Abstract
The binding domain of the chicken leptin receptor [chLBD (chicken leptin-binding domain)], subcloned from the full-size chicken leptin receptor and prepared in an Escherichia coli system, was subjected to site-directed mutagenesis to identify the amino acids involved in leptin binding. A total of 22 electrophoretically pure, >90% monomer-containing mutants were expressed, refolded and purified. The effects of the mutations were tested by the ability to form complexes with ovine leptin, and the kinetic parameters of interaction were determined by surface plasmon resonance. Six mutants were used to determine whether mutations of several amino acids that differ between chLBD and mammalian LBDs will affect affinity: none showed any such effect, except the mutant A105D (Ala105→Asp), which exhibited some decrease in affinity. Surface plasmon resonance analysis identified six mutants in which binding activity was totally abolished (F73A, Y14A/F73A, V76A/F77A, L78A/L79A, V76A/F77A/L78A/L79A and A105D/D106V) and six mutants (Y14A, R41A, R41A/S42A/K43A, V103A, V135A/F136A and F136A) in which affinity for the hormone was reduced, mainly by increased dissociation rates. Gel-filtration experiments indicated the formation of a 1:1 ovine or human leptin–chLBD complex with a molecular mass of approx. 41 kDa. Gel-filtration experiments yielded 1:1 complexes with those mutants in which affinity had decreased, but not with the six mutants, which had totally lost their binding capacity. Modelling the leptin–chLBD complex indicated that the binding domain of the latter is located mainly in the L3 loop, which contributes nine amino acid residues interacting with leptin. Contact-surface analysis identified the residues having the highest contribution to the recognition site to be Phe73, Phe77 and Leu79.
Keywords: chicken leptin receptor, immunoglobulin-like domain (IGD), leptin, leptin binding domain (LBD), MON105 cells, surface plasmon resonance (SPR)
Abbreviations: chLBD, chicken leptin-binding domain; chLep, chicken leptin; CM5, carboxymethyl dextran; ECD, extracellular domain; EDC, N-ethyl-N′-(3-dimethylaminopropyl)-carbodi-imide hydrochloride; hLBD, human LBD; hLep, human leptin; IB, inclusion body; IGD, immunoglobulin-like domain; NHS, N-hydroxysuccinimide; oLep, ovine leptin; RT, retention time; SPR, surface plasmon resonance; WT, wild-type
INTRODUCTION
Leptin, the product of the ob gene, has been reported to suppress appetite by regulating satiety-centre activities in the brain via its receptor (LEPR) and to affect body weight [1]. However, further studies have shown that leptin receptors are also expressed in many other tissues [2–6] and have suggested that leptin is involved in more diverse biological functions than previously thought. A leptin tertiary-structure solution revealed its connection to the long-chain cytokine superfamily [7]. Although the tertiary structure of the leptin receptor has not yet been determined, its amino acid sequence analysis shows a high similarity to receptors of the class I cytokine receptor family, such as the receptors for growth hormone, G-CSF (granulocyte colony-stimulating factor), interleukin-6 and erythropoietin. The receptors from this family share multiple similar domains in their ECD (extracellular domain), such as C2, CK and F3. Like the G-CSF receptor, the leptin receptor has two repeats of the CK-F3 domain, suggesting it to be the ligand-binding site [8–10]. A study performed by Fong et al. [11] localized the LBD (leptin-binding domain) to the membrane-proximal CK-F3 (∼200 amino acids) in the leptin receptor ECD. However, recent data have shown that the binding of leptin to its receptor more closely resembles the interaction of interleukin-6 with its receptor [12], and the IGD (immunoglobulin-like domain) located between the distal and proximal CK-F3 domains appears to be essential for productive dimerization or tetramerization of the leptin receptor [13]. However, binding to the receptor was not affected by removal of the IGD [13], and alanine mutagenesis of leptin's site III that interacts with IGD abolishes the leptin-inducible receptor activation but does not effect binding [14,15].
Recently, we subcloned, expressed, purified and characterized the LBD of hLep (human leptin) receptor [16]. This LBD is capable of forming a 1:1 complex with leptin, and the binding constants of this short part of the leptin receptor ECD are in the nanomolar range, similar or somewhat lower than that of the full-length membrane-embedded receptor. In the present study, a similar approach was followed to prepare the recombinant LBD of chicken leptin receptor [chLBD (chicken LBD)] and to characterize its binding capacities relative to hLBD (human LBD), in order to provide an additional aspect to the interaction-site-mapping study of both leptin and its receptor. Since chLBD is more easily prepared than its human analogue but interacts with mammalian leptins with similar affinity, we have used this protein for site-directed mutagenesis aimed at the identification of residues important for its interaction with leptin.
MATERIALS AND METHODS
Materials
oLeps (ovine leptins) and hLeps were prepared in our laboratory as described previously [17,18]. pMon3403 expression vector and MON105 cells (strain of Escherichia coli cells) were provided by Monsanto (St. Louis, MO, U.S.A.). Restriction enzymes used in the molecular biology experiments were obtained from Fermentas (Vilnius, Lithuania) and New England Biolabs (Beverly, MA, U.S.A.). DNA primers were ordered from Gibco BRL, NV Life Technologies S.A. (Ghent, Belgium). RPMI 1640 medium, interleukin-3, nalidixic acid and MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide, also known as Thiazolyl Blue] were purchased from Sigma (St. Louis, MO, U.S.A.), fetal calf serum from Biolab (Jerusalem, Israel) and pTrc 99A expression vector, Superdex 75 HR 10/30 column and Q-Sepharose from Pharmacia LKB Biotechnology AB (Uppsala, Sweden). A research-grade CM5 (carboxymethyl dextran) sensor chip, NHS (N-hydroxysuccinimide), EDC [N-ethyl-N′-(3-dimethylaminopropyl)-carbodi-imide hydrochloride], ethanolamine/HCl and HBS-EP running buffer (10 mM Hepes, 150 mM NaCl, 3.4 mM EDTA and 0.005%, v/v, surfactant P20 at pH 7.4) were purchased from Biacore AB (Uppsala, Sweden). All other chemicals were of analytical grade.
Preparation of chLBD expression plasmid
A DNA insert encoding the chLBD fragment, consisting of amino acids 420–626 of the chLep (chicken leptin) receptor, was prepared by PCR using the following primers: the 5′-sense primer, 5′-AAAACCATGGCGATTGATGTGAATATCAATATC-3′, containing an NcoI restriction site (underlined), and the 3′-antisense primer, 5′-CCCAAGCTTTCAATCTTTTACAGCTGCATA-3′, containing a stop codon (boldface) followed by a HindIII restriction site (underlined). The resulting PCR product was digested with NcoI/HindIII, purified and cloned into the pMon3403 expression vector, predigested with the same restriction enzymes. The expression plasmid was then transformed into E. coli MON105 expression cells.
Preparation of chLBD mutants
To prepare the chLBD mutants, the DNA insert encoding the WT (wild-type) LBD, subcloned into pTrc 99A or pMon3401 expression vector, was used as starting material. The insert was modified with the Stratagene Quik Change mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.) according to the manufacturer's instructions, using two complementary primers (Table 1). The primers were designed to contain base changes (marked in boldface) to obtain the respective mutations, but still conserve the appropriate amino acid sequence, and to modify a specific restriction site (underlined) for colony screening. The procedure included 18 PCR cycles using Pfu polymerase. The mutated construct was then digested with DpnI restriction enzyme, which is specific to methylated and hemi-methylated DNA (target sequence: 5′-Gm6ATC-3′), in order to digest the template and select for mutations containing synthesized DNA. The plasmids were then transfected into XL1 competent cells. Five colonies of each mutant were screened for mutation, using the specifically designed restriction site, and revealed at least 80% efficiency. Two colonies of each mutant were sequenced and they were confirmed to contain the mutation but no unwanted misincorporation of nucleotides. The inserts, coding for ovine and chicken mutated leptins, were then removed from the pTrc 99A plasmid, ligated to the pMon3401 vector between the NcoI and HindIII restriction sites and transfected into MON105 competent cells.
Table 1. Primers used for the preparation of chLBD mutants.
In the primers, S stands for a sense primer and A for an antisense primer; all mutations are in boldface. Modified restriction sites are underlined.
| Primer | Primer sequence | Modified restriction site |
|---|---|---|
| Y14A-5 | S 5′-GTGAAACTGATGGCGCCTTAACTAAAATGACTTGCAG-3 | NarI |
| Y14A-3 | A 5′-CTGCAAGTCATTTTAGTTAAGGCGCCATCAGTTTCAC-3′ | NarI |
| R41A-5 | S 5′-GGGGAGTTCCTTGCAGTTAAGATATCACGCGAGCAAAATTTATTG-3′ | EcoRV |
| R41A-3 | A 5′-CAATAAATTTTGCTCGCGTGATATCTTAACTGCAAGGAACTCCCC-3′ | EcoRV |
| S42A-5 | S 5′-GGGAGTTCCTTGCAGTTAAGATATCACAGGGCCAAAATTTATTG-3′ | EcoRV |
| S42A-3 | A 5′-CAATAAATTTTGGCCCTGTGATATCTTAACTGCAAGGAACTCCC-3′ | EcoRV |
| K43A-5 | S 5′-GTTCCTTGCAGTTAAGATACCACAGGAGCGCTATTTATTGTTC-3′ | Eco47III |
| K43A-3 | A 5′-GAACAATAAATAGCGCTCCTGTGGTATCTTAACTGCAAGGAAC-3′ | Eco47III |
| R41S42K43A-5 | S 5′-GGAGTTCCTTGCAATTGAGATACCACGCGGCCGCTATTTATTGTTC-3′ | MunI |
| R41S42K43A-3 | A 5′-GAACAATAAATAGCGGCCGCGTGGTATCTCAATTGCAAGGAACTCC-3′ | MunI |
| F73A-5 | S 5′-GAGTGCACAGCTCAGCCTGTTTTTCTTTTATCCGGACATACC-3′ | BspEI |
| F73A-3 | A 5′-GGTATGTCCGGATAAAAGAAAAACAGGCTGAGCTGTGCACTC-3′ | BspEI |
| V76AF77A-5 | S 5′-CACATTTCAGCCTGCTGCTCTTTTATCCGGATATACCATGTGG-3′ | BspEI |
| V76AF77A-3 | A 5′-CCACATGGTATATCCGGATAAAAGAGCAGCAGGCTGAAATGTGC-3′ | BspEI |
| L78AL79A-5 | S 5′-GCACATTTCAGCCTGTTTTTGCTGCATCCGGATATACCATGTGG-3′ | BspEI |
| L78AL79A-3 | A 5′-CCACATGGTATATCCGGATGCAGCAAAAACAGGCTGAAATGTGC-3′ | BspEI |
| V76AF77AL78AL79A-5 | S 5′-GCACATTTCAGCCTGCTGCAGCTGCATCTGGATATACCATG-3′ | PstI |
| V76AF77AL78AL79A-3 | A 5′-CATGGTATATCCAGATGCAGCTGCAGCAGGCTGAAATGTGC-3′ | PstI |
| V103A-5 | S 5′-GAATCCTCACCAACATGTGTCGCTCCAGCAGATGTG-3′ | NspI |
| V103A-3 | A 5′-CACATCTGCTGGAGCGACACATGTTGGTGAGGATTC-3′ | NspI |
| V103L-5 | S 5′-CAACTTGTGTCCTTCCAGCTGATGTGGTGAAGCC-3′ | PvuII |
| V103L-3 | A 5′-GGCTTCACCACATCAGCTGGAAGGACACAAGTTG-3′ | PvuII |
| A105D-5 | S 5′-CTTGTGTCGTTCCGGATGATGTGGTGAAGCCACTG-3′ | BspEI |
| A105D-3 | A 5′-CAGTGGCTTCACCACATCATCCGGAACGACACAAG-3′ | BspEI |
| A105D106V-5 | S 5′-CTTGTGTCGTTCCAGACGTCGTGGTGAAGCCACTG-3′ | AatII |
| A105D106V-3 | A 5′-CAGTGGCTTCACCACGACGTCTGGAACGACACAAG-3′ | AatII |
| D106S-5 | S 5′-CTTGTGTCGTTCCAGCTAGCGTGGTGAAGCCACTGCCTCCC-3′ | NheI |
| D106S-3 | A 5′-GGGAGGCAGTGGCTTCACCACGCTAGCTGGAACGACACAAG-3′ | NheI |
| V135A-5 | S 5′-GGGCTGCTGAACGTTAGCTGGACAAACCCCGCGTTTACAAATGATG-3′ | AclI |
| V135A-3 | A 5′-CATCATTTGTAAACGCGGGGTTTGTCCAGCTAACGTTCAGCAGCCC-3′ | AclI |
| F136A-5 | S 5′-CAAACCCCGTGGCTACAAATGATGACCTCAAGTTTCAGATCC-3′ | AflII(–) |
| F136A-3 | A 5′-GGATCTGAAACTTGAGGTCATCATTTGTAGCCACGGGGTTTG-3′ | AflII(–) |
| V135AF136A-5 | S 5′-CGTGAGCTGGACAAACCCCGCGGCTACAAATGATGACC-3′ | SacII |
| V135AF136A-3 | A 5′-GGTCATCATTTGTAGCCGCGGGGTTTGTCCAGCTCACG-3′ | SacII |
| D140N-5 | S 5′-GTGTTTACAAATGATAACCTTAAATTTCAGATCCGG-3′ | AflII(–) |
| D140N-3 | A 5′-CCGGATCTGAAATTTAAGGTTATCATTTGTAAACAC-3′ | AflII(–) |
| K142Q-5 | S 5′-GTTTACAAATGATGACCTGCAGTTTCAGATCCGG-3′ | PstI |
| K142Q-3 | A 5′-CCGGATCTGAAACTGCAGGTCATCATTTGTAAAC-3′ | PstI |
| L162Y-5 | S 5′-CACATGGGAGCTCTATGAAGTTTATAGCGTACCAACAAGATCAG-3′ | SacI |
| L162Y-3 | A 5′-CTGATCTTGTTGGTACGCTATAAACTTCATAGAGCTCCCATGTG-3′ | SacI |
| L191A-5 | S 5′-CAGATCCGCTGTAGAGCCCTGGATGGCGCAGGCTACTGG-3′ | PstI(–) |
| L191A-3 | A 5′-CCAGTAGCCTGCGCCATCCAGGGCTCTACAGCGGATCTG-3′ | PstI(–) |
Expression, refolding and purification of chLBD and its mutants
The recombinant chLBD (or the respective chLBD mutant) with an extra methionine–alanine at the N-terminus was expressed upon nalidixic acid induction (50 μg/ml) in a 10 litre culture of MON105 cells, transformed with the appropriate expression plasmid, grown in 20×2.5 litre flasks in Terrific Broth medium (12 g/l protein hydrolysate, 24 g/l yeast extract, 9.2 g/l dipotassium hydrogen phosphate, 2.2 g/l potassium dihydrogen phosphate and 8 ml/l glycerol) at 37 °C to an attenuance D600 of 0.9. After an additional 4 h of incubation, the cells were harvested by 10 min centrifugation at 10000 g and frozen at −20 °C. The bacterial pellet from 10 litre of bacterial culture was thawed on ice and resuspended in lysis buffer (10 mM Tris/HCl and 10 mM EDTA, pH 8). IBs (inclusion bodies) were then prepared as described previously [16] and frozen. Subsequently, IBs obtained from 5 litres of bacterial culture were solubilized in 600 ml of 4.5 M urea and 40 mM Tris, containing 50 mM cysteine. The pH value of the solution was adjusted to 11.5 by NaOH. After 2 h of stirring at 4 °C, 3 vol. of 0.67 M arginine was added to a final concentration of 0.5 M and stirred overnight. The next morning, the solution was transferred to dialysis tubes and dialysed against 10 litres of 10 mM Tris/HCl (pH 9) for 60 h, with external solution exchange every 6–10 h. The protein was then applied at maximal flow rate (400–500 ml/h) to a Q-Sepharose column (30 ml bead volume), preequilibrated with the same buffer. The breakthrough fraction, which contained no LBD, was discarded, and the absorbed protein was eluted in a stepwise manner (50, 100, 150 and 200 mM NaCl in 10 mM Tris/HCl, pH 9). Fractions of 50 ml were collected and protein concentration was determined by measuring A280.
Determination of purity and monomer content
SDS/PAGE was carried out by the method of Laemmli [19] in a 15% (w/v) polyacrylamide gel under reducing and non-reducing conditions. The gel was stained with Coomassie Brilliant Blue R. Gel-filtration chromatography was performed on a Superdex™ 75 HR 10/30 column with 0.2 ml aliquots of the Q-Sepharose column-eluted fraction using TN buffer (25 mM Tris/HCl and 150 mM NaCl, pH 8).
Determination of CD spectra
The CD spectra (in millidegrees) were measured with an AVIV model 62A DS CD spectrometer (AVIV, Lakewood, NJ, U.S.A.) using a 0.020 cm rectangular QS Hellma cuvette. The absorption spectra were measured with an AVIV model 17DS UV–visible IR spectrophotometer using a 1.000 cm QS cuvette and correction for light scattering. Collection of the data and calculation of the results were carried out as described previously [16].
Determination of complex stoichiometry
Complexes between chLBD and oLep or hLep were prepared at various molar concentrations in TN buffer. The final concentrations of the proteins in the 1:1 ratio were 10 μM. After a 20 min incubation at room temperature (25 °C), 200 μl aliqouts were applied to a Superdex™ 75 HR 10/30 column preequilibrated with TN buffer. To determine the molecular mass of the complex, the column was calibrated with several pure proteins.
Binding assays
Radiolabelled 125I-hLep served as a ligand, and human and ovine non-labelled leptins served as competitors. The experiments were conducted using recombinant chLBD. Each tube contained 200 μl of reaction buffer (12.5 mM sodium barbiturate, 0.1%, w/v, BSA, 7.5 mM EDTA, 150 mM NaCl and 0.1%, v/v, Triton X-100, pH 8.6), 100 μl of 125I-hLep (180000–220000 c.p.m.) and 100 μl of leptin solutions (providing 0–5000 ng/tube) in the reaction buffer. The reaction was started by the addition of 100 μl of LBD or LBD mutants (20 ng). The tubes were incubated for 24 h at room temperature, then the leptin–LBD complex was precipitated by adding 250 μl of 1% (w/v) bovine immunoglobulins and 500 μl of 20% (w/v) poly(ethylene glycol). After thorough mixing, the tubes were incubated for 20 min at 4 °C and centrifuged at 12000 g for 15 min at 4 °C. The supernatant was carefully aspirated, and the precipitates were counted in a Kontron γ-counter. hLep was iodinated by a method described previously for the iodination of human growth hormone [20].
Kinetic measurements of chLBD–leptin interactions
All experiments were performed at 25 °C using SPR (surface plasmon resonance) methodology. The kinetics and equilibrium constants for the interactions between human and chicken recombinant LBD and oLep and hLep were determined using the Biacore 3000 system. Leptin (human or ovine) was immobilized in a flow cell of a research-grade CM5 sensor chip using amine-coupling chemistry [21]. The immobilization steps were carried out at a flow rate of 5 μl/min in HBS-EP buffer. The surface was activated for 7 min with a mixture of 0.05 M NHS and 0.2 M EDC. Leptin was injected at a concentration of 50 μg/ml in 10 mM acetate (pH 3.5) until the desired level [1000 RU (resonance units)] was achieved. Ethanolamine (1 M, pH 8.5) was injected for 7 min to block the remaining activated groups. A control surface was prepared by activating the carboxy groups and then blocking the activated groups by ethanolamine as described in [21,29]. For the binding studies, the hLBD or chLBD, resuspended in HBS-EP buffer, was passed at different concentrations (31.25, 62.5, 125 and 250 nM) through three flow cells (carrying hLep, ovLep or a control) at the rate of 30 μl/min. The surface was regenerated after each interaction with a 10 μl pulse of 10 mM glycine buffer (pH 2). The experiments were controlled by the kinetics Wizard of the Biacore control software, which automatically corrects for refractive-index changes and non-specific binding by subtraction of the responses obtained for the control surface from the data obtained for the interactions between LBD and leptin. The resultant binding curves were fitted to the association and dissociation phases at all LBD concentrations simultaneously using Biacore evaluation software. In all cases, the best fit was obtained for a simple bimolecular interaction (Langmuir model).
To evaluate the binding kinetics in a different way, we performed additional experiments in which the chLBD was immobilized and hLep was in the mobile phase. Thus chLBD was amino-linked to the CM5 chip; the pH range within which leptin-binding capacity is preserved is very narrow, and after a number of trials below pH 6, we chose an optimum pH of 5.7 using 5 mM maleate buffer. The experimental conditions were: flow 5 μl/min, 8 min activation with an EDC/NHS mixture, 8 min injection of 2 μM monomeric LBD solution and 8 min ethanolamine neutralization. An immobilization level of up to 1500 RU was consistently obtained under these conditions. As no chemical agent was suitable for the dissociation of leptin from chLBD while ensuring good preservation and reproducibility of leptin docking, an overnight simple wash, at a flow rate of 50 μl/min, was used to eliminate bound leptin.
RESULTS
Purification and characterization of chLBD and its mutants
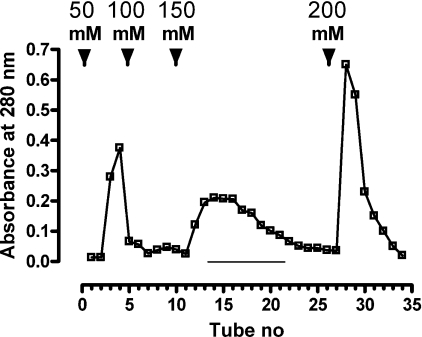
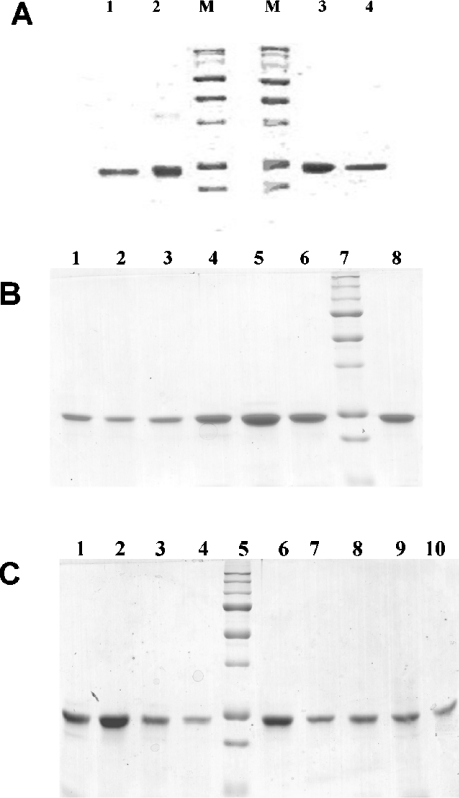
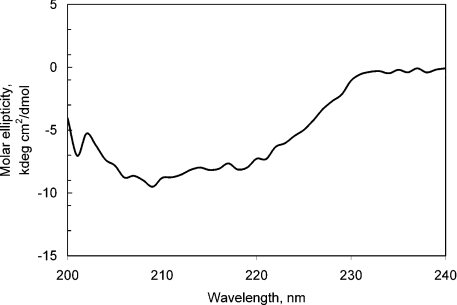
The recombinant LBD representing the membrane-proximal repetition of the CK-F3 domain (419–626 amino acids) of the chLep receptor ECD was subcloned by PCR using appropriate primers on the cDNA template that was cloned by Horev et al. [22]. The protein was expressed in E. coli MON105 cells. Upon induction, chLBD was expressed in high amounts and, after 4 h of fermentation, represented the major protein fraction of the bacteria, which appeared as a main band in the IBs, as detected by SDS/PAGE (results not shown). IBs were purified and solubilized as described in the Materials and methods section. After refolding, the protein was purified by one-step anion-exchange chromatography on a Q-Sepharose column. The fractions were eluted by increasing concentrations of NaCl (Figure 1) and every third tube was tested for LBD appearance by gel filtration on a Superdex™ 75 HR 10/30 column; fractions containing monomers were subsequently pooled (see Figure 2). Only the fraction eluted with 150 mM NaCl contained over 95% monomeric protein with 5% dimers; fractions eluted with higher NaCl concentrations contained mostly oligomers. The molar absorbance coefficient of chLBD at 280 nm was calculated by Pace et al. [23] to be 50390 M−1·cm−1 or 2.11 for 1 mg/ml. To achieve a protein concentration of approx. 0.5 mg/ml, the pool was Amicon-concentrated, filter-sterilized and stored at 4 °C. Under those conditions, the purified chLBD retained its monomeric structure as well as its ability to form a complex with leptin for up to 1 year. The monomeric profile and molecular chLBD mass were also determined by SDS/PAGE, showing only one band of approx. 24 kDa in the presence of reducing agent (the calculated value for Met-chLBD is 23.891 kDa): this matched the result obtained when recombinant human LBD, previously prepared in our laboratory, was tested under the same conditions (Figure 3A, lanes 3 and 4 respectively). In the absence of reducing agent, a small amount of dimers could be seen in chLBD (Figure 3A, lane 2), in agreement with the gel-filtration results. The dimers probably appeared due to the formation of S–S intermolecular links. The yield of the monomeric fraction (average of six preparations) varied between 15 and 25 mg from 5 litres of bacterial culture. The results of the CD analysis at neutral pH are presented in Figure 4. The secondary-structure calculations revealed the contents of α-helices, β-strands and β-turns to be similar to those observed for hLBD (Table 2 and results not shown).
Figure 1. Purification of chLBD extracted from IBs and refolded on a Q-Sepharose column.
The column (2.5 cm×7 cm) was equilibrated with 10 mM Tris/HCl (pH 9.0) at 4 °C. The dialysed solution of refolded protein was applied to the column at a rate of 400 ml/h. Elution was carried out using a discontinuous NaCl gradient in the same buffer at 400 ml/h, and 50 ml fractions were collected. Protein concentration was determined by measuring absorbance at 280 nm. Every third tube was analysed for chLBD content by gel filtration in a Superdex™ 75 HR column. Fractions eluted with 150 mM NaCl (underlined) contained monomeric protein and were pooled.
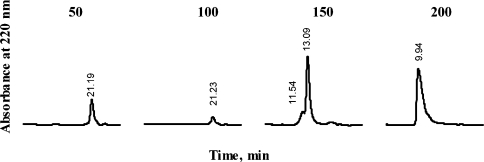
Figure 2. Gel-filtration analysis of the pools eluted with increasing concentrations of NaCl (in mM) from a Q-Sepharose column.
Figure 3. SDS/PAGE (12% polyacrylamide) of recombinant chLBD and selected mutants.
(A) Lanes 1 and 4, hLBD; lanes 2 and 3, chLBD; lanes M, molecular mass standards [250, 150, 100, 75 (strong band), 50 (strong band), 37, 25 (strong band) and 20 kDa]. Lanes 1 and 2 are without and lanes 3 and 4 with 2-mercaptoethanol. (B) Lane 1, chLBD F136A; lane 2, chLBD V135AF136A; lane 3, chLBD V103L; lane 4, chLBD D140N; lane 5, chLBD K142Q; lane 6, chLBD L162Y; lane 7, molecular mass standards [250, 150, 100, 75 (strong band), 50 (strong band), 37, 25 (strong band), 20 and 15 kDa]; and lane 8, WT chLBD. (C) Lane 1, chLBD Y14A; lane 2, chLBD L191A; lane 3, chLBD L78A/L79A; lane 4, chLBD R41A; lane 5, molecular mass standards [250, 150, 100, 75 (strong band), 50 (strong band), 37, 25 (strong band), 20 and 15 kDa]; lane 6, WT chLBD; lane 7, chLBD S42A; lane 8, chLBD K43A; lane 9, chLBD R41A/S42A/K43A; and lane 10, chLBD V135A. All proteins in (B, C) are with 2-mercaptoethanol.
Figure 4. CD spectra of purified recombinant chicken LBD in 20 mM KH2PO4 buffer (pH 7.5).
Table 2. Secondary structure of recombinant chLBD and hLBD at neutral pH.
Results are given as means±S.D. Errors only arose from uncertainty in the fitting of the experimental CD spectrum by the set of standard protein CD spectra in the CONTIN program. Errors in both the CD measurements and the protein concentration determination were not included. hLBD values are previously published values [16].
| Secondary structure (%) | chLBD | hLBD |
|---|---|---|
| α-Helix | 11±1.0 | 6.6±0.4 |
| β-Strands | 31±2.1 | 37±1.2 |
| β-Turns | 10±1.6 | 25±1.0 |
| The remainder | 48±3.3 | 31±1.6 |
chLBD mutants were prepared in a similar manner and their purity was verified by both SDS/PAGE (Figures 3B and 3C) and gel filtration on a Superdex 75 column, which resulted in a main peak of monomers (90–95%) and a small amount of dimers (results not shown). In cases in which the relative amount of dimer was higher, the monomeric fraction was further isolated by preparative gel filtration using a Superdex 75 (2.6 cm×60 cm) column. Their CD spectra were similar to the WT chLBD (results not shown). Similar to WT chLBD, the mutants could be stored at 4 °C as sterile solutions (0.4–0.5 mg/ml) for at least 6 months without undergoing any change in their monomeric content or in their capacity to interact with leptin.
Binding experiments
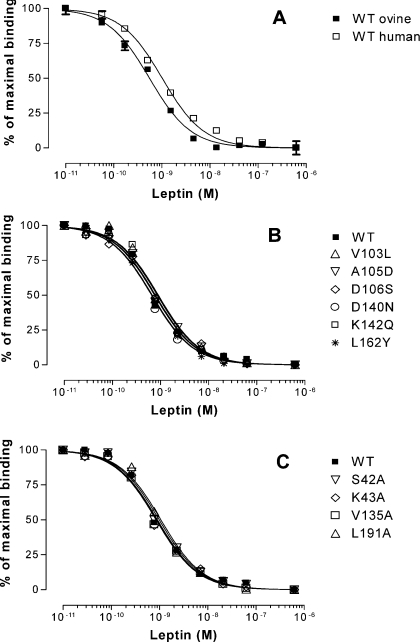
The affinity of non-mutated chLBD for hLep and oLep was determined in competition studies using 125I-hLep as a ligand. Unlabelled hLep and oLep dose-dependently inhibited binding of the radioactive ligand to the chLBD with IC50 values of 1.02 and 0.66 nM respectively, and maximal displacement of the 125I-hLep occurred in the presence of approx. 100 nM hLep (Figure 5A). All chLBD mutants aimed at humanizing chLBD were also tested for binding of 125I-hLep using the same amount of LBD (20 ng) for each tube and oLep as competitor. The results presented in Figure 5(B) show no significant difference as compared with non-mutated (WT) chLBD. The respective IC50 values for WT chLBD and the following mutants [V103L (Val103→Leu), A105D, D106S, D140N, K142Q and L162Y] were (in nM) 0.75, 0.90, 0.79, 0.76, 0.65, 0.86 and 0.77. In additional experiments, the binding capacity of WT chLBD was compared with mutants aimed at lowering the affinity: S42A, K43A, V135A and L191A (Figure 5C). The respective IC50 values (in nM) were comparable with those obtained in the two former experiments: 0.90, 0.98, 0.87, 0.85 and 1.07. Like in the former experiment, these differences were not statistically significant and all were within the 95% confidence limit. In contrast, the following mutants (Y14A/F73A, F73A, V76A/F77, L78A/L79A, V76A/F77/L78A/L79A and A105D/D106V) did not bind specifically any 125I-hLep even when their concentration in the tube was elevated to 200 ng. The binding of 125I-hLep to the mutants (20 ng or more per tube) Y14A, R41A, R41A/S42A/K43A, V103A, F136A and V135A/F136A was very low compared with WT chLBD, making quantitative comparison not feasible because of the high non-specific values resulting from the large quantity of mutated LBDs. Therefore more detailed studies were performed using SPR (see below).
Figure 5. Competition of unlabelled hLep or oLep with 125I-hLep (180000–220000 c.p.m./tube) for binding to purified chLBD or chLBD mutants.
(A) Comparison of hLep and oLep capacity to compete with 125I-hLep. The respective EC50 values for hLep and oLep were 1.02 and 0.66 nM but the differences were within 95% confidence intervals and statistically not significant. The variability was low, so the S.E.M. values are in most cases not seen. (B) Comparison of oLep capacity to compete with 125I-hLep for binding to chLBD or its mutants aimed at humanizing the chLBD or (C) with mutants aimed at decreasing the interaction of chLBD with leptin. Results are presented as means±S.E.M. for three experiments in (A) and as the mean for one experiment (in triplicates) in (B, C). Because of small differences in the maximal specific binding values obtained with WT chLBD and different mutants, which varied between 12 and 15%, the specific bindings obtained in each experiment were normalized to 100%. For other details, see text.
Detection of chLBD–leptin or mutant-chLBD–leptin complexes by gel filtration
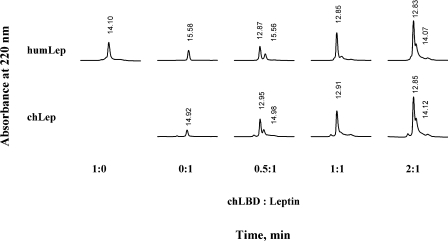
To characterize the binding stoichiometry between leptin and chLBD or its mutants, the two were mixed in different molar ratios and separated by gel filtration through an analytical Superdex 75 column to determine the molecular mass of the binding complex under non-denaturing conditions. The experiments were performed using a constant concentration of 10 μM ovine or human recombinant leptin and 5, 10 or 20 μM chLBD (Figure 6). Both species (ovine and human) of leptin bound the chLBD in a 1:1 ratio. This stoichiometry was determined by the appearance of a single peak when the components were mixed at the same molar ratio and the appearance of an additional peak when one of them was in excess. The molecular mass calculation, based on the corresponding peak RT (retention time), was approx. 41 kDa. It should be noted that gel-filtration experiments even yielded 1:1 complexes in mutants in which affinity had decreased (see results of the SPR analysis below) and the results were comparable with those shown in Figure 6. This probably results from the fact that the gel-filtration experiments were carried out with concentrations of 10 μM. In contrast, no complex formation was detected with mutants in which the ability to interact with leptin had been totally abolished (F73A, Y14A/F73A, V76A/F77A, L78A/L79A, V76A/F77A/L78A/L79A and A105D/D106V), as also shown by SPR analysis.
Figure 6. Gel-filtration analysis of complexes between chLBD and hLep (humLEP) or oLep (chLep) on a Superdex™ 75 HR 10/30 column.
Complex formation was achieved by 20 min incubation at room temperature in TN buffer using various molar ratios; 200 μl aliquots of the mixture were applied to the column, preequilibrated with the same buffer. The initial hormone concentration (10 μM) was constant in all cases. The column was developed at 0.8 ml/min and calibrated with BSA [66 kDa, RT=11.47 min], ovine placental lactogen (23 kDa, RT=14.5 min) and lysozyme (14 kDa, RT=21.45 min). Protein concentration in the eluate was monitored by measuring the absorbance at 220 nm.
SPR determination of the interaction between chLBD or its mutants and leptin
To characterize further the binding capacities of chLBD with leptin, the SPR technique using leptin immobilized on a sensor chip was employed. oLep or hLep was immobilized on the sensor-chip surface by amine-coupling, and binding of chLBD was determined. Binding of recombinant hLBD, previously prepared by us, was examined simultaneously on the same sensor chip. The most acceptable interactions were obtained from comparison with a 1:1 theoretical model using χ2 analysis. The calculated data are presented in Table 3. The affinity of hLep for hLBD was comparable with chLBD and the differences in kinetics and thermodynamic constants were not significant. The thermodynamic dissociation constant (Kd), kinetic dissociation constant (koff) and kinetic association constant (kon) values obtained for non-mutated oLep were comparable with those obtained for the interaction with hLep (for comparison, see Table 4, first row). To verify the interaction of chLBD with the respective leptins, the SPR was repeated ‘in reverse’ by immobilizing the LBD to the sensor-chip surface and using the leptin in the mobile phase. Different concentrations of hLep were successively injected on to the LBD chip, in the 50–1000 nM range at pH 7.3. Qualitatively, hLep exhibited rapid association and slow dissociation, both monophasic (see Table 3). There were no differences in the kon values but the koff values were 2.5–6-fold lower. However, the 1:1 stoichiometry of the interaction was not changed.
Table 3. Kinetic constants measured by SPR for complex formation of hLBD and LBD with hLep.
Values presented are means±S.E.M.
| Immobilized ligand | Injected ligand | kon (mol−1·s−1×105) | koff (s−1×10−3) | Kd (M×10−8) |
|---|---|---|---|---|
| hLep | hLBD | 1.20±0.23 | 1.85±0.30 | 1.54 |
| chLBD | 1.83±0.95 | 4.25±3.40 | 2.32 | |
| chLBD | hLep | 1.68±0.30 | 0.74±0.23 | 0.44 |
Table 4. Kinetic constants (means±S.E.M.) measured by SPR for complex formation of chLBD mutants with immobilized oLep.
The S.E.M. values for the kon and koff values were calculated using the values obtained from their respective calculations in four or five concentrations of chLBD or chLBD mutants in each experiment. The numbering of amino acids is that of chLBD and the numbers in parentheses indicate the numbering in the full-size receptor (see also Figure 7).
| Mutant | kon (mol−1·s−1×105) | koff (s−1×10−3) | Kd (M×10−8) | χ2 |
|---|---|---|---|---|
| Wild-type* | 2.65±0.55 | 4.58±0.12 | 1.72±0.38 | 1.57–5.31 |
| Mutations aimed at humanizing the chLBD | ||||
| V103L (531)† | 2.94±0.43 | 6.65±0.67 | 2.26 | 0.8–2.97 |
| A105D (533) | 1.38±0.33 | 10.4±0.98 | 7.54 | 0.78 |
| D106S (534)† | 4.27±0.65 | 7.94±0.89 | 1.86 | 1.14–6.9 |
| D140N (568) | 1.50±0.22 | 5.19±0.63 | 3.34 | 1.88 |
| K142Q (570) | 1.85±0.28 | 6.67±0.86 | 3.59 | 2.78 |
| L162Y (590) | 2.07±0.46 | 5.54±0.66 | 2.66 | 1.88 |
| Mutations aimed at decreasing the interaction of chLBD with leptin | ||||
| Y14A (442) | 1.75±0.23 | 14.0±1.73‡ | 8.41 | 3.31 |
| Y14A/F73A (442/551) | No binding | |||
| R41A (469) | 1.05±0.33 | 64.0±9.55 | 63.8 | 0.42 |
| S42A (470) | 3.14±0.54 | 4.55±0.54 | 1.45 | 1.06 |
| K43A (471) | 2.08±0.32 | 7.35±0.80 | 3.53 | 1.06 |
| R41A/S42A/K43A (469–471) | 0.59±0.23 | 29.0±4.41 | 50.2 | 0.29 |
| F73A (501) | No binding | |||
| V76A/F77A (504/505) | No binding | |||
| L78A/L79A (506/507) | No binding | |||
| V76A/F77A/L78A/L79A (504–507) | No binding | |||
| V103A (531) | 0.42±0.16 | 10.80±1.93 | 25.7 | 1.24 |
| A105D/D106V (533/5343) | No binding | |||
| V135A (563)† | 4.59±0.66 | 5.55±0.73 | 1.21 | 1.32–4.52 |
| F136A (564) | 1.95±0.23 | 59.0±7.88 | 30.3 | 0.42 |
| V135A/F136A (563/564) | 3.48±0.49 | 87.0±9.97 | 28.7 | 0.48 |
| L191A (619)* | 3.93±0.70 | 5.78±0.67 | 1.47 | 1.97–4.40 |
* The results are means for three experiments. The respective S.E.M. values for kon, koff and Kd were 0.55, 0.12 and 0.38. Lower χ2 values indicate better fit.
† Average of two experiments.
‡ Values that differ by >3-fold from the WT LBD are shown in boldface.
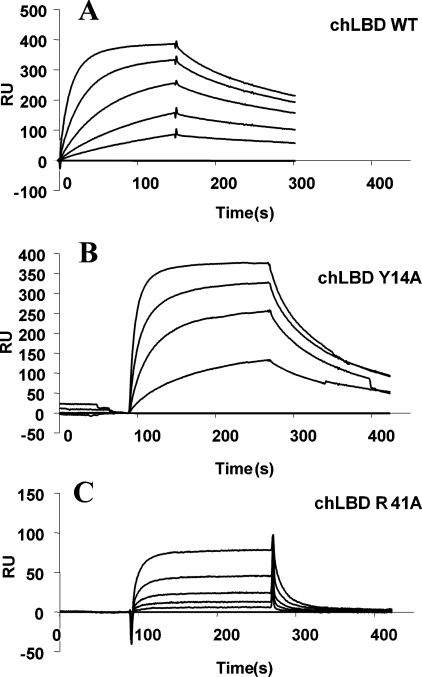
To test the effect of mutagenesis on the affinity of chLBD for the mutants, we used the system in which leptin or leptin mutants are immobilized: as in the reverse situation, very long periods were required to remove the leptin bound to immobilized chLBD (see the Materials and methods section). Thus chLBD or its mutants were used in the mobile phase and the results are summarized in Table 4. Mutagenesis aimed at determining whether mutations of several amino acids that differ between chLBD and mammalian LBDs will affect affinity clearly showed the lack of such an effect (see the upper part of Table 4), except for mutant A105D in which a slight decrease in affinity was observed. In contrast, alanine-scanning mutagenesis was helpful in identifying six residues in which the mutations resulted in total loss of binding capacity (Table 4 and Figure 7). It should be noted that four residues are identical and two (Val76 and Val103 in birds and, respectively, Ile76 and Leu103 in mammals) are similar in all compared species, hinting at their general importance for leptin binding. Mutations of four additional amino acids caused a 5–37-fold decrease in the Kd value, resulting in three mutants (Y14A, R41A and F136A) with faster dissociation and two cases (R41A/S42A/K43A and V103A) with both faster dissociation and slower association (see Table 4 and Figure 7). Sensograms of three typical cases are shown in Figure 8, demonstrating such differences in SPR analysis.
Figure 7. Sequence alignment of chicken, turkey, human, mouse and rat LBD.
Amino acids designated with ‘–’ are identical with those of chLBD. The sequence begins at amino acid 429. Mutated residues are in boldface and residues in which the mutation reduced or abolished the affinity towards oLep are underlined. The missing amino acid in chicken and turkey receptor is marked as ‘.’.
Figure 8. Interaction of oLep covalently linked to CM-dextran with chLBD (A), chLBD Y14A (B) and chLBD R41A (C).
After oLep immobilization, serial dilutions (2-fold) of each LBD (starting at 250 nM) were injected for 150–180 s at the rate of 30 μl/min. Then the chip was washed with HBS-EP buffer for 150 s. For other details, see text.
DISCUSSION
In the present study, the minimal binding domain of the chLep receptor (chLBD) was subcloned and recombinantly prepared in an E. coli system. The resultant protein sequence differs slightly from its counterpart in the previously published chLep receptor sequence ([22,24]; GenBank® accession no. AF169827 versus accession no. AB033383 respectively), and the subcloned chLBD has the Ile residue at position 1 (after Met-Ala) instead of Val. This residue was inserted by the 5′-primer that was also used for subcloning hLBD. The rest of the chLBD sequence is similar to the Ohkubo et al. [24] sequence, excluding the residues at positions 63 and 95, where Phe and Leu were exchanged with Leu and Phe. On the basis of this comparison, we concluded that there is no significant difference between the chLBD cloned in this work and the equivalent part of the chLep receptor reported previously [24].
The recombinant chLBD was purified to a homogeneity of more than 95% monomeric protein, as shown by SDS/PAGE and gel-filtration analyses. The yield was relatively low, averaging 15–25 mg per 5 litre fermentation, but the ease of preparation made it feasible to produce enough material for structural and functional studies. The secondary structure of the chLBD was similar to that of hLBD and the ECD of other cytokine receptors, indicating proper folding [25–27].
The binding capacities of hLBD compared with full-length membrane-embedded hLep receptor were evaluated in a previous study [16]. Similar to hLBD, the pure chLBD was able to form a stable complex with leptin in a 1:1 stoichiometric ratio, as revealed by gel-filtration experiments and SPR analysis. Comparison of the SPR-determined binding kinetics of chLBD and hLBD showed no significant differences. In a vice versa situation, wherein chLBD was immobilized and hLep was used as a soluble ligand, the Kd of the interaction was approx. 6-fold lower than that in the reverse situation. The change in the affinity did not result from a different kon value but was due to slower dissociation (see Table 3). Although we have no explanation for this phenomenon, it may result from the fact that immobilized leptin has less flexibility to bind LBD as compared with the opposite situation, wherein the chLBD was immobilized. A similar discrepancy has also been observed in a study of the interaction between the growth hormone receptor ECD and growth hormone [28,29].
The SPR analysis identified six mutants in which binding was completely abolished (F73A, Y14A/F73A, V76A/F77A, L78A/L79A, V76A/F77A/L78A/L79A and A105D/D106V) and six mutants (Y14A, R41A, R41A/S42A/K43A, V103A, F136A and V135A/F136A) in which affinity towards the hormone was reduced, mainly by increased dissociation rates. The results obtained by SPR were also verified by binding experiments in solution using 125I-hLep as a ligand and oLep as competitor. All mutants that did not show significant decreases in the affinity for the ligand in SRP experiments did not differ from chLBD in the binding assay as well (see Figures 5B and 5C) and the six mutants in which the binding was abolished (see Table 4) could not bind specifically 125I-hLep. It should be noted that despite the fact that the random mobilization technique was used, the stoichiometry analysis indicated 1:1 molar ratio of the ligand–chLBD complexes, similar to the gel-filtration experiments. Furthermore, even though not all ligand molecules were homogenously immobilized, the comparison of the binding properties of the WT versus mutated chLBD was valid as also verified by binding experiments in solutions, using the radioactive ligand.
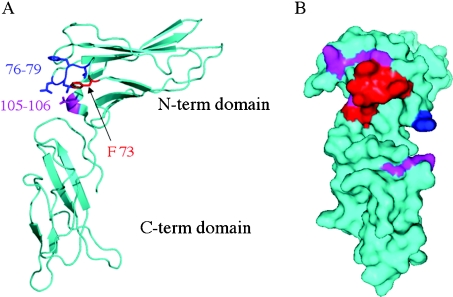
Based on our previous homology model of human LBD, we modelled the chLBD–oLep complex and examined the binding determinants with reference to the affinities of the chLBD mutations. The model of the chLBD–oLep complex indicates a site-1 contact surface between the receptor's binding domain and the hormone. The binding determinants consist of six binding regions, which are in consensus with other cytokine receptor complexes [25–27,30,31]. The six regions consist of five extended loop segments (L1, L2, L3, L5 and L6) and the short interdomain linker (L4). Extensive mutational analysis indicated the contribution of the chLBD loops to the binding properties. In chLBD, there are select amino acid residues that, upon mutagenesis, completely abolish hormone binding. These residues are located in the two binding segments L3 and L4. The L3 loop contributes nine amino acid residues that interact with leptin. Single and multiple mutations in the L3 region indicate that it is the most crucial segment for leptin binding. In this context, the L3 loop contains two Phe residues at positions 73 and 77, which are extended towards the ligand. A point mutation, F73A, abolishes leptin binding. In addition, the double mutants V76A/F77A and L78A/L79A, as well as the resultant quadruple combination, result in complete loss of leptin binding (Table 4). Based on these data, we conclude that the L3 loop contains several critical residues, which are essential for hormone binding (Figure 9A). Contact-surface analysis [32] indicated that the residues with the highest contribution to the recognition site are Phe73, Phe77 and Leu79. These results are in full agreement with the mutagenesis data. Available data on cytokine-receptor-binding determinants identify a few residues that are essential to ligand binding [33], and these usually include aromatic residues. In growth hormone receptor, prolactin receptor and erythropoietin receptor, L3 contributes an essential aromatic residue; the second residue is donated by another loop from the D2 domain. In chLBD, both essential aromatic residues are donated by the L3 loop and, in addition, Leu79 may also play an important role in ligand binding.
Figure 9. inding determinants of chLBD with oLep.
(A) Tube presentation of the chLBD indicating the segments that, upon mutations, abolished binding towards oLep completely. These segments include Phe73 located in L3 (red), residues 76–79 from L3 (blue) and residues 105–106 located in L4 that links the two domains (magenta). (B) Surface presentation of chLBD where the molecule is rotated by 90° clockwise along the vertical axis with reference to the orientation on the left-hand side. The colour code indicates the effect of the mutations on oLep binding. Red, magenta and blue colours designate no binding, substantial and moderate reductions in binding affinities respectively according to the results displayed in Table 4.
The L4 region makes an important contribution to ligand recognition in chLBD. Mutants V103A and A105D reduce affinity for leptin by factors of seven and four respectively. Conversely, the D106S mutation maintains an affinity for oLep that is similar to that of the WT. However, the double-mutant A105D/D106V does not show any binding to the hormone. This is quite unexpected, since our model indicates that there is very little contact area between the three residues in L4 and the ligand. L4 is located in the linker between the D1 and D2 domains and stabilizes their mutual orientation. We thus assume that the combined mutation in the L4 helical segment disrupts correct folding in this critical region, resulting in a different D1–D2 orientation, which will not permit any ligand binding (Figure 9A).
The chLBD mutation data also indicate that the L5 region contributes to hormone binding and that the double-mutant V135A/F136A shows reduced affinity for leptin. L5 has been shown to contribute critical residues in the binding of other members of the cytokine receptor superfamily, such as growth hormone and prolactin receptors. In addition, Tyr14, located in the L1 (Figure 9B) loop, makes a minor contribution to the binding since the Y14A mutation has almost no effect on the affinity for the hormone. Several other mutations that have minor effects on hormone binding (Figure 9B) are part of the critical multiple mutations that abolish binding.
Acknowledgments
A.G. was supported by the Israeli Science Foundation (grant no. 594/02).
References
- 1.Friedman J. M., Halaas J. L. Leptin and the regulation of body weight in mammals. Nature (London) 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 2.Cioffi J. A., Shafer A. W., Zupancic T. J., Smith-Gbur J., Mikhail A., Platika D., Snodgrass H. R. Novel B219/OB receptor isoforms: possible role of leptin in hematopoiesis and reproduction. Nat. Med. 1996;2:585–589. doi: 10.1038/nm0596-585. [DOI] [PubMed] [Google Scholar]
- 3.Emilsson V., Liu Y. L., Cawthorne M. A., Morton N. M., Davenport M. Expression of the functional leptin receptor mRNA in pancreatic islets and direct inhibitory action of leptin on insulin secretion. Diabetes. 1997;46:313–316. doi: 10.2337/diab.46.2.313. [DOI] [PubMed] [Google Scholar]
- 4.Hoggard N., Hunter L., Duncan J. S., Williams L. M., Trayhurn P., Mercer J. G. Leptin and leptin receptor mRNA and protein expression in the murine fetus and placenta. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11073–11078. doi: 10.1073/pnas.94.20.11073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glasow A., Haidan A., Hilbers U., Breidert M., Gillespie J., Scherbaum W. A., Chrousos G. P., Bornstein S. R. Expression of Ob receptor in normal human adrenals: differential regulation of adrenocortical and adrenomedullary function by leptin. J. Clin. Endocrinol. Metab. 1998;83:4459–4466. doi: 10.1210/jcem.83.12.5337. [DOI] [PubMed] [Google Scholar]
- 6.Briscoe C. P., Hanif S., Arch J. R., Tadayyon M. Leptin receptor long-form signalling in a human liver cell line. Cytokine. 2001;14:225–229. doi: 10.1006/cyto.2001.0871. [DOI] [PubMed] [Google Scholar]
- 7.Zhang F., Basinski M. B., Beals J. M., Briggs S. L., Churgay L. M., Clawson D. K., DiMarchi R. D., Furman T. C., Hale J. E., Hsiung H. M., et al. Crystal structure of the obese protein leptin-E100. Nature (London) 1997;387:206–209. doi: 10.1038/387206a0. [DOI] [PubMed] [Google Scholar]
- 8.Livnah O., Stura E. A., Middleton S. A., Johnson D. L., Jolliffe L. K., Wilson I. A. Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science. 1999;283:987–990. doi: 10.1126/science.283.5404.987. [DOI] [PubMed] [Google Scholar]
- 9.Wells J. A., De Vos A. M. Hematopoietic receptor complexes. Annu. Rev. Biochem. 1996;65:609–634. doi: 10.1146/annurev.bi.65.070196.003141. [DOI] [PubMed] [Google Scholar]
- 10.Aritomi M., Kunishima N., Okamoto T., Kuroki R., Ota Y., Morikawa K. Atomic structure of the GCSF-receptor complex showing a new cytokine-receptor recognition scheme. Nature (London) 1999;401:713–717. doi: 10.1038/44394. [DOI] [PubMed] [Google Scholar]
- 11.Fong T. M., Huang R. R., Tota M. R., Mao C., Smith T., Varnerin J., Karpitskiy V. V., Krause J. E., Van der Ploeg L. H. Localization of leptin binding domain in the leptin receptor. Mol. Pharmacol. 1998;53:234–240. doi: 10.1124/mol.53.2.234. [DOI] [PubMed] [Google Scholar]
- 12.Muller-Newen G. The cytokine receptor gp130: faithfully promiscuous. Science STKE 2003. 2003. p. PE40. [DOI] [PubMed]
- 13.Zabeau L., Defeau D., Van der Heyden J., Iserentant H., Vandekerckhove J., Tavernier J. Functional analysis of leptin receptor activation using a Janus kinase/signal transducer and activator of transcription complementation assay. Mol. Endocrinol. 2004;18:150–161. doi: 10.1210/me.2003-0078. [DOI] [PubMed] [Google Scholar]
- 14.Peelman F., Van Beneden K., Zabeau L., Iserentant H., Ulrichts P., Defeau D., Verhee A., Catteeuw D., Elewaut D., Tavernier J. Mapping of the leptin binding sites and design of a leptin antagonist. J. Biol. Chem. 2004;279:41038–41046. doi: 10.1074/jbc.M404962200. [DOI] [PubMed] [Google Scholar]
- 15.Niv-Spector L., Gonen-Berger D., Gourdou I., Biener E., Gussakowsky E. E., Benomar Y., Ramanujan K. V., Taouis M., Herman B., Callebout I., et al. Identification of the hydrophobic strand in the A-B loop of leptin as major binding site III: implications for large-scale preparation of potent recombinant human and ovine leptin antagonists. Biochem. J. 2005 doi: 10.1042/BJ20050457. doi:10.1042/BJ20050457. [DOI] [PMC free article] [PubMed]
- 16.Sandowski Y., Raver N., Gussakovsky E. E., Shochat S., Dym O., Livnah O., Rubinstein M., Krishna R., Gertler A. Subcloning, expression, purification, and characterization of recombinant human leptin-binding domain. J. Biol. Chem. 2002;277:46304–46309. doi: 10.1074/jbc.M207556200. [DOI] [PubMed] [Google Scholar]
- 17.Gertler A., Simmons J., Keisler D. H. Large-scale preparation of biologically active recombinant ovine obese protein (leptin) FEBS Lett. 1998;442:137–140. doi: 10.1016/s0014-5793(97)01613-x. [DOI] [PubMed] [Google Scholar]
- 18.Raver N., Vardy E., Livnah O., Devos R., Gertler A. Comparison of R128Q mutations in human, ovine, and chicken leptins. Gen. Comp. Endocrinol. 2002;126:52–58. doi: 10.1006/gcen.2001.7766. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Gertler A., Ashkenazi A., Madar Z. Binding sites of human growth hormone and ovine and bovine prolactins in the mammary gland and the liver of lactating dairy cow. Mol. Cell. Endocrinol. 1984;34:51–57. doi: 10.1016/0303-7207(84)90158-8. [DOI] [PubMed] [Google Scholar]
- 21.Johnsson B., Lofas S., Lindquist G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem. 1991;198:268–277. doi: 10.1016/0003-2697(91)90424-r. [DOI] [PubMed] [Google Scholar]
- 22.Horev G., Einat P., Aharoni T., Eshdat Y., Friedman-Einat M. Molecular cloning and properties of the chicken leptin-receptor (CLEPR) gene. Mol. Cell. Endocrinol. 2000;162:95–106. doi: 10.1016/s0303-7207(00)00205-7. [DOI] [PubMed] [Google Scholar]
- 23.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohkubo T., Tanaka M., Nakashima K. Structure and tissue distribution of chicken leptin receptor (cOb-R) mRNA. Biochim. Biophys. Acta. 2000;1491:303–308. doi: 10.1016/s0167-4781(00)00046-4. [DOI] [PubMed] [Google Scholar]
- 25.De Vos A. M., Ultsch M., Kossiakoff A. A. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 26.Somers W., Ultsch M., De Vos A. M., Kossiakoff A. A. The X-ray structure of a growth hormone-prolactin receptor complex. Nature (London) 1994;372:478–481. doi: 10.1038/372478a0. [DOI] [PubMed] [Google Scholar]
- 27.Elkins P. A., Christinger H. W., Sandowski Y., Sakal E., Gertler A., de Vos A. M., Kossiakoff A. A. Ternary complex between placental lactogen and the extracellular domain of the prolactin receptor. Nat. Struct. Biol. 2000;7:808–815. doi: 10.1038/79047. [DOI] [PubMed] [Google Scholar]
- 28.Cunningham B. C., Wells J. A. Comparison of a structural and a functional epitope. J. Mol. Biol. 1993;234:554–563. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]
- 29.Gertler A., Grosclaude J., Strasburger C. J., Nir S., Djiane J. Real-time kinetic measurements of the interactions between lactogenic hormones and prolactin-receptor extracellular domains from several species support the model of hormone-induced transient receptor dimerization. J. Biol. Chem. 1996;271:24482–24491. doi: 10.1074/jbc.271.40.24482. [DOI] [PubMed] [Google Scholar]
- 30.Livnah O., Stura E. A., Johnson D. L., Middleton S. A., Mulcahy L. S., Wrighton N. C., Dower W. J., Jolliffe L. K., Wilson I. A. Functional mimicry of a protein hormone by a peptide agonist: the EPO receptor complex at 2.8 A. Science. 1996;273:464–471. doi: 10.1126/science.273.5274.464. [DOI] [PubMed] [Google Scholar]
- 31.Syed R. S., Reid S. W., Li C., Cheetham J. C., Aoki K. H., Liu B., Zhan H., Osslund T. D., Chirino A. J., Zhang J., et al. Efficiency of signalling through cytokine receptors depends critically on receptor orientation. Nature (London) 1998;395:511–516. doi: 10.1038/26773. [DOI] [PubMed] [Google Scholar]
- 32.Jones S., Thronton J. M. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clackson T., Wells J. A. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]