Abstract
PrPC [normal cellular PrP (prion-related protein)] is a glycosylphosphatidylinositol-linked cell-surface glycoprotein that is expressed primarily by cells of the central and peripheral nervous system and the lymphoreticular system. During prion disease, PrPC undergoes structural modification to PrPSc (abnormal disease-specific conformation of PrP). The appearance of prion infectivity and PrPSc within different peripheral lymphoid tissue sites during natural scrapie infection in sheep is suggestive of haematogenic dissemination. For this to occur, blood cells may harbour or carry disease-associated PrP and in doing so present altered conformations of PrP on their cell-surface. In the present study, we show that changes in PrP epitope expression, or accessibility, can be detected on peripheral blood mononuclear cells during the course of experimental scrapie in susceptible sheep. Peripheral blood mononuclear cells isolated from VRQ homozygous lambs inoculated orally with scrapie were probed with either N- or C-terminal-specific anti-PrP monoclonal antibodies and analysed by flow cytometry. During the progression of scrapie, significant alterations were seen in the exposure of particular cell-surface PrP epitopes. These modifications included increased accessibility to N-terminal regions of the PrP molecule, to the region between β-strand-2 and residue 171, and to the C-terminal region of helix-3. Increased accessibility in the globular C-terminal domain of PrP occurred in the vicinity of tyrosine dimers, which are believed to have increased solvent exposure in disease-associated PrP. We suggest that the alterations in anti-PrP monoclonal antibody recognition of cell-surface PrP on blood cells from scrapie-infected sheep are indicative of structural changes within this molecule that may be relevant to prion disease.
Keywords: blood cell, cell-surface, prion disease, prion-related protein (PrP), secondary structure, transmissible spongiform encephalopathies
Abbreviations: FDC, follicular dendritic cell; MVV, maedi-visna virus; PBMC, peripheral blood mononuclear cell; p.i., post-inoculation; PK, proteinase K; PrP, prion-related protein; PrPC, normal cellular PrP; PrPSc, abnormal disease-specific conformation of PrP; Prp nomenclature, amino acid residue numbers refer to the ovine PrP sequence
INTRODUCTION
Prion diseases, such as scrapie in sheep, BSE (bovine spongiform encephalopathy) in cattle and CJD (Creutzfeldt–Jakob disease) in humans, are transmissible chronic neurodegenerative disorders characterized by the accumulation of PrPSc [the abnormal disease-specific conformation of PrP (prion-related protein)], an abnormal isomer of the host protein PrPC (normal cellular PrP). The protein-only hypothesis postulates that the transmissible prion agent consists solely of proteinaceous material [1]. Consequently, it is proposed that PrPSc forms part, or all, of the infectious prion agent and is responsible for the modification of PrPC. During the preclinical phase of prion disease induced by peripheral inoculation, prion infectivity and PrPSc can be detected within peripheral lymphoid tissue. At these sites, PrPSc depositions may be found in TBMs (tingible body macrophages) and FDCs (follicular dendritic cells), although replication of the infectious prion agent appears to be sustained by FDCs [2,3]. The temporal appearance of PrPSc in the lymphoreticular system, the PNS (peripheral nervous system) and eventually the CNS (central nervous system) of prion-infected individuals is indicative of spread by the nervous system [4,5]. This is supported by extensive studies in transgenic and gene-knockout mice, which demonstrate that neuroinvasion occurs principally via the PNS [6–9].
In natural scrapie of sheep, the main portal of entry of the infectious agent is believed to be the oral route [10]. The early appearance of infectivity [11,12] and PrPSc [4,13] in aggregated intestinal lymphoid tissue after oral exposure to TSE (transmissible spongiform encephalopathy)-infected material has identified gut-associated lymphoid tissue as the most probable site of intestinal uptake of prions. The investigation of naturally and orally exposed lambs to scrapie has indicated that the earliest site of peripheral lymphoid tissue uptake was to the dome regions of ileal Peyer's patches [14]. However, the rapid accumulation of PrPSc in lymphoid nodules of other gut and non-gut lymphoid tissues implied lymphatic and haematogenous dissemination of the scrapie agent within the host [15]. Haematogenous dissemination of infectivity between different lymphoreticular sites is an accepted feature of the preclinical phase of prion disease in experimental rodent scrapie following non-neural inoculation [16]. Consequently, peripheral blood is considered to be a possible reservoir of prion infectivity in scrapie-infected sheep and other TSE-affected individuals. It has been shown that prion disease can be transmitted through transfusion of whole blood, or buffy coat, from natural scrapie-infected, or BSE-experimentally infected sheep, into recipient sheep [17,18]. Similarly, prion infectivity has been detected in whole blood and in buffy coat from mice infected with a human-derived strain of variant CJD during both the preclinical and clinical phases of the disease [19]. The presence of detectable infectivity in blood of experimentally infected animals has reinforced concerns that human blood supplies may be contaminated with prion infectivity [20].
Circulating PBMCs (peripheral blood mononuclear cells) of sheep [21,22] and other species [23] express PrPC on their cell surface, which may serve as a substrate for conversion to a diseaseassociated form of PrP. In such a scheme, sheep PBMCs may harbour or carry disease-associated PrP and contribute to the deposition of PrPSc on resident lymphoid tissue cells such as FDCs. Polymorphisms in the ovine PrP protein at amino acid residues 136, 154 and 171 are associated with variation in susceptibility to natural scrapie. VRQ (Val136Arg154Gln171) or ARQ (Ala136-Arg154Gln171) animals show susceptibility to scrapie, whilst those that express ARR (Ala136Arg154Arg171) show resistance [24,25]. We have recently shown that PrPC expressed on the surface of ovine PBMCs displays conformational variation between scrapie-susceptible and -resistant genotypes, and between different susceptible allelic variants [26]. This indicates that ovine PrPC expressed on the surface of blood cells is capable of structural flexibility and has provided the basis for an investigation into the presence of disease-associated PrP on the surface of blood cells during prion disease. These disease-associated forms of PrP may be distinct from PK (proteinase K)-resistant PrPSc, which has not been readily detected on the surface of ovine blood cells [21,27], and may serve as surrogate markers of ongoing prion disease.
In the present study, we have used anti-PrP monoclonal antibodies and flow cytometry to probe the conformation of ovine PBMCs' cell-surface PrP during the progression of experimental scrapie infection in orally inoculated VRQ homozygous lambs. During the course of scrapie infection, significant changes were seen in the reactivity of PBMCs with both N- and C-terminal-specific anti-PrP monoclonal antibodies. There was increased accessibility to N-terminal regions of the prion protein and decreased accessibility to central portions of the molecule, including the region around residue 171. During the progression of scrapie, significant alterations were seen in the exposure of particular cell-surface PrP epitopes. These changes, which were indicative of modifications of cell-surface PrP epitope exposure, included increased accessibility to N-terminal regions of the PrP molecule, to the region between β-strand-2 and residue 171, and to the C-terminal region of helix-3. Increased accessibility in the globular C-terminal domain of PrP occurred in the vicinity of tyrosine dimers, which are believed to have increased solvent exposure in disease-associated PrP. Information on the presence of altered forms of PrP on the surface of blood cells from scrapie-infected sheep will be relevant to the understanding and diagnosis of prion diseases.
MATERIALS AND METHODS
Purification of recombinant and truncated PrP peptides
Full-length ovine PrP–VRQ (residues 25–232) or truncated ovine PrP-ARR or ovine PrP–VRQ (residues 89–233) was generated as described previously [26]. Recombinant PrP proteins were verified by MS to confirm the correct protein sequence and the presence of a disulphide bond.
Generation of monoclonal antibodies
Anti-PrP monoclonal antibodies were generated as described previously [26]. The N-terminal-specific anti-PrP monoclonal antibodies T164, T188 and T325 were produced from Prnpo/o (Prnp-knockout) mice immunized with murine brain homogenate and the C-terminal-specific anti-PrP monoclonal antibodies A516, V468 and V26 were produced from Prnpo/o mice immunized with ovine recombinant PrP [26].
Epitope mapping of monoclonal antibodies
The epitope specificity of the anti-PrP monoclonal antibodies was determined by the PEPSCAN method described previously [28]. Briefly, synthetic 15-mer peptides, overlapping by 14 amino acids, covering residues 25–232 of ovine PrP (190 peptides in total, including allelic variants) were covalently linked to polyethylene mini-PEPSCAN cards and screened by PEPSCAN-based ELISA. The PrP peptide-coated mini-PEPSCAN cards were reacted for 16 h at 4 °C with individual anti-PrP monoclonal antibodies routinely at 10 μg/ml in blocking solution that contained 5% (v/v) horse serum and 5% (w/v) ovalbumin. After washing, anti-PrP monoclonal antibodies were detected by rabbit-anti-mouse peroxidase using 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulphonic acid) and H2O2 as substrate. Colour development of the ELISA was quantified with a charge-coupled-device camera and processing system using the Optimas version 6.5 image-processing software package. Further epitope mapping of the N-terminal-specific monoclonal antibodies was carried out with a series of overlapping 21-mer peptides (comprising residues 25–46, 36–57, 47–68, 58–79, 69–90 and 80–101) of ovine PrP.
Western-blot analysis of full-length ovine recombinant PrP–VRQ
For Western-blot analysis of full-length ovine recombinant PrP–VRQ, the protein was subjected to SDS/PAGE under reducing conditions (200 ng of protein/track) and subsequently transferred on to nitrocellulose membranes by semi-dry blotting. Membranes were blocked with TBS-T (10 mM Tris/HCl, pH 7.8, 100 mM NaCl and 0.05% Tween 20) containing 5% (w/v) non-fat milk and then incubated with neat cell-culture supernatants at 20 °C for 2 h. This was followed by incubation with goat anti-mouse IgG-horseradish peroxidase (Sigma, catalogue no. A-2304) at 1:2000 dilution in 1% (w/v) non-fat milk in TBS-T at 20 °C for 2 h. PrP bands were detected by ECL® (enhanced chemiluminescence; Amersham Biosciences).
Direct immunoassay
Full-length ovine recombinant PrP–VRQ was coated at 200, 40, 8, 1.6 and 0 ng/well in 96-well flat-bottomed ELISA plates overnight at 4 °C. All dilutions were tested in triplicate. Excess protein was removed and wells were blocked with block buffer consisting of 2% (w/v) BSA and 0.05% sodium azide in PBS for 1.5 h at 20 °C with shaking. Plates were washed four times with wash buffer (PerkinElmer, Warrrington, Cheshire, U.K., catalogue no. 1244-114; concentrate diluted 25-fold in distilled water) and the relevant biotinylated anti-PrP monoclonal antibody was added at 50 ng/well diluted in assay buffer (PerkinElmer, catalogue no. 1244-111) and incubated for 1 h at 20 °C with shaking. Plates were washed four times with wash buffer followed by incubation with Europium-labelled streptavidin (PerkinElmer, catalogue no. 1244-360) at 50 ng/well for 1 h at 20 °C with shaking. Plates were washed eight times with wash buffer followed by the addition of enhancement solution (PerkinElmer, catalogue no. 1244-105). Plates were incubated for 5 min at 20 °C with shaking and the fluorescence was measured in a time-resolved fluorimeter (PerkinElmer, Victor™). Results were analysed and the means minus the control readings were plotted to give recombinant PrP–VRQ in ng/well against fluorescent counts/s.
MovS6 VRQ cell culture for immunofluorescence staining
MovS6 cells [29] were grown in Dulbecco's modified Eagle's medium (Sigma, catalogue no. D-5796) supplemented with Ham's F-12 with L-glutamine (Cambrex Bio Science, Verviers, Belgium, catalogue no. BE12-615F) containing 200 units/ml of penicillin, 100 μg/ml of streptomycin and 8% (v/v) heat-inactivated FCS (foetal calf serum). Cells were removed from the flasks using trypsin-free versene just before reaching confluence and were washed three times in FACS buffer (PBS containing 1% heat-inactivated FCS supplemented with 0.1% sodium azide) before immunofluorescence staining. Cell-surface phenotype was assessed using aliquots of 1×106 cells incubated with monoclonal antibody cell-culture supernatants or normal mouse serum at 1:1000 as control, for 20 min at 4 °C followed by three washes in FACS buffer and incubation with goat anti-mouse IgG-biotin (Sigma, catalogue no. B-7264) at 1:1000 for 20 min at 4 °C. Cells were washed three times with FACS buffer and subsequently incubated with 0.25 μg of streptavidin-phycoerythrin (Pharmingen, BD UK, London, U.K., catalogue no. 554061) for 20 min at 4 °C. Finally, cells were washed three times with FACS buffer and analysed for cell-surface fluorescence using a FACSCalibur® (Becton Dickinson, Mountain View, CA, U.S.A.). For each sample 10000 cells were analysed with dead cells excluded on the basis of forward and side light scatter.
Sheep inoculations
Scrapie-free VRQ homozygous Cheviot lambs were derived by embryo transfer from ewes of U.K. origin. Lambs were born and inoculated with scrapie within a purpose built experimental facility isolated from other livestock. Six lambs, at 1–2 weeks of age, were dosed orally with 1 g of undiluted scrapie-infected sheep brain homogenate placed on the back of the tongue. The homogenate was prepared from brains of scrapie-infected VRQ and ARQ homozygous or heterozygous sheep. Inoculated lambs were maintained under normal sheep husbandry conditions and monitored for clinical signs of scrapie. Three animals were culled at 6 months post-scrapie inoculation and the three remaining animals were maintained until they displayed unequivocal signs of scrapie, at which point they were killed. Scrapie infection was confirmed in all clinical cases by the presence of disease-specific PrP in brain sections and by typical vacuolar pathology in the brain stem.
Blood sampling
Peripheral blood was collected by jugular venepuncture from scrapie-inoculated sheep, or from age- and sex-matched New Zealand-derived scrapie-free sheep. Scrapie-inoculated lambs were bled immediately before prion challenge and subsequently at monthly intervals post-scrapie inoculation. Blood was also collected from sheep persistently infected with MVV (maedi-visna virus). Blood was either collected in EDTA tubes or allowed to clot and was subsequently transported on ice and stored overnight at 4 °C before the preparation of PBMCs.
Isolation of sheep PBMCs for immunofluorescence staining
A buffy coat was prepared from EDTA-treated blood by centrifugation at 733 g for 20 min at 21 °C and the harvested cells were layered on to NycoPrep™ animal (density 1.077 g/ml; osmolarity 265 mosmol) and centrifuged at 600 g for 15 min at 21 °C. Mononuclear cells were recovered from the density medium interface and washed three times with FACS buffer. Cell-surface phenotype was assessed using aliquots of 1×106 cells incubated with monoclonal antibody culture supernatant, or normal mouse serum at 1:1000 as control, for 20 min at 4 °C followed by three washes in FACS buffer and incubation with goat anti-mouse IgG-biotin (Sigma, catalogue no. B-7264) at 1:1000 or goat anti-mouse IgG1-biotin (Caltag Laboratories, Burlingame, CA, U.S.A., catalogue no. M32115) at 1:500, for 20 min at 4 °C. Cells were washed three times with FACS buffer and subsequently incubated with 0.25 μg of streptavidin-phycoerythrin for 20 min at 4 °C. Cells were finally washed three times with FACS buffer and analysed for cell-surface fluorescence using a FACSCalibur®. For each sample 10000 cells were analysed with dead cells excluded on the basis of forward and side light scatter.
Statistical analysis
Statistical analysis of the data was performed using the two-tailed Student t test (paired-samples).
RESULTS
Reactivity of anti-PrP monoclonal antibodies with recombinant ovine PrP
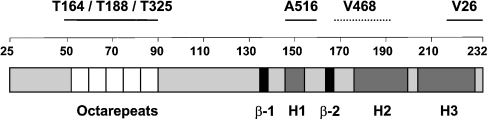
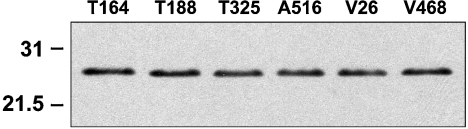
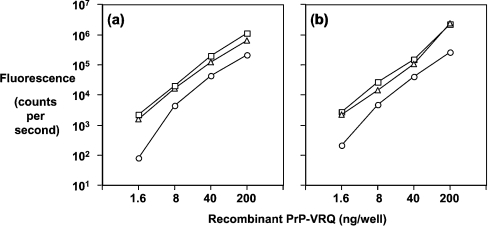
We have utilized a panel of anti-PrP monoclonal antibodies [26] to probe the conformation of the ovine prion protein on the surface of blood cells from VRQ homozygous sheep during experimental scrapie disease. The specificity of the anti-PrP monoclonal antibodies has been investigated by epitope mapping using truncated ovine PrP and a panel of overlapping 15-mer peptides that spanned the whole length of full-length ovine PrP [26], and a series of overlapping 21-mer peptides spanning residues 25–101 of ovine PrP. The monoclonal antibodies T164, T188 and T325 all reacted with full-length ovine recombinant PrP (residues 25–232) but not a peptide comprising residues 89–233. This indicated that T164, T188 and T325 were N-terminal-specific but to date these monoclonal antibodies have not shown any reactivity during epitope mapping using overlapping 15-mers. However, using 21-mer peptides that spanned residues 25–101 of ovine PrP, all three monoclonal antibodies were shown to react within the amino acid sequence 47–90, which includes the octapeptide repeat region of PrP. Monoclonal antibodies A516, V26 and V468 all reacted with full-length (residues 25–232) and truncated VRQ (residues 89–233). In epitope mapping studies, monoclonal antibody A516 reacted most strongly with a peptide comprising residues 145–156 around the C-terminal region of helix-1. Monoclonal antibody V26 reacted with the amino acid sequence CITQYQRESQAYYQRG that consisted of residues 217–232 of ovine PrP and was located in the extreme C-terminal region of helix-3. Further indirect evidence that V26 reacted in this region of PrP was the fact that this monoclonal antibody failed to react with murine recombinant PrP but showed enhanced reactivity with bovine recombinant PrP (results not shown). Ovine, murine and bovine PrP show amino acid differences in this region of the molecule that may account for this genotypic variation in V26 reactivity. Monoclonal antibody V468 has not shown any reactivity in epitope mapping to date, although its epitope appears to be influenced by residue 171 since this monoclonal antibody has little reactivity with PrP-ARR [26]. Figure 1 shows the epitope location of the N- and C-terminal-specific anti-PrP monoclonal antibodies presented in this study. Figure 2 showed that all the monoclonal antibodies reacted equally well with full-length ovine recombinant PrP–VRQ (residues 25–232) when tested by Western blotting. Similarly, all the monoclonal antibodies reacted with ovine recombinant PrP–VRQ when measured by direct immunoassay as shown by the data in Figure 3. These data show that all monoclonal antibodies were capable of recognizing the VRQ allelic variant of ovine prion protein.
Figure 1. Schematic diagram showing the epitope location of N- and C-terminal-specific anti-PrP monoclonal antibodies.
β represents a β-strand and H represents a helix. Numbers represent amino acid residues 25–232.
Figure 2. Western-blot analysis of monoclonal antibodies with full-length ovine recombinant PrP–VRQ.
Full-length ovine recombinant PrP–VRQ (200 ng/lane) was analysed by Western blotting using the following anti-PrP monoclonal antibodies: T164 (lane 1); T188 (lane 2); T325 (lane 3); A516 (lane 4); V26 (lane 5); and V468 (lane 6). Molecular-mass standards (kDa) are shown on the left-hand side.
Figure 3. Detection of ovine recombinant PrP by direct immunoassay.
ELISA plates were coated with 5-fold dilutions of full-length ovine recombinant PrP–VRQ starting from 200 ng/well as described in the Materials and methods section. Biotinylated anti-PrP monoclonal antibodies were used at 50 ng/well and the results are shown as the mean counts/s minus the background for triplicate wells. (a) T164 (○), T188 (□) or T325 (△); (b) A516 (○), V26 (□) or V468 (△).
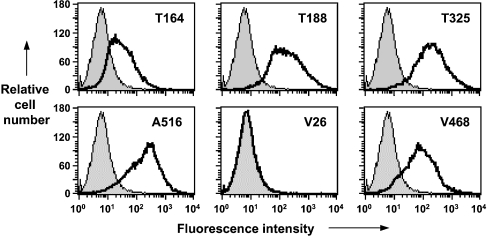
Reactivity of anti-PrP monoclonal antibodies with MovS6 VRQ cells
The N- and C-terminal-specific anti-PrP monoclonal antibodies were tested for their reactivity with cell-surface ovine PrPC. Monoclonal antibodies were reacted with MovS6 cells and analysed by flow cytometry. MovS6 cells are a continuously growing cell line derived from dorsal root ganglia of ovine PrP–VRQ transgenic mice crossed with mice expressing SV40T antigen [29] that express glycosylated PrP–VRQ similar to that seen in sheep brain material. The data in Figure 4 (upper panels) show that the flow cytometric profiles for MovS6 cells reacted with the N-terminal-specific monoclonal antibodies T164, T188 and T325. All three N-terminal-specific monoclonal antibodies showed a similar monophasic profile, although the percentage of T164-positive cells was reduced compared with that seen with monoclonal antibodies T188 or T325. Figure 4 also shows the reactivity of the C-terminal-specific monoclonal antibodies A516, V26 and V468 with MovS6 cells (lower panels). Monoclonal antibodies A516 and V468 reacted with similar percentages of MovS6 cells and with similar fluorescence intensity. However, monoclonal antibody V26 failed to react with cell-surface PrPC despite the fact that it showed reactivity with recombinant protein. The epitope for V26 is located at the extreme C-terminal region of ovine PrP (residues 217–232). The failure of monoclonal antibody V26 to react with this region of PrP suggested that the epitope for this monoclonal antibody was buried or obscured in the native cell-surface molecule under normal conditions. Other anti-PrP monoclonal antibodies such as V47, which bind to a similar epitope as V26, reacted in a similar manner (results not shown).
Figure 4. Reactivity of N- and C-terminal-specific anti-PrP monoclonal antibodies with MovS6 cells.
Representative flow cytometry profiles of MovS6 cells are shown following reactivity with T164, T188 or T325 (upper panels), and A516, V26 or V468 (lower panels). Shaded peak represents control fluorescence; black line represents T164, T188, T325, A516, V26 and V468 fluorescence.
Increased expression of N-terminal PrP epitopes by PBMCs from scrapie-infected sheep
We have previously shown that PBMCs from scrapie-free VRQ homozygous sheep expressed high levels of PrP on their cell surface, which could be detected by N-terminal-specific anti-PrP monoclonal antibodies [26]. We subsequently investigated whether these cells presented altered forms of prion protein on their cell surface as detected by flow cytometry during the progression of scrapie disease using the panel of anti-PrP monoclonal antibodies described here.
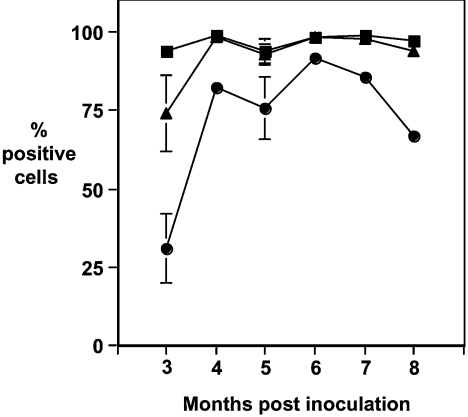
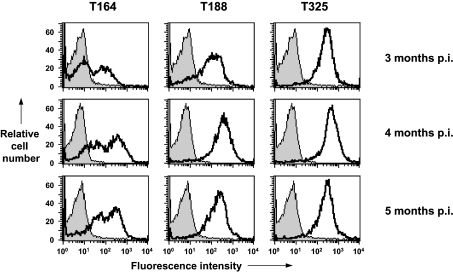
Figure 5 shows the reactivity of PBMCs from scrapie-infected sheep with anti-PrP monoclonal antibodies that are reactive with the N-terminal region of the prion protein. The data from months 3–8 p.i. (post-inoculation) are shown. T164, T188 and T325 all have specific epitopes within amino acid residues 47–90 of ovine PrP. Whilst these anti-PrP monoclonal antibodies recognize epitopes within the N-terminal octapeptide region of ovine PrP, they showed variation in their reactivity with blood cells from scrapieinfected sheep. At 3 months post-scrapie inoculation (Figure 6), T325 reacted with virtually all of the PBMCs from scrapie-infected sheep, whilst T188 and T164 reacted with less cells. T164 reacted with less than 40% of PBMCs at this time point. The failure of T164 and T188 to react with all of the PBMCs at this early time point in the disease process may reflect presentation of PrP on some cells where the specific epitopes for these monoclonal antibodies are buried or obscured by homotypic or heterotypic protein interaction. Alternatively, the epitopes for these monoclonal antibodies may be lost through truncation of blood cell PrP. The absence of T164 and T188 reactivity with PBMCs from scrapie-infected sheep was not sustained throughout the course of the incubation period. At subsequent time points, there was an increase in the number of T164- and T188-reactive PBMCs (Figure 6). At 5 months p.i. and at later time points, the percentage of cells reactive with each of the N-terminal monoclonal antibodies approached that seen with T325, although the percentage of T164-reactive cells was always less than the percentage of T188- or T325-positive cells. At later time points, as the percentage of T164-positive cells increased, it became clear that there were T164-high and T164-low positive cells. In contrast, T325 showed a uniform single reactivity profile with virtually all of the PBMCs binding this monoclonal antibody at all of the time points analysed.
Figure 5. Percentage of PBMCs from scrapie-infected sheep reactive with N-terminal-specific anti-PrP monoclonal antibodies.
Ovine PBMCs from VRQ homozygous sheep were collected at various time points following oral inoculation with scrapie brain material and analysed by flow cytometry with anti-PrP monoclonal antibodies T164 (●), T188 (▲) or T325 (■) as described in the Materials and methods section. n=6 for months 3–5 p.i.; n=3 for months 6–7 p.i.; and n=2 for month 8 p.i. Results are shown as mean±S.D.
Figure 6. Reactivity of N-terminal-specific anti-PrP monoclonal antibodies with PBMCs from VRQ homozygous scrapie-infected sheep.
Representative flow cytometry profiles of ovine PBMCs collected at 3–5 months post-scrapie inoculation following reactivity with T164, T188 or T325. Profiles shown are representative of 6 out of 6 sheep for each anti-PrP monoclonal antibody. Shaded peak represents control fluorescence; black line represents T164, T188 and T325 fluorescence.
Variation in C-terminal PrP epitope accessibility on PBMCs from scrapie-infected sheep
It has been shown that ligand binding events at the N-terminal region of PrP may influence conformational changes or binding events elsewhere in the molecule [30–32]. Furthermore, we have shown that genotypically encoded N-terminal variation in ovine PBMCs cell-surface PrPC expression occurs concomitantly with C-terminal region variations [26]. This supports the suggestion that structural differences in the N-terminal region of the molecule can have conformational effects in the C-terminal portion. Consequently, we investigated the expression of C-terminal PrP epitopes on blood cells from scrapie-infected sheep to correlate any change that occurred with C-terminal-reactive anti-PrP monoclonal antibodies.
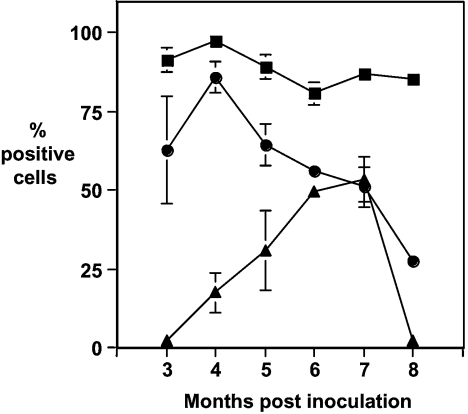
Figure 7 shows the percentage of PBMCs from scrapie-infected sheep reactive with monoclonal antibodies A516, V468 and V26. These monoclonal antibodies were generated against truncated ovine PrP that lacked amino acid residues 25–88 and are therefore reactive in the C-terminal portion of the molecule [26]. At all of the time points investigated, monoclonal antibody A516 showed reactivity with the majority of PBMCs from scrapie-infected sheep. In contrast, monoclonal antibodies V26 and V468 showed variation in their reactivity with PBMCs from scrapie-infected sheep. V26 failed to show significant reactivity with PBMCs at the earliest time points measured. However, at subsequent time points, there was a progressive increase in the percentage of cells reactive with this monoclonal antibody. At 7 months p.i., approx. 50% of the PBMCs reacted with monoclonal antibody V26. In contrast, monoclonal antibody V468 showed a reciprocal reactivity to that seen with V26 during the course of the experiment. The percentage of V468-positive cells decreased from approx. 75 to 25% over the course of the experiment.
Figure 7. Percentage of PBMCs from scrapie-infected sheep reactive with C-terminal-specific anti-PrP monoclonal antibodies.
Ovine PBMCs from VRQ homozygous sheep were collected at various time points following oral inoculation with scrapie brain material and analysed by flow cytometry with anti-PrP monoclonal antibodies V468 (●), V26 (▲) or A516 (■) as described in the Materials and methods section. n=6 for months 3–5 p.i.; n=3 for months 6–7 p.i.; and n=2 for month 8 p.i. Results are shown as means±S.D.
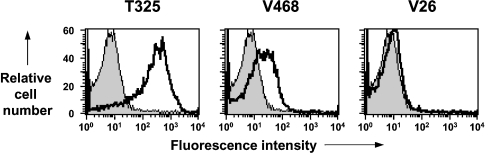
The changes in epitope exposure by PrP expressed on the surface of ovine PBMCs during scrapie disease do not appear to be due to non-specific changes in PrP expression. As a control for disease specificity, we investigated the reactivity of PBMCs prepared from sheep with a persistent virus infection (MVV) with anti-PrP monoclonal antibodies used here. PBMCs from MVV-infected sheep did not show the same changes in cell-surface PrP as those from scrapie-infected sheep, in particular no reactivity was seen with monoclonal antibody V26 (Figure 8).
Figure 8. Reactivity of anti-PrP monoclonal antibodies with PBMCs from sheep persistently infected with MVV.
Representative flow cytometry profiles of ovine PBMCs collected from sheep persistently infected with MVV. Profiles shown are representative of 8 out of 8 sheep for each anti-PrP monoclonal antibody shown. Shaded peak represents control fluorescence; black line represents T325, V468 and V26 fluorescence.
PrP expression by PBMCs from age-matched scrapie-free or scrapie-infected sheep
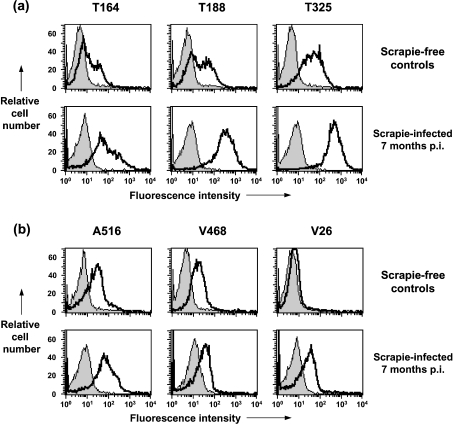
To exclude the possibility that the changes in PrP expression seen on the surface of blood cells isolated from scrapie-infected sheep were not simply an age-related phenomenon, N- and C-terminal FACS staining of PBMCs from age-matched New Zealand-derived scrapie-free VRQ homozygous sheep was investigated. Figure 9 shows representative FACS profiles of PBMCs from scrapie-free and scrapie-infected sheep stained with either N- (Figure 9a) or C-terminal-specific (Figure 9b) anti-PrP monoclonal antibodies. Figure 9(a) shows that there were fewer PBMCs from scrapie-free control sheep reactive with the N-terminal monoclonal antibodies T164 and T188 compared with PBMCs from scrapie-infected animals. T325 showed similar reactivity with PBMCs from both scrapie-free and scrapie-infected animals, although the scrapie-free controls showed lower fluorescence intensity. Figure 9(b) shows that the C-terminal monoclonal antibody V26 had little, if any, reactivity with PBMCs from scrapie-free sheep but showed significant reactivity with PBMCs from age-matched scrapie-infected sheep. Monoclonal antibodies A516 and V468 showed similar reactivity with PBMCs from both scrapie-free and scrapie-infected animals.
Figure 9. Reactivity of N- and C-terminal-specific anti-PrP monoclonal antibodies with PBMCs from VRQ homozygous terminal scrapie-infected sheep and age-matched scrapie-free controls.
Representative flow cytometry profiles of ovine PBMCs collected at either 7 months post-scrapie inoculation, or from age-matched scrapie-free control sheep are shown for (a) N-terminal-specific anti-PrP monoclonal antibodies T164, T188 and T325; (b) C-terminal-specific anti-PrP monoclonal antibodies A516, V468 and V26. Shaded peak represents control fluorescence; black line represents T164, T188, T325, A516, V468 and V26 fluorescence. n=3 for scrapie-infected sheep; n=4 for age-matched scrapie-free controls.
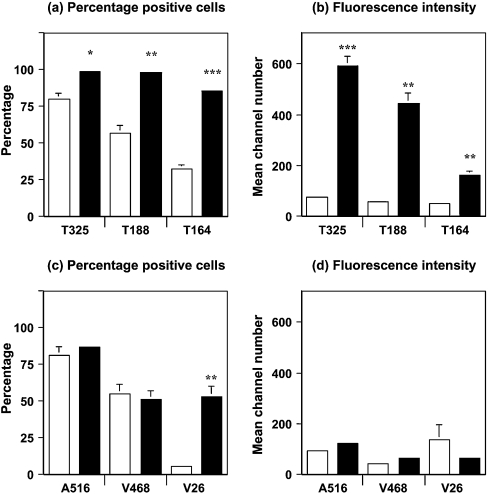
Figure 10 shows the quantification of the N- and C-terminal FACS analysis of PBMCs from scrapie-free and scrapie-infected sheep. Figure 10(a) shows that the percentage of PBMCs from scrapie-infected sheep reactive with N-terminal monoclonal antibodies was significantly greater than that seen with PBMCs from scrapie-free sheep. PBMCs from scrapie-infected sheep showed a significantly higher percentage of cells reactive with T325 compared with age-matched scrapie-free controls (P<0.05). This trend was also seen when the PBMCs were stained with T188 (P<0.01) or T164 (P<0.001). In addition, the fluorescence intensity of all three N-terminal-specific monoclonal antibodies was significantly greater with PBMCs from scrapie-infected sheep compared with cells from scrapie-free animals as shown in Figure 10(b). PBMCs from scrapie-infected sheep gave a significantly higher mean channel number with T325 compared with age-matched scrapie-free controls (P<0.001). This trend was also seen when the PBMCs were stained with T188 or T164 (P<0.01). Figure 10(c) shows that the percentage of PBMCs from scrapie-infected sheep reactive with monoclonal antibody V26 was significantly greater than that seen with PBMCs from scrapie-free sheep (P<0.01).
Figure 10. Quantification of N- and C-terminal PrP flow cytometric analysis.
(a) Percentage of positive cells (means±S.D.) and (b) mean channel number (means±S.D.) are shown for PBMCs from scrapie-infected sheep at 7 months p.i. (black bar) (n=3) and age-matched scrapie-free control sheep (white bar) (n=4) stained with N-terminal-specific anti-PrP monoclonal antibodies T325, T188 and T164. (c) Percentage of positive cells (means±S.D.) and (d) mean channel number (means±S.D.) are shown for PBMCs from scrapie-infected sheep at 7 months p.i. (black bar) (n=3) and age-matched scrapie-free control sheep (white bar) (n=4) stained with C-terminal-specific anti-PrP monoclonal antibodies A516, V468 and V26. Statistical analysis of the data in Figure 10 (comparison with scrapie-free equivalent): *P<0.05, **P<0.01 and ***P<0.001.
Collectively, these data indicate that during the progression of disease in scrapie-infected sheep, there are modifications in epitope exposure at the N- and C-terminal regions of PrP expressed on the surface of blood cells.
DISCUSSION
We have previously shown that conformational variation of ovine PrPC on the surface of PBMCs can be analysed by the use of anti-PrP monoclonal antibodies and flow cytometry [26]. In this study, we have investigated whether changes in PrP epitope expression occur on blood cells isolated from sheep during experimental scrapie. To do so, we have orally inoculated VRQ homozygous lambs with scrapie-infected brain material and subsequently collected PBMCs for PrP analysis at various time points post-prion challenge.
Three different monoclonal antibodies, T164, T188 and T325, which all have epitopes located within amino acid residues 47–90 of ovine PrP (A. M. Thackray and R. Bujdoso, unpublished work), revealed scrapie-induced changes in the N-terminal region of PBMCs cell-surface prion protein. Monoclonal antibody T164 showed the most noticeable change in reactivity with PBMCs from scrapie-infected sheep. This monoclonal antibody reacted with very few PBMCs harvested shortly after scrapie challenge but reacted with progressively more cells collected at later time points. A similar trend, but to a lesser extent, was seen with monoclonal antibody T188. The changes in N-terminal-specific monoclonal antibody reactivity is indicative of PBMCs from scrapie-infected sheep expressing PrP with structural modifications in, or around, the copper-binding domain. These changes would appear to be quite localized and did not involve all of the N-terminal region since monoclonal antibody T325 reacted with a similar percentage of cells at all of the time points analysed.
Changes in the C-terminal region of cell-surface PrP on PBMCs from scrapie-infected sheep were evident as seen by the differential reactivity of monoclonal antibodies specific for epitopes in the globular domain of the protein. Monoclonal antibody V26 reacted with a progressively increasing percentage of cells during the course of scrapie disease. The epitope for this monoclonal antibody is located in the C-terminal region of helix-3. Changes in the region between β-strand-2 and residue 171 of cell-surface PrP were indicated by monoclonal antibodies that bind to this region of PrP. Monoclonal antibodies 968 and 683 react with the epitopes YYRPVD (amino acid residues 165–170) and PVDQY (amino acid residues 168–172) respectively and showed reactivity with cells from scrapie-infected sheep (results not shown). Further changes in the region around residue 171 were indicated by monoclonal antibody V468, which recognizes an epitope that includes, or is influenced by, residue 171 [26], and reacted with progressively fewer cells during the course of scrapie disease. It is interesting to note that monoclonal antibodies V26, 683 and 968 reacted with PBMCs from scrapie-infected sheep but do not bind to native cell-surface PrPC expressed by PBMCs from scrapie-free animals (data presented here and [26]). It is also noteworthy that the epitopes of V26 and those of 683 and 968 are located in the vicinity of YYQ (amino acid residues 228–230) and YYR (amino acid residues 165–167) motifs respectively. These motifs are considered to be buried or obscured in normal PrPC but exposed in disease-associated PrP [33]. This would suggest that structural modifications do occur in cell-surface PrP expressed by PBMCs during scrapie disease. These modifications may be similar, although possibly not as extensive as those seen in disease-associated PrP that accumulates on more long-lived cells such as FDCs in lymphoid tissue and neurons in CNS tissue. The changes in epitope exposure by PrP expressed on the surface of ovine PBMCs during scrapie disease does not appear to be due to non-specific effects. Cells prepared from age-matched scrapie-free sheep did not show the same PrP expression profiles nor did cells from sheep with a persistent MVV infection; in particular, no reactivity was seen with monoclonal antibody V26. In addition, the changes in PrP expression did not appear to be due to the appearance of a unique cell type in peripheral blood since there was no significant change in the scatter profiles of PBMCs during the progression of scrapie disease. Secondly, there was no significant difference in the number of PBMCs in the blood from scrapie-infected and scrapie-free sheep (results not shown).
Our results are consistent with the appearance of altered conformers of PrP on the surface of blood cells from scrapie-infected sheep. Indirect evidence in support of this was found in [46], where it is shown through the use of Raman spectroscopy that a membrane preparation from scrapie-infected sheep blood cells has a significantly higher β-sheet content compared with normal cells. As PrPC conversion to PrPSc involves an increase in β-sheet content, blood cell PrP is considered to be the candidate protein responsible for the increased β-sheet content within the membrane fraction. However, whilst scrapie-infected sheep blood, including buffy coat, does contain prion infectivity as shown by intraspecies blood transfusions [17,18], the presence of PK-resistant PrPSc in the blood of scrapie-infected sheep remains debatable [34]. The protein-only hypothesis predicts that prion infectivity and PK-resistant PrPSc are congruent. The simplest explanation for the apparent dichotomy with respect to the blood of scrapie-infected sheep is that the level of PK-resistant PrPSc in this fluid is below the detection level of the presently available methods. Alternatively, disease-associated forms of PrP present on the surface of blood cells may exist, which are distinct from PK-resistant PrPSc. Other forms of disease-associated PrP, distinct from classical PrPSc, have been described such as PK-sensitive PrPSc [35,36]. Since the conversion of PrPC into PrPSc is believed to be an unfolding process that involves intermediate structures, it is plausible to assume that such forms of PrP may exist on the surface of cells that do not readily present or accumulate the end-stage PK-resistant PrPSc. With respect to lymphocytes, this may be a consequence of the relatively short life span of these cells compared with long-lived cells such as FDCs. Cells other than FDCs that are able to accumulate PrPSc are lymphoid tissue TBMs, and do so possibly as a consequence of their phagocytic activity. Whether these cells are able to enter the blood stream and are a source of prion infectivity still remains un-established. In this regard, it is interesting to note that immunohistochemical staining of ovine lymphoid tissue does not show the presence of PrPSc-positive lymphocytes, suggesting these cells do not acquire PrPSc by passive means as they migrate through lymph nodes. This may be due to the fact that these cells do not display the phagocytic activity of lymphoid macrophages.
A key issue to be addressed is what factor(s) induce the phenotypic changes in PrP expressed on the surface of ovine PBMCs. PrPC is proposed to be converted into PrPSc by autocatalytic conversion induced by PrPSc [1]. For this mechanism to occur in the peripheral blood of scrapie-infected sheep and affect the majority of cells present, as indicated by the FACS profiles seen here, it is reasonable to assume that significant levels of circulating PrPSc should be present. Since PrPSc is not readily detectable in scrapie-infected sheep blood, other mechanisms may be considered. One alternative may be an alteration in the blood level of PrP ligands such as metal ions, during the course of scrapie disease. Metal ions such as copper or manganese have been shown to bind to the N-terminus of PrP and cause structural modifications within the protein [37–44]. Ligand binding at the N-terminal region of PrP results in subsequent modification of epitopes in C-terminal regions of the protein. We have previously shown that mice undergoing prion disease are characterized by the presence of an increased level of manganese in peripheral blood during preclinical and clinical disease [45]. If the same occurs in the peripheral blood of scrapie-infected sheep, then the increased level of blood manganese may be capable of modifying the normal metal ion content of cell-surface PrP, thereby inducing conformational changes in the protein.
Our results are compatible with the presentation of novel conformers of PrP on the surface of sheep blood cells during the progression of prion disease. We have considered that these novel forms of PrP are related to scrapie disease and now we need to establish what their relationship is to other altered conformations of this protein, such as PrPSc. We are currently investigating whether the modifications seen in epitope exposure of cell-surface PrP during scrapie disease described here are accompanied by changes in epitope expression by serum or plasma PrP and if they correlate with prion infectivity.
Acknowledgments
This work was supported in part by funds from Defra (Department for Environment, Food and Rural Affairs). We thank the VLA, Weybridge (Addlestone, Surrey, U.K.) for the kind supply of ovine tissues and normal and scrapie-infected sheep blood. We thank Dr B. Blacklaws, Dr S. Budhia and Dr C. Barbezange (University of Cambridge) for the kind supply of maedi-visna virus-infected sheep blood. We thank Dr H. Laude (INRA, France) for the MovS6 cells.
References
- 1.Prusiner S. B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 2.McBride P. A., Eikelenboom P., Kraal G., Fraser H., Bruce M. E. PrP protein is associated with follicular dendritic cells of spleens and lymph nodes in uninfected and scrapie-infected mice. J. Pathol. 1992;168:413–418. doi: 10.1002/path.1711680412. [DOI] [PubMed] [Google Scholar]
- 3.Aguzzi A. Prions and the immune system: a journey through gut, spleen, and nerves. Adv. Immunol. 2003;81:123–171. doi: 10.1016/s0065-2776(03)81004-0. [DOI] [PubMed] [Google Scholar]
- 4.Beekes M., McBride P. A. Early accumulation of pathological PrP in the enteric nervous system and gut-associated lymphoid tissue of hamsters orally infected with scrapie. Neurosci. Lett. 2000;278:181–184. doi: 10.1016/s0304-3940(99)00934-9. [DOI] [PubMed] [Google Scholar]
- 5.van Keulen L. J., Schreuder B. E., Vromans M. E., Langeveld J. P., Smits M. A. Pathogenesis of natural scrapie in sheep. Arch. Virol. Suppl. 2000;16:57–71. doi: 10.1007/978-3-7091-6308-5_5. [DOI] [PubMed] [Google Scholar]
- 6.Glatzel M., Heppner F. L., Albers K. M., Aguzzi A. Sympathetic innervation of lymphoreticular organs is rate limiting for prion neuroinvasion. Neuron. 2001;31:25–34. doi: 10.1016/s0896-6273(01)00331-2. [DOI] [PubMed] [Google Scholar]
- 7.Lasmezas C. I., Cesbron J. Y., Deslys J. P., Demaimay R., Adjou K. T., Rioux R., Lemaire C., Locht C., Dormont D. Immune system-dependent and -independent replication of the scrapie agent. J. Virol. 1996;70:1292–1295. doi: 10.1128/jvi.70.2.1292-1295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Race R., Oldstone M., Chesebro B. Entry versus blockade of brain infection following oral or intraperitoneal scrapie administration: role of prion protein expression in peripheral nerves and spleen. J. Virol. 2000;74:828–833. doi: 10.1128/jvi.74.2.828-833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baldauf E., Beekes M., Diringer H. Evidence for an alternative direct route of access for the scrapie agent to the brain bypassing the spinal cord. J. Gen. Virol. 1997;78:1187–1197. doi: 10.1099/0022-1317-78-5-1187. [DOI] [PubMed] [Google Scholar]
- 10.Hadlow W. J., Kennedy R. C., Race R. E. Natural infection of Suffolk sheep with scrapie virus. J. Infect. Dis. 1982;146:657–664. doi: 10.1093/infdis/146.5.657. [DOI] [PubMed] [Google Scholar]
- 11.Kimberlin R. H., Walker C. A. Pathogenesis of scrapie in mice after intragastric infection. Virus Res. 1989;12:213–220. doi: 10.1016/0168-1702(89)90040-3. [DOI] [PubMed] [Google Scholar]
- 12.Kimberlin R. H., Hall S. M., Walker C. A. Pathogenesis of mouse scrapie. Evidence for direct neural spread of infection to the CNS after injection of sciatic nerve. J. Neurol. Sci. 1983;61:315–325. doi: 10.1016/0022-510x(83)90165-x. [DOI] [PubMed] [Google Scholar]
- 13.Maignien T., Lasmezas C. I., Beringue V., Dormont D., Deslys J. P. Pathogenesis of the oral route of infection of mice with scrapie and bovine spongiform encephalopathy agents. J. Gen. Virol. 1999;80:3035–3042. doi: 10.1099/0022-1317-80-11-3035. [DOI] [PubMed] [Google Scholar]
- 14.Heggebo R., Press C. M., Gunnes G., Lie K. I., Tranulis M. A., Ulvund M., Groschup M. H., Landsverk T. Distribution of prion protein in the ileal Peyer's patch of scrapie-free lambs and lambs naturally and experimentally exposed to the scrapie agent. J. Gen. Virol. 2000;81:2327–2337. doi: 10.1099/0022-1317-81-9-2327. [DOI] [PubMed] [Google Scholar]
- 15.Heggebo R., Press C. M., Gunnes G., Ulvund M. J., Tranulis M. A., Lsverk T. Detection of PrPSc in lymphoid tissues of lambs experimentally exposed to the scrapie agent. J. Comp. Pathol. 2003;128:172–181. doi: 10.1053/jcpa.2002.0625. [DOI] [PubMed] [Google Scholar]
- 16.Scott J. R. Scrapie pathogenesis. Br. Med. Bull. 1993;49:778–791. doi: 10.1093/oxfordjournals.bmb.a072646. [DOI] [PubMed] [Google Scholar]
- 17.Hunter N., Foster J., Chong A., McCutcheon S., Parnham D., Eaton S., MacKenzie C., Houston F. Transmission of prion diseases by blood transfusion. J. Gen. Virol. 2002;83:2897–2905. doi: 10.1099/0022-1317-83-11-2897. [DOI] [PubMed] [Google Scholar]
- 18.Houston F., Foster J. D., Chong A., Hunter N., Bostock C. J. Transmission of BSE by blood transfusion in sheep. Lancet. 2000;356:999–1000. doi: 10.1016/s0140-6736(00)02719-7. [DOI] [PubMed] [Google Scholar]
- 19.Cervenakova L., Yakovleva O., McKenzie C., Kolchinsky S., McShane L., Drohan W. N., Brown P. Similar levels of infectivity in the blood of mice infected with human-derived vCJD and GSS strains of transmissible spongiform encephalopathy. Transfusion. 2003;43:1687–1694. doi: 10.1046/j.0041-1132.2003.00586.x. [DOI] [PubMed] [Google Scholar]
- 20.Llewelyn C. A., Hewitt P. E., Knight R. S., Amar K., Cousens S., Mackenzie J., Will R. G. Possible transmission of variant Creutzfeldt-Jakob disease by blood transfusion. Lancet. 2004;363:417–421. doi: 10.1016/S0140-6736(04)15486-X. [DOI] [PubMed] [Google Scholar]
- 21.Herrmann L. M., Davis W. C., Knowles D. P., Wardrop K. J., Sy M. S., Gambetti P., O'Rourke K. I. Cellular prion protein is expressed on peripheral blood mononuclear cells but not platelets of normal and scrapie-infected sheep. Haematologica. 2001;86:146–153. [PubMed] [Google Scholar]
- 22.Holada K., Simak J., Risitano A. M., Maciejewski J., Young N. S., Vostal J. G. Activated platelets of patients with paroxysmal nocturnal hemoglobinuria express cellular prion protein. Blood. 2002;100:341–343. doi: 10.1182/blood.v100.1.341. [DOI] [PubMed] [Google Scholar]
- 23.Holada K., Vostal J. G. Different levels of prion protein (PrPc) expression on hamster, mouse and human blood cells. Br. J. Haematol. 2000;110:472–480. doi: 10.1046/j.1365-2141.2000.02158.x. [DOI] [PubMed] [Google Scholar]
- 24.Clouscard C., Beaudry P., Elsen J. M., Milan D., Dussaucy M., Bounneau C., Schelcher F., Chatelain J., Launay J. M., Laplanche J. L. Different allelic effects of the codons 136 and 171 of the prion protein gene in sheep with natural scrapie. J. Gen. Virol. 1995;76:2097–2101. doi: 10.1099/0022-1317-76-8-2097. [DOI] [PubMed] [Google Scholar]
- 25.Goldmann W., Hunter N., Smith G., Foster J., Hope J. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J. Gen. Virol. 1994;75:989–995. doi: 10.1099/0022-1317-75-5-989. [DOI] [PubMed] [Google Scholar]
- 26.Thackray A. M., Yang S., Wong E., Fitzmaurice T. J., Morgan-Warren R. J., Bujdoso R. Conformational variation between allelic variants of cell-surface ovine prion protein. Biochem. J. 2004;381:221–229. doi: 10.1042/BJ20040351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrmann L. M., Baszler T. V., Knowles D. P., Cheevers W. P. PrP(Sc) is not detected in peripheral blood leukocytes of scrapie-infected sheep: determining the limit of sensitivity by immunohistochemistry. Clin. Diagn. Lab. Immunol. 2002;9:499–502. doi: 10.1128/CDLI.9.2.499-502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slootstra J. W., Puijk W. C., Ligtvoet G. J., Langeveld J. P., Meloen R. H. Structural aspects of antibody-antigen interaction revealed through small random peptide libraries. Mol. Divers. 1996;1:87–96. doi: 10.1007/BF01721323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Archer F., Bachelin C., Andreoletti O., Besnard N., Perrot G., Langevin C., Le Dur A., Vilette D., Baron-Van Evercooren A., Vilotte J. L., et al. Cultured peripheral neuroglial cells are highly permissive to sheep prion infection. J. Virol. 2004;78:482–490. doi: 10.1128/JVI.78.1.482-490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thackray A. M., Madec J. Y., Wong E., Morgan-Warren R., Brown D. R., Baron T., Bujdoso R. Detection of bovine spongiform encephalopathy, ovine scrapie prion-related protein (PrPSc) and normal PrPc by monoclonal antibodies raised to copper-refolded prion protein. Biochem. J. 2003;370:81–90. doi: 10.1042/BJ20021280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li R., Liu T., Wong B. S., Pan T., Morillas M., Swietnicki W., O'Rourke K., Gambetti P., Surewicz W. K., Sy M. S. Identification of an epitope in the C terminus of normal prion protein whose expression is modulated by binding events in the N terminus. J. Mol. Biol. 2000;301:567–573. doi: 10.1006/jmbi.2000.3986. [DOI] [PubMed] [Google Scholar]
- 32.Matsunaga Y., Peretz D., Williamson A., Burton D., Mehlhorn I., Groth D., Cohen F. E., Prusiner S. B., Baldwin M. A. Cryptic epitopes in N-terminally truncated prion protein are exposed in the full-length molecule: dependence of conformation on pH. Proteins. 2001;44:110–118. doi: 10.1002/prot.1077. [DOI] [PubMed] [Google Scholar]
- 33.Paramithiotis E., Pinard M., Lawton T., LaBoissiere S., Leathers V. L., Zou W. Q., Estey L. A., Lamontagne J., Lehto M. T., Kondejewski L. H., et al. A prion protein epitope selective for the pathologically misfolded conformation. Nat. Med. 2003;9:893–899. doi: 10.1038/nm883. [DOI] [PubMed] [Google Scholar]
- 34.Schmerr M. J., Jenny A. L., Bulgin M. S., Miller J. M., Hamir A. N., Cutlip R. C., Goodwin K. R. Use of capillary electrophoresis and fluorescent labeled peptides to detect the abnormal prion protein in the blood of animals that are infected with a transmissible spongiform encephalopathy. J. Chromatogr. A. 1999;853:207–214. doi: 10.1016/s0021-9673(99)00514-2. [DOI] [PubMed] [Google Scholar]
- 35.Safar J., Cohen F. E., Prusiner S. B. Quantitative traits of prion strains are enciphered in the conformation of the prion protein. Arch. Virol. Suppl. 2000;16:227–235. doi: 10.1007/978-3-7091-6308-5_22. [DOI] [PubMed] [Google Scholar]
- 36.Safar J., Wille H., Itri V., Groth D., Serban H., Torchia M., Cohen F. E., Prusiner S. B. Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 37.Whittal R. M., Ball H. L., Cohen F. E., Burlingame A. L., Prusiner S. B., Baldwin M. A. Copper binding to octarepeat peptides of the prion protein monitored by mass spectrometry. Protein Sci. 2000;9:332–343. doi: 10.1110/ps.9.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown D. R., Qin K., Herms J. W., Madlung A., Manson J., Strome R., Fraser P. E., Kruck T., von Bohlen A., Schulz-Schaeffer W., et al. The cellular prion protein binds copper in vivo. Nature (London) 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 39.Hornshaw M. P., McDermott J. R., Candy J. M., Lakey J. H. Copper binding to the N-terminal tandem repeat region of mammalian and avian prion protein: structural studies using synthetic peptides. Biochem. Biophys. Res. Commun. 1995;214:993–999. doi: 10.1006/bbrc.1995.2384. [DOI] [PubMed] [Google Scholar]
- 40.Miura T., Hori-i A., Mototani H., Takeuchi H. Raman spectroscopic study on the copper(II) binding mode of prion octapeptide and its pH dependence. Biochemistry. 1999;38:11560–11569. doi: 10.1021/bi9909389. [DOI] [PubMed] [Google Scholar]
- 41.Aronoff-Spencer E., Burns C. S., Avdievich N. I., Gerfen G. J., Peisach J., Antholine W. E., Ball H. L., Cohen F. E., Prusiner S. B., Millhauser G. L. Identification of the Cu2+ binding sites in the N-terminal domain of the prion protein by EPR and CD spectroscopy. Biochemistry. 2000;39:13760–13771. doi: 10.1021/bi001472t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burns C. S., Aronoff-Spencer E., Dunham C. M., Lario P., Avdievich N. I., Antholine W. E., Olmstead M. M., Vrielink A., Gerfen G. J., Peisach J., et al. Molecular features of the copper binding sites in the octarepeat domain of the prion protein. Biochemistry. 2002;41:3991–4001. doi: 10.1021/bi011922x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stockel J., Safar J., Wallace A. C., Cohen F. E., Prusiner S. B. Prion protein selectively binds copper(II) ions. Biochemistry. 1998;37:7185–7193. doi: 10.1021/bi972827k. [DOI] [PubMed] [Google Scholar]
- 44.Brown D. R., Hafiz F., Glasssmith L. L., Wong B. S., Jones I. M., Clive C., Haswell S. J. Consequences of manganese replacement of copper for prion protein function and proteinase resistance. EMBO J. 2000;19:1180–1186. doi: 10.1093/emboj/19.6.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thackray A. M., Knight R., Haswell S. J., Bujdoso R., Brown D. R. Metal imbalance and compromised antioxidant function are early changes in prion disease. Biochem. J. 2002;362:253–258. doi: 10.1042/0264-6021:3620253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carmona P., Monleon E., Monzon M., Badiola J. J., Monreal J. Raman analysis of prion protein in blood cell membranes from naturally affected scrapie sheep. Chem. Biol. 2004;11:759–764. doi: 10.1016/j.chembiol.2004.04.005. [DOI] [PubMed] [Google Scholar]