Abstract
In the present work, the effect of Na+ binding on the conformational, stability and molecular recognition properties of thrombin was investigated. The binding of Na+ reduces the CD signal in the far-UV region, while increasing the intensity of the near-UV CD and fluorescence spectra. These spectroscopic changes have been assigned to perturbations in the environment of aromatic residues at the level of the S2 and S3 sites, as a result of global rigidification of the thrombin molecule. Indeed, the Na+-bound form is more stable to urea denaturation than the Na+-free form by ∼2 kcal/mol (1 cal≡4.184 J). Notably, the effects of cation binding on thrombin conformation and stability are specific to Na+ and parallel the affinity order of univalent cations for the enzyme. The Na+-bound form is even more resistant to limited proteolysis by subtilisin, at the level of the 148-loop, which is suggestive of the more rigid conformation this segment assumes in the ‘fast’ form. Finally, we have used hirudin fragment 1–47 as a molecular probe of the conformation of thrombin recognition sites in the fast and ‘slow’ form. From the effects of amino acid substitutions on the affinity of fragment 1–47 for the enzyme allosteric forms, we concluded that the specificity sites of thrombin in the Na+-bound form are in a more open and permissible conformation, compared with the more closed structure they assume in the slow form. Taken together, our results indicate that the binding of Na+ to thrombin serves to stabilize the enzyme into a more open and rigid conformation.
Keywords: allostery, coagulation, hirudin, limited proteolysis, Na+, thrombin
Abbreviations: ARA, synthetic analogue of the hirudin fragment 1–47 containing the substitutions Val1→Ala, Ser2→Arg and Tyr3→Ala; ARW, synthetic analogue of the fragment 303–316 of thermolysin, with the sequence AASVRQAWDAVGVK; Bip, L-4,4′-biphenylalanine; ChCl, choline chloride; FPR, D-Phe-Pro-Arg-p-nitroanilide; βNal, L-2-naphthylalanine; PDB, Protein DataBank; PEG, poly(ethylene glycol); pGndPhe, p-guanidino-L-phenylalanine; pNA, p-nitroanilide; RP, reverse-phase; TFA, trifluoroacetic acid
INTRODUCTION
Thrombin plays a key role at the interface between coagulation, inflammation and cell growth [1], and exerts either procoagulant or anticoagulant functions in haemostasis [2]. The procoagulant role entails conversion of fibrinogen into fibrin and platelet activation, whereas the anticoagulant role involves the activation of Protein C [3]. The most effective modulator of thrombin function in solution is Na+, which triggers the transition of the enzyme from an anticoagulant (‘slow’) form to a procoagulant (‘fast’) form [4–6]. The Na+-bound (fast) form displays procoagulant properties, since it cleaves more specifically fibrinogen and protease-activated receptor, whereas the Na+-free (slow) form is anticoagulant because it retains the normal activity toward Protein C, but is unable to promote acceptable hydrolysis of procoagulant substrates [5,6]. The importance of Na+ in thrombin function is highlighted by the fact that under physiological conditions the two forms occur almost equally, and that natural thrombin variants with compromised Na+ binding result into bleeding phenotypes [6].
The effect of Na+ binding on thrombin function is allosteric in nature [5,7], and several crystal structures of the enzyme, with and without Na+ bound, have been reported [8–11]. The structures of the pseudo wild-type thrombin mutant [Arg77a→Ala (where the lower case letter here and elsewhere in paper refers to the residue present in the insertion loops of thrombin, that are absent in chymotrypsin)], reported by Di Cera and co-workers [8,9], in the presence [Protein DataBank (PDB) code, 1SG8] or absence (PDB code, 1SGI) of Na+, displayed only small changes in the side-chain orientation of Ser195 in the active site, Asp189 in the S1 site, Glu192 and Asp222 on the protein surface, and some rearrangement of the water molecules filling the S1 site. The binding of Na+ also increased the average crystallographic B factor of thrombin, suggesting a higher conformational flexibility in the fast form [9]. Conversely, the structures of the fast (1JOU_AB and 1JOU_CD) and slow (1JOU_EF) form reported by Huntington and Esmon [10], based on the inactive Ser195→Ala mutant, all contained a Na+ ion bound and showed significant differences at the level of the S2 and S3 sites, which in the slow form protrude on to the protein surface and limit the access to the catalytic pocket. In particular, the apolar cavity of the S3 site is restricted by protrusion of Trp215, possibly caused by reorientation of the underlying Phe227 and Cys168–Cys182 disulphide bond. Similar changes are also observed in the structure of the Glu217→Lys mutant (PDB code, 1RD3), which does not bind Na+ and has been proposed to capture the essential features of the slow form [11]. Other conformational changes occur in the Na+-binding site, in the 148-loop and at the level of the exosite I. In particular, the salt bridges anchoring the Na+ site to the 148-loop (Glu146–Arg221a) and to the 166–171 helix (Lys185–Glu164) are disrupted in this mutant structure, with a consequent partial unfolding of the Na+ site and collapse of the 148-loop on to the groove leading to the catalytic pocket. However, it may be difficult to discriminate between the structural changes pertaining to the slow form of the wild-type enzyme and those caused by charge reversal (Glu→Lys substitution) in the Na+-binding site. Furthermore, all the putative structures of the fast and slow forms reported so far display numerous contacts between thrombin monomers in the crystal lattice [8–11], and therefore crystal packing effects cannot be ruled out.
In the present study we have investigated the conformational, stability and molecular recognition properties of thrombin in solution by combining several different spectroscopic and biochemical techniques, under temperature and salt conditions in which the enzyme predominantly (>90%) exists in the fast (0.2 M NaCl) or in the slow [0.2 M ChCl (choline choride)] form [4,5]. Our results demonstrate that Na+ binding stabilizes the enzyme in a more open and rigid conformation and that this effect is specific to Na+.
EXPERIMENTAL
Reagents
The chromogenic substrate FPR (D-Phe-Pro-Arg-p-nitroanilide) was synthesized as previously described [12]. The peptide ARW (synthetic analogue of the fragment 303–316 of thermolysin, with the sequence AASVRQAWDAVGVK) was obtained from Neosystem Laboratories (Strasburg, France). Nα-Fmoc (fluoren-9-ylmethoxycarbonyl)-protected amino acids, solvents and reagents for peptide synthesis were purchased from Applied Biosystems (Foster City, CA, U.S.A.) or Bachem AG (Bubendorf, Switzerland). Proteases, reagents for electrophoresis, buffers and organic solvents were of analytical grade and obtained from Fluka.
Preparation of thrombin samples
Aliquots (0.4–0.7 mg) of commercial human α-thrombin from Haematologic Technologies (Essex Junction, VT, U.S.A.) were loaded on to a fast-flow HiTrap column (1.6 cm×2.5 cm) from Pharmacia (Uppsala, Sweden), eluted at a flow rate of 1 ml/min with 5 mM Tris/HCl buffer, pH 8.0, containing 0.1% (w/v) PEG [poly(ethylene glycol)]-8000 and 0.2 M chloride salt, as indicated. Alternatively, 5 mM Bis-Tris buffer, pH 6.5, was used. The absorbance of the effluent was recorded at 280 nm. The purity of thrombin preparations (∼98%) was established by SDS/PAGE (12% acrylamide gel) and RP (reverse-phase)-HPLC on a C4 analytical column from Vydac (Hesperia, CA, U.S.A.). MS analysis of thrombin samples, carried out on a Mariner instrument from Perseptive Biosystems (Stafford, TX, U.S.A.), gave a mass value of 36030±3 a.m.u. (atomic mass units). Freshly prepared thrombin samples were immediately used for subsequent experiments.
Spectroscopic techniques
Measurements were carried out at least in duplicate in the correct buffer and the final spectrum resulted from subtraction of the corresponding baseline. Temperature correction was applied for Tris buffer.
Peptide and protein concentration was determined by UV absorbance at 280 nm on a Lambda-2 spectrophotometer from PerkinElmer (Cupertino, CA, U.S.A.) using molar absorption coefficients (ϵ) obtained experimentally, as for thrombin [13], or calculated on the basis of the amino acid composition [14] (see Supplementary Table 1S, http://www.BiochemJ.org/bj/390/bj3900485add.htm). CD spectra were recorded on a Jasco (Tokyo, Japan) model J-810 spectropolarimeter. Far-UV spectra were recorded in a 1-mm cell, at a scan-speed of 10 nm/min, with a response time of 16 s, and were the average of four accumulations. Near-UV spectra were recorded in a 1-cm cell, at a scan-speed of 50 nm/min, with a response time of 2 s, and were the average of 16 accumulations. Fluorescence spectra were recorded on a PerkinElmer spectrofluorimeter model LS-50B. Spectra were taken at a scan speed of 120 nm/min by exciting the protein samples (equilibrated for 10 min in the correct buffer) at 280 nm, using an excitation/emission slit of 5 nm.
Urea-mediated denaturation of human α-thrombin and bovine α-chymotrypsin was carried out at 25 °C, at pH 8.0 and 6.5, in the presence of 0.2 M chloride salts, after 30-min incubation in the correct buffer, as described in [15]. Spectra were recorded at a scan speed of 240 nm/min, with an excitation/emission slit of 5 nm and resulted from the average of four accumulations, after baseline subtraction. Reversibility of the denaturation process was evaluated by measuring the recovery of the fluorescence intensity or λmax value upon 20-fold dilution of enzyme stock solutions (3 μM) in 7 M urea with non-denaturing buffer at either pH 8.0 or 6.5. When denaturation was only partially reversible (i.e. at pH 8.0), the value of ([urea]1/2) was estimated as reported [16]. When the unfolding was fully reversible (i.e. at pH 6.5), the data were analysed within the framework a two-state process [15].
Limited proteolysis
Time-course kinetic analyses of proteolysis reaction of human α-thrombin in 5 mM Tris/HCl buffer (pH 8.0)/0.1% PEG-8000 were carried out in duplicate at 5 and 25 °C (±0.2 °C) under identical and carefully controlled experimental conditions, in the fast (0.2 M NaCl) and slow (0.2 M ChCl) form. To a solution (600 μl) of thrombin (14 μM) were added 20–40 μl of subtilisin stock solution (0.55 μM) in the same buffer containing 5 mM CaCl2. At fixed time intervals, acid-trapped aliquots (35 μg) were analysed by SDS/PAGE (12% acrylamide gel) and by RP-HPLC on C4 Vydac analytical column, eluted with a linear acetonitrile/0.078% TFA (trifluoroacetic acid) gradient from 25 to 35% in 5 min and from 35 to 40% in 20 min, at a flow rate of 0.8 ml/min. The chemical identity of the material eluted from the RP-HPLC column was established by MS and by N-terminal sequence analysis carried out on a model 477A protein sequenator (Applied Biosystems). At each time point, the concentration of α-thrombin in solution was estimated by integrating the area under the chromatographic peak corresponding to the intact enzyme, and the data were analysed under pseudo-first-order conditions [17] using the computer program Microcal Origin 6.0. As a control, the effect of salt on the amidolytic activity of subtilisin was studied by incubating the enzyme (3 nM) at 5 and 25 °C with the synthetic peptide ARW (30 μM), under fast and slow conditions. Acid-trapped aliquots were analysed by RP-HPLC on a C18 Vydac column, eluted with a linear acetonitrile/0.078% TFA gradient from 5 to 40% in 25 min, at a flow rate of 0.8 ml/min.
Synthesis and anti-thrombin activity of hirudin 1–47 analogues
Hirudin analogues were synthesized, allowed to refold and purified as described previously [18,19]. The inhibitory potency of the synthetic analogues was determined by measuring the release of pNA (p-nitroaniline) from FPR (20 μM), as described previously [12,20]. In the case of slow-tight binders [i.e. Ser2→Arg and Ser2→pGndPhe (p-guanidino-L-phenylalanine)], thrombin (Calbiochem) was first incubated for 1 h with the inhibitor and then the reaction was started by addition of FPR.
Computational methods
The mutated side-chains were modelled essentially without bump on the structure of hirudin–thrombin complex (PDB code, 4HTC) [21], which was crystallized in the presence of NaCl, using the program Insight-II (Biosym Inc.). Cavity volumes were calculated by the computer program CASTp (http://cast.engr.uic.edu/cast1/), using a water probe radius of 1.4 Å. ASA (accessible surface area) and B factor calculations were carried out using a program available at http://molbio.info.nih.gov/structbio/basic.html. Strain energy of disulphide bonds were calculated as described in [22].
RESULTS AND DISCUSSION
Conformational and stability properties of thrombin allosteric forms
The effect of univalent cations having different sizes (Ch+>K+>Na+>Li+) on the conformation and stability of thrombin was investigated by numerous spectroscopic techniques at 25 °C and 0.2 M chloride salt, as indicated. Choline was used as an ‘inert’ cation to control ionic strength [23], since it is too bulky to interact with the enzyme. Significant differences were observed in the far- and near-UV CD spectra, fluorescence spectra and stability to urea (see below), whereas second-derivative UV spectroscopy [24], quenching of Trp fluorescence [25] and ANS (8-anilinonaphthalene-l-sulphonic acid)-binding experiments [26] (see Supplementary Figures 3S, 4S and 5S respectively, http://www.BiochemJ.org/bj/390/bj3900485add.htm) indicated that Na+ binding does not appreciably alter the solvent accessibility of aromatic amino acids and the surface hydrophobicity of thrombin. As a control, similar experiments were conducted in parallel on α-chymotrypsin (a serine protease displaying high structural similarity with thrombin, but lacking Na+-binding properties [27,28]) and on model compound solutions of aromatic amino acids. In both cases, the spectroscopic signal was found to be insensitive to the salt-type (see also Supplementary Figures 1S and 2S, http://www.BiochemJ.org/bj/390/bj3900485add.htm).
Conformational analysis
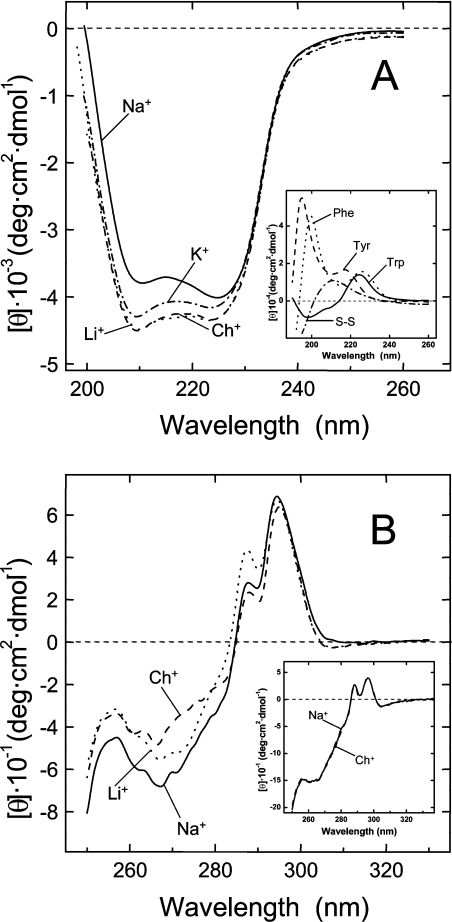
The shape of the far-UV CD spectrum of human α-thrombin resembles that of a protein possessing significant helical content (Figure 1A). However, the two minima centred at 210 and 225 nm are red-shifted by 3–5 nm relative to the bands typical of the helical conformation, occurring at 220–222 nm and 208 nm [29]. In addition, the intensity of the CD bands is unrealistically low for a polypeptide chain with substantial helical structure. Indeed, the crystallographic structure of thrombin [28] yields 14% α-helix and 27% β-sheet. These observations suggest that the far-UV CD spectrum of thrombin may be affected by the contribution of aromatic amino acids and disulphide bonds in the far-UV region. With this in mind, the spectra of cystine and suitable models of Phe, Tyr and Trp are shown in Figure 1(A, inset). Such effects become more important for those proteins displaying low signal intensity and are most prominent in systems where aromatic groups are clustered in the protein structure [29]. Of note, three aromatic/hydrophobic clusters can be clearly identified in thrombin, comprising the S2 and S3 specificity sites [28] (see also the legend to Figure 5). Compared with choline, Na+ binding to thrombin reduces the intensity and alters the shape of the CD spectrum. Interestingly, this effect is specific to Na+ (Na+>K+>Li+≈Ch+) and reflects conformational changes likely to occur in the environment of some aromatic amino acids clustered within the S2 and S3 sites of thrombin. However, all the putative structures of the fast and slow forms reported so far do not display any change in the secondary structure content of the enzyme [8–11].
Figure 1. Far-UV (A) and near-UV (B) CD spectra of thrombin in the presence of univalent cations.
All spectra were recorded at 25 °C in 5 mM Tris/HCl buffer, pH 8.0, containing 0.1% PEG-8000, in the presence of different chloride salts (0.2 M), as indicated. (A) CD spectra of thrombin in the far-UV region were taken at a protein concentration of 5 μM. Inset: far-UV spectra of cystine and Nα-acetyl-amide derivatives of Tyr, Phe and Trp in the same buffer, without salt added. Ellipticity value for model compounds solutions is expressed as molar ellipticity. (B) CD spectra of thrombin in the near-UV region were taken at a protein concentration of 30 μM. The spectrum recorded in the presence of KCl is intermediate between that obtained with LiCl and NaCl and, for the sake of clarity, it has been omitted. Inset: near-UV spectra of α-chymotrypsin (30 μM), in the presence of NaCl (——) or ChCl (------).
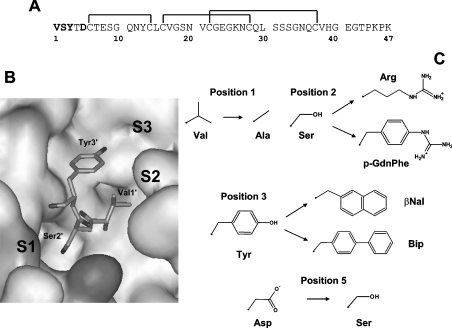
Figure 5. Amino acid substitutions introduced in the N-terminal domain (residues 1–47) of hirudin.
(A) Amino acid sequence of fragment 1–47 from hirudin HM2 (hirudin variant isolated from the leech Hirudinaria manillensis). The positions mutated are indicated in bold and disulphide bonds by the lines joining the cysteine residues. (B) Representation of the interaction of the N-terminal tripeptide of hirudin fragment 1–47 with thrombin (4HTC) [21]. The approximate localization of the amino acids defining the specifitity sites (S1, S2, and S3) on thrombin structure is indicated. Val1′ of hirudin is orientated towards the S2 site; Ser2′ covers the S1 site; Tyr3′ occupies the apolar S3 site. Three aromatic/hydrophobic clusters can be clearly identified in thrombin structure (1PPB) [28]: cluster I comprises the S2 site and is formed by Tyr94, Trp96, Pro60c, Tyr60a, and Trp60d; cluster II lines the S3 site and is formed by Trp215, Phe227, Cys168–Cys182 and Val163, the latter being in contact with Tyr225 in the Na+ site; cluster III is close to cluster II and formed by residues Phe181, Phe199 and Tyr228. (C) Amino acid replacements introduced into the hirudin fragment 1–47.
The near-UV spectrum of thrombin (Figure 1B) displays extensive detailed structure between 282 and 305 nm, assigned to the contribution of Trp residues, and between 260 and 280 nm, where the absorption of Phe and Tyr is predominant and masks the contribution of the four disulphide bonds [30]. In the presence of Na+, the CD signal is enhanced throughout the wavelength range explored, suggesting that cation binding induces local environmental asymmetry and conformational rigidification of the aromatic chromophores. Reorientation of the disulphide bridge of Cys168–Cys182, reported by Huntington and co-workers [10,11] (see the Introduction section), can also influence the CD spectrum. Interestingly, the presence of LiCl only slightly increases the CD signal in the 250–280 nm range, in agreement with the lower affinity (∼20-fold) of this cation for thrombin relative to Na+ [31].
Binding of Na+ to thrombin enhances the fluorescence intensity by ∼18% (Figure 2), with no change in the λmax value at 342 nm, in agreement with previous results [32] and compatible with a global rigidification of the enzyme in the Na+-bound form [25], even though specific local interactions (i.e. reorientation of the Phe227–Trp215 pair [10,11]) can also influence thrombin fluorescence. As already observed in the far-UV CD, the effect of univalent cations on thrombin fluorescence is specific for Na+ and follows the order Na+≫K+>Li+≈Ch+ (Figure 2).
Figure 2. Fluorescence spectra of thrombin in the presence of monovalent cations.
Fluorescence spectra were recorded at 25 °C in 5 mM Tris/HCl buffer, pH 8.0, containing 0.1% PEG-8000, in the presence of different chloride salts (0.2 M), as indicated. Thrombin samples (2 ml, 0.15 μM), prepared by 40-fold dilution of a stock solution with the proper buffer, were excited at 280 nm. The spectrum of denatured thrombin in 6 M guanidino-HCl is also shown. Inset: fluorescence spectra of α-chymotrypsin (0.15 μM) recorded under fast (0.2 M NaCl) and slow (0.2 M ChCl) conditions.
Stability studies
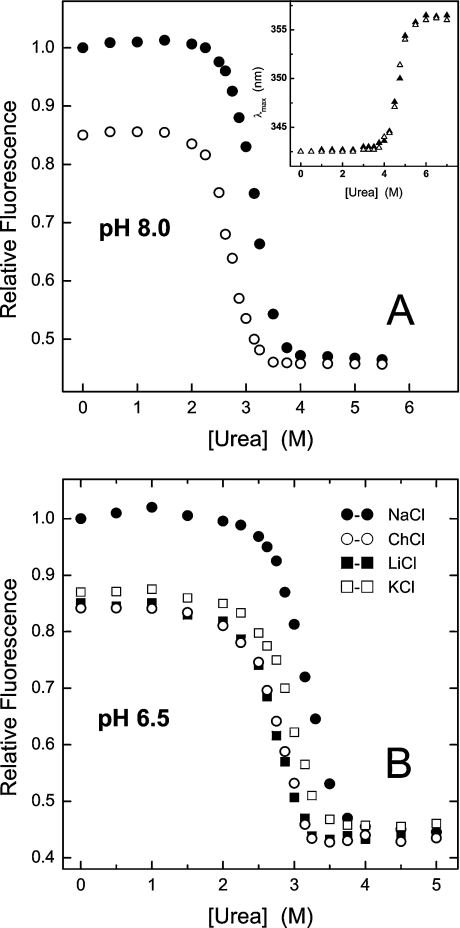
Thrombin denaturation experiments were carried out at pH 8.0 and 6.5 (Figures 3A and 3B respectively). In both cases, a single sharp transition was observed, suggestive of a highly co-operative unfolding process [15]. At pH 8.0 the concentration of urea inducing the 50% effect, [urea]1/2, on the fluorescence signal (i.e. intensity at 342 nm or λmax shift) was estimated as 2.7 M for the slow form and 3.2 M for the fast form. As a control, the stability of chymotrypsin remained constant after exchanging ChCl with NaCl (Figure 3A, inset). Thrombin denaturation was only 60% reversible at pH 8.0, whereas full reversibility was observed when the λmax shift was considered. This effect is probably caused by the increased autolytic degradation of the enzyme at pH 8.0, where thrombin is maximally active, and/or by the sparing solubility of the unfolded enzyme at this pH, quite close to its isoelectric point (pI=7.6) [13]. Hence, we carried out urea denaturation experiments at pH 6.5, where thrombin autolysis is minimized and its solubility much increased [13] (Figure 3B). Notably, at this pH the unfolding was completely reversible (>95%) and denaturation data were analysed within the framework of a two-state model, as previously reported for α-chymotrypsin [33]. The values of [urea]1/2 were identical to those obtained at pH 8.0, and the thermodynamic stability (ΔGU) of the fast and slow forms were estimated as 10.3±0.8 and 8.2±0.5 kcal·mol−1 (1 cal≡4.184 J) respectively. As expected for a protein undergoing a two-state reversible unfolding transition [34], the free energy change of Na+ binding to thrombin at 25 °C (ΔGb=−2.2 kcal·mol−1) [32] was completely channelled into stabilization of the fast form (ΔΔGU=ΔGUslow−ΔGUfast=−2.1 kcal·mol−1), in keeping with the notion that metal ions stabilize proteins by exclusively binding to the folded state [34]. Figure 3(B) clearly demonstrates that the stabilizing effect of cation binding is specific for Na+ and follows the order Na+>K+>Li+≈Ch+, identical to the affinity order of univalent cations for thrombin (Na+>K+>Li+) [31,32].
Figure 3. Effect of univalent cations on the urea stability of thrombin at pH 8.0 and 6.5.
(A) Measurements were carried out at 25 °C in 5 mM Tris/HCl buffer, pH 8.0, containing 0.1% PEG-8000, in the presence of 0.2 M NaCl (●) or ChCl (○). Fluorescence measurements were carried out by exciting the samples (2 ml, 0.15 μM) at 280 nm and recording the intensity at 342 nm as a function of urea concentration. The value of [urea]1/2 was estimated as 2.7 M for the Na+-free (slow) form and 3.2 M for the Na+-bound (fast) form. Inset: urea-induced denaturation of bovine α-chymotrypsin under fast (▲) and slow (△) conditions. Because the fluorescence change of chymotrypsin unfolding at 342 nm was too small (<20%) to be accurately measured, the λmax shift was taken as a probe of denaturation [15]. An identical value of 4.7 M was estimated for [urea]1/2, either in the presence of NaCl or ChCl. (B) Thrombin denaturation was carried out at 25 °C in 5 mM Bis-Tris, pH 6.5, containing 0.1% PEG-8000, in the presence of 0.2 M chloride salt, as indicated. A value of [urea]1/2 was estimated as 2.7 M with ChCl (○) or LiCl (■); 2.9 M in the presence of KCl (□); 3.2 M in the presence of NaCl (●).
Kinetic studies in solution indicated substantial conformational heterogeneity of the slow form [35], and the crystal structures of thrombin allosteric forms proposed by Huntington and co-workers [10,11] provide evidence for a significant Na+-binding-coupled structural rearrangement. In this view, the stabilizing effect of cation binding is likely to originate from the balance of opposing forces, including electrostatic coupling and cation desolvation, side-chain packing and conformational ordering. Conformational energy calculations [22] (see Supplementary Table 2S, http://www.BiochemJ.org/bj/390/bj3900485add.htm) indicate that the strain energy (E) of the Cys168–Cys182 disulphide bond in the hypothetical fast form (1JOU_AB) is significantly higher (E=3.8–4.0 kcal/mol) than that estimated for the slow form (1JOU_EF; 1RD3) (E=2.0–2.2 kcal/mol) [10,11].
Probing the conformational flexibility of thrombin allosteric forms by limited proteolysis
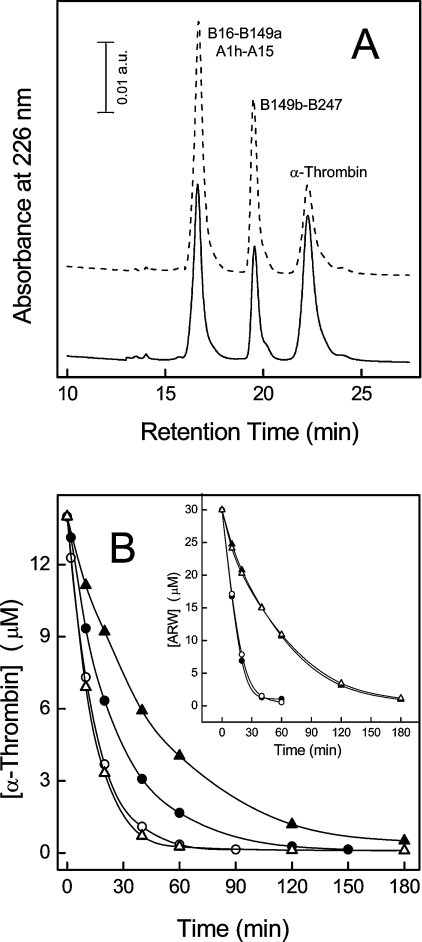
Limited proteolysis reaction is a very sensitive probe of protein structure and chain flexibility [36], and therefore, to unravel possible differences in the conformation and dynamics of thrombin allosteric forms, we compared the susceptibility of thrombin to proteolysis by subtilisin at 5 and 25 °C, under slow and fast conditions. Proteolysis reactions were analysed by RP-HPLC (Figure 4A) and SDS/PAGE (results not shown), whereas the chemical identity of the proteolytic fragments was established by MS and N-terminal sequence analysis. Our results indicate that subtilisin specifically cleaves the fast and slow form of thrombin at the same level, between Ala149a and Asn149b in the 148-loop, in agreement with previous results [37] and consistent with the higher flexibility of this segment in the enzyme structure [28].
Figure 4. Limited proteolysis of thrombin allosteric forms by subtilisin.
(A) RP-HPLC analysis of the proteolysis reaction of thrombin with subtilisin at 25 °C, in the fast (—) and slow (---) form, using a subtilisin/thrombin ratio of 1:1000 (w/w). The numbers near the chromatographic peaks identify the proteolytic fragments, according to the chymotrypsinogen numbering. (B) Time-course kinetics of the limited proteolysis reaction of thrombin allosteric forms. The reaction was conducted at 5 and 25 °C using subtilisin/thrombin ratios of 1:500 and 1:1000 respectively. Continuous lines were drawn using the equation describing the pseudo-first-order kinetics [17], with best-fit parameters: fast form at 25 °C (●), s=0.037 μM−1·s−1; slow form at 25 °C (○), s=0.062 μM−1·s−1; fast form at 5 °C (▲), s=0.009 μM−1·s−1; slow form at 5 °C (△), s=0.031 μM−1·s−1. Typically, the error on the determination of s values was <5%. The extent of autolytic degradation of thrombin in the absence of subtilisin was estimated by RP-HPLC analysis and found to be negligible (<1%) after 3 h. Inset: proteolysis reaction of the ARW peptide, used as a control. The reaction was conducted at 5 and 25° at a subtilisin/ARW molar ratio of 1:10000 in the presence of 0.2 M NaCl or ChCl. The concentration of uncleaved ARW was determined by RP-HPLC. Best-fit parameters values are: fast form at 25 °C (●), s=0.370 μM−1·s−1; slow form at 25 °C (○), s=0.350 μM−1·s−1; fast form at 5 °C (▲), s=0.097 μM−1·s−1; slow form at 5 °C (△), s=0.102 μM−1·s−1.
The 148-loop is strategically positioned between the Na+-binding site and exosite I in thrombin [28] and is crucial for fibrinogen recognition [38]. It is characterized by high intrinsic conformational heterogeneity [10,11,21,28,39], but it is still unclear whether the different structural modes observed for the 148-loop represent real responses to different ligands or if they result from crystal-packing effects. Analysis of the progress curves reported in Figure 4(B) indicates the specificity constant (s) for the hydrolysis of thrombin by subtilisin at 25 °C is increased by almost two times in the slow form relative to the fast form, suggesting that the Na+-bound form of the enzyme is more resistant to proteolysis. This effect is more pronounced (4-fold higher) at 5 °C, where the Na+-bound form is even more populated (Kd=5 mM) [35]. By using the ARW peptide, which is fully unfolded in solution (V. De Filippis, unpublished work), we have verified that the proteolytic activity of subtilisin was independent of the salt type (i.e. NaCl or ChCl) (Figure 4B, inset). These results indicate that the different susceptibility of thrombin allosteric forms to subtilisin do reflect the different conformation that the 148-loop assumes in the two forms and provide clear-cut evidence that upon Na+ binding this loop becomes more conformationally rigid.
The inverse relation existing between susceptibility to proteolysis and urea stability of thrombin allosteric forms suggests that rigidification induced by Na+ binding is not restricted to the 148-loop, but instead is a more widely distributed property of the enzyme in the Na+-bound form, reflecting the general rigidity–stability relationship observed in other (metal-binding) proteins [34,40].
Structural mapping of thrombin recognition sites in the fast and slow form
The N-terminal domain 1-47 of hirudin (Figure 5A) extensively interacts with thrombin recognition sites (Figure 5B) and, like intact hirudin [5,41] and fibrinogen [4,5], binds preferentially to the fast form of the enzyme. Therefore, it may serve as a reliable structural probe for the transition of thrombin from the slow into the fast form. In the present study, we have extended our previous work [12] by introducing, at the N-terminal end of fragment 1–47, natural (Ala and Arg) and non-natural [pGndPhe, βNal (L-2-naphthylalanine) and Bip (L-4,4′-biphenylalanine)] amino acids having different side-chain volume, hydrophobicity and charge properties (Figure 5C).
Hirudin analogues were synthesized, purified to homogeneity, and characterized with respect to their chemical identity and functional properties, as described previously [18,19]. The results of the thrombin-binding experiments (Table 1) can be summarized as follows: first, alanine replacement at either position 1 or position 3 reduces affinity almost exclusively for the fast form; secondly, side-chain enlargement at position 3 with bulky and hydrophobic amino acids (i.e. βNal and Bip) strongly enhances affinity for both forms, but preferentially for the fast form; thirdly, electrostatic perturbation of the primary specificity site S1, by Ser2→Arg or Ser2→pGndPhe exchange, enhances affinity preferentially for the slow form; fourthly, disruption of the Asp5′–Arg221a interaction reduces binding strength to a similar extent (∼1 kcal/mol) under both fast and slow conditions, suggesting that this link exists in the hirudin–thrombin complex either in the presence or absence of Na+; fifthly, the effects of cumulative amino acid exchanges in the analogue ARA (synthetic analogue of the hirudin fragment 1–47 containing the substitutions Val1→Ala, Ser2→Arg and Tyr3→Ala) are additive for the fast form, whereas strong deviation from additivity is observed for the slow form; strikingly, replacement of even a single amino acid (Ala3→Bip) can change the affinity of hirudin fragment 1–47 for thrombin more than 1.3×104 times.
Table 1. Thermodynamic data for the binding of synthetic analogues of hirudin fragment 1–47 to the fast and slow form of thrombin.
Thrombin inhibitory potency of the synthetic analogues was determined as detailed elsewhere [12,20].
| Fast form | Slow form | |||||
|---|---|---|---|---|---|---|
| Analogue | Kd (nM) | ΔΔGb (kcal/mol)* | Kd (nM) | ΔΔGb (kcal/mol) | ΔGc (kcal/mol)† | r‡ |
| Tyr3 (wild-type) | 42±0.5 | – | 1460±20 | – | −2.10 | 35 |
| Val1→Ala | 630±7 | 1.61 | 4080±40 | 0.61 | −1.10 | 6.5 |
| Ser2→Arg§ | 1.7±0.02 | −1.90 | 12±2 | −2.84 | −1.16 | 7.0 |
| Ser2→pGndPhe | 2.5±0.1 | −1.67 | 16±2 | −2.67 | −1.10 | 6.4 |
| Tyr3→Ala§ | 2650±50 | 2.46 | 3450±90 | 0.51 | −0.15 | 1.3 |
| Tyr3→βNal§ | 1.1±0.1 | −2.14 | 94±4 | −1.64 | −2.64 | 85 |
| Tyr3→Bip | 0.2±0.01 | −3.17 | 12±1 | −2.84 | 2.43 | 60 |
| Asp5→Ser | 238±3 | 1.02 | 5500±90 | 0.83 | −1.86 | 23 |
| ARA | 650±17 | 1.62 | 2100±30 | 0.21 | −0.70 | 3.2 |
* ΔΔGb is the difference in the free energy change of binding to thrombin between the synthetic analogue (ΔGb*) and the natural fragment (ΔGbwt), ΔΔGb=ΔGb*−ΔGbwt. A negative value of ΔΔGb indicates that the mutated species binds thrombin more tightly than the natural fragment. Errors are ±0.1 kcal/mol or less.
† ΔGc is the free energy of coupling to thrombin, measured as ΔGc=ΔGb, fast−ΔGb, slow [23]. The value of ΔGc is negative if the inhibitor binds to the fast form with higher affinity than to the slow form.
‡ r is the ratio Kd(slow)/Kd(fast) and gives a value for the binding selectivity of the analogue for the fast form.
§ For these analogues, thrombin inhibition data were determined in this study for a second time and found in close agreement with those reported in our previous work [12].
Reasonable explanations for the effects of Ala replacement can be found in the lower hydrophobicity of Ala relative to Val and Tyr, and in the loss of numerous van der Waals contacts in the fast form, but not in the slow form. Hence, thrombin binding data indicate that the specificity sites of the enzyme in the Na+-bound (fast) form are in a more open and accessible conformation compared with the more closed structure they assume in the Na+-free (slow) form. Also consistent with our model, are the effects of the replacement of Ser2 with Arg or pGndPhe, whose long charged side-chain is expected to facilitate penetration of the inhibitor into thrombin recognition sites, to a greater extent in the case of the more closed slow form than in the case of the fast form of the enzyme, which is already accessible for binding. Finally, the non-additive behaviour observed for the binding of ARA to the slow form of thrombin, but not for the fast form, indirectly reflects the higher conformational heterogeneity of the slow form and is in keeping with the notion that co-operativity is more frequent in proteins characterized by high flexibility and low intrinsic stability [42].
Conclusions
By combining several different spectroscopic and biochemical tools, we have shown that Na+ binding specifically promotes a transition in thrombin, mainly involving the specificity sites of the enzyme and the 148-loop, from a more closed and flexible conformation (i.e. the slow form) to a more open and rigid one (i.e. the fast form). This is in agreement with the notion that Na+ binding enhances thrombin activity preferentially for the more sterically demanding substrates, such as fibrinogen and protease-activated receptor 1 [5], related to the procoagulant functions of the enzyme.
Even though it is difficult to identify a unique mechanism by which Na+ binding is transduced into changes in thrombin structure, we speculate that conformational ordering at the Na+ site may propagate to the flanking regions (i.e. the 148-loop and 171-helix) through the formation of specific salt bridges (i.e. Glu146–Arg221a and Glu164–Lys185) which, in turn, can ‘freeze’ the 148-loop and the S3 site into a more open and rigid conformation. In addition, reorientation of the Cys168–Cys182 disulphide bridge may represent a key element in the on–off mechanism by which the anticoagulant (slow) form is switched to the procoagulant (fast) form, only after the binding of Na+. This model is compatible with the crystal structures reported by Huntington and co-workers showing that the total cavity volume of the fast form is reduced by ∼200 Å3 in the slow form [10,11] and that the two salt bridges stabilizing the fast form are broken in the slow form [11]. Futhermore, Ala replacement of Glu146 or Arg221a stabilizes thrombin in the slow form [43]. Instead, even extensive perturbation in the 148-loop, generated by segment deletion (i.e. the mutant ΔAla149a–Lys149e) [38] or autolytic cleavage (i.e. γ-thrombin) [32] (not involving Glu146), does not alter the Na+ sensitivity of the enzyme.
A key aspect emerging from our work is the structural plasticity of thrombin, since perturbation at different sites can evoke very similar changes in the enzyme conformation. Indeed, as observed for Na+ binding, interaction of the hirudin peptide 54–65 to the exosite I enhances the affinity of thrombin for substrates and inhibitors [20,35,37], induces very similar changes in the far-UV CD [44] and fluorescence spectra [35,37], and even increases the resistance of the 148-loop to proteolysis [37]. Similar rigidification of the autolysis loop has been reported upon binding of PPACK (D-Phe-Pro-Arg-chloromethylketone) to the enzyme active site [37,45]. In conclusion, our results can contribute to elucidate the structural basis of thrombin allostery and may have important implications in the study of other Na+-regulated coagulative proteases.
Online data
Acknowledgments
This work was supported by a grant (PRIN-2003) from the Italian Ministry of University and Scientific Research to V.D.F. We thank Professor Angelo Fontana and Dr Daniele Dalzoppo for critical reading of the manuscript, and Dr Raimondo De Cristofaro for helpful comments. The support of Dr Olmetta Iadicicco is also gratefully acknowledged.
References
- 1.Minami T., Sugiyama A., Wu S. O., Abid R., Kodama T., Aird W. C. Thrombin and phenotypic modulation of the endothelium. Arterioscler. Thromb. Vasc. Biol. 2004;24:41–53. doi: 10.1161/01.ATV.0000099880.09014.7D. [DOI] [PubMed] [Google Scholar]
- 2.Griffin J. H. The thrombin paradox. Nature (London) 1995;378:337–338. doi: 10.1038/378337a0. [DOI] [PubMed] [Google Scholar]
- 3.Esmon C. T. The Protein C pathway. Chest. 2003;124:26S–32S. doi: 10.1378/chest.124.3_suppl.26s. [DOI] [PubMed] [Google Scholar]
- 4.Dang Q. D., Vindigni A., Di Cera E. An allosteric switch controls the procoagulant and anticoagulant activities of thrombin. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5977–5981. doi: 10.1073/pnas.92.13.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Cera E., Dang Q. D., Ayala Y. M. Molecular mechanisms of thrombin function. Cell Mol. Life Sci. 1997;53:701–730. doi: 10.1007/s000180050091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Cera E. Thrombin interactions. Chest. 2003;124:11S–17S. doi: 10.1378/chest.124.3_suppl.11s. [DOI] [PubMed] [Google Scholar]
- 7.Di Cera E., Guinto E. R., Vindigni A., Dang Q. D., Ayala Y. M., Wuyi M., Tulinsky A. The Na+ binding site of thrombin. J. Biol. Chem. 1995;270:22089–22092. doi: 10.1074/jbc.270.38.22089. [DOI] [PubMed] [Google Scholar]
- 8.Pineda A. O., Savvides S. N., Waksman G., Di Cera E. Crystal structure of the anticoagulant slow form. J. Biol. Chem. 2002;277:40177–40180. doi: 10.1074/jbc.C200465200. [DOI] [PubMed] [Google Scholar]
- 9.Pineda A. O., Carrell C. J., Bush L. A., Prasad S., Caccia S., Chen Z. W., Mathews F. S., Di Cera E. Molecular dissection of Na+ binding to thrombin. J. Biol. Chem. 2004;279:31842–31853. doi: 10.1074/jbc.M401756200. [DOI] [PubMed] [Google Scholar]
- 10.Huntington J. A., Esmon C. The molecular basis of thrombin allostery revealed by a 1.8-Å structure of the ‘slow’ form. Structure. 2003;11:469–479. doi: 10.1016/s0969-2126(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 11.Carter W. J., Myles T., Gibbs C. S., Leung L. L., Huntington J. A. Crystal structure of anticoagulant thrombin variant E217K provides insights into thrombin allostery. J. Biol. Chem. 2004;279:26387–26394. doi: 10.1074/jbc.M402364200. [DOI] [PubMed] [Google Scholar]
- 12.De Filippis V., Colombo G., Russo I., Spadari B., Fontana A. Probing hirudin–thrombin interaction by incorporation of noncoded amino acids and molecular dynamics simulation. Biochemistry. 2002;43:1537–1550. doi: 10.1021/bi0203482. [DOI] [PubMed] [Google Scholar]
- 13.Fenton J. W., II, Fasco M., Stackrow A. B. Human thrombins: production, evaluation, and properties of α-thrombin. J. Biol. Chem. 1977;252:3587–3598. [PubMed] [Google Scholar]
- 14.Gill S. G., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 15.Eftink M. R. The use of fluorescence methods to monitor unfolding transitions in proteins. Biophys. J. 1994;66:482–501. doi: 10.1016/s0006-3495(94)80799-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Stasio E., Bizzarri P., Misiti F., Pavoni E., Brancaccio A. A fast, accurate procedure to collect and analyse unfolding fluorescence signal: the case of dystroglycan. Biophys. Chem. 2004;107:197–211. doi: 10.1016/j.bpc.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Copeland R. A. Enzymes. New York: Wiley-VHC Inc.; 2000. Kinetics of single-substrate enzyme reactions; pp. 109–145. [Google Scholar]
- 18.De Filippis V., Vindigni A., Altichieri L., Fontana A. Core domain of hirudin from leech Hirudinaria manillensis: chemical synthesis, purification and characterization of a Trp3 analog of fragment 1–47. Biochemistry. 1995;34:9552–9564. doi: 10.1021/bi00029a032. [DOI] [PubMed] [Google Scholar]
- 19.De Filippis V., Quarzago D., Vindigni A., Di Cera E., Fontana A. Synthesis and characterization of more potent analogues of hirudin fragment 1–47 containing non-natural amino acids. Biochemistry. 1998;37:13507–13515. doi: 10.1021/bi980717n. [DOI] [PubMed] [Google Scholar]
- 20.Ayala J. M., Vindigni A., Nayal M., Spolar R. S., Record M. T., Jr, Di Cera E. Thermodynamic investigation of hirudin binding to the slow and fast forms of thrombin: evidence for folding transitions in the inhibitor and protease coupled to binding. J. Mol. Biol. 1995;253:787–798. doi: 10.1006/jmbi.1995.0591. [DOI] [PubMed] [Google Scholar]
- 21.Rydel T. J., Tulinsky A., Bode W., Huber R. Refined structure of the hirudin–thrombin complex. J. Mol. Biol. 1991;221:583–601. doi: 10.1016/0022-2836(91)80074-5. [DOI] [PubMed] [Google Scholar]
- 22.Katz B. A., Kossiakoff A. The crystallographically determined structures of atypical strained disulfides engineered into subtilisin. J. Biol. Chem. 1986;261:15480–15485. [PubMed] [Google Scholar]
- 23.Ayala Y., Di Cera E. Molecular recognition by thrombin. Role of the slow→fast transition, site-specific ion binding energetics and thermodynamic mapping of structural components. J. Mol. Biol. 1994;235:733–746. doi: 10.1006/jmbi.1994.1024. [DOI] [PubMed] [Google Scholar]
- 24.Ragone R., Colonna G., Balestrieri C., Servillo L., Irace G. Determination of tyrosine exposure in proteins by second-derivative spectroscopy. Biochemistry. 1984;23:1871–1875. doi: 10.1021/bi00303a044. [DOI] [PubMed] [Google Scholar]
- 25.Lakowicz J. R. 2nd ed. New York: Kluwer Academic/Plenum; 1999. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 26.Cardamone M., Puri M. K. Spectrofluorimetric assessment of the surface hydrophobicity of proteins. Biochem. J. 1992;282:589–593. doi: 10.1042/bj2820589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsukada H., Blow D. M. Structure of α-chymotrypsin refined at 1.68-Å resolution. J. Mol. Biol. 1985;184:703–711. doi: 10.1016/0022-2836(85)90314-6. [DOI] [PubMed] [Google Scholar]
- 28.Bode W., Turk D., Karshikov A. The refined 1.9-Å X-ray crystal structure of D-Phe-Pro-Arg-chloromethylketone-inhibited human α-thrombin: Structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure–function relationships. Protein Sci. 1992;1:426–471. doi: 10.1002/pro.5560010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brahms S., Brahms J. Determination of protein secondary structure in solution by vacuum ultraviolet circular dichroism. J. Mol. Biol. 1980;138:149–178. doi: 10.1016/0022-2836(80)90282-x. [DOI] [PubMed] [Google Scholar]
- 30.Strickland E. H. Aromatic contributions to circular dichroism spectra of proteins. CRC Crit. Rev. Biochem. 1974;3:113–175. doi: 10.3109/10409237409105445. [DOI] [PubMed] [Google Scholar]
- 31.Prasad S., Wright K. J., Banerjee Roy D., Bush L. A., Cantwell A. M., Di Cera E. Redesigning the monovalent cation specificity of an enzyme. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13785–13790. doi: 10.1073/pnas.2333109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wells C. M., Di Cera E. Thrombin is a Na+-activated enzyme. Biochemistry. 1992;31:11721–11730. doi: 10.1021/bi00162a008. [DOI] [PubMed] [Google Scholar]
- 33.Greene R. F., Pace N. C. Urea and guanidine hydrochloride denaturation of ribonuclease, lysozyme, α-chymotrypsin, and β-lactoglobulin. J. Biol. Chem. 1974;249:5388–5393. [PubMed] [Google Scholar]
- 34.Arnold F. H., Zhang J.-H. Metal-mediated protein stabilization. Trends Biotech. 1994;12:189–192. doi: 10.1016/0167-7799(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 35.Lai M.-T., Di Cera E., Shaker J. A. Kinetic pathway for the slow–fast transition of thrombin. J. Biol. Chem. 1997;272:30275–30282. doi: 10.1074/jbc.272.48.30275. [DOI] [PubMed] [Google Scholar]
- 36.Fontana A., Polverino De Laureto P., De Filippis V., Scaramella E., Zambonin M. Probing the partly folded states of proteins by limited proteolysis. Structure Fold. Des. 1997;2:17–26. doi: 10.1016/S1359-0278(97)00010-2. [DOI] [PubMed] [Google Scholar]
- 37.Parry M. A. A., Stone S. R., Hofsteenge J., Jackman M. P. Evidence for common structural changes in thrombin induced by active-site or exosite binding. Biochem. J. 1993;290:665–670. doi: 10.1042/bj2900665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dang Q. D., Sabetta M., Di Cera E. Selective loss of fibrinogen clotting in a loop-less thrombin. J. Biol. Chem. 1997;272:19649–19651. doi: 10.1074/jbc.272.32.19649. [DOI] [PubMed] [Google Scholar]
- 39.Vijayalakshmi J., Padmanabhan K. P., Mann K. G., Tulinsky A. The isomorphous structures of prethrombin-2, hirugen-, and PPACK-thrombin: changes accompanying activation and exosite binding to thrombin. Protein Sci. 1994;3:2254–2271. doi: 10.1002/pro.5560031211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang K. E. S., Dill K. A. Native protein fluctuations: the conformational-motion temperature and the inverse correlation of protein flexibility with protein stability. J. Biomol. Struct. Dyn. 1998;16:397–411. doi: 10.1080/07391102.1998.10508256. [DOI] [PubMed] [Google Scholar]
- 41.De Filippis V., Russo I., Vindigni A., Di Cera E., Salmaso S., Fontana A. Incorporation of noncoded amino acids into the N-terminal domain 1–47 of hirudin yields a highly potent and selective thrombin inhibitor. Protein Sci. 1999;8:2213–2217. doi: 10.1110/ps.8.10.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luque I., Freire E. Structural stability of binding sites: consequences for binding affinity and allosteric effects. Proteins. 2000;4(Suppl.):63–71. doi: 10.1002/1097-0134(2000)41:4+<63::aid-prot60>3.3.co;2-y. [DOI] [PubMed] [Google Scholar]
- 43.Dang Q. D., Guinto E. R., Di Cera E. Rational engineering of activity and specificity in a serine protease. Nat. Biotechnol. 1997;15:146–149. doi: 10.1038/nbt0297-146. [DOI] [PubMed] [Google Scholar]
- 44.Mao S. J., Yates M. T., Owen T. J., Krstenansky J. L. Interaction of hirudin with thrombin: identification of a minimal binding domain of hirudin that inhibits clotting activity. Biochemistry. 1988;27:8170–8173. doi: 10.1021/bi00421a027. [DOI] [PubMed] [Google Scholar]
- 45.Croy C. H., Koeppe J. R., Bergqvist S., Komives E. A. Allosteric changes observed in thrombin upon active site occupation. Biochemistry. 2004;43:5246–5255. doi: 10.1021/bi0499718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.