Abstract
NDH (NADH-quinone oxidoreductase)-1 complexes in cyanobacteria have specific functions in respiration and cyclic electron flow as well as in active CO2 uptake. In order to isolate NDH-1 complexes and to study complex–complex interactions, several strains of Thermosynechococcus elongatus were constructed by adding a His-tag (histidine tag) to different subunits of NDH-1. Two strains with His-tag on CupA and NdhL were successfully used to isolate NDH-1 complexes by one-step Ni2+ column chromatography. BN (blue-native)/SDS/PAGE analysis of the proteins eluted from the Ni2+ column revealed the presence of three complexes with molecular masses of about 450, 300 and 190 kDa, which were identified by MS to be NDH-1L, NDH-1M and NDH-1S respectively, previously found in Synechocystis sp. PCC 6803. A larger complex of about 490 kDa was also isolated from the NdhL-His strain. This complex, designated ‘NDH-1MS’, was composed of NDH-1M and NDH-1S. NDH-1L complex was recovered from WT (wild-type) cells of T. elongatus by Ni2+ column chromatography. NdhF1 subunit present only in NDH-1L has a sequence of -HHDHHSHH- internally, which appears to have an affinity for the Ni2+ column. NDH-1S or NDH-1M was not recovered from WT cells by chromatography of this kind. The BN/SDS/PAGE analysis of membranes solubilized by a low concentration of detergent indicated the presence of abundant NDH-1MS, but not NDH-1M or NDH-1S. These results clearly demonstrated that NDH-1S is associated with NDH-1M in vivo.
Keywords: affinity purification, CO2 uptake, cyanobacteria, NAD(P)H dehydrogenase, NDH-1 complex, thylakoid membrane
Abbreviations: BN, blue-native; β-DM, n-dodecyl β-D-maltoside; Cm, chloramphenicol; ESI MS/MS, electrospray ionization tandem MS; His-tag, histidine tag; His6, hexahistidine; MALDI-TOF, matrix-assisted laser-desorption–ionization time-of-flight; PSI, Photosystem I; Synechocystis 6803, Synechocystis sp. PCC 6803; T. elongatus, Thermosynechococcus elongatus BP-1; WT, wild-type
INTRODUCTION
The NDH-1 complex is a proton-pumping NADH-quinone oxidoreductase, also called Complex I [1]. In all organisms, the NDH-1 complexes form a characteristic L-shaped structure with a membrane domain and a peripheral domain composed of various numbers of hydrophobic and hydrophilic subunits [2–4]. The bacterial NDH-1 complexes, because of their relative structural simplicity and easy gene manipulation, are valuable models with which to study the corresponding complexes in eukaryotes [5]. In particular, the NDH-1 complexes of cyanobacteria closely resemble the NDH-1 complexes discovered in the chloroplasts of green plants, and thus present a unique opportunity to investigate the NDH-1 complexes specific to photosynthetic organisms. The NDH-1 complexes in cyanobacteria and higher plants function both in respiration and PSI (Photosystem I)-cyclic electron flow [6–11]. Additionally, cyanobacterial NDH-1 complexes are involved in inorganic-carbon-concentrating mechanisms [6,12–17]. Thus the NDH-1 complexes have multiple functions and play a crucial role in the acclimation and survival strategy of photosynthetic organisms under a variety of stress conditions [18,19].
In cyanobacteria and chloroplasts, there are 11 genes ndhA–ndhK encoding homologues of Escherichia coli NDH-1 subunits (NuoA–NuoD and NuoH–NuoN), whereas homologous genes encoding the soluble subunits NuoE–NuoG, which belong to the catalytic domain of E. coli NDH-1, have not yet been found [20–22]. Four additional subunits (NdhL–NdhO) have recently been identified in Synechocystis 6803 (Synechocystis sp. PCC 6803) [23,24], and three of them (NdhM–NdhO) also in higher plants [25]. Further, and unique to cyanobacteria, the ndhD and ndhF genes comprise small families. There are five ndhD and three ndhF genes in T. elongatus, and six and three respectively in Synechocystis 6803 (CyanoBase, the genome database for cyanobacteria; http://www.kazusa.or.jp/cyano/cyano.html). Diversity of the NdhD and NdhF subunits makes possible the multitude of functions of NDH-1 complexes in cyanobacteria [14,26]. Both reverse genetic [12,13,17,26] and functional proteomic studies [27] have demonstrated that the NDH-1 complexes containing NdhD1(D2) and NdhF1 are involved in PSI-cyclic electron flow and cellular respiration, whereas the NDH-1 complexes containing other NdhD/F members function in CO2 uptake. Proteomic studies have also been helpful in elucidating the overall structure and subunit composition of different NDH-1 complexes in Synechocystis 6803 [23,24,27]. We found three distinct NDH-1 complexes in the thylakoid membrane of Synechocystis 6803: NDH-1L, containing NdhD1 and NdhF1 and involved in respiration; NDH-1M, lacking NdhD/NdhF subunits and associated with cyclic electron flow; and NDH-1S, composed of NdhD3/F3, CupA and Sll1735 and being essential for active CO2 uptake [27].
Attempts to isolate intact NDH-1 complexes from cyanobacteria or plant chloroplasts have met severe difficulties, owing to the fragility of the enzyme and the low quantity present in the thylakoid membrane [23,25,28,29]. Therefore it is crucial to search for organisms with stable thylakoid membrane complexes. In order to investigate the composition, assembly and interaction of different NDH-1 complexes, we selected a thermophilic cyanobacterium, T. elongatus, and constructed a number of strains in which a His6 (hexahistidine) tag was added to C-termini of various subunits of NDH-1 complexes. Here we show that the strains that have a His-tag on NdhL and CupA were successfully used to isolate two types of NDH-1 complexes, namely NDH-1L and NDH-1MS, and provide the first evidence, at the protein level, that the functional distinction among NDH-1 complexes resides in the distal part of the membrane module containing the specific NdhD and NdhF subunits.
EXPERIMENTAL
NdhL-His and CupA-His strains of T. elongatus: construction and growth
The following primers were used to amplify the DNA containing cupA (ElcupA1 and ElcupA2) and ndhL (ElndhL1 and ElndhL2) and the downstream regions of these genes (ElcupA3 and ElcupA4, ElndhL3 and ElndhL4). ELcupA2 and ElndhL2 contain the sequences encoding six histidine residues. In addition, the primers for amplification of cupA and ndhL contain an SacI site and those for amplification of the downstream regions contain an SphI site. ELcupA1: 5′-GGG GAG CTC ACC AGT GCT CAG ATC GT-3′; ELcupA2: 5′-GGG GAG CTC CTA GTG ATG ATG ATG ATG ATG CAA ATA ATT GGG ATC CT-3′; ELcupA3: 5′-GGG GCA TGC GGA GTT GTA GGG CAA GG-3′; ELcupA4: 5′-GGG GCA TGC CAC CGA TCG GTT CGC CG-3′; ELndhL1: 5′-GGG GAG CTC GCC ATT GGC TTG GAT CG-3′; ELndhL2: 5′-GGG GAG CTC CTA GTG ATG ATG ATG ATG ATG GGA ATT GAG GGA GCG CG-3′ (+228 to +212); ELndhL3: 5′-GGG GCA TGC GTG CTA TGC GCC GCA TT-3′; ELndhL4: 5′-GGG GCA TGC CAC CGG TTT CTG GAT TC-3′. After the Cm (chloramphenicol) cassette [30] was inserted into the HincI site of pUC18 vector, the PCR products containing the CupA-His- or NdhL-His-encoding region and the downstream region were inserted into the SacI and SphI sites respectively after digestion with the restriction enzymes. The sequencing of the inserted PCR products confirmed that there was no mutation. Cells of wild-type or tll2230 disruptant mutant of T. elongatus were transformed with the above constructs according to the procedure described by Iwai et al. [31] and then grown on BG-11 [32] plates containing 3.4 μg/ml Cm at 45 °C in the light under high-CO2 conditions. We have also constructed strains by adding a His-tag to the C-termini of NdhB, NdhD1 and NdhE using the same method as that described above. However, His-tagged proteins could not be detected from these strains (results not shown).
For the isolation of protein complexes, T. elongatus WT, NdhL-His and CupA-His strains were grown in BG-11 medium at 45 °C under 50 μmol of photons·s−1·m−2 in 1-litre batch cultures under gentle agitation at high CO2 (4.5% CO2 in air) or low CO2 (the level in air). The mutant strains were grown in the presence of antibiotic.
Isolation and solubilization of thylakoid membrane
Thylakoid membranes were isolated [33] and suspended in buffer A [25 mM Bistris (pH 7.0)/20% (w/v) glycerol/1 mM Pefabloc] at a final concentration of 10 μg of protein/μl. The suspension was supplemented with an equal volume of buffer A containing 0.5% β-DM (n-dodecyl β-D-maltoside) or 2% digitonin, and incubated for 10 min on ice. Insoluble material was removed by centrifugation at 28 000 g for 30 min.
Purification of NdhL- and CupA-containing complexes
NdhL- and CupA-containing complexes were purified using Ni2+-affinity chromatography at 4 °C. The column (Chelating Sepharose™ High Performance; Amersham Biosciences) was saturated with 0.1 M NiSO4 and equilibrated with buffer B [25 mM Bistris (pH 7.0)/20% (w/v) glycerol/1 mM Pefabloc/0.02% β-DM]. Solubilized thylakoids (50 mg of protein) were filtered through a 0.45-μm-pore-size membrane (Schleicher & Schuell MicroScience GmbH, Dassel, Germany), and applied on to the 1 ml Ni2+-affinity chromatography column at a flow rate of 1 ml/min. After 1.5 h of sample circulation, the column was washed with buffer B containing 10 mM imidazole. His-tagged complexes were eluted with a 10–200 mM imidazole gradient in buffer B (40 ml) to optimize the elution conditions. Afterwards, 150 mM imidazole was adopted for routine experiments. The eluted complexes were concentrated by using the Microcon YM-50 (Millipore) system, washed with buffer B and stored at −80 °C for further analysis.
Electrophoresis and immunoblotting
Solubilized thylakoids (150 μg of protein) mixed with a 1/40th volume of BN (blue-native)-gel sample buffer [34] and purified samples (7 μg of protein) were loaded on 4.5–11% (w/v) polyacrylamide gradient BN-gel in a Hoefer Mighty Small mini-vertical unit. Electrophoresis was run at 4 °C for 5 h [34]. The BN-gel lane was solubilized and loaded on to a 1-mm-thick SDS/14%-(w/v)-polyacrylamide/6 M urea gel [27]. After electrophoresis, the proteins were visualized by silver staining. Immunoblotting was performed as described by Zhang et al. [27] using the CDP-Star® Chemiluminescent Detection Kit (New England Biolabs). The NdhF3 and NdhJ antibodies were as described previously [27]. Supersignal West HisProbe Kit (Pierce) was used to detect His-tagged proteins.
Identification of proteins by MALDI-TOF (matrix-assisted laser-desorption–ionization time-of-flight) MS and ESI MS/MS (electrospray ionization tandem MS)
Silver-stained protein spots were excised from gels and digested with trypsin or CNBr [35]. The digests were analysed by MALDI-TOF MS and ESI MS/MS as described by Battchikova et al. [24].
Bioinformatic tools
The sequence information was consulted at NCBI (http://www.ncbi.nlm.nih.gov/) and CyanoBase. Sequence similarity searches were performed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). FASTA (http://fasta.bioch.virginia.edu/) was used for sequence comparison. Protein sequences were characterized using TMHMM (http://www.cbs.dtu.dk/services/TMHMM/), ClustalW (http://www.ebi.ac.uk/clustalw/) and Mascot (www.matrixscience.com).
RESULTS
Purification of NDH-1 complexes by Ni2+ affinity chromatography
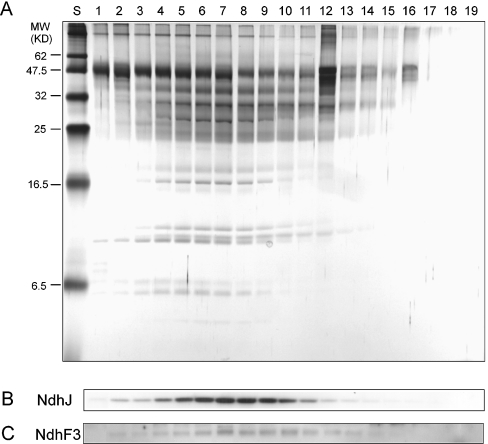
Solubilized thylakoid membrane extracts isolated from WT, NdhL-His and CupA-His cells, grown under high- or low-CO2 conditions, were applied on to a Ni2+ column. Optimal imidazole concentration for elution of the target complexes was determined using a gradient of imidazole (10–200 mM) in the elution buffer. The elution fractions were subjected to SDS/PAGE. The silver-stained gel and immunoblots probed with NdhJ and NdhF3 antibodies are shown in Figure 1. The results demonstrated that both the NdhF3- and NdhJ-containing complexes were co-eluted in fractions 2–14, with a peak at fraction 7 (Figures 1B and 1C). We also noticed that some proteins were eluted out earlier than target complexes, with the peak at fraction 5. Similar results were obtained with the CupA-His strain (results not shown). Therefore 150 mM imidazole was routinely used in further experiments.
Figure 1. SDS/PAGE profiles of fractions containing His-tagged NDH-1 complexes eluted from Ni2+ affinity column.
The NDH-1 complexes were solubilized from the NdhL-His strain grown at low CO2 concentration, and 19 fractions were eluted from Ni2+ column by an increasing imidazole gradient from 10 to 200 mM. Eluted fractions were concentrated to 100 μl and subjected to SDS/PAGE. (A) Silver-stained SDS/PAGE gels: lane S, molecular-mass standards; lanes 1–19, elution fractions of gradient immidazole. (B) Immunoblot probed with anti-NdhJ antibody. (C) Immunoblot probed with anti-NdhF3 antibody.
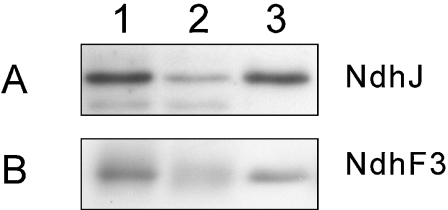
The efficiency of the Ni2+ affinity column was estimated by immunoblotting with NdhJ and NdhF3 antibodies (Figure 2). About 80% of the target protein was bound to the column, and about 90% of the bound material could be recovered from the column. Finally, approx. 0.12 mg of protein was obtained from 50 mg of thylakoid proteins.
Figure 2. Efficiency of the Ni2+ column in the purification of NDH-1 complexes.
(A) Complexes were isolated from the NdhL-His strain using the Ni2+ affinity column, separated by SDS/PAGE, electroblotted to a PVDF membrane and probed with anti-NdhJ antibody. In (B) the CupA-His strain was used and the membrane was probed with anti-NdhF3 antibody. Thylakoid proteins equivalent to 50 mg were solubilized and applied to the Ni2+ affinity column. Equal relative volumes were loaded on the wells. Lane 1, initial sample; lane 2, unbound material; lane 3, eluted material.
Analysis and subunit identification of the NdhL- and CupA-containing complexes
Membrane protein complexes isolated by Ni2+ affinity chromatography were further analysed by two-dimensional BN/SDS/PAGE. Different numbers of protein complexes were resolved depending on the strains and growth conditions, as shown in silverstained gels (Figures 3A–3C). Subunit composition of the complexes was analysed by MS (Tables 1 and 2).
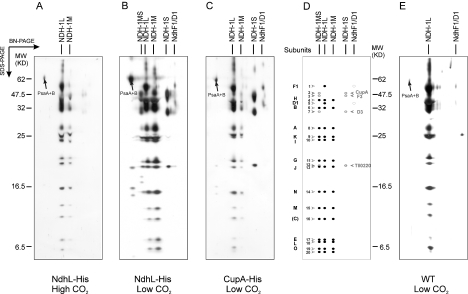
Figure 3. Two-dimensional gels of NDH-1 complexes purified on the Ni2+ affinity column.
Thylakoid membranes were isolated, solubilized with 0.25% β-DM, and applied to the Ni2+ affinity column. Eluted complexes were subjected to BN/SDS/PAGE, and the gels were stained with silver (A, B, C and E): (A) NdhL-His strain grown at high CO2; (B) NdhL-His strain grown at low CO2; (C) CupA-His strain grown at low CO2; (D) scheme of NDH-1 subunits; (E) WT grown at low CO2. The identify of the spots is based on MS analysis and as indicated in Tables 1 and 2.
Table 1. Subunits of NDH-1 from T. elongatus identified by MALDI-TOF.
The spot number corresponds to that in Figure 3. ORF, the open reading frame in CyanoBase. M, molecular mass. Experimental molecular-masses were estimated by comparison of the migration of the respective proteins with that of molecular-mass markers. MALDI-TOF MS results are presented as the ratios of peptide masses providing the identity with the total amount of peptides used in the search, percentages of coverage for full-length proteins and score values according to Mascot.
| M (kDa) | |||||||
|---|---|---|---|---|---|---|---|
| Spot no. | NDH-1 subunit | ORF | Theoretical | Experimental | Peptides matched/total | Cover (%) | Score |
| 2 | CupA | tlr0906 | 51.0 | 49 | 18/24 | 43 | 277 |
| 4 | NdhH | tlr1288 | 45.5 | 41 | 20/26 | 57 | 209 |
| 9 | NdhK | tlr0705 | 25.9 | 24.5 | 6/9 | 26 | 85 |
| 10 | NdhI | tlr0668 | 22.9 | 24 | 12/16 | 55 | 138 |
| 12 | Tll0220 | tll0220 | 15.8 | 20.2 | 4/7 | 47 | 76 |
| 13 | NdhJ | tlr1430 | 19.4 | 20 | 12/17 | 83 | 187 |
| 14 | NdhN | tlr1130 | 16.6 | 15.5 | 7/9 | 57 | 110 |
| 15 | NdhM | tll0447 | 12.6 | 13 | 5/6 | 45 | 73 |
Table 2. Subunits of NDH-1 from T. elongatus identified by ESI MS/MS.
The spot number corresponds to that in Figure 3. ORF, the open reading frame in CyanoBase. Mtheo, values of theoretical molecular masses (in kDa). Experimental molecular masses (Mexp, in kDa), were estimated by comparison of the migration of the respective proteins with that of molecular-mass markers. The number of transmembrane helices (Mem) was predicted by TMHMM. ESI MS/MS results are presented as total scores and percentages of coverage for full-length proteins followed by sequences of peptides (up to five per protein) with observed m/z values, charge state of peptides and scores according to Mascot. Proteins were digested with trypsin, except NdhF1, which was treated with CNBr. C-terminal methionine in CNBr-cleaved peptides was converted into homoserine lactone and shown by an asterisk. Miscleavage is shown as (1) and oxidation of methionine as (o).
| Spot no. | NDH-1 subunit | ORF | Mtheo | Mexp | Mem | Total score | Cover (%) | Sequence | Observed peptide mass, charge/score | [M+H]+ theoretical |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NdhF1 | tll0720 | 72.1 | 55 | 16 | 436 | 26 | VAAGVFLIARM* | 550.31, 2+, /59 | 1099.66 |
| LFLGSGSVIHSM* | 600.29, 2+, /49 | 1199.64 | ||||||||
| GVGAYSAGLFHLM* | 637.80, 2+, /47 | 1274.66 | ||||||||
| EGVVGHNPDLAQDM* | 717.30, 2+, /55 | 1433.67 | ||||||||
| EGPTPISALIHAATM* | 487.57, 3+, /52 | 1460.78 | ||||||||
| 3 | NdhF3 | tlr0904 | 66.4 | 46 | 16 | 291 | 13 | ITAWFDR | 454.74, 2+, /31 | 908.47 |
| ATVVGVVDMISR | 623.84, 2+, /43 | 1246.69 | ||||||||
| TFCLVWGGEVKPMTAR | 617.98, 3+, /45 | 1851.92 | ||||||||
| TFVDGTGNAFGVVTLLGGDR | 998.52, 2+, /121 | 1996.02 | ||||||||
| 5 | NdhD1 | tll0719 | 56.2 | 38 | 12 | 405 | 14 | MGGYALIR | 440.75, 2+, /51 | 880.47 |
| ELVEHEALVDAEPR | 803.92, 2+, /84 | 1606.81 | ||||||||
| THTLILEEMGGVGQK | 538.30, 3+, /76 | 1612.84 | ||||||||
| LLTQIYDATTGQVIAR | 881.99, 2+, /86 | 1762.97 | ||||||||
| 6 | NdhB | tll0045 | 55.2 | 32 | 14 | 202 | 7 | AAGFALAIR | 445.28, 2+, /48 | 889.53 |
| EPQEMSEAVR | 588.28, 2+, /49 | 1175.54 | ||||||||
| TGTDQISEYAGLYQK | 837.42, 2+, /70 | 1673.80 | ||||||||
| 7 | NdhD3 | tlr0905 | 53.9 | 30.5 | 14 | 168 | 4 | LGAYGLVR | 424.76, 2+, /47 | 848.50 |
| TNELVATLAK | 530.32, 2+, /47 | 1059.61 | ||||||||
| ELNVLNGLLNPLR | 732.94, 2+, /75 | 1464.86 | ||||||||
| 8 | NdhA | tlr0667 | 41.3 | 27 | 6 | 188 | 13 | YSLLGGLR | 439.77, 2+, /41 | 878.53 |
| IDQLLDLGWK | 600.84, 2+, /68 | 1200.68 | ||||||||
| IGPEYIGPLGILAPLADGLK | 1004.09, 2+, /55 | 2007.18 | ||||||||
| 11 | NdhG | tlr0669 | 21.6 | 21 | 5 | 128 | 25 | ETYTPVPGR | 510.27, 2+, /32 | 1019.54 |
| QGGAAVVSLGVFALLTK | 816.00, 2+, /73 | 1630.99 | ||||||||
| ELVLEPEPILGEEVVPPLELPERPR | 950.54, 2+, /22 | 2849.61 | ||||||||
| 16 | NdhC | tlr1429 | 15.1 | 11 | 3 | 32 | 21 | VAIPRLR | 412.74, 2+, /4 | 824.55 |
| LTTYESGMEPIGGAWIQFNVR | 790.37, 3+, /28 | 2369.16 | ||||||||
| 17 | NdhE | tlr0670 | 11.2 | 8 | 3 | 112 | 12 | FNLLK | 317.69, 2+, /36 | 634.39 |
| DTVDMEKFNLLK(1) | 484.90, 3+, /11 | 1452.74 | ||||||||
| DTVDM(o)EKFNLLK(1) | 734.80, 3+, /43 | 1468.74 | ||||||||
| 18 | NdhL | tsr0706 | 8.6 | 7.5 | 2 | 32 | 12 | WYVASSFER | 572.75, 2+, /32 | 1144.54 |
| 19 | NdhO | tsl0017 | 7.9 | 6 | 0 | 208 | 68 | GEVVDIR | 394.20, 2+, /36 | 787.43 |
| LDQLEVAQ | 458.24, 2+, /24 | 915.48 | ||||||||
| YPPYLFEGR | 571.26, 2+, /43 | 1141.57 | ||||||||
| FPVPTPTVWLR | 656.85, 2+, /47 | 1312.74 | ||||||||
| LANSLEALASDHR | 466.23, 2+, /58 | 1396.72 |
Two complexes, of molecular mass about 450 and 300 kDa, interacting with Ni2+ column, were recovered from NdhL-His strain grown at high CO2. They were analogous to NDH-1L and NDH-1M complexes of Synechocystis 6803 [31]. From cells of the same strain grown at low CO2, four major complexes were isolated. Besides NDH1-L and NDH-1M, we observed a small complex (190 kDa) analogous to NDH-1S of Synechocystis 6803, and a large complex (490 kDa) not detected earlier in Synechocystis. The new complex contained subunits of both NDH-1M and NDH-1S, and was therefore named ‘NDH-1MS’. Only three complexes, NDH-1L, NDH-1M and NDH-1S, were recovered from CupA-His strain grown at low CO2. The NDH-1MS complex was not recovered and the amount of NDH-1M was less than that from NdhL-His strain grown under similar conditions (Figure 3C).
An additional complex of 1000 kDa, present in the gels to various extents, appeared to represent the PSI trimer as contamination (Figures 3A–3C and 3E). This complex was eluted from the Ni2+ column at low imidazole concentration (Figure 1) and could be largely eliminated (Figure 3E).
To investigate the possibility of unspecific binding of native proteins to the Ni2+ column, WT cells grown at low CO2 were also analysed. We observed that NDH-1L, but not NDH-1M or NDH-1S, was effectively recovered from the Ni2+ column without any His-tag modification (Figure 3E). We next tested whether NDH-1L from WT Synechocystis 6803 could also be recovered from Ni2+ column, but this was not the case (results not shown).
In the NDH-1L complex, 15 subunits were identified (Tables 1 and 2), including the hydrophilic components (NdhH–NdhO) and the membrane proteins (NdhA, NdhB, NdhC, NdhD1, NdhE, NdhF1, NdhG and NdhL). The NDH-1M complex contained the same 13 single copy ndh gene products but lacked the NdhD and NdhF subunits. The NDH-1S complex contained NdhD3, NdhF3, CupA and Tll0220, similar to those in Synechocystis 6803 [24,27]. The NDH-1MS complex comprised all the subunits of NDH-1M and NDH-1S and was distinctly characterized by its high molecular mass together with the presence of a clearly distinguishable CupA protein in the complex (Figure 3B).
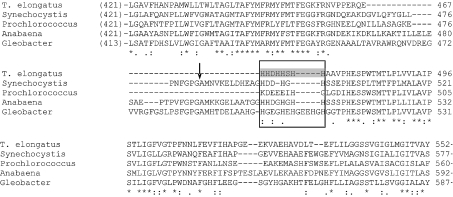
It should be noted that NdhF1 was the largest subunit of NDH-1L of T. elongatus. No proteolysis of this subunit was observed, in contrast with NdhF1 of Synechocystis 6803, which was proteolytically processed between Gly482 and Ala483 [27,28]. The multiple alignment [36] of NdhF1 sequences from various cyanobacteria showed that the sequence of T. elongatus, as well as some other cyanobacteria, lacks the proteolytic site present in Synechocystis 6803 (Figure 4).
Figure 4. Sequence comparison of the NdhF1 proteins from cyanobacteria.
Multiple sequence alignment of the NdhF1 protein on the region surrounding the proteolytic site and the probable Ni2+-affinity-column-binding site was performed by ClustalW. Sequences of the following organisms were used: T. elongatus (T. elongatus BP-1), Tll0720; Synechocystis (Synechocystis sp. PCC 6803), Slr0844; Prochlorococcus (Prochlorococcus marinus SS120), Pro0172; Anabaena (Anabaena sp. PCC 7120), Alr3956; Gleobacter (Gleobacter violaceus PCC 7421), Glr0218. The arrow indicates the proteolytic site in Synechocystis 6803 NdhF1 protein. The box indicates the probable Ni2+-affinity-column-binding site, and the grey shading shows the histidine-rich region in T. elongatus.
Fragility of the NDH-1MS supercomplex
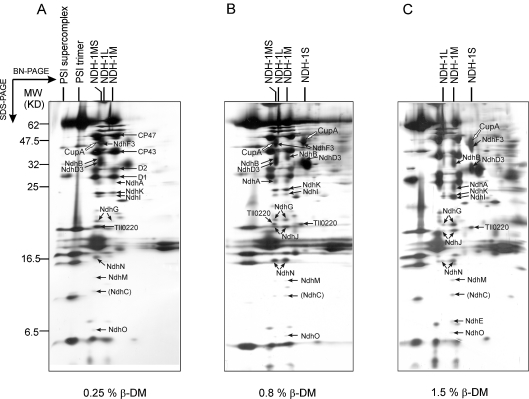
Only a small amount of NDH-1MS was recovered under conditions for complex isolation, suggesting the fragility of this complex. To demonstrate the presence of the NDH-1MS supercomplex in vivo, the WT thylakoid membranes were solubilized with a series of detergent concentrations, followed by immediate separation of the protein complexes by BN/SDS/PAGE. Preliminary experiments had shown that the membrane complexes of T. elongatus could be solubilized at a lower detergent concentration than those from Synechocystis 6803. The presence of the NDH-1MS complex in T. elongatus was analysed from thylakoids of low-CO2-grown WT cells solubilized with β-DM (0.1–1.5%). The lowest concentration capable of solubilizing the thylakoid protein complexes was 0.25%. At this concentration, the supercomplex NDH-1MS was the main form of the NDH-1 complexes, and almost no dissociated subcomplexes (NDH-1M and NDH-1S) were observed (Figure 5A). β-DM concentrations of 0.8% and 1.5% dissociated approximately half and nearly all of the supercomplexes respectively (Figures 5B and 5C). Digitonin, which has been considered to be a milder detergent than β-DM and suitable for other supercomplex studies [37–39], was also tested at different concentrations (0.5–2.0%). With 1.0% digitonin, the pattern obtained was similar to that obtained on solubilization of the membrane by 0.25% β-DM (results not shown). These results clearly demonstrated that the NDH-1M and NDH-1S complexes in vivo form an NDH-1MS supercomplex.
Figure 5. Stability of NDH-1MS from WT T. elongatus cells grown at low CO2.
Thylakoids were solubilized with (A) 0.25% β-DM, (B) 0.8% β-DM, or (C) 1.5% β-DM, protein complexes were separated by two-dimensional BN/SDS/PAGE and the gels were stained with silver. The white arrow shows the CupA protein in the NDH-1 complex that is dominating under the given detergent condition. The major NDH-1 subunits as well as the PSII (Photosystem II) subunits D1, D2, CP43 and CP47 are indicated by black arrows.
DISCUSSION
Selection of the subunit for His tagging is crucial for isolation of NDH-1 complexes
Cyanobacterial NDH-1 complexes have attracted much attention because of their multiple functions [6,7,12–16,26], but the understanding of their structure is still in its infancy. The recent discovery of different NDH-1 complexes opened the era in which the structure–function relationships of these complexes could be studied [23,24,27,34]. However, a thorough investigation of NDH-1 complexes has been hampered by their low abundance and fragility. We therefore took an approach involving the His-tagging of subunits of NDH-1 complexes of a thermophilic cyanobacterium, namely T. elongatus. Out of five constructed strains (NdhB-His, NdhD1-His, NdhE-His, NdhL-His and CupA-His), His-tagged proteins were present only in NdhL-His and CupA-His. Failure to detect His-tagged proteins on other strains suggested that the membrane subunits (NdhB, NdhD1 and NdhE) are not stably integrated into NDH-1 complexes when six histidine residues are added and become rapidly degraded. Reverse-genetic studies in Synechocystis 6803 showed that NdhL is an essential subunit for the functioning of NDH-1 complexes [40]. On the other hand, the deletion of NdhL, in contrast with other hydrophobic subunits, does not seriously hamper the assembly of NDH-1 complexes [24]. It is therefore conceivable that the NdhL subunit is located at the edge of the NDH-1 hydrophobic membrane domain, and introducing a His-tag to NdhL subunit would not critically affect the assembly and function of the NDH-1 complexes. Indeed, no growth retardation of NdhL-His strain has been recorded, although the different functions of various NDH-1 complexes were not specifically tested. The insertion of His-tag into the CupA subunit also allowed normal growth of cells independently of the CO2 conditions, but it seems that the His-tag in CupA hampered the stability of the NDH-1MS complex (see below).
NDH-1 complexes isolated from NdhL-His and CupA-His strains
Isolation of the NDH-1MS supercomplex from the NdhL-His strain confirms the co-operation of NDH-1M and NDH-1S in CO2 uptake [27]. In addition to NDH-1MS, considerable amounts of NDH-1M and NDH-1S were co-purified from NdhL-His (Figure 3B). Since NDH-1S does not itself bind to the column (Figure 3E), it can be ‘rescued’ from the NdhL-His strain only if associated with NDH-1M during membrane solubilization and chromatography. Thus NDH-1MS bound to the Ni2+ column was partially dissociated into NDH-1M and NDH-1S, most probably during the elution step. These results imply a fragile structure for the supercomplex NDH-1MS.
Only one type of NDH-1 complex, namely NDH-1MS, which is specialized in respect of inducible CO2 uptake [15,16], was expected to be isolated from the CupA-His strain. However, three complexes (NDH-1L, NDH-1M and NDH-1S) were rescued by affinity chromatography and no NDH-1MS was isolated from low-CO2-grown cells of this strain (Figure 3C). In addition, the amount of NDH-1M isolated from CupA-His strain was less than that isolated from NdhL-His strain. This suggested that Histagging of the hydrophilic CupA subunit destabilized the NDH-1MS supercomplex. Indeed, a better integrity of NDH-1MS was achieved by tagging the hydrophobic subunit NdhL. Severe destabilization of the NDH-1 complex also occurred when the His-tag was fused to the hydrophilic NdhH subunit for isolation of the NDH-1(L) complex from tobacco (Nicotiana tabacum) thylakoids [25].
A considerable amount of intact NDH-1L was rescued from the NdhL-His strain, but, unexpectedly, also from the CupA-His strain (Figures 3A–3C). This, however, does not indicate an interaction between NDH-1L and NDH-1S, since NDH-1L was also rescued from WT T. elongatus by Ni2+ column chromatography (Figure 3E). We found that the NdhF1 of T. elongatus contains a histidine-rich region, namely -H468HDHHSHH475- (Figure 4). Analysis of the sequence of this subunit using TMHMM v. 2.0 [41], predicted that the histidine-rich region is located on the cytoplasmic side, and thus appears to have an affinity for the Ni2+ column. This might have enabled one to efficiently rescue NDH-1L from WT T. elongatus, this being consistent with the failure to rescue NDH-1L from WT Synechocystis 6803, whose NdhF1 does not contain as rich a histidine region as does T. elongatus NdhF1 (Figure 4).
Structural and functional diversity of cyanobacterial NDH-1 complexes resides on different NdhD/F subunits
NDH-1 complexes and their homologues are widespread enzymes in eubacteria, cyanobacteria and eukaryotes [42]. In contrast with the mitochondrial complex I involved in respiration, the NDH-1 complexes in cyanobacteria and in the chloroplasts of green plants have an additional function in PSI-cyclic electron transport [7,9,25] and also in CO2 uptake in cyanobacteria [6]. To acquire this functional diversity, cyanobacteria have developed different types of NDH-1 complexes. This was achieved by allowing different types of NdhD/F-containing complexes to associate with a basal complex, NDH-1M, to form NDH-1L and NDH-1MS. In addition to these complexes, the presence of an NdhD4/NdhF4/CupB-containing complex, participating in constitutive CO2 uptake and having a structure similar to NDH-1MS, was highly expected, but could not be identified in the present study. This is puzzling, since reverse genetics and functional studies have shown considerable fluxes of CO2 through such a putative NDH-1 complex [15,16]. Moreover, attempts to find the NdhD4/NdhF4/CupB-containing (sub)complex or individual subunits in purified plasma membranes have failed (N. Battchikova, P. Zhang and E.-M. Aro, unpublished work), leaving the identity and location of this complex still to be discovered.
Model structure of the NDH-1 complexes
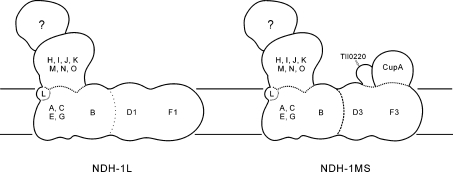
On the basis of the discovery of the NDH-1MS complex and because of the possibility of limited dissociation of NdhD1/NdhF1 from the NDH-1L complex, as shown in the previous studies on NDH-1 complexes of Synechocystis 6803 and E. coli [23,43], it is most likely that the hydrophobic subunits of the NdhF/NdhD family are located at a distal part of the membrane arm in the NDH-1 complex (Figure 6). The other subunits, representing NDH-1M, are likely to form a relatively independent structure with a membrane domain composed of NdhA, NdhB, NdhC, NdhE, NdhG and NdhL and a hydrophilic domain composed of NdhH–NdhO (Figure 6). Upon assembly of the NDH-1M complex with the NdhD1/NdhF1 subcomplex, a respiratory NDH-1L complex is formed, whereas upon assembly with the NDH-1S complex an inducible CO2-uptake complex (NDH-1MS) is formed. It is conceivable that, upon sudden changes in the CO2 level, which generally induces opposite changes in the contents of the NDH-1MS and NDH-1L complexes [27], the cyanobacterial cells can flexibly use the NDH-1M subcomplex and induce the synthesis/degradation of only the distal arm of the complex containing the NdhD and NdhF subunits.
Figure 6. Model of subunit arrangement in the NDH-1L and NDH-1MS complexes.
The question mark indicates a still unknown subcomplex corresponding to the electron input module.
Acknowledgments
We thank the Academy of Finland, the Nordic Joint Committee for Agricultural Research (grant to E. M. A.), the U.S. National Science Foundation and the Department of Energy (grants to T. O. and H. B. P.) for financial support. The Proteomics and Mass Spectrometry Unit in the Turku Center for Biotechnology is thanked for technical advice in protein analysis.
References
- 1.Yagi T., Matsuno-Yagi A. The proton-translocating NADH-quinone oxidoreductase in the respiratory chain: the secret unlocked. Biochemistry. 2003;42:2266–2274. doi: 10.1021/bi027158b. [DOI] [PubMed] [Google Scholar]
- 2.Guenebaut V., Vincentelli R., Mills D., Weiss H., Leonard K. R. Three-dimensional structure of NADH-dehydrogenase from Neurospora crassa by electron microscopy and conical tilt reconstruction. J. Mol. Biol. 1997;265:409–418. doi: 10.1006/jmbi.1996.0753. [DOI] [PubMed] [Google Scholar]
- 3.Grigorieff N. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 Å in ice. J. Mol. Biol. 1998;277:1033–1046. doi: 10.1006/jmbi.1998.1668. [DOI] [PubMed] [Google Scholar]
- 4.Matsuno-Yagi A., Yagi T. Introduction: complex I – an L-shaped black box. J. Bioenerg. Biomembr. 2001;33:155–157. doi: 10.1023/a:1010702116348. [DOI] [PubMed] [Google Scholar]
- 5.Weidner U., Geier S., Ptock A., Friedrich T., Leif H., Weiss H. The gene locus of the proton-translocating NADH:ubiquinone oxidoreductase in Escherichia coli. Organization of the 14 genes and relationship between the derived proteins and subunits of mitochondrial complex I. J. Mol. Biol. 1993;233:109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- 6.Ogawa T. A gene homologous to the subunit-2 gene of NADH dehydrogenase is essential to inorganic carbon transport of Synechocystis PCC 6803. Proc. Natl. Acad. Sci. U.S.A. 1991;88:4275–4278. doi: 10.1073/pnas.88.10.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mi H., Endo T., Schreiber U., Ogawa T., Asada K. Electron donation from cyclic and respiratory flows to the photosynthetic intersystem chain is mediated by pyridine nucleotide dehydrogenase in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol. 1992;33:1233–1237. [Google Scholar]
- 8.Shikanai T., Endo T. Physiological function of a respiratory complex, NAD(P)H dehydrogenase in chloroplasts: dissection by chloroplast reverse genetics. Plant Biotech. 2000;17:79–86. [Google Scholar]
- 9.Joet T., Cournac L., Peltier G., Havaux M. Cyclic electron flow around photosystem I in C3 plants. In vivo control by the redox state of chloroplasts and involvement of the NADH-dehydrogenase complex. Plant Physiol. 2002;128:760–769. doi: 10.1104/pp.010775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peltier G., Cournac L. Chlororespiration. Annu. Rev. Plant Biol. 2002;53:523–550. doi: 10.1146/annurev.arplant.53.100301.135242. [DOI] [PubMed] [Google Scholar]
- 11.Munekage Y., Hashimoto M., Miyake C., Tomizawa K.-I., Endo T., Tasaka M., Shikanai T. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature (London) 2004;429:579–582. doi: 10.1038/nature02598. [DOI] [PubMed] [Google Scholar]
- 12.Ohkawa H., Sonoda M., Katoh H., Ogawa T. The use of mutants in the analysis of the CCM in cyanobacteria. Can. J. Bot. 1998;76:1025–1034. [Google Scholar]
- 13.Price G. D., Klughammer B., Ludwig M., Badger M. R. The functioning of the CO2 concentrating mechanism in several cyanobacterial strains: a review of general physiological characteristics, genes, proteins and recent advances. Can. J. Bot. 1998;76:973–1002. [Google Scholar]
- 14.Klughammer B., Sültemeyer D., Badger M. R., Price G. D. The involvement of NAD(P)H dehydrogenase subunits, NdhD3 and NdhF3, in high-affinity CO2 uptake in Synechococcus sp. PCC 7002 gives evidence for multiple NDH-1 complexes with specific roles in cyanobacteria. Mol. Microbiol. 1999;32:1316–1332. doi: 10.1046/j.1365-2958.1999.01457.x. [DOI] [PubMed] [Google Scholar]
- 15.Shibata M., Ohkawa H., Kaneko T., Fukuzawa H., Tabata S., Kaplan A., Ogawa T. Distinct constitutive and low CO2-induced CO2 uptake systems in cyanobacteria: genes involved and their phylogenetic relationship with homologous genes in other organisms. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11789–11794. doi: 10.1073/pnas.191258298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda S., Badger M. R., Price G. D. Novel gene products associated with NdhD3/D4-containing NDH-1 complexes are involved in photosynthetic CO2 hydration in the cyanobacterium, Synechococcus sp. PCC7942. Mol. Microbiol. 2002;43:425–435. doi: 10.1046/j.1365-2958.2002.02753.x. [DOI] [PubMed] [Google Scholar]
- 17.Ogawa T., Kaplan A. Inorganic carbon acquisition systems in cyanobacteria. Photosynth. Res. 2003;77:105–115. doi: 10.1023/A:1025865500026. [DOI] [PubMed] [Google Scholar]
- 18.Figge R. M., Cassier-Chauvat C., Chauvat F., Cerff R. Characterization and analysis of an NAD(P)H dehydrogenase transcriptional regulator critical for the survival of cyanobacteria facing inorganic carbon starvation and osmotic stress. Mol. Microbiol. 2001;39:455–468.. doi: 10.1046/j.1365-2958.2001.02239.x. [DOI] [PubMed] [Google Scholar]
- 19.Martin M., Casano L. M., Zapata J. M., Guera A., del Campo E. M., Schmitz-Linneweber C., Maier R. M., Sabater B. Role of thylakoid Ndh complex and peroxidase in the protection against photo-oxidative stress: fluorescence and enzyme activities in wild-type and ndhF-deficient tobacco. Physiol. Plant. 2004;122:443–452. [Google Scholar]
- 20.Kaneko T., Sato S., Kotani H., Tanaka A., Asamizu E., Nakamura Y., Miyajima N., Hirosawa M., Sugiura M., Sakamoto S., et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 21.Rasmusson A. G., Heiser V., Zabaleta E., Brennicke A., Grohmann L. Physiological, biochemical and molecular aspects of mitochondrial complex I in plants. Biochim. Biophys. Acta. 1998;1364:101–111. doi: 10.1016/s0005-2728(98)00021-8. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura Y., Kaneko T., Sato S., Ikeuchi M., Katoh H., Sasamoto S., Watanabe A., Iriguchi M., Kawashima K., Kimura T., et al. Complete Genome Structure of the Thermophilic Cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res. 2002;9:123–130. doi: 10.1093/dnares/9.4.123. [DOI] [PubMed] [Google Scholar]
- 23.Prommeenate P., Lennon A. M., Markert C., Hippler M., Nixon P. J. Subunit composition of NDH-1 complexes of Synechocystis sp. PCC 6803: identification of two new ndh gene products with nuclear encoded homologues in the chloroplast Ndh complex. J. Biol. Chem. 2004;279:28165–28173. doi: 10.1074/jbc.M401107200. [DOI] [PubMed] [Google Scholar]
- 24.Battchikova N., Zhang P., Rudd S., Ogawa T., Aro E.-M. Identification of NdhL and Ssl1690 (NdhO) in NDH-1L and NDH-1M complexes of Synechocystis sp. PCC 6803. J. Biol. Chem. 2005;280:2587–2595. doi: 10.1074/jbc.M410914200. [DOI] [PubMed] [Google Scholar]
- 25.Rumeau D., Becuwe-Linka N., Beyly A., Louwagie M., Garin J., Peltier G. New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid Ndh complex functioning in higher plants. Plant Cell. 2005;17:219–232. doi: 10.1105/tpc.104.028282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohkawa H., Pakrasi H. B., Ogawa T. Two types of functionally distinct NAD(P)H dehydrogenases in Synechocystis sp. strain PCC6803. J. Biol. Chem. 2000;275:31630–31634. doi: 10.1074/jbc.M003706200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang P., Battchikova N., Jansen T., Appel J., Ogawa T., Aro E.-M. Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp. PCC 6803. Plant Cell. 2004;16:3326–3340. doi: 10.1105/tpc.104.026526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger S., Ellersiek U., Kinzelt D., Steinmüller K. Immunopurification of a subcomplex of the NAD(P)H-plastoquinone-oxidoreductase from the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 1993;326:246–250. doi: 10.1016/0014-5793(93)81800-f. [DOI] [PubMed] [Google Scholar]
- 29.Matsuo M., Endo T., Asada K. Properties of the respiratory NAD(P)H dehydrogenase isolated from the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol. 1998;39:263–267. doi: 10.1093/oxfordjournals.pcp.a029366. [DOI] [PubMed] [Google Scholar]
- 30.Elhai J., Wolk C. P. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmid that allows cloning into long polylinkers. Gene. 1988;68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 31.Iwai M., Katoh H., Katayama M., Ikeuchi M. Improved genetic transformation of the thermophilic cyanobacterium Thermosynechococcus elongatus sp. BP-1. Plant Cell Physiol. 2004;45:171–175. doi: 10.1093/pcp/pch015. [DOI] [PubMed] [Google Scholar]
- 32.Williams J. K. G. Construction of specific mutations in PSII photosynthetic reaction center by genetic engineering. Methods Enzymol. 1988;167:766–778. [Google Scholar]
- 33.Gombos Z., Wada H., Murata N. The recovery of photosynthesis from low-temperature photoinhibition is accelerated by the unsaturation of membrane lipids: a mechanism of chilling tolerance. Proc. Natl. Acad. Sci. U.S.A. 1994;91:8787–8791. doi: 10.1073/pnas.91.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herranen M., Battchikova N., Zhang P., Graf A., Sirpiö S., Paakkarinen V., Aro E.-M. Towards functional proteomics of membrane protein complexes in Synechocystis sp. PCC 6803. Plant Physiol. 2004;134:470–481. doi: 10.1104/pp.103.032326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Montfort B. A., Doeven M. K., Canas B., Veenhoff L. M., Poolman B., Robillard G. T. Combined in-gel tryptic digestion and CNBr cleavage for the generation of peptide maps of an integral membrane protein with MALDI-TOF mass spectrometry. Biochim. Biophys. Acta. 2002;1555:111–115. doi: 10.1016/s0005-2728(02)00264-5. [DOI] [PubMed] [Google Scholar]
- 36.Higgins D. G., Thompson J. D., Gibson T. J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 37.Cline K., Mori H. Thylakoid ΔpH-dependent precursor proteins bind to a cpTatC–Hcf106 complex before Tha4-dependent transport. J. Cell Biol. 2001;154:719–729. doi: 10.1083/jcb.200105149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schägger H. Respiratory chain supercomplexes of mitochondria and bacteria. Biochim. Biophys. Acta. 2002;1555:154–159. doi: 10.1016/s0005-2728(02)00271-2. [DOI] [PubMed] [Google Scholar]
- 39.Heinemeyer J., Eubel H., Wehmhöner D., Jänsch L., Braun H.-P. Proteomic approach to characterize the supramolecular organization of photosystems in higher plants. Phytochemistry. 2004;65:1683–1692. doi: 10.1016/j.phytochem.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 40.Ogawa T. Identification and characterization of the ictA/ndhL gene product essential to inorganic carbon transport of Synechocystis PCC 6803. Plant Physiol. 1992;99:1604–1608. doi: 10.1104/pp.99.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Bol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 42.Friedrich T., Steinmüller K., Weiss H. The proton-pumping respiratory complex I of bacteria and mitochondria and its homologue in chloroplasts. FEBS Lett. 1995;367:107–111. doi: 10.1016/0014-5793(95)00548-n. [DOI] [PubMed] [Google Scholar]
- 43.Holt P. J., Morgan D. J., Sazanov L. A. The location of NuoL and NuoM subunits in the membrane domain of the Escherichia coli Complex I. Implications for the mechanism of proton pumping. J. Biol. Chem. 2003;278:43114–43120. doi: 10.1074/jbc.M308247200. [DOI] [PubMed] [Google Scholar]