Abstract
GSH synthesis occurs through a two-step enzymatic reaction driven by GCL (glutamate–cysteine ligase; made up of catalytic and modifying subunits) and GSS (glutathione synthetase). In humans, oxidative stress regulates GCL expression in an antioxidant response element-dependent manner via Nrf2 [NFE (nuclear factor erythroid)-related factor 2]. In the rat, GSS and GCL are regulated co-ordinately by oxidative stress, and induction of GSS further increases GSH synthetic capacity. Transcriptional regulation of the human GSS has not been examined. To address this, we have cloned and characterized a 2.2 kb 5′-flanking region of the human GSS. The transcriptional start site is located 80 nt upstream of the translation start site. The human GSS promoter efficiently drove luciferase expression in Chang cells. Overexpression of either Nrf1 or Nrf2 induced the GSS promoter activity by 130 and 168% respectively. Two regions homologous to the NFE2 motif are demonstrated to be important for basal expression of human GSS, as mutation of these sites reduced the promoter activity by 66%. Nrf1, Nrf2 and c-Jun binding to these NFE2 sites under basal conditions was demonstrated using chromatin immunoprecipitation assays. In summary, two NFE2 sites in the human GSS promoter play important roles in the basal expression of GSS and, similar to the GCL subunits, the human GSS gene expression is also regulated by Nrf2.
Keywords: Chang cells, cloning, 5′-flanking region, glutathione synthetase, NFE2, Nrf1
Abbreviations: AP-1, activating protein-1; ARE, antioxidant response element; C/EBP, CCAAT/enhancer binding protein site; ChIP, chromatin immunoprecipitation; CNC, cap-n-collar; EMSA, electrophoretic mobility-shift assay; FBS, fetal bovine serum; GCL, glutamate–cysteine ligase; GCLC, GCL catalytic; GCLM, GCL modifying; GSS, glutathione synthetase; HCC, hepatocellular carcinoma; MEM, minimal essential medium; NF-κB, nuclear factor κB; NFE2, nuclear factor erythoid 2; Nrf1, NFE-related factor 1; SP-1, stimulating protein-1; STAT, signal transducer and activator of transcription; TBH, t-butylhydroquinone
INTRODUCTION
GSH is formed from L-glutamate, L-cysteine and glycine in a two-step enzymatic reaction catalysed by GCL (glutamate–cysteine ligase) and GSS (glutathione synthetase) [1]. GSH is a major antioxidant in host defence, synthesized in large part by the liver, and is important for phase II detoxification and elimination. It is important for maintenance of cellular thiol/disulphide redox potential and has been associated with signal transduction, gene expression and apoptosis [1]. Increased levels of GSH are found in proliferating hepatocytes and HCC (hepatocellular carcinoma) [2] and has been associated with drug-resistant tumour cell lines and tumour cells from patients with tumours resistant to drug therapy [3,4]. The first step in GSH synthesis is widely accepted to be rate-limiting. As a consequence, regulation of the GCL enzymes [both GCLC (GCL catalytic) and GCLM (GCL modifying) subunits] has been extensively examined [5–9]. AREs (antioxidant response elements) in the human promoter region have been implicated in the regulation of both GCL subunits by agents such as β-NF (β-napthaflavone) and TBH (t-butylhydroquinone) [8–10]. Furthermore, the CNC-bZip family (where CNC stands for cap-n-collar) has been suggested to play a role in ARE responsiveness in GCL and other antioxidant gene expressions, mediated in part by increased binding of NFE2 (nuclear factor erythroid 2)-related nuclear factors (Nrf1 and Nrf2) [9,11–17].
There are lines of evidence that suggest a more prominent role for GSS in GSH regulation than previously recognized. In response to surgical trauma, decreased GSS activity with no significant change in GCL activity is related to a reduced GSH level in skeletal muscle [18]. Additionally, increased rat GSS and GCLC expression further enhanced GSH synthesis above that observed with increased GCLC expression alone [7]. Using the rat promoter, we recently demonstrated co-ordinated regulation of rat GCL and GSS by AP-1 (activating protein-1) in response to TBH [6]. Generally, in the rat, agents that induced both catalytic and modifying subunits of GCL also induced GSS expression. An exception to this was liver regeneration following two-thirds partial hepatectomy, which induced GSS and the catalytic, but not the modifying, subunit of GCL [7,19]. Much less is known about the regulation of the human GSS gene. We reported that GSS is transcriptionally induced in human HCC [2]. Recently, we demonstrated a decreased steady-state mRNA level for both GCLC and GSS in patients with acute alcoholic hepatitis, possibly contributing to the decreased hepatic GSH level observed in these patients [20]. Deficiency in GSS in humans results in a build-up of γ-glutamylcysteine, which is converted into 5-oxoproline. This has been associated with metabolic acidosis, haemolytic anaemia and central nervous system damage [21,22]. Thus a better understanding of transcriptional regulation of human GSS is important. Nrf1 and Nrf2 are demonstrated regulators of human and mouse GCLC and GCLM and mouse GSS [9,11,17], however whether they also regulate human GSS is unknown. To address this, we have cloned and characterized a 2.2 kb 5′-flanking region of the human GSS gene. This work presents basal activity of the human GSS promoter and investigates a potential role for Nrf1 and Nrf2 transcription factors in the regulation of human GSS. Furthermore, we identify two regions within the promoter that are important for basal activity.
MATERIALS AND METHODS
Cloning of the 5′-flanking region of the human GSS gene
To identify the promoter region of the human GSS gene, a primer (CH1, 5′-AGCTGCTGTTTATCCTGCAAGAG-3′) corresponding to +22 to +44 (numbered according to the translational start site) of the human GSS cDNA (GenBank® accession no. BC007927) was used for PCR amplification (Advantage 2 PCR Enzyme system; BD Biosciences Clontech, Palo Alto, CA, U.S.A.) of a human genomic library (Genome Walker, Clontech) prepared by DraI, EcoRV, PvuII or SspI restriction digests. This initial PCR was followed by a second round of PCR using the nested primer supplied with the Genome Walker kit and the genespecific nested primer (GCH2, 5′-CATCCCAACACTAGTTCGCCTTTC-3′) corresponding to −21 to +3 of the human GSS cDNA. A specific 729 bp fragment from the DraI library was isolated by gel electrophoresis and cloned into TOPO pCR2.1 (Invitrogen) and subsequently used to transform Top10F′ competent cells (Invitrogen). A positive clone was identified and sequenced. This sequence was blasted against human chromosome 20 to identify the promoter region of the human GSS gene. Oligonucleotide primers designed to correspond to −46/−65 bp (T7 primer, 5′-TCGCGCCGCTACCCAGGCTC-3′) of the promoter sequence and from the human genome sequence (5′-GACATCAGGAAACATTGTTCTG-3′), 2151 bp upstream from this site, were used to PCR-amplify a 2170 bp region of the GSS promoter (−2216/−46 bp). This fragment was isolated by gel electrophoresis, cloned into the pCR2.1 TA vector (Invitrogen) and sequenced using the ABI Prism dRhodamine Terminator Cycle Sequencer performed by the Microchemical Core Facility (Norris Comprehensive Cancer Center, Keck School of Medicine). This 2.2 kb 5′-flanking region of the human GSS was cloned into the KpnI/XhoI site of promoterless pGL3-enhancer vector creating the recombinant plasmid −2216/−46 hGSS-LUC (where LUC stands for luciferase).
Using 5′-RACE (5′-rapid amplification of cDNA ends) analysis to determine the transcriptional start site in Chang cells
Chang cells (human liver cell line) were obtained from the Cell Culture Core (USC Research Center for Liver Diseases) and grown according to instructions provided by A.T.C.C. (Manassas, VA, U.S.A.). The GeneRacer kit (Invitrogen) was used to determine the 5′-end/transcriptional start site of the human GSS mRNA. Total cellular RNA was isolated from Chang cells using the TRIzol® method. Subsequently, mRNA was isolated using an Oligotex column (Qiagen, Valencia, CA, U.S.A.). Using a genespecific primer (5′-CTCAGCAATACTCCCTCAGCCAG-3′), gene-race-ready cDNA was prepared with the GeneRacer kit. Finally, 5′-cDNA ends were amplified using the GeneRacer 5′-primer and the nested gene-specific primer CH1 (corresponding to +22 to +44 of the human GSS cDNA). The PCR amplification parameters were 2 min at 94 °C, 35 cycles of 30 s at 94 °C followed by 30 s at 63 °C and 30 s at 72 °C, and a final extension for 10 min at 72 °C. Resulting fragments were isolated on a 3% (w/v) agarose gel, cloned into the pCR2.1 vector (Invitrogen), used to transform Top10 competent cells (Invitrogen) and sequenced. The sequence resulting from CH1 priming was confirmed using a second primer, GCH2 (corresponding to −21 to +3 of the human GSS cDNA), to repeat the experiment.
Construction of 5′-deletion constructs
The 2.2 kb fragment in the sense orientation upstream of the luciferase coding sequence of the pGL3-enhancer vector (Promega, Madison, WI, U.S.A.) is the construct that contains the longest 5′-flanking sequence (−2216/−46) employed in the transfection assay. A −1686/−46 5′-deletion construct was constructed by PCR amplification of HepG2 genomic DNA with the gene-specific primers T4 (5′-CCACTGATTTACGATCACTAGGAG-3′) and T7, followed by PCR amplification with the nested primers T5 (5′-TCTGTTCGCTTCGCTCCATCTC-3′) and T7. The resulting fragment was inserted into the pCR2.1 cloning vector. This 1.7 kb 5′-region was subcloned into the KpnI/XhoI site of the promoterless pGL3 enhancer vector, creating the recombinant plasmid −1686/−46 hGSS-LUC. A 1073 bp 5′-deletion construct was created by restriction digestion of the −1686/−46 hGSSpCR2.1 cloning vector by XhoI and SacI, generating a 1073 bp fragment. This 1073 bp fragment was ligated into the pGL3-enhancer vector at the XhoI/SacI site. A −685/−46 5′-deletion construct was generated by PCR amplification of HepG2 genomic DNA with GCH2 and T4 primers, followed by nested PCR using T7 and T1 (5′-CTGCCCCAGATCCTGATGTAAAC-3′) primers. The resulting fragment was ligated into the pCR2.1 cloning vector, sequenced and subcloned into the pGL3-enhancer vector at the SacI/XhoI site. A −340/−46 5′-deletion construct was created by PCR amplification of HepG2 genomic DNA with GCH2 and T4 primers, followed by nested PCR using T8 (5′-CCATCTTCCACTAGTCGTCGTC-3′) and T7 primers. The resulting fragment was cloned into the pCR2.1 cloning vector, sequenced and subcloned into the pGL3-enhancer vector at the KpnI/XhoI site. For analysis of promoter activity in Chang cells overexpressing Nrf1 or Nrf2, the −2216/−46 promoter construct was subcloned from the pGL3-enhancer vector into the pGL3-basic vector at the KpnI/NcoI site. Correct orientation of each deletion construct was confirmed by restriction digest analysis.
Mutagenesis of proximal and distal NFE2 sites in the human GSS promoter
The proximal and distal NFE2 sites within the full-length human GSS promoter-sequencing construct were mutated using the GeneTailor site-directed mutagenesis system (Invitrogen), according to the manufacturer's instructions using primers designed to replace the TCAGC core sequence with GAGAC. Transformants were identified by sequence analysis. Finally, the 2.2 kb insert containing both mutations was subcloned into the KpnI/XhoI site of the promoter-less pGL3-enhancer vector. Correct orientation was confirmed by restriction digest analysis.
Analysis of promoter constructs in Chang cell culture
Relative basal transcriptional activity of the pGL3-enhancer/GSS promoter fragments was studied using Chang cells (2.9×105 cells·ml−1·well−1 in 12-well tissue culture plates) transiently transfected with GSS promoter-luciferase constructs for 20 h using the Superfect transfection reagent (Qiagen). Cells were washed with PBS and using SuperFect reagent were transfected with 50 ng·ml−1·well−1 SV-40 (simian virus 40) driven Renilla luciferase (control for transfection efficiency) and 1 μg·ml−1·well−1 empty pGL3 enhancer or molar equivalent of pGL3 enhancer containing human GSS 5′-deletion constructs. Cells were incubated with the transfection complexes for 2.5 h, washed with PBS, the medium was replaced with 10% (v/v) FBS (fetal bovine serum)/MEM (minimal essential medium) and incubated at 37 °C under normal growth conditions for 20 h. Cell lysates were harvested with passive lysis buffer (part #E194A; Promega). Luciferase activity was measured using the dual luciferase assay system (Promega) and relative luciferase activity was calculated as the ratio of sample luciferase activity to Renilla luciferase activity. Values are expressed as fold pGL3-control.
The modulation of GSS promoter activity by Nrf1 or Nrf2 transcription factors was determined by measuring the luciferase activity driven by the pGL3-basic −2216/−46 GSS promoter construct in the presence of 1.5 μg/ml empty expression vector (pEF1αCV) or molar equivalent of Nrf1 or Nrf2 expression vectors (gifts from Dr J. Chan, University of California, Irvine, CA, U.S.A.) in a 12-well tissue culture plate for 24 h. SV-40 Renilla plasmid was included at 2–37 ng/well as a control for transfection efficiency. Relative luciferase activity was calculated as above. The empty expression vector was prepared by excising the Nrf1 insert from the Nrf1 expression vector (pEF1Nrf1) by restriction digestion with XbaI/KpnI. Resulting overhangs were filled in by a Klenow reaction and the vector was re-ligated. Transient trasfection was performed as above.
The effect of mutation of the NFE2 sites within the GSS promoter was determined by measuring the luciferase activity driven by the pGL3-enhancer −2216/−46 GSS promoter construct with both sites containing a 4 bp mutation. Relative luciferase activity was calculated as above.
Northern-blot analysis
Total cellular RNA (20 μg) was run on a formaldehyde–agarose (1%) gel at 80 V. The RNA was subsequently transferred on to a Duralon-UV membrane (Stratagene, La Jolla, CA, U.S.A.) overnight at room temperature (25 °C) in 20×SSC (where 2×SSC is 0.15 M NaCl and 0.015 M sodium citrate). The RNA was cross-linked to the membrane by UV and the membrane was stored at 4 °C in 6×SSC until probed for GSS and β-actin. Probes (50 ng) were labelled with [32P]dCTP in a Klenow reaction with 10×decamers (Ambion, Austin, TX, U.S.A.) for 1 h at 37 °C and denatured at 90 °C for 5 min before hybridization with the RNA membrane. The membrane was prehybridized for 2 h at 42 °C with Ultrahyb buffer (ULTRAhyb ultrasensitive hybridization buffer #8670; Ambion) and subsequently hybridized with labelled probe overnight at 42 °C. The membrane was washed twice for 5 min with 2×SSC and 0.1% SDS, twice for 15 min with 0.1×SSC and 0.1% SDS and exposed to autoradiograph film. Values are determined by densitometry and normalized to β-actin and are expressed as percentage of control (transfected with control vector).
EMSA (electrophoretic mobility-shift assay)
Nuclear proteins isolated from Chang cells (5–15 μg) were incubated at room temperature for 10 min with binding buffer (Gel Shift Assay Core System #E3050; Promega) and 100× unlabelled probe (for competitor), followed by 20 min at room temperature with 200000 c.p.m. [γ-32P]ATP-labelled probe. Reactions were run on a 6% (w/v) non-denaturing acrylamide gel at 200 V in 0.5×TBE (where 1×TBE is 45 mM Tris/borate/1 mM EDTA) at room temperature and the gel was subsequently dried and exposed to autoradiograph film.
ChIP (chromatin immunoprecipitation) analysis
ChIP in Chang cells was performed using the ChIP assay kit (Upstate Biotechnology, Lake Placid, NY, U.S.A.). The manufacturer's instructions were followed except for the extension of the incubation period for collecting antibody histone complexes with salmon-sperm DNA/Protein A–agarose slurry to 3 h at 4 °C. Antibodies against Nrf1, Nrf2 and c-Jun (Santa Cruz Biotechnology, New York, NY, U.S.A.) were used for immunoprecipitation. PCR amplification was used to assay the immunoprecipitates using nested primers specific for the sites of interest. Primers for the distal NFE2 site included 5′-GACATCAGGAAACATTGTTCTG-3′ and 5′-GAGACGGAGTTTTCGTTGTTG-3′ and nested primers 5′-CTGGGAATAACCAGACACCTA-3′ and 5′-CAGGTTCAAGCAATTCTCCTG-3′, producing a 333 bp amplicon. Primers for the proximal NFE2 site included the T7 primer and 5′-GAATGCTGAAGTCAGGACTGTTTG-3′ and the nested primers 5′-TCGATGCGACCCAGTGCGCTT-3′ and 5′CTGGGTTCTAATACAGGCTCAG-3′, producing a 403 bp amplicon. These primers were chosen to flank the NFE2 sites and to yield amplicons within the size range recommended by the ChIP kit manufacturer (250–750 bp). Input starting material was used as a positive control and a no antibody immunoprecipitation was used as a negative control.
Statistical analysis
Results are expressed as means±S.E.M. Statistical analysis was performed using ANOVA followed by Fisher's test for multiple comparisons, or Student's t test for paired comparisons. For changes in mRNA levels, ratios of GSS to β-actin densitometric values were compared; P<0.05 was considered to be statistically significant.
RESULTS
Cloning and sequencing of the 5′-flanking region of the human GSS
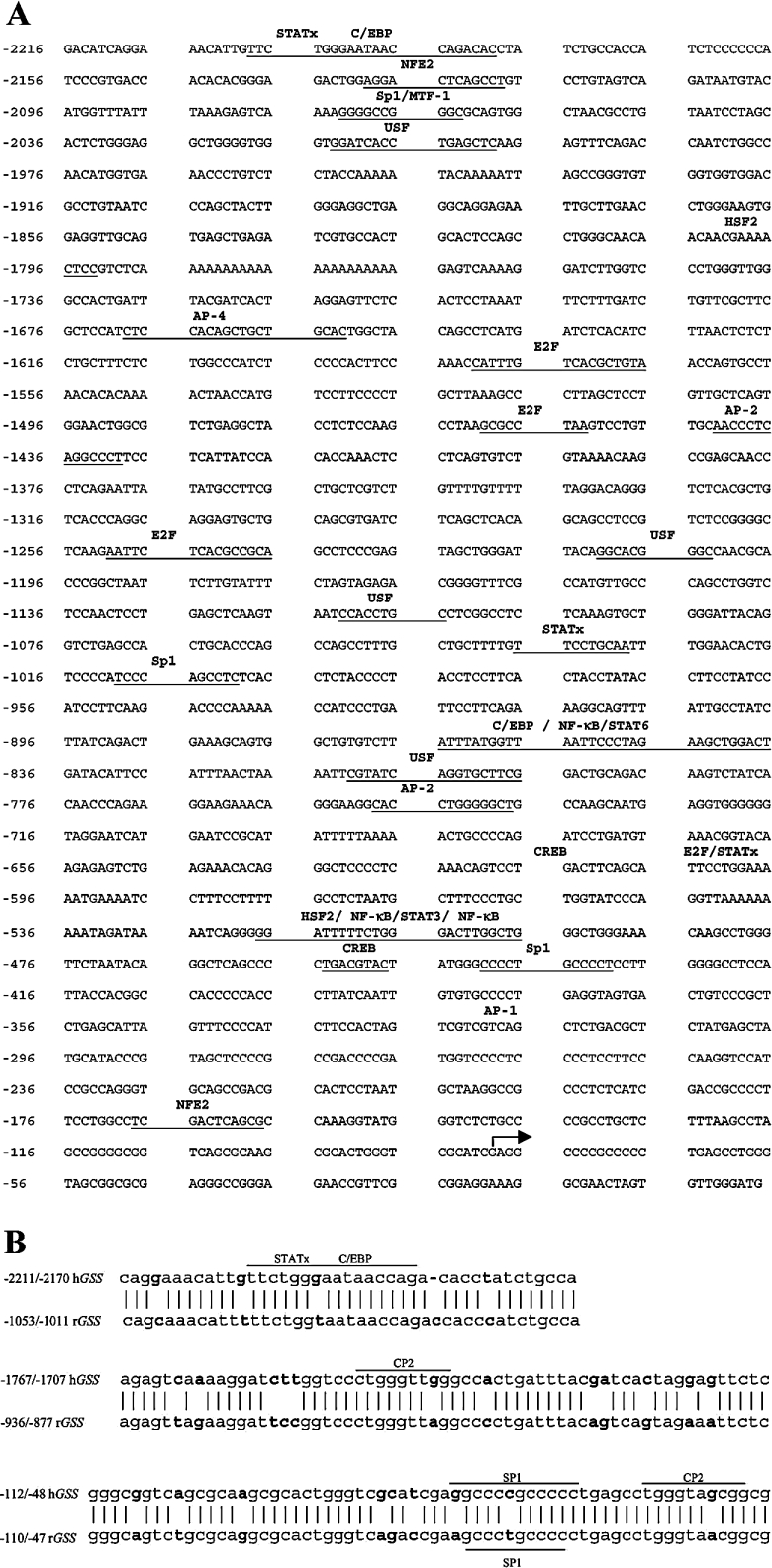
The sequence of the 2.2 kb 5′-flanking region of the human GSS (GenBank® accession no. AY184210) is shown in Figure 1(A). Using the MultAlin program for alignment of DNA sequences (Pôle Bio Informatique Lyonnais), we compared the human GSS promoter reported here with the rat GSS promoter (GenBank® accession no. AF333982). Analysis revealed that 40.85% of the sequences are 90% or more conserved, 51.18% are 50–90% conserved and 7.89% are less than 50% conserved. TFSearch search (http://www.cbrc.jp/research/db/TFSEARCH.html) and MatInspector databases (http://www.genomatix.de/cgi-bin/eldorado/main.pl) were used to analyse the promoter sequence for potential transcription factor-binding sites. These sites are indicated in Figure 1(A) and include several potential binding sites for STATs (signal transducers and activators of transcription), SP-1 (stimulating protein-1), NFE2, several NF-κB (nuclear factor κB) sites, AP-1, AP-2 and AP-4, several C/EBPs (CCAAT/enhancer binding protein sites), HSF-2 (heat shock factor-2) and CREB (cyclic AMP-responsive element binding protein 1) sites. Comparison of the rat and human GSS promoters using NCBI blast analysis (Figure 1B) indicated only three areas that shared a high degree of homology (>80%). These regions included −2211/−2170, −1767/−1707 and −112/−48 of human GSS corresponding to −1053/−1011, −936/−877 and −110/−47 of the rat GSS respectively. Putative transcription factor-binding sites present in these regions include STATs, SP-1, C/EBP and two CP2 sites.
Figure 1. 5′-Flanking region of human GSS.
(A) Sequence of the 5′-flanking region of human GSS. Two primers (see the Materials and methods section) were used for 5′-RACE to determine the transcriptional start site, indicated by the forward arrow. TFSearch and MatInspector databases identified potential transcription factor-binding sites, denoted in bold above the sequence and underlined. (B) Comparison of the human and rat GSS promoter using NCBI blast analysis indicated only three areas that shared a high degree of homology (>80%). These regions included −2211/−2170, −1767/−1707 and −112/−48 of human GSS corresponding to −1053/−1011, −936/−877 and −110/−47 of the rat GSS respectively. Putative transcription factor-binding sites present in these regions are shown.
Transcriptional start site
The transcriptional start site was determined by cloning products of 5′-RACE using cDNA from Chang cells, followed by sequencing. Two different primers corresponding to +22 to +44 and −21 to +3 of the human GSS cDNA were used and yielded products beginning from 80 nt upstream of the translation start site (Figure 1A, indicated by forward arrow). Using AceView (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/), we compared our experimentally determined transcriptional start site with all publicly available mRNAs and expressed sequence tags on the genome sequence. Of these, the start site determined in our study is identical to GSS variants a, d and h (as identified by AceView). Four other variants also showed homology with our sequence, however these sequences included an additional 3, 10 and 19 bp further 5′ of our start site.
Human GSS promoter activity in Chang cells
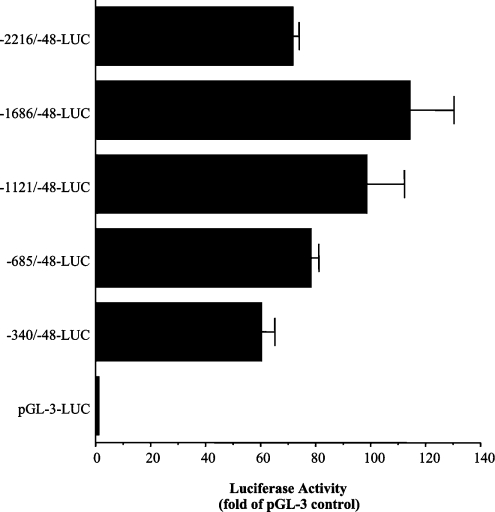
To assess basal GSS promoter activity, Chang cells were transiently transfected for 20 h with 5′-sequential deletion constructs of the human GSS promoter (Figure 2) cloned into the promoterless pGL3 enhancer luciferase reporter plasmid. The relative luciferase activity as compared with samples transfected with reporter vector alone was determined. The smallest construct (−340/−48-LUC) that contains a strongly GC-rich region, an AP-1 and NFE2 site efficiently drove luciferase activity. Increased activity was observed with each sequential construct up to the −1686/−48-LUC construct. A repressor region appears to be present between the −1686 and −2216 nt region of the promoter, as promoter activity decreased with the full-length promoter (−2216/−48-LUC) construct.
Figure 2. Human GSS promoter activity in Chang cells.
Relative luciferase activity (expressed as fold pGL3-control) in Chang cells 20 h post-transfection with 1 μg·ml−1·well−1 human GSS 5′-deletion promoter constructs in 10% FBS/MEM. Values represent an average of three experiments performed in quadruplicate.
Nrf1 or Nrf2 overexpression enhances expression of human GSS in Chang cells
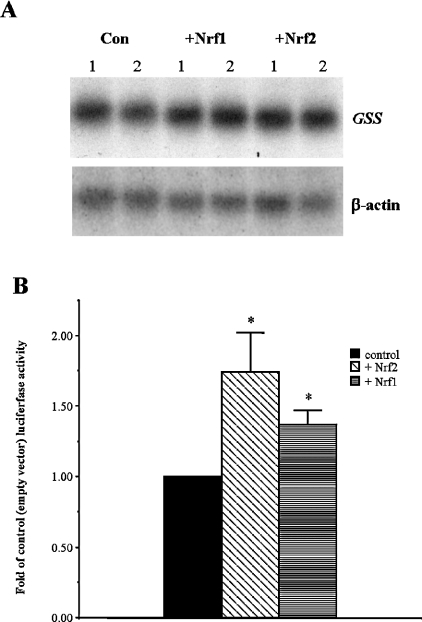
Nrf1 and Nrf2 have been demonstrated to be important regulators of GCL and mouse GSS [9,11]. To determine whether Nrf1 or Nrf2 plays a role in the regulation of the human GSS gene, we assessed the effect of overexpression of these transcription factors on human GSS (Figure 3). Chang cells were co-transfected with the −2216/−48 GSS pGL3 basic construct and an empty expression vector or an expression vector coding for either Nrf1 or Nrf2 transcription factor. We observed enhanced promoter activity when cells overexpressed either Nrf1 (130% of control) or Nrf2 (168% of control) (Figure 3B). We determined that the enhancement resulting from overexpression of these transcription factors was real and not simply an effect seen with the artificial luciferase assay, as we also observed enhanced steady-state mRNA levels of the endogenous GSS by 140% in Chang cells (Figure 3A).
Figure 3. Nrf1 or Nrf2 overexpression enhances expression of human GSS in Chang cells.
(A) Effect of Nrf1 or Nrf2 overexpression on human GSS steady-state mRNA levels in Chang cells. Chang cells were transfected for 24 h with Nrf1, Nrf2 or empty expression plasmid (pEFαCV) and 20 μg of total cellular RNA probed for GSS and β-actin mRNA by Northern-blot analysis. (B) Quantification of fold-increase of full-length GSS promoter activity following 24 h transfection of Chang cells with Nrf1, Nrf2 or pEFαCV expression plasmid. Relative luciferase was calculated and is expressed as fold-control. Values represent an average of five and seven experiments performed in triplicate for Nrf1 and Nrf2 overexpression respectively.
Importance of two NFE2 sites for basal human GSS promoter activity
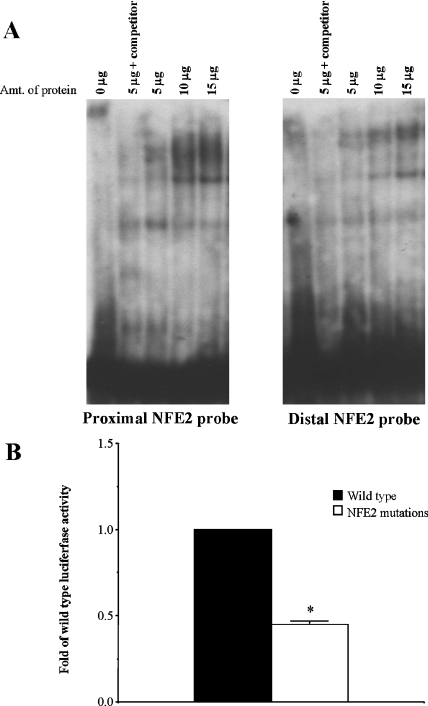
There are two NFE2 sites in the human GSS promoter that can potentially bind Nrf1 and Nrf2. From EMSA analysis, we observed binding of Chang nuclear proteins to probes for both the proximal and distal NFE2 sites present in the promoter (Figure 4A). Mutations to the proximal and distal NFE2 sites of the promoter construct from 5′-GACTCAGC-3′ to 5′-GACGAGAC-3′ resulted in decreased promoter activity (44% of control) following transfection of Chang cells (Figure 4B), providing further confirmation that these sites are important for the regulation of human GSS.
Figure 4. A role for NFE2 sites in basal human GSS promoter activity.
(A) EMSA demonstrating binding of Chang nuclear proteins (5, 10 and 15 μg) to both the proximal and distal NFE2 sites. Specificity of binding is determined by competition of binding with 100-fold excess unlabelled probe. (B) Mutagenesis of proximal and distal NFE2 sites within the full-length promoter decreases basal promoter activity by 66%. Relative luciferase activity was measured following 24 h transfection with wild-type full-length GSS and full-length GSS promoter with NFE2 site mutations. Values are expressed as fold-wild-type luciferase activity and represent three experiments performed in triplicate. *P<0.05 versus wild-type construct.
Transcription factor binding to the two NFE2 sites
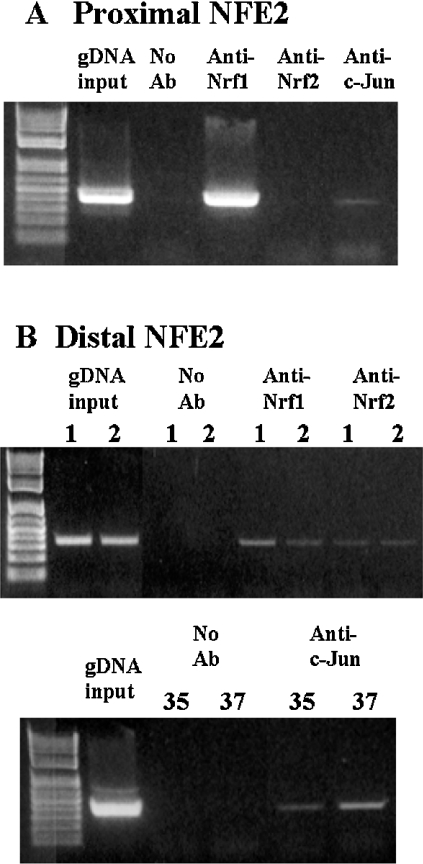
To see if Nrf1 and Nrf2 bind to the GSS promoter in the native chromosomal DNA, we examined binding of Nrf1 and Nrf2 to these two NFE2 sites of the human GSS promoter using ChIP analysis. Since Nrf1 and Nrf2 have been demonstrated to interact with Jun proteins, we also investigated possible binding of c-Jun to these sites. Figure 5 shows that Nrf1 and c-Jun bind to both NFE2 sites while Nrf2 binds only to the distal NFE2 site.
Figure 5. ChIP analysis of the NFE2 sites of the human GSS promoter.
(A) Proximal NFE2 site. PCR products from amplification of the NFE2 site following immunoprecipitation with antisera against Nrf1, Nrf2 or c-Jun demonstrate Nrf1 and c-Jun binding but not Nrf2 binding to the proximal NFE2 site. Input genomic DNA (gDNA input) was used as a positive control and a no antibody immunoprecipitation (no Ab) was used as a negative control. A representative Figure from two experiments is shown. (B) Distal NFE2 site. PCR products from amplification of the NFE2 site following immunoprecipitation with antisera against Nrf1, Nrf2 or c-Jun demonstrate that Nrf1, Nrf2 and c-Jun all bind to the distal NFE2 site. Numbers above the Figure for Nrf1 and Nrf2 represent two different experiments, numbers above the Figure for c-Jun (bottom panel) represent number of cycles. These are representative figures for three experiments.
DISCUSSION
In the rat, GCL and GSS expression are regulated in a similar fashion [6,7]. Generally, agents that induced both subunits of GCL also induced GSS expression, with the exception of partial hepatectomy that specifically induced GSS and the catalytic, but not the modifying, subunit of GCL [7,19]. Differences in the regulation of the rat and human GCL promoters have been documented. Whereas ARE and AP-1 sites are implicated in the expression of human GCL (both subunits), induction of the rat GCLC and GSS occurs independently of AREs and no ARE has been identified within the cloned fragment of these promoters. AP-1 is required for the basal and the TBH-mediated induction in expression of rat GCL subunits and GSS [6]. In contrast, TBH induces the human GCL in an ARE-dependent manner [10]. Regulation of the human GSS is essentially unknown except for our previous work showing increased expression following thioacetamide treatment and in HCC [2,7]. Based on the fact that agents that up-regulate both rat GCL subunits also up-regulate rat GSS, we speculate that human GSS and GCL are also regulated in a similar manner. To address this, we have cloned and characterized a 2.2 kb 5′-flanking region of the human GSS. Since Nrf1 and Nrf2 have been shown to be the key transcription factors for the regulation of the human GCL subunits, we also examined the role of these transcription factors in the regulation of the human GSS.
The transcriptional start site is located 80 nt upstream of the translation start site. Analysis of the 5′-flanking human GSS sequence revealed that it does not share a great degree of homology to the rat promoter overall. There are three regions with >80% homology between the human and rat GSS promoter, which contain putative transcription factor-binding sites for STATs, C/EBP and CP-2. C/EBP is a liver-enriched transcription factor demonstrated to increase following partial hepatectomy [23]. STATs regulate expression of genes controlling proliferation, survival, differentiation, development, and tumourigenesis [24]. CP2 is a ubiquitously expressed transcription factor involved in globin gene transcription [25]. Several other potential binding sites within the full-length human GSS promoter were identified by the MatInspector and TFSearch databases including SP-1, NF-κB, AP-1, AP-2, AP-4, HSF-2 and CREB. NF-κB and AP-1 are redox-sensitive transcription factors that regulate expression of antioxidant enzymes [26].
We demonstrate that the human GSS promoter efficiently drove luciferase expression in transiently transfected Chang cells. Maximal promoter activity was achieved with the −1686/−48 GSS construct. The 5′-flanking region of the human GSS contains two sites with a high degree of homology to the NFE2p45 binding element [27]. NFE2p45 is the first identified protein in the CNC-basic leucine zipper family and is specifically expressed in erythroid cells [12,28,29]. Further CNC-basic leucine zipper family members were identified by a yeast expression system that encodes proteins that bind to the tandem NFE2/AP-1-like cis-elements [also referred to as MARE (maf recognition element)] in the β-globin locus control region [30,31]. Two NFE2 related factors (Nrf1 and Nrf2) also bind to the NFE2/AP-1 site motif but are more ubiquitously expressed [11,12,30]. The two NFE2p45 sites in the human GSS promoter share homology with MARE [32], ARE [10] and TRE (phorbol-12-o-tetradecanoate-13-acetate response element) [27]. Of the two sites, one is located proximally to the transcription start site (within 100 bp of the start site), and the other is found at the distal end of the promoter. These sites caught our attention because of the homology to ARE. Nrf1 and Nrf2 have been suggested to be important in the regulation of components of the phase II detoxification mechanism and GSH homoeostasis through the ARE/EpRE (electrophile response element) sites [11,13,33]. Mice deficient in the Nrf1 and Nrf2 transcription factors exhibit decreased GSH, GCL and GSS levels consistent with predisposition of these animals to liver injury and oxidative stress [11,17,34]. Nrf1 and Nrf2 have been demonstrated to act by binding as heterodimers with small maf proteins or Jun (c-Jun, JunB and JunD) to effect ARE-mediated expression of the GCL, human NAD(P)H quinone oxidoreductase and glutathione S-transferase genes [9,11,12,15,16]. However, the mechanism of action and complete list of binding partners for these transcription factors is still not completely described. We next examined whether Nrf1 and Nrf2 regulate the human GSS expression. Overexpression of Nrf1 or Nrf2 in Chang cells resulted in enhanced expression of endogenous GSS steady-state mRNA levels and increased promoter activity in Chang cells to 130 and 168% of controls respectively. These modest levels of induction could be due to a constitutively high level of basal GSS expression in Chang cells. As Nrf1 overexpression reported in the literature induces GCLM approx. 2-fold above control [11], the small induction of GSS we observe with overexpression of Nrf1 and Nrf2 is not surprising.
To determine whether these two NFE2 sites were important for basal expression, we investigated whether they were able to bind to nuclear proteins from untreated Chang cells. By EMSA we observed binding to both the proximal and distal NFE2 sites. The nuclear proteins from the untreated Chang cells bound to the proximal NFE2 site, producing a smear in addition to a clear band, suggesting that a number of protein–DNA complexes may have formed. Proteins bound to the distal site also produced several bands, indicating that more than one protein or complex formed as well. Using ChIP analysis, we demonstrated that Nrf1 and c-Jun bind to both NFE2 sites of the human GSS promoter, whereas Nrf2 binds to only the distal NFE2 site. To further establish the importance of these sites, mutagenesis of the distal and proximal NFE2 binding sites resulted in decreased promoter activity, 44% when compared with controls. The results suggest that although the NFE2 sites are not the only elements involved in basal expression (as activity was not completely abolished), they are important for basal expression of human GSS.
In summary, we characterized for the first time the 5′-flanking region of the human GSS gene. We found modest induction of gene expression or promoter activity by Nrf1 or Nrf2. Finally, we identified NFE2 sites within the promoter to be important for basal promoter activity and identified Nrf1, Nrf2 and c-Jun proteins binding to these NFE2 sites within the proximal and distal promoter.
Acknowledgments
This work was supported by National Institutes of Health (Bethesda, MD, U.S.A.) grant DK-45334. Chang cells were provided by the Cell Culture Core of the USC Research Center for Liver Diseases (P30 DK48522).
References
- 1.Sies H. Glutathione and its role in cellular functions. Free Radicals Biol. Med. 1999;27:916–921. doi: 10.1016/s0891-5849(99)00177-x. [DOI] [PubMed] [Google Scholar]
- 2.Huang Z. Z., Chen C., Zeng Z., Yang H., Oh J., Chen L., Lu S. C. Mechanism and significance of increased glutathione level in human hepatocellular carcinoma and liver regeneration. FASEB J. 2001;15:19–21. doi: 10.1096/fj.00-0445fje. [DOI] [PubMed] [Google Scholar]
- 3.Lee F. Y., Siemann D. W., Sutherland R. M. Changes in cellular glutathione content during adriamycin treatment in human ovarian cancer – a possible indicator of chemosensitivity. Br. J. Cancer. 1989;60:291–298. doi: 10.1038/bjc.1989.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masters J. R., Thomas R., Hall A. G., Hogarth K., Matheson E. C., Cattan A. R., Lohrer H. Sensitivity of testis tumour cells to chemotherapeutic drugs: role of detoxifying pathways. Eur. J. Cancer. 1996;32A:1248–1253. doi: 10.1016/0959-8049(96)00033-0. [DOI] [PubMed] [Google Scholar]
- 5.Yang Y., Dieter M. Z., Chen Y., Shertzer H. G., Nebert D. W., Dalton T. P. Initial characterization of the glutamate-cysteine ligase modifier subunit Gclm(−/−) knockout mouse. Novel model system for a severely compromised oxidative stress response. J. Biol. Chem. 2002;277:49446–49452. doi: 10.1074/jbc.M209372200. [DOI] [PubMed] [Google Scholar]
- 6.Yang H., Zeng Y., Lee T. D., Yang Y., Ou X., Chen L., Haque M., Rippe R., Lu S. C. Role of AP-1 in the coordinate induction of rat glutamate-cysteine ligase and glutathione synthetase by tert-butylhydroquinone. J. Biol. Chem. 2002;277:35232–35239. doi: 10.1074/jbc.M203812200. [DOI] [PubMed] [Google Scholar]
- 7.Huang Z. Z., Yang H., Chen C., Zeng Z., Lu S. C. Inducers of γ-glutamylcysteine synthetase and their effects on glutathione synthetase expression. Biochim. Biophys. Acta. 2000;1493:48–55. doi: 10.1016/s0167-4781(00)00156-1. [DOI] [PubMed] [Google Scholar]
- 8.Mulcahy R. T., Wartman M. A., Bailey H. H., Gipp J. J. Constitutive and β-naphthoflavone-induced expression of the human γ-glutamylcysteine synthetase heavy subunit gene is regulated by a distal antioxidant response element/TRE sequence. J. Biol. Chem. 1997;272:7445–7453. doi: 10.1074/jbc.272.11.7445. [DOI] [PubMed] [Google Scholar]
- 9.Wild A. C., Moinova H. R., Mulcahy R. T. Regulation of γ-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J. Biol. Chem. 1999;274:33627–33636. doi: 10.1074/jbc.274.47.33627. [DOI] [PubMed] [Google Scholar]
- 10.Erickson A. M., Nevarea Z., Gipp J. J., Mulcahy R. T. Identification of a variant antioxidant response element in the promoter of the human glutamate-cysteine ligase modifier subunit gene. J. Biol. Chem. 2002;277:30730–30737. doi: 10.1074/jbc.M205225200. [DOI] [PubMed] [Google Scholar]
- 11.Kwong M., Kan Y. W., Chan J. Y. The CNC basic leucine zipper factor, Nrf1, is essential for cell survival in response to oxidative stress-inducing agents. Role for Nrf1 in γ-gcsL and gss expression in mouse fibroblasts. J. Biol. Chem. 1999;274:37491–37498. doi: 10.1074/jbc.274.52.37491. [DOI] [PubMed] [Google Scholar]
- 12.Jaiswal A. K. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radicals Biol. Med. 2004;36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 13.Kwak M.-K., Itoh K., Yamamoto M., Kensler T. W. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol. Cell. Biol. 2002;22:2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nioi P., McMahon M., Itoh K., Yamamoto M., Hayes J. D. Identification of a novel Nrf2-mediated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeyapaul J., Jaiswal A. K. Nrf2 and c-Jun regulation of antioxidant response element (ARE)-mediated expression and induction of γ-glutatmylcysteine synthetase heavy subunit gene. Biochem. Pharmacol. 2000;59:1433–1439. doi: 10.1016/s0006-2952(00)00256-2. [DOI] [PubMed] [Google Scholar]
- 16.Venugopal R., Jaiswal A. K. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene. 1998;17:3145–3156. doi: 10.1038/sj.onc.1202237. [DOI] [PubMed] [Google Scholar]
- 17.Chan J. Y., Kwong M. Impaired expression of glutathione synthetic enzyme genes in mice with targeted deletion of the Nrf2 basic-leucine zipper protein. Biochim. Biophys. Acta. 2000;1517:19–26. doi: 10.1016/s0167-4781(00)00238-4. [DOI] [PubMed] [Google Scholar]
- 18.Luo J. L., Hammarqvist F., Andersson K., Wernerman J. Surgical trauma decreases glutathione synthetic capacity in human skeletal muscle tissue. Am. J. Physiol. 1998;275:E359–E365. doi: 10.1152/ajpendo.1998.275.2.E359. [DOI] [PubMed] [Google Scholar]
- 19.Huang Z. Z., Li H., Cai J., Kuhlenkamp J., Kaplowitz N., Lu S. C. Changes in glutathione homeostasis during liver regeneration in the rat. Hepatology. 1998;27:147–153. doi: 10.1002/hep.510270123. [DOI] [PubMed] [Google Scholar]
- 20.Lee T. D., Sadda M. R., Mendler M. H., Bottiglieri T., Kanel G., Mato J. M., Lu S. C. Abnormal hepatic methionine and glutathione metabolism in patients with alcoholic hepatitis. Alcohol. Clin. Exp. Res. 2004;28:173–181. doi: 10.1097/01.ALC.0000108654.77178.03. [DOI] [PubMed] [Google Scholar]
- 21.Huang C. S., He W., Meister A., Anderson M. E. Amino acid sequence of rat kidney glutathione synthetase. Proc. Natl. Acad. Sci. U.S.A. 1995;92:1232–1236. doi: 10.1073/pnas.92.4.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Z. Z., Habib G. M., Rhead W. J., Gahl W. A., He X., Sazer S., Lieberman M. W. Mutations in the glutathione synthetase gene cause 5-oxoprolinuria. Nat. Genet. 1996;14:361–365. doi: 10.1038/ng1196-361. [DOI] [PubMed] [Google Scholar]
- 23.Diehl A. M., Yang S. Q. Regenerative changes in C/EBPα and C/EBPβ expression modulate binding to the C/EBP site in the c-fos promoter. Hepatology. 1994;19:447–456. [PubMed] [Google Scholar]
- 24.Sánchez A., Nagy P., Thorgeirsson S. S. STAT-3 activity in chemically-induced hepatocellular carcinoma. Eur. J. Cancer. 2003;39:2093–2098. doi: 10.1016/s0959-8049(03)00393-9. [DOI] [PubMed] [Google Scholar]
- 25.Zhou W., Zhao Q., Sutton R., Cumming H., Wang X., Cerruti L., Hall M., Wu R., Cunningham J. M., Jane S. M. The role of p22 NF-E4 in human globin gene switching. J. Biol. Chem. 2004;18:26227–26232. doi: 10.1074/jbc.M402191200. [DOI] [PubMed] [Google Scholar]
- 26.Catani M. V., Savini I., Duranti G., Caporossi D., Ceci R., Sabatini S., Avigliano L. Nuclear factor κB and activating protein 1 are involved in differentiation-related resistance to oxidative stress in skeletal muscle cells. Free Radicals Biol. Med. 2004;37:1024–1036. doi: 10.1016/j.freeradbiomed.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 27.Itoh K., Igarashi K., Hayashi N., Nishizawa M., Yamamoto M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol. Cell. Biol. 1995;15:4184–4193. doi: 10.1128/mcb.15.8.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrews N. C., Erdjument-Bromage H., Davidson M. B., Tempst P., Orkin S. H. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature (London) 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- 29.Mignotte V., Wall L., de Boer E., Grosveld F., Romeo P. H. Two tissue-specific factors bind the erythroid promoter of the human porphobilinogen deaminase gene. Nucleic Acids Res. 1989;17:37–54. doi: 10.1093/nar/17.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan J. Y., Han X.-L., Kan Y. W. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc. Natl. Acad. Sci. U.S.A. 1993;90:11371–11375. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moi P., Chan K., Asunis I., Cao A., Kan Y. W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the β-globin locus control region. Proc. Natl. Acad. Sci. U.S.A. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeda H., Nishi S., Sakai M. Transcription factor Nrf2/MafK regulates rat glutathione S-transferase P gene during hepatocarcinogenesis. Biochem. J. 2004;380:515–521. doi: 10.1042/BJ20031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morito N., Yoh K., Itoh K., Hirayama A., Koyama A., Yamamoto M., Takahashi S. Nrf2 regulates the sensitivity of death receptor signals by affecting intracellular glutathione levels. Oncogene. 2003;22:9275–9281. doi: 10.1038/sj.onc.1207024. [DOI] [PubMed] [Google Scholar]
- 34.Chan K., Han X.-D., Kan Y. W. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4611–4616. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]