
- 2.Contag, C. H., A. Ehrnst, J. Duda, A. Bohlin, S. Lindgren, and J. Mullins. 1997. Mother-to-infant transmission of human immunodeficiency virus type 1 involving five envelope sequence subtypes. J. Virol. 71:1292–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delwart, E. L., H. Pan, H. Sheppard, D. Wolpert, A. Neumann, B. Korber, and J. Mullins. 1997. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J. Virol. 71:7498–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delwart, E. L., H. Sheppard, B. Walker, J. Goudsmit, and J. Mullins. 1994. HIV-1 evolution in vivo tracked by DNA heteroduplex mobility assays. J. Virol. 68:6672–6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delwart, E. L., E. Shpaer, J. Louwagie, F. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. Mullins. 1993. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science 262:1257–1261. [DOI] [PubMed] [Google Scholar]
- 6.Essajee, S. M., H. Pollack, G. Rochford, I. Oransky, K. Krasinski, and W. Borkowsky. 2000. Early changes in quasispecies repertoire in HIV-infected infants: correlation with disease progression. AIDS Res. Hum. Retrovir. 16:1945–1953. [DOI] [PubMed] [Google Scholar]
- 7.Fast, P. E., and M. Walker. 1993. Human trials of experimental AIDS vaccines. AIDS 7:S147–159. [PubMed] [Google Scholar]
- 8.Ganeshan, S., R. Dickover, B. Korber, Y. Bryson, and S. Wolinsky. 1997. human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J. Virol. 71:663–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambert, J. S., J. McNamara, S. Katz, et al. 1998. Safety and immunogenicity of HIV recombinant envelope vaccines in HIV-infected infants and children. National Institutes of Health-sponsored Pediatric AIDS Clinical Trials group (ACTG 218). J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 19:451–461. [DOI] [PubMed] [Google Scholar]
- 10.Liu, S.-L., T. Schacker, L. Musey, D. Shriner, M. McElrath, L. Corey, and J. Mullins. 1997. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J. Virol. 71:4282–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panther, L. A., L. Tucker, C. Xu, R. Tuomala, J. Mullins, and D. Anderson. 2000. Genital tract human immunodeficiency virus type 1 (HIV-1) shedding and inflammation and HIV-1 env diversity in perinatal HIV-1 transmission. J. Infect. Dis. 181:555–563. [DOI] [PubMed] [Google Scholar]
- 12.Redfield, R. R., D. Birx, N. Ketter, E. Tramont, V. Polonis, C. Davis, J. Brundage, G. Smith, S. Johnson, and A. Fowler. 1991. A phase I evaluation of the safety and immunogenicity of immunization with recombinant gp160 in patients with early HIV infection. N. Engl. J. Med. 324:1677–1684. [DOI] [PubMed] [Google Scholar]
- 13.Vigano, A., D. Bricalli, D. Trabattoni, D. Salvaggio, A. Ruzzante, M. Barbi, G. Di Sanzo, N. Principi, and M. Clerici 1998. Immunization with both T cell-dependent and T cell-independent vaccines augments HIV viral load secondarily to stimulation of tumor necrosis factor alpha. AIDS Res. Hum. Retroviruses 14:727–734. [DOI] [PubMed] [Google Scholar]
- 14.Wolinsky, S. M., B. Korber, A. Neumann, M. Daniels, K. Kunstman, A. Whetsell, M. Furtado, Y. Cao, D. Ho, and J. Safrit. 1996. Adaptive evolution of HIV-1 during the natural course of infection. Science 272:537–542. [DOI] [PubMed] [Google Scholar]
- 15.Zagury, D., J. Bernard, A. Hallbreich, B. Bizzini, C. Carelli, A. Achour, M. Defer, J. Bertho, K. Lanneval, and J. Zagury. 1992. One-year follow-up of vaccine therapy in HIV-infected immune-deficient individuals — a new strategy. J. AIDS 5:676–681. [PubMed] [Google Scholar]