Abstract
Heparan sulphate (HS) is a ubiquitous constituent of the extracellular matrix that is required for the biological activity of circulating soluble and insoluble extracellular ligands. GLCE (D-glucuronyl C5-epimerase), an enzyme responsible for the epimerization of D-glucuronic acid into L-iduronic acid of HS, endows the nascent polysaccharide chain with the ability to bind to growth factors and cytokines. In order to examine the mechanism of regulation of GLCE expression, the functional organization of the human GLCE gene promoter has been investigated. Studies utilizing stepwise deleted and site-directed mutagenized promoter constructs have shed light on the functional relevance of two cis-acting binding elements for the β-catenin–TCF4 complex (where TCF4 stands for T-cell factor 4) that are located in the enhancer region of the promoter. The ability of the putative binding sequences to bind the β-catenin–TCF4 complex has been confirmed through electrophoretic mobility-shift and supershift analyses. We have found that, in a set of human colon carcinoma cell lines, the expression of GLCE correlates with the degree of activation of the β-catenin–TCF4 transactivation complex. Furthermore, the ectopic expression of β-catenin–TCF4 in cells that constitutively express low levels of the transactivation complex produces a significant increase of GLCE transcript level and, at the same time, enhances the rate of D-glucuronic acid epimerization in HS. The data obtained are consistent with the idea that the β-catenin–TCF4 transactivation pathway plays a major role in modulating GLCE expression, thus contributing to the regulation of HS biosynthesis and its structural organization.
Keywords: β-catenin–TCF4, colon carcinogenesis, glucuronic acid (GlcA), heparan sulphate, iduronic acid, Wnt signalling
Abbreviations: APC, adenomatous polyposis coli protein; Fz, Frizzled; GlcA, glucuronic acid; GLCE, D-glucuronyl C5-epimerase; HS, heparan sulphate; IdoA, iduronic acid; RT, reverse transcriptase; TCF, T-cell factor; UTR, untranslated region
INTRODUCTION
Heparan sulphate (HS) is a negatively charged polysaccharide present at the cell surface and is required for the biological activity of circulating soluble and insoluble extracellular ligands [1]. HS is required for efficient signalling through the FGF (fibroblast growth factor), the Wnt, the BMP (bone morphogenetic protein) and the Hedgehog pathways, which all play key roles during embryonic development and in the etiology of major diseases [2,3]. The biological activity of HS is dependent on the degree of sulphation and conformational freedom of the polysaccharide chain. HS is synthesized through a series of steps whose regulation is still poorly understood [4]. A key modification of the nascent polysaccharide chain is carried out in the Golgi system by the GLCE (D-glucuronyl C5-epimerase) enzyme, which converts GlcA (glucuronic acid) into IdoA (iduronic acid). GlcA epimerization releases the conformational constraints of the polysaccharide chain, allowing the access of ligands to specific regions of HS [5–7]. IdoA residues are more frequently 2-O-sulphated than GlcA residues, resulting in the appearance of clustered, highly charged domains along the glycosaminoglycan chain [8]. IdoA content may also affect the rate of HS degradation by the action of heparanase-like enzymes [9]. Transgenic mice lacking GLCE are not viable, corroborating the idea that the enzyme introduces a key modification in the HS structure that is necessary for its biological activity [10]. In order to elucidate how the expression of the epimerase is regulated, in the present study, we have cloned and functionally characterized the human GLCE gene promoter. The results show that two β-catenin–TCF4 cis-acting elements (where TCF4 stands for T-cell factor 4), located approx. 1.3 kb upstream of the translation start site, are functionally relevant in modulating the gene expression. We furthermore demonstrate that GLCE expression and its enzymatic activity are enhanced when the β-catenin–TCF4 complex is overexpressed in colon carcinoma cells. The findings support the conclusion that a key step of HS biosynthesis is under the control of the Wnt/APC (adenomatous polyposis coli protein)/β-catenin axis, an oncogenic pathway that is dysregulated in a large proportion of human epithelial tumours such as colon carcinomas [11].
EXPERIMENTAL
Cloning of the human GLCE promoter
RNA obtained from HSF, HeLa and HCT116 cells was used to identify the gene transcriptional start sites through 5′-primer extension experiments. For this purpose, an oligonucleotide of the sequence 5′-GAAGGAGACAGCAGAGAAGGGGTGGCT-3′, complementary to a region flanking the 3′-end of the GLCE putative translational start site, was end-labelled with [γ-32P]ATP. After purification on a spin column, 10 pmol (∼25000 c.p.m.) of the 32P-probe was hybridized with 2 μg of total RNA and reverse transcription was initiated by the addition of 20 units of Qiagen Sensiscript. After 30 min of incubation at 37 °C, the extension product was analysed on 6% (w/v) polyacrylamide/8 M urea sequencing gel, and the bands corresponding to the extension products were evidenced by autoradiography. The sequence of the GLCE gene promoter was retrieved from the human genome database using the sequence of the 5′-UTR (5′-untranslated region) as query. The putative gene promoter was amplified using pooled human genomic DNA as the template and primers of the sequences 5′-AGGCAGCTACTAAGGGAAGAGATGTCAAGCAT-3′ and 5′-CTCGGCAAGGAAGCGCTTGTGTGAGGAGA-3′. The product of expected size 1.9 kb was cloned in pGEM-T vector and sequenced.
Reporter vectors and promoter deletion and mutation constructs
The pGL3-C5:+23/−1893 reporter vector encoding firefly luciferase was generated by standard molecular biology techniques by retrieving the GLCE promoter from the pGEM-T plasmid by KpnI/SalI restriction digestion. pGL3-C5:+23/−1417 and pGL3-C5:+23/−419 were generated by taking advantage of unique BstXI and AgeI sites present in the promoter sequence and by digestion of the vector with KnpI followed by religation of the blunt-ended linearized constructs. pGL3-C5:+23/−1191 was generated by retrieving the BamHI/NcoI insert from pGL3-C5:+23/−1417 followed by ligation at the BglII/NcoI sites of pGL3. MUT-pGL3-C5:+23/−1417 was obtained by mutagenesis of the putative β-catenin–TCF4-binding sites (where TCF4 is T-cell factor 4) of sequences CTTTGTT and TTCAAAG using a Stratagene Quik Change kit and complementary primers of the sequence 5′-GAGATTGGGTGGCTGTGTTCTGTTCACAGGATGGGCGAG-3′ (the mutated sites are underlined) corresponding to the −1281/−1319 bp region of the GLCE promoter. For the assay of the reporter activity, cells in 12-well plates were transfected with 1 μg of the firefly luciferase-reporter vector and 0.05 μg of phRL-SV40 (where SV40 stands for simian virus 40) encoding the Renilla luciferase to correct for the transfection efficiency. Lipofectamine™ was used as the transfection agent. Where indicated, cells were also co-transfected with 0.1 or 1 μg/ml pcDNA3.0-human β-catenin and 1 μg/ml pHR-human TCF4 expression vectors. A TOP-FLASH vector (kindly provided by Dr B. Vogelstein, Johns Hopkins University, Baltimore, MD, U.S.A.) was utilized to monitor the activity of the β-catenin–TCF4 transactivation complex in different cell lines [12]. Cells were collected 24 h after transfection and the luciferase activity was measured using a dual-luciferase assay kit (Promega, Madison, WI, U.S.A.).
Electrophoretic mobility-shift assay
Nuclear extracts from SW480 cells were prepared by extraction using an NE-PER kit (Pierce). Before use, the nuclear extracts were dialysed at 4 °C overnight against 20 mM Hepes (pH 7.9), 75 mM KCl and 0.1% Na2-EDTA. A double-stranded oligonucleotide of the sequence 5′-TGGCTTTGTTCTGTTCAAAGGATG-3′ corresponding to the −1287/−1311 bp regions of the GLCE promoter was 32P-labelled with T4-polynucleotide kinase [12] and 10 pmol (∼15000 c.p.m.) was incubated with 5 μg of cell nuclear proteins in 20 mM Hepes (pH 7.9) buffer containing 75 mM KCl, 0.1% Na2-EDTA and 0.5 μg of poly(dI-dC)·(dI-dC) in a final volume of 50 μl for 120 min at room temperature (25 °C). Binding specificity was assessed by incubation in the presence of 30-fold excess of unlabelled oligonucleotide. For the supershift assay, 0.5 μg of antibody was added to the incubation mixture. DNA–protein hybrids were analysed on a 6% non-denaturing polyacrylamide gel (acrylamide/bisacrylamide, 39:1) in 25 mM Tris and 200 mM glycine buffer, and the bands were visualized by autoradiography.
Transcript level assay by semiquantitative RT (reverse transcriptase)–PCR
Cellular RNA was extracted in TRI Reagent and 1 μg was reverse-transcribed using the Qiagen Sensiscript kit and oligo-dT primers. First-strand cDNA was quantified by PCR amplification, stopping the reaction after 25 cycles in order to avoid limitation in the reagents availability. The generated GLCE cDNA was quantified by agarose electrophoresis. Gels were photographed and the propidium iodide-stained bands were quantified by densitometric scanning.
Epimerase enzymatic assay
Control and β-catenin–TCF4-transfected cells cultured in 35 mm wells were washed with PBS and collected by scraping using a rubber policeman into 300 μl of ice-cold 0.25 M sucrose and 20 mM Tris/HCl buffer (pH 7.4) supplemented with 100 μM NaF, 2 mM Na3VO4, 10 mM PMSF, 500 μM 4-(2-aminoethyl)benzenesulphonyl fluoride, 150 nM aprotinin and 1 μM leupeptin. Cells were lysed by sonication while held on ice using a Bronson sonifier fitted with a microtip and the preparation was stored at −70 °C until use. Protein concentration was assessed using a bicinchoninic acid-based assay (Pierce). The substrate for the epimerase enzymatic assay consisted of modified bacterial N-acetyl-heparosan prepared essentially as described by Hagner-McWhirter et al. [13]. For this purpose, the bacterial strain O10:K5:H4 (culture Bi 8337-41 of [14]) was obtained from the Escherichia coli repository at the Pennsylvania State University. For the metabolic radiolabelling of the capsular polysaccharide, the bacteria were grown in 5 ml of Luria–Bertani medium containing 0.5% glucose at 37 °C overnight. The culture (100 μl) was then added to 15 ml of Luria–Bertani medium without glucose and incubated in the presence of 250 μCi of D-[5-3H]glucose (10 Ci/mmol) for 24 h at 37 °C. After centrifugation, the supernatant was combined with 0.75 ml of 1 M sodium acetate buffer (pH 4.0) and the solution was cycled at a flow rate of 1 ml/min through a 1 ml prepacked DEAE-FF column (Amersham Biosciences) equilibrated in 50 mM sodium acetate and 50 mM NaCl (pH 4.0). After 30 min, the column was washed with 10 ml of the same buffer and the bound K5 polysaccharide was eluted using a stepwise NaCl gradient (0.05–1.0 M) in 50 mM sodium acetate buffer (pH 4.0) at a flow rate of 1 ml/min by increasing the NaCl concentration in 0.2 M steps every 5 min. The peak of radioactivity eluting at the 0.40 M NaCl step was pooled and desalted on a Sephadex G25 1 cm×30 cm column. N-deacetylation of the 3H-labelled K5 polysaccharide was carried out by treatment with hydrazine/hydrazine sulphate at 96 °C for 5 h. After desalting, the samples were freeze-dried and treated with 0.25 M HIO3 to eliminate the residual hydrazides. N-sulphation was achieved by treatment with trimethylamine–sulphotrioxide complex (Sigma) adjusted to pH 9.5 with NaHCO3 at 56 °C for 24 h. After desalting, the uronic acid concentration in the sample was assessed by a carbazol assay [15]. The 3H-sulphated heparosan produced had a specific activity of 6.1×106 c.p.m./mg of uronic acid. The enzymatic assay, performed essentially as described by Crawford et al. [16], is based on the release of 3H2O from [C5-3H]GlcA units present in the polysaccharide [17]. Briefly, the reactions were set up by combining 200 μg of cell proteins with 40 nmol of labelled substrate in 200 μl of 25 mM Hepes buffer (pH 7.0) containing 0.1% Triton X-100. Samples depleted of enzymatic activity for the determination of the background radioactivity were prepared by preheating the cell extract at 100 °C for 10 min. After 2 h of incubation at 37 °C, the reactions were stopped by the addition of 50 μl of 125 mM sodium acetate (pH 4.0) and 125 mM LiCl. After centrifugation, the supernatant was loaded on to a 1 ml DEAE-FF prepacked column connected to an FPLC injector and pump and equilibrated with 50 mM sodium acetate (pH 4.0) and 50 mM NaCl. The radioactivity recovered in the initial 3 ml of the flow-through volume and corresponding to freed 3H2O was measured in a Wallac 1219 Rackbeta liquid-scintillation counter.
RESULTS
Cloning of the human GLCE gene promoter
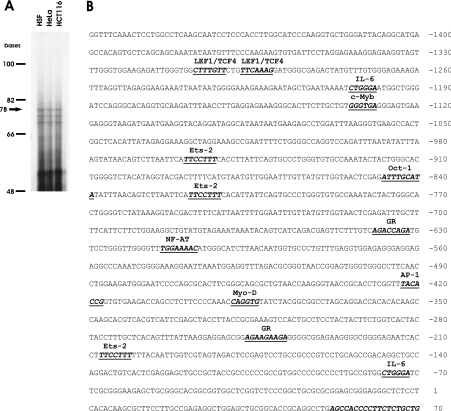
Preliminary information on the GLCE promoter was obtained by a BLAST search of the human genome database utilizing the published sequence of the murine Glce gene 5′-UTR as a query. This procedure gave results that putatively identified the 5′-UTR of the human gene. To confirm that this region was part of the transcribed gene product, RT–PCR was used to specifically amplify and clone a 2082 bp cDNA fragment encompassing the 5′-end and the stop codon of the putative gene transcript. DNA sequencing of the cloned product confirmed its identity (GenBank® accession no. AY635582). In order to identify conclusively the translational start sites of the gene, 5′-primer extension experiments were performed. A primer annealing to the 5′-UTR of the gene was utilized to reverse-transcribe RNA obtained from HSF, HeLa and HCT116 cells. The results (Figure 1A) enabled us to locate the major transcriptional start site of the gene 229 bp upstream of the translational start site. The sequence of the putative human GLCE gene promoter was retrieved from the human genome database by BLAST search utilizing the sequence of the gene 5′-UTR as query. The identified genomic region was amplified, cloned in pGEM-T vector and sequenced.
Figure 1. Structural analysis of the GLCE promoter.
(A) Identification of the transcriptional start site of the human GLCE gene. The arrow points to the largest transcript generated by an RT from a primer anchored in the 5′-UTR of the gene. (B) Mapping of the cis-acting transcriptional factor binding sites on the human GLCE gene promoter. Only transcriptional factor-binding sites that are conserved at the same position in the human and mouse promoter sequences are indicated. The acronyms used to identify the ligands are: c-myb, protoncogene, required for G1/S transition; Lef1–TCF4, β-catenin–TCF4 complex; NFAT, nuclear factor of activated T-cells [activated by CAML (calcium-modulator cyclophilin ligand)]. A part of the sequence (+52/+70), complementary to the oligonucleotide used as the primer to identify the transcriptional start sites, is also highlighted.
Functional characterization of the human GLCE promoter
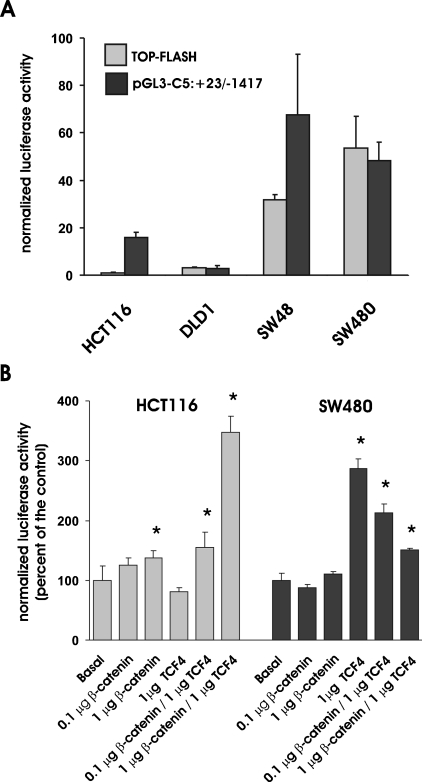
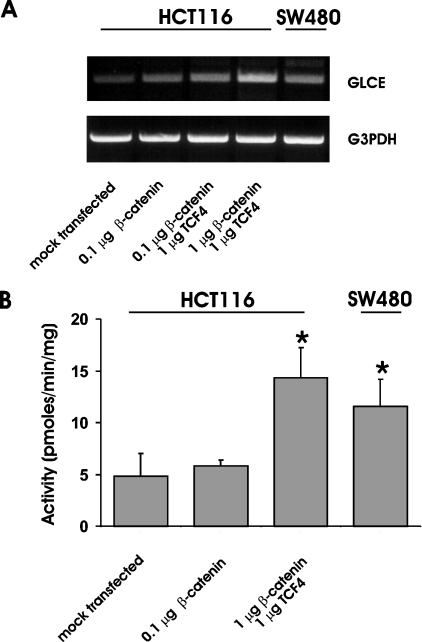
In silico analysis of the promoter organization was carried out by querying the TESS database (www.cbil.upenn.edu/tess/index). The same analysis was carried out on the sequence of the mouse Glce gene promoter retrieved from the mouse genome database BLAST searched with the 5′-UTR sequence of the gene [16]. The alignment of the human and murine gene promoters allowed the identification of cis-acting binding elements that are conserved in the two species on the basis of their sequences and their relative distance from the transcriptional start site (Figure 1B). In addition to binding domains for transcriptional factors that may be relevant for the constitutive expression of the gene {Oct-1 [octamer binding transcription factors, positively interacts with GR (glucorticoid receptor nuclear factor)] and MyoD} or that act as mediators of different signalling [Ets-2 (protoncogene with mitogenic and oncogenic activities; functionally interacts with AP-1 and c-Myb), GR and IL-6 (interleukin-6; interacts with GR)] and oncogenic [AP-1 (activator protein-1; composed of Jun-Jun or Jun-Fos dimers) and c-Myb] pathways, the GLCE promoter contains a conserved β-catenin/TCF4 cis-acting element that is a potential target of the HS-dependent Wnt signalling pathway. In particular, both the human and murine gene promoters harbour a tandem of canonical β-catenin–Lef1–TCF4 cis-acting binding elements of sequences CTTTGTT and TTCAAAG [18] located approx. 1.3 kb upstream of the transcriptional start site. Because deregulation of the β-catenin–TCF4 gene transactivation plays a key role in the pathogenesis of colon carcinomas and the signalling pathways have been well characterized in this cellular context, the function of the identified sites was investigated in a set of colon carcinoma cell lines. HCT116, DLD-1, SW48 and SW480 cells have been previously characterized with respect to the activation of the β-catenin–TCF4 pathway [12,19–22]. Before use in the present study, their status was further confirmed by utilizing TOP-FLASH, a reporter vector in which luciferase expression is driven by tandem repeats of the Lef1–TCF4 canonical consensus sequence [22] (Figure 2A). In order to examine the correlation between the activity of the GLCE promoter and the status of activation of the β-catenin–TCF4 complex, the cells were transfected with pGL3-C5:+23/−1417. The activity of the GLCE promoter reporter was significantly higher in cells with elevated β-catenin–TCF4 transactivation activity (i.e. SW48 and SW480) than in those with a low activity level (HCT116 and DLD-1; Figure 2A). In order to examine the effect of elevation of the ectopic expression of the β-catenin–TCF4 complex on the GLCE gene transactivation, HCT116 and SW480 cells were transfected with increasing doses of β-catenin and TCF4 expression vectors. HCT116 cells constitutively express low levels of β-catenin and TCF4 [12]. As expected, in these cells, the ectopic expression of β-catenin has little effect on the transactivation of the GLCE promoter unless the transcriptional activator TCF4 that complexes with β-catenin in the nucleus is also present in excess (Figure 2B). On the contrary, in SW480 cells, which constitutively express high levels of the β-catenin–TCF4 complex, the reporter activity was not affected by β-catenin and was in fact suppressed when both β-catenin and TCF4 were overexpressed as observed for other genes that are targets of the β-catenin–TCF4 transactivation complex when the complex is present in excess [12,23]. In order to assess whether transactivation of the GLCE promoter results in increased GLCE expression as well as enzymatic activity, HCT116 cells were transfected with increasing doses of β-catenin and TCF4 expression vectors. Semiquantitative analysis of the GLCE transcript in HCT116 cells revealed a positive dose-dependent effect of β-catenin–TCF4 (Figure 3A), consistent with the results of the reporter vector experiments seen previously (Figure 2B). In addition, the epimerase enzymatic activity was increased in those HCT116 cells expressing a high level of the β-catenin–TCF4 complex, consistent with the idea that GLCE protein level is affected (Figure 3B). GLCE transcript level was constitutively elevated in SW480 cells and the epimerase activity was significantly higher than that measured in untransfected HCT116 cells. These results support the conclusion that, in colon carcinoma cells, the β-catenin–TCF4 transactivation pathway is an important modulator of the GLCE expression and the rate of HS GlcA epimerization.
Figure 2. GLCE transcriptional activity in human colon carcinoma cells expressing different levels of β-catenin and TCF4.
(A) Transcriptional activity of the TOP-FLASH and the pGL3-C5:+23/−1417 reporter vectors in different human colon carcinoma cell lines with constitutively low (HCT116 and DLD-1) or elevated (SW48 and SW480) activity level of the β-catenin–TCF4 transactivation complex. Cells in 12-well plates were co-transfected with 1 μg/ml of the reporter vectors and 0.1 μg/ml phRL-SV40 to normalize values to the transfection efficiency. Luciferase activity was assayed 24 h later. Results shown are the means±S.D. (n=3) of the normalized luciferase values. (B) Effects of β-catenin and TCF4 on the transcriptional activity of pGL3-C5:+23/−1417 in HCT116 and SW480 cells. Cells were transfected with 1 μg/ml of the GLCE promoter reporter vector together with various combinations of the β-catenin and TCF4 expression vectors as indicated. Results shown are the means±S.D. (n=3) of the normalized luciferase values. *P<0.05 versus cells transfected with pGL3-C5:+23/−1417 alone (basal group).
Figure 3. Effect of the ectopic expression of β-catenin and TCF4 on GLCE activity.
(A) Semiquantitative RT–PCR assay of GLCE transcript level in HCT116 and SW480 cells. In order to examine the effect of the ectopic expression of β-catenin–TCF4, HCT116 cells were transfected with increasing amounts of human β-catenin and human TCF4 expression vectors. (B) GLCE enzymatic activity in cells expressing different levels of the β-catenin–TCF4 complex. For the enzymatic assay, the substrate composed of 3H-sulphated heparosan (200 μM) was incubated for 2 h at 37 °C with 200 μg of cell extract, and the 3H2O liberated as a result of GlcA conversion into IdoA was isolated from the labelled substrate by ion-exchange chromatography. Results shown are the means±S.D. for three independent experiments.
The GLCE gene promoter β-catenin–TCF4 cis-acting elements are functionally active
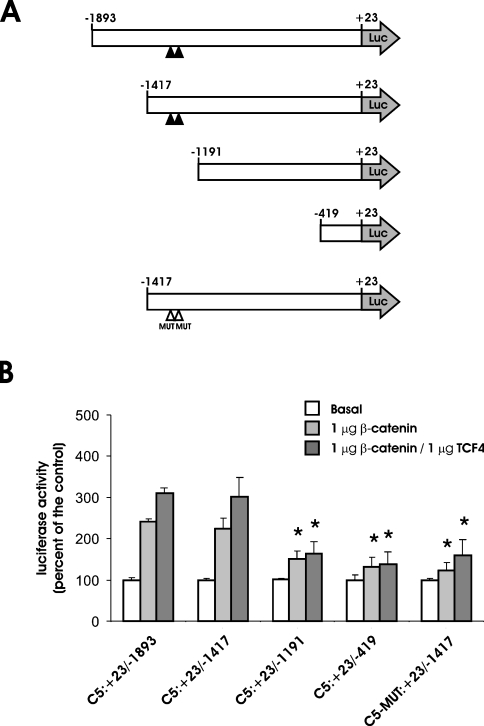
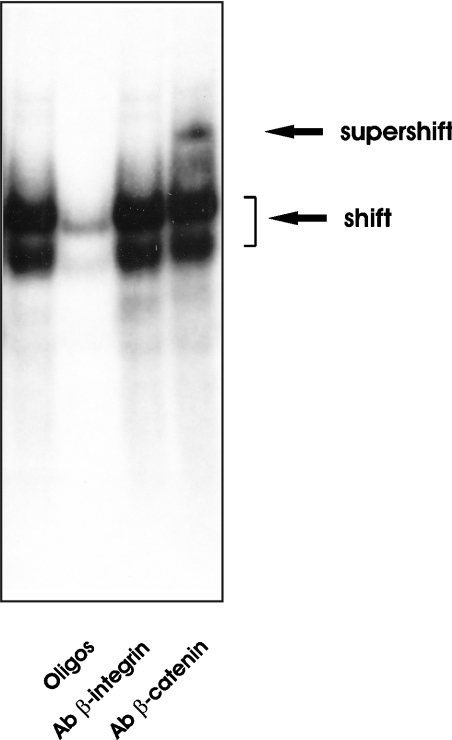
In order to confirm the direct dependence of the observed effect on the presence of the β-catenin–TCF4 cis-acting binding elements, two new constructs were tested in HCT116 cells: pGL3-C5:+23/−1191, in which 226 bp of the promoter region harbouring the putative binding sites had been deleted, and pGL3-C5-MUT:+23/−1417, in which the binding sequence had been mutated (Figure 4A). Either deletion or single-base mutation of the consensus sequence resulted in a significant loss of responsiveness to the elevation of β-catenin or β-catenin–TCF4 level (Figure 4B). This result supports the conclusion that the two conserved sites are functionally relevant in modulating GLCE transcription. The ability of the putative binding sequences to bind the β-catenin–TCF4 complex was confirmed through electrophoretic mobility-shift and supershift analyses (Figure 5). When a 21 bp labelled double-stranded oligonucleotide of the sequence corresponding to that of the putative β-catenin–TCF4 cis-acting binding element of the GLCE promoter was incubated with nuclear extract from SW480 cells, the formation of DNA–protein complexes could be evidenced by gel electrophoresis. The size of the complex increases when anti-β-catenin antibody was also added, confirming the presence of the transcriptional factor in the complexes formed by the labelled oligonucleotide. The formation of the DNA–protein complex was inhibited when 30-fold excess of cold oligonucleotide was added and the supershifted complex did not form after the addition of an irrelevant antibody (anti-β-integrin), thus validating the oligonucleotide–DNA binding specificity.
Figure 4. Identification and functional characterization of the β-catenin/TCF4 cis-acting binding elements present in the GLCE promoter.
(A) Schematic representation of the various reporter constructs utilized in the present study. Filled arrowheads indicate the position of the β-catenin–TCF4 cis-acting binding sites. Mutated sites are identified with empty arrowheads. (B) Effects of β-catenin and TCF4 overexpression in HCT116 cells on the transcriptional activity of the series of GLCE promoter reporter vectors shown on the left. Cells in 12-well plates were transfected with the reporter vector alone (1 μg/ml pGL3-C5:+23/−1417) together with β-catenin or β-catenin + TCF4 expression vectors (1 μg/ml each). Luciferase activity was assayed 24 h later and the values were corrected for the transfection efficiency. Results shown are the means±S.D. (n=9) of the normalized luciferase activity. *P<0.05 versus cells transfected with pGL3-C5:+23/−1893 alone (Basal group).
Figure 5. Protein–DNA complex formation between β-catenin/TCF4 and the corresponding binding domain of the GLCE promoter.
γ-32P-labelled oligonucleotides of the sequence corresponding to the putative β-catenin–TCF4 binding site of the GLCE promoter were incubated with a nuclear extract from SW480 cells, and the DNA–protein complex formed was analysed on 6% non-denaturing polyacrylamide gel. After electrophoresis, bands were visualized by autoradiography. For the competition binding, 30-fold excess of unlabelled oligonucleotides were included in the incubation mixture (lane 2). Two shifted bands corresponding respectively to DNA–TCF4 (lower band) and DNA:TCF4/β-catenin (upper band) complexes were detected [29]. For the supershift analysis, 0.5 μg of an irrelevant monoclonal antibody (anti-β-integrin, lane 3) or anti β-catenin antibody (lane 4) were added.
DISCUSSION
In this study, we present evidence that GLCE, encoding a key enzyme in HS biosynthesis, is a target of the β-catenin–TCF4 transactivation complex, the terminal effector of the Wnt/APC/β-catenin signalling pathway. The fact that ectopic expression in cells of β-catenin alone or together with TCF4 enhances the GLCE promoter transcriptional activity and increases the gene transcript level, resulting in increased enzymatic activity, supports the conclusion that the pathway is functionally significant in modulating the gene expression in vivo, resulting in enhanced conversion of HS GlcA into IdoA. We furthermore demonstrate that the transactivation effect is mediated through two conserved β-catenin–TCF4 cis-acting binding elements located in the enhancer region of the gene promoter.
The gene encoding the HS epimerase is represented only once in the mammalian genome, implying that HS uronic acid epimerization is ultimately dependent on GLCE expression. Limited information is currently available regarding how GLCE activity is regulated. The HS chain modification produced by this enzyme is an irreversible process in vivo [24] and occurs after the nascent polysaccharide chain has undergone GlcNAc N-deacetylation and N-sulphation but before glucosamine residues are 6-O- and 3-O-sulphated [25,26]. GLCE physically interacts with other enzymes involved in HS biosynthesis, leading to the formation of multimeric enzymatic complexes that may perform sequentially related functions. The association of GLCE with the 2-O-sulphotransferase, in particular, positively affects the epimerase stability and mediates its translocation to the Golgi [27]. The availability of a suitable HS substrate at the site where GLCE is localized can also, in principle, affect the overall rate of HS epimerization. In this context, specific domains of the core protein to which the HS chain is attached may route the nascent proteoglycan to Golgi subcompartments where GLCE is located [28]. The relative impact of these various mechanisms on the overall rate of GlcA epimerization remains to be assessed fully. Conceivably, the GLCE expression level is a major determinant of the enzymatic activity. In the present study, we found that a positive correlation exists between the epimerase activity and the GLCE transcript level. In addition, our results indicate that agents like β-catenin and TCF4, capable of affecting GLCE gene transcription, are important modulators of the rate of HS GlcA epimerization in cells.
β-Catenin acts as a transcriptional cofactor by migrating to the nucleus and associating with members of a family of DNA-binding proteins known as TCFs, of which TCF4 is expressed in the intestinal epithelium [29]. The targets of β-catenin–TCF4 transactivation include development-related genes activated through the Wingless/Wnt signalling pathway [11]. Many genes relevant for colorectal tumour formation and progression have been identified as being transcriptionally activated by the β-catenin–TCF4 complex. Some are relevant for growth control and cell cycling (SMC3, c-myc, cyclin D1, c-Jun, fra-1, gastrin and ITF2), some are implicated in cell survival (Id2 and MDR1) and some are implicated in tumour invasion and metastasis (matrilysin and VEGF) [12,21,22,30–37]. GLCE is the first gene involved in glycosaminoglycan biosynthesis that has been identified as the target for β-catenin. Two conserved transcriptional binding sites for β-catenin–TCF4 are present in the human and mouse GLCE promoters. They are located in tandem arrangement in the enhancer region of the promoter. This location is not unusual in promoters that are targets of the β-catenin–TCF4 complex and it has been previously observed in the c-myc and VEGF gene promoters [22,33].
Wnts are secreted proteins that function in numerous developmental processes in vertebrates and invertebrates [38]. Wnt proteins bind to the Fz (Frizzled) receptors and LRP5/6 co-receptors [39] and, by stabilizing the critical mediator β-catenin, initiate a complex signalling cascade that plays an important role in regulating cell proliferation and differentiation. It has been proposed that proteoglycan-bound HS modulates Wg signalling through two general mechanisms. HS can either protect the Wnt protein from being degraded or reduce the range of Wnt ligand diffusion, thereby affecting the local concentration of the Wnt ligand that is available to bind to the Fz receptors [40]. HS proteoglycans also act as co-receptors that directly facilitate the formation of Wnt/Fz signalling complexes [41]. The different models of activation of Wnt signalling by HS predict that changes in HS composition and/or quantity can significantly affect the morphogen activity in vivo. In this context, our finding that the expression of a key enzyme in HS biosynthesis is a target of the β-catenin-dependent transactivation pathway raises the possibility that HS biosynthesis and β-catenin-mediated signalling are interdependent. In particular, the deregulation of the Wnt/APC/β-catenin transactivation pathway, by enhancing HS protein-binding efficiency, may engage a Wnt-dependent autocrine loop that can also intensify the signalling of other mitogenic and angiogenic heparin-binding factors, such as FGF (fibroblast growth factor), PDGF (platelet-derived growth factor) and VEGF (vascular endothelial growth factor) [1]. This functional loop may be particularly relevant in the initiation and progression of colon tumours where loss of heterozygosity of the APC or the β-catenin gene leads to constitutive activation of the β-catenin–TCF4 transactivation pathway. Currently, there is no documentation of any mutation or amplification of genes encoding Wnt ligands or receptors linked to human cancer. On the other hand, changes in HS composition have been identified in transformed colon cells during the transition from colon adenoma to carcinoma [42] or during cell differentiation [43]. In particular, the HS IdoA-2-O-sulphate content is affected, suggesting that either the activity of GLCE or that of the 2-O-sulphotransferase or both is specifically affected during cell transformation. Recently, evidence has been presented that syndecan-1, a major cell HS proteoglycan, is required to create a β-catenin–TCF responsive cell population during mammary development [44].
In conclusion, we provide evidence supporting the idea that GlcA epimerization, a step in HS biosynthesis affecting the polysaccharide activity and specificity, is a target of the β-catenin–TCF4 transactivation pathway. A large proportion of human epithelial tumours, including almost all colon tumours, are known to be initiated by dysregulation of the β-catenin metabolism. Since differentiation and transformation of colonic epithelial cells involve changes in composition of HS, we postulate that transcriptional regulation of GLCE is of significance in mediating the oncogenic potential of β-catenin, leading to the expansion of a clonal population of adenomatous cells.
Acknowledgments
This work was supported by a National Institutes of Health grant (RO1-CA82290) to G.G.
References
- 1.Rapraeger A. C. In the clutches of proteoglycans: how does heparan sulfate regulate FGF binding? Chem. Biol. 1995;2:645–649. doi: 10.1016/1074-5521(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 2.Ornitz D. M. FGFs, heparan sulfate and FGFRs: complex interactions essential for development. BioEssays. 2000;22:108–112. doi: 10.1002/(SICI)1521-1878(200002)22:2<108::AID-BIES2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 3.Bernfield M., Gotte M., Park P. W., Reizes O., Fitzgerald M. L., Lincecum J., Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 4.Esko J. D., Selleck S. B. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 5.Mulloy B., Forster M. J. Conformation and dynamics of heparin and heparan sulfate. Glycobiology. 2000;10:1147–1156. doi: 10.1093/glycob/10.11.1147. [DOI] [PubMed] [Google Scholar]
- 6.Ferro D. R., Provasoli A., Ragazzi M., Casu B., Torri G., Bossennec V., Perly B., Sinay P., Petitou M., Choay J. Conformer populations of L-iduronic acid residues in glycosaminoglycan sequences. Carbohydr. Res. 1990;195:157–167. doi: 10.1016/0008-6215(90)84164-p. [DOI] [PubMed] [Google Scholar]
- 7.Inoue Y., Inouye Y., Nagasawa K. Conformational equilibria of the L-iduronate residue in non-sulphated di-, tetra- and hexa-saccharides and their alditols derived from dermatan sulphate. Biochem. J. 1990;265:533–538. doi: 10.1042/bj2650533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casu B., Lindahl U. Structure and biological interactions of heparin and heparan sulfate. Adv. Carbohydr. Chem. Biochem. 2001;57:159–206. doi: 10.1016/s0065-2318(01)57017-1. [DOI] [PubMed] [Google Scholar]
- 9.Bai X., Bame K. J., Habuchi H., Kimata K., Esko J. D. Turnover of heparan sulfate depends on 2-O-sulfation of uronic acids. J. Biol. Chem. 1997;272:23172–23179. doi: 10.1074/jbc.272.37.23172. [DOI] [PubMed] [Google Scholar]
- 10.Li J. P., Gong F., Hagner-Mcwhirter A., Forsberg E., Abrink M., Kisilevsky R., Zhang X., Lindahl U. Targeted disruption of a murine glucuronyl C5-epimerase gene results in heparan sulfate lacking L-iduronic acid and in neonatal lethality. J. Biol. Chem. 2003;278:28363–28366. doi: 10.1074/jbc.C300219200. [DOI] [PubMed] [Google Scholar]
- 11.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 12.Ghiselli G., Coffee N., Munnery C. E., Koratkar R., Siracusa L. D. The cohesin SMC3 is a target for the beta-catenin/TCF4 transactivation pathway. J. Biol. Chem. 2003;278:20259–20267. doi: 10.1074/jbc.M209511200. [DOI] [PubMed] [Google Scholar]
- 13.Hagner-McWhirter A., Hannesson H. H., Campbell P., Westley J., Roden L., Lindahl U., Li J. P. Biosynthesis of heparin/heparan sulfate: kinetic studies of the glucuronyl C5-epimerase with N-sulfated derivatives of the Escherichia coli K5 capsular polysaccharide as substrates. Glycobiology. 2000;10:159–171. doi: 10.1093/glycob/10.2.159. [DOI] [PubMed] [Google Scholar]
- 14.Orskov I., Orskov F., Jann B., Jann K. Serology, chemistry, and genetics of O and K antigens of Escherichia coli. Bacteriol. Rev. 1977;41:667–710. doi: 10.1128/br.41.3.667-710.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bitter T., Muir H. M. A modified uronic acid carbazole reaction. Anal. Biochem. 1962;4:330–334. doi: 10.1016/0003-2697(62)90095-7. [DOI] [PubMed] [Google Scholar]
- 16.Crawford B. E., Olson S. K., Esko J. D., Pinhal M. A. Cloning, Golgi localization, and enzyme activity of the full-length heparin/heparan sulfate-glucuronic acid C5-epimerase. J. Biol. Chem. 2001;276:21538–21543. doi: 10.1074/jbc.M100880200. [DOI] [PubMed] [Google Scholar]
- 17.Lindahl U., Jacobsson I., Hook M., Backstrom G., Feingold D. S. Biosynthesis of heparin. Loss of C-5 hydrogen during conversion of D-glucuronic to L-iduronic acid residues. Biochem. Biophys. Res. Commun. 1976;70:492–499. doi: 10.1016/0006-291x(76)91073-1. [DOI] [PubMed] [Google Scholar]
- 18.van De W. M., Oosterwegel M., Dooijes D., Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakita K., Tetsu O., McCormick F. A mammalian two-hybrid system for adenomatous polyposis coli-mutated colon cancer therapeutics. Cancer Res. 2001;61:854–858. [PubMed] [Google Scholar]
- 20.Leung J. Y., Kolligs F. T., Wu R., Zhai Y., Kuick R., Hanash S., Cho K. R., Fearon E. R. Activation of AXIN2 expression by beta-catenin-T cell factor. A feedback repressor pathway regulating Wnt signaling. J. Biol. Chem. 2002;277:21657–21665. doi: 10.1074/jbc.M200139200. [DOI] [PubMed] [Google Scholar]
- 21.Rockman S. P., Currie S. A., Ciavarella M., Vincan E., Dow C., Thomas R. J., Phillips W. A. Id2 is a target of the beta-catenin/T cell factor pathway in colon carcinoma. J. Biol. Chem. 2001;276:45113–45119. doi: 10.1074/jbc.M107742200. [DOI] [PubMed] [Google Scholar]
- 22.He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., da Costa L. T., Morin P. J., Vogelstein B., Kinzler K. W. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 23.Fodde R., Smits R., Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer. 2001;1:55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 24.Hagner-Mcwhirter A., Li J. P., Oscarson S., Lindahl U. Irreversible glucuronyl C5-epimerization in the biosynthesis of heparan sulfate. J. Biol. Chem. 2004;279:14631–14638. doi: 10.1074/jbc.M313760200. [DOI] [PubMed] [Google Scholar]
- 25.Jacobsson I., Lindahl U., Jensen J. W., Roden L., Prihar H., Feingold D. S. Biosynthesis of heparin. Substrate specificity of heparosan N-sulfate D-glucuronosyl 5-epimerase. J. Biol. Chem. 1984;259:1056–1063. [PubMed] [Google Scholar]
- 26.Backstrom G., Hook M., Lindahl U., Feingold D. S., Malmstrom A., Roden L., Jacobsson I. Biosynthesis of heparin. Assay and properties of the microsomal uronosyl C-5 epimerase. J. Biol. Chem. 1979;254:2975–2982. [PubMed] [Google Scholar]
- 27.Pinhal M. A., Smith B., Olson S., Aikawa J., Kimata K., Esko J. D. Enzyme interactions in heparan sulfate biosynthesis: uronosyl 5-epimerase and 2-O-sulfotransferase interact in vivo. Proc. Natl. Acad. Sci. U.S.A. 2001;98:12984–12989. doi: 10.1073/pnas.241175798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seidler D. G., Breuer E., Grande-Allen K. J., Hascall V. C., Kresse H. Core protein dependence of epimerization of glucuronosyl residues in galactosaminoglycans. J. Biol. Chem. 2002;277:42409–42416. doi: 10.1074/jbc.M208442200. [DOI] [PubMed] [Google Scholar]
- 29.Morin P. J., Sparks A. B., Korinek V., Barker N., Clevers H., Vogelstein B., Kinzler K. W. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 30.Tetsu O., McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature (London) 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 31.Yamada T., Takaoka A. S., Naishiro Y., Hayashi R., Maruyama K., Maesawa C., Ochiai A., Hirohashi S. Transactivation of the multidrug resistance 1 gene by T-cell factor 4/beta-catenin complex in early colorectal carcinogenesis. Cancer Res. 2000;60:4761–4766. [PubMed] [Google Scholar]
- 32.Crawford H. C., Fingleton B. M., Rudolph-Owen L. A., Goss K. J., Rubinfeld B., Polakis P., Matrisian L. M. The metalloproteinase matrilysin is a target of beta-catenin transactivation in intestinal tumors. Oncogene. 1999;18:2883–2891. doi: 10.1038/sj.onc.1202627. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X., Gaspard J. P., Chung D. C. Regulation of vascular endothelial growth factor by the Wnt and K-ras pathways in colonic neoplasia. Cancer Res. 2001;61:6050–6054. [PubMed] [Google Scholar]
- 34.Kolligs F. T., Nieman M. T., Winer I., Hu G., Van M. D., Feng Y., Smith I. M., Wu R., Zhai Y., Cho K. R., et al. ITF-2, a downstream target of the Wnt/TCF pathway, is activated in human cancers with beta-catenin defects and promotes neoplastic transformation. Cancer Cell. 2002;1:145–155. doi: 10.1016/s1535-6108(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 35.Koh T. J., Bulitta C. J., Fleming J. V., Dockray G. J., Varro A., Wang T. C. Gastrin is a target of the beta-catenin/TCF-4 growth-signaling pathway in a model of intestinal polyposis. J. Clin. Invest. 2000;106:533–539. doi: 10.1172/JCI9476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mann B., Gelos M., Siedow A., Hanski M. L., Gratchev A., Ilyas M., Bodmer W. F., Moyer M. P., Riecken E. O., Buhr H. J., et al. Target genes of beta-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc. Natl. Acad. Sci. U.S.A. 1999;96:1603–1608. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shtutman M., Zhurinsky J., Simcha I., Albanese C., D'Amico M., Pestell R., Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. U.S.A. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perrimon N., Bernfield M. Specificities of heparan sulphate proteoglycans in developmental processes. Nature (London) 2000;404:725–728. doi: 10.1038/35008000. [DOI] [PubMed] [Google Scholar]
- 39.Tamai K., Semenov M., Kato Y., Spokony R., Liu C., Katsuyama Y., Hess F., Saint-Jeannet J. P., He X. LDL-receptor-related proteins in Wnt signal transduction. Nature (London) 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 40.Hacker U., Lin X., Perrimon N. The Drosophila sugarless gene modulates Wingless signaling and encodes an enzyme involved in polysaccharide biosynthesis. Development. 1997;124:3565–3573. doi: 10.1242/dev.124.18.3565. [DOI] [PubMed] [Google Scholar]
- 41.Lin X., Perrimon N. Dally cooperates with Drosophila Frizzled 2 to transduce Wingless signalling. Nature (London) 1999;400:281–284. doi: 10.1038/22343. [DOI] [PubMed] [Google Scholar]
- 42.Jayson G. C., Lyon M., Paraskeva C., Turnbull J. E., Deakin J. A., Gallagher J. T. Heparan sulfate undergoes specific structural changes during the progression from human colon adenoma to carcinoma in vitro. J. Biol. Chem. 1998;273:51–57. doi: 10.1074/jbc.273.1.51. [DOI] [PubMed] [Google Scholar]
- 43.Molist A., Romaris M., Lindahl U., Villena J., Touab M., Bassols A. Changes in glycosaminoglycan structure and composition of the main heparan sulphate proteoglycan from human colon carcinoma cells (perlecan) during cell differentiation. Eur. J. Biochem. 1998;254:371–377. doi: 10.1046/j.1432-1327.1998.2540371.x. [DOI] [PubMed] [Google Scholar]
- 44.Liu B. Y., Kim Y. C., Leatherberry V., Cowin P., Alexander C. M. Mammary gland development requires syndecan-1 to create a beta-catenin/TCF-responsive mammary epithelial subpopulation. Oncogene. 2003;22:9243–9253. doi: 10.1038/sj.onc.1207217. [DOI] [PubMed] [Google Scholar]