Abstract
Phototropin is a blue light photoreceptor for tropic responses, relocation of chloroplasts, and stomata opening in plants. Phototropin has two chromophoric domains named light-oxygen-voltage-sensing (LOV) 1 and 2 in the N-terminal half, and a serine/threonine (Ser/Thr) protein kinase motif in the C-terminal half. Concerning the kinase activity of phototropin, only autophosphorylation has been detected so far. However, we found that phototropin can phosphorylate a protein other than phototropin itself. Bacterially expressed Arabidopsis phototropin 2 kinase domain (KD) with GST-tag showed a constitutive kinase activity on casein, a common in vitro substrate of Ser/Thr protein kinase. By using this in vitro assay system, the roles of each LOV domain were studied. Addition of LOV2 to KD (GST-L2-KD) inhibits the kinase activity that is canceled by light. This light activation of kinase disappeared on introduction of a mutation blocking photochemical reaction in the LOV2 domain. Accordingly, LOV2 domain acts as a major light-regulated molecular switch of casein phosphorylation. Interestingly, isolated LOV2 from the KD still binds to the KD in a light-dependent manner and functions in similar ways, indicating the role of LOV2 domain as an inhibitor of the kinase activity in the substrate phosphorylation. LOV1, in contrast, contributes little to the photoactivation in GST-L1-L2-KD; however, it acts as an attenuator of the light activation of the kinase by LOV2.
Keywords: photoregulation, photoreceptor, phosphorylation, LOV domain
Light in the wavelength region from UV to far-red is an important stimulus for plants that precisely regulates developmental and cell motility processes. To sense the light conditions including intensity, quality, and direction, plants have acquired three major photoreceptors (1): phytochrome, cryptochrome, and phototropin.
Phototropin (2, 3), originally identified as a photoreceptor for tropic responses (4), has also been found to mediate chloroplast relocations (5–7), stomatal opening (8), leaf expansion (9), and rapid inhibition of hypocotyl elongation (10). Most plants have two isoforms of phototropin, named phot1 and phot2 (2). In Arabidopsis thaliana (At), phot1 and phot2 share tropic responses and also one of the chloroplast relocation responses, photoaccumulation, depending on fluence rate of light (6). Photoavoidance response, on the other hand, is mediated by only phot2 (5). In contrast, stomatal opening is regulated redundantly by both phot1 and phot2 (8).
The phototropins are composed of 900–1,000 amino acid residues and two prosthetic flavin mononucleotide (FMN) molecules (2, 3) (Fig. 1A Top). The N-terminal half has a pair of FMN-binding domains with ≈100 residues designated light-oxygen-voltage-sensing (LOV) 1 and 2 (11), and the C-terminal half forms a serine/threonine (Ser/Thr) kinase domain connected to the LOV2 domain with a linker region (Fig. 1 A).
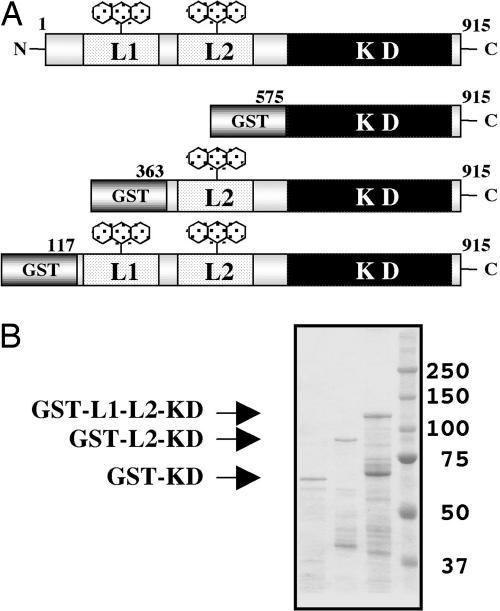
Fig. 1.
Expression of Atphot2 recombinant polypeptides in Escherichia coli. (A) Schematic drawing of an Atphot2 domain structure and three expression constructs of N-terminal deletions, GST-KD (1–574 deletion), GST-L2-KD (1–362 deletion), and GST-L1-L2-KD (1–116 deletion), respectively. KD, kinase domain; L1, LOV1; L2, LOV2. (B) SDS/PAGE of the three recombinant polypeptides purified by glutathione-Sepharose. GST-KD (leftmost lane), GST-L2-KD (second lane from the left), and GST-L1-L2-KD (third lane from the left) were stained by Coomassie brilliant blue. Molecular masses (kDa) of the markers (right lane) are indicated to the right of the gel.
Upon absorption of blue light, FMN in the two LOV domains undergoes a unique cyclic photochemical reaction through formation and breakage of a covalent bond (12) with a highly conserved cysteine found in all of the LOV domains of phototropin-like proteins (13–15). Crystal structures of the Chlamydomonas phot-LOV1 (16) and Adiantum phytochrome 3 (phy3)-LOV2 (17, 18) have revealed very small conformational changes in the vicinity of the chromophore on formation of the protein–FMN adduct, whereas Fourier transform infrared studies have shown secondary-structural changes in phy3-LOV2. Recently, an NMR study has reported light-dependent structural changes in a part of the linker region in oat phot1 (19), and a small-angle x-ray scattering study has proposed a relative movement of the linker region to the LOV2 domain in Arabidopsis phot1 (20). These changes are hypothesized to lead the activation of the kinase domain upon adduct formation (21).
Phototropin was originally found as a protein phosphorylated rapidly on light-illumination in a membrane fraction. Since then, autophosphorylation of phototropin has been established both in vivo (22) and in vitro (4). The autophosphorylation sites reside between the LOV1 and LOV2 domains and also in the N-terminal region (23, 24). A rescue experiment with a phototropin-deficient Arabidopsis mutant has revealed that photoactive LOV2 domains play a predominant role in activating autophosphorylation by light in both phot1 and phot2 (25). These studies indicate a correlation between the autophosphorylation of phototropin and phototropic responses; however, its involvement in the signal transduction is still unclear. Furthermore, no kinase activity of phototropin, except for autophosphorylation, has been detected so far.
In the present study, we searched for substrates for the kinase activity of Atphot2 in common substrates of Ser/Thr kinase and found that casein can be a good substrate. This observation that phototropin can phosphorylate proteins other than phototropin itself is unique. Using the in vitro phosphorylation system, we investigated the role of LOV1 and LOV2 in regulation of the kinase activity by light and found that LOV2 acts as a light-regulated molecular switch of constitutively active kinase domain and that LOV1 attenuates the light sensitivity of the switch, indicating distinct roles for the two LOV domains.
Materials and Methods
Construction of Expression Vectors. By using cDNA of Atphot2 as a template, required DNA fragments with appropriate restriction sites were synthesized with PCR and oligonucleotide primers. Amplified DNA was isolated, digested, and cloned into a pGEX4T1 bacterial expression vector (Amersham Pharmacia Biosciences) as a translational fusion to the glutathione S-transferase (GST). The following polypeptides, fused with GST in the N terminus, were prepared: GST-KD (575–915), GST-L2-KD (363–915), GST-L1-L2-KD (117–915), GST-L1 (117–265), and GST-L2 (363–500). In addition to these wild-type polypeptides, their non-adduct-forming homologues were prepared by mutating the adduct-forming Cys to Ala. The homologues are GST-L2(C426A)-KD [GST-L2(C/A)-KD], GST-L1(C170A)-L2-KD [GST-L1(C/A)-L2-KD], GST-L1-L2(C426A)-KD [GST-L1-L2(C/A)-KD], and GST-L1(C170A)-L2(C426A)-KD [GST-L1(C/A)-L2(C/A)-KD]. The kinase-inactive mutant of GST-KD was prepared by replacement of an aspartate residue essential to the kinase activity by asparagine (26) in subdomain VII of the KD and designated as GST-KD(D720N). The amino acid substitutions were introduced by using a Quick Change site-directed mutagenesis kit (Stratagene), following the instructions, and verified by DNA sequencing with a CEQ2000XL DNA analysis system (Beckman Coulter).
Expression and Purification of Recombinant Proteins. The E. coli JM109 strain transformed by each expression plasmid was grown at 37°C in LB medium containing 50 μg·ml-1 ampicillin until OD600 reached 0.3, and then incubated with 0.1 mM isopropyl β-d-thiogalactopyranoside for 20 h at 20°C in darkness. The following purification procedures were carried out at 0–4°C under dim red light. Bacteria were collected by centrifugation and suspended in the extraction buffer containing 50 mM Tris·HCl (pH 7.8), 100 mM NaCl, 1 mM EGTA, 10% glycerol, and 1 mM phenylmethylsulfonyl fluoride (PMSF). The cells were lysed by sonication, and the supernatant was mixed with glutathione-Sepharose 4B (Amersham Pharmacia Biosciences). The resin was washed with the extraction buffer. The GST-fusion proteins were eluted with 10 mM reduced glutathione in the extraction buffer and stored at -80°C until use. For the purification of GST-free LOV1 and LOV2, bacteria were collected by centrifugation, washed with PBS, and resuspended in PBS containing 1 mM PMSF. The cells were lysed by sonication, and the supernatant was mixed with glutathione-Sepharose 4B. After washing the resin with PBS, the LOV polypeptides were cleaved from GST tag with thrombin, which leaves a linker of five amino acid residues, Gly-Ser-Pro-Glu-Phe, to the N terminus of each LOV construct. The dissociated polypeptides from the gel were purified further by gel filtration on a Sephacryl S-100 HR column (Amersham Pharmacia Biosciences), equilibrated and eluted with 100 mM NaCl/25 mM Tris·HCl/1 mM Na2EGTA (pH 7.8). Purity of the polypeptides was examined with Coomassie brilliant blue and anti-GST IgG staining of SDS/PAGE gels.
In Vitro Phosphorylation Assay. Phosphorylation assay was performed under dim red light. KD-containing polypeptides were incubated with casein (dephosphorylated for protein kinase assay, Sigma) at 30°C in a kinase reaction mixture containing 30 mM Tris·HCl (pH 7.8), 100 mM NaCl, 1 mM Na2EGTA, 10% glycerol, 10 mM MgCl2, 20 μM ATP, and 37 kBq of [γ-32P]ATP. The reaction was terminated by the addition of a concentrated SDS/PAGE sample buffer followed by boiling for 3 min. Then, the samples were run on SDS/PAGE, and molecular masses of the bands were estimated by their positions revealed by Coomassie brilliant blue staining. Phosphorylation of the bands was visualized with imaging plates and a bioimaging analyzer (Fuji). The analyzer also quantified each band in the autoradiogram. Light-dependent phosphorylation was measured with GST-L2-KD, GST-L1-L2-KD, and their photoinactive homologues and GST-KD in the presence of purified LOV polypeptides by giving a mock irradiation or irradiating with white light. Fluence-response of the light-induced kinase activity was studied with GST-KD, GST-L2-KD, and GST-L1-L2-KD by irradiating with white light supplied with a combination of a fluorescent tube and a variable power supply for 10 min at different fluence rates. Fluence rates of the light were measured with a spectroradiometer LI-1800 (Li-Cor, Lincoln, NE).
GST Pull-Down Assay. GST or GST-KD was charged to glutathione-Sepharose 4B resin according to the manufacturer's protocol. The resin was incubated for 1 h at 4°C in binding buffer containing 25 mM Tris·HCl (pH 7.8), 100 mM NaCl, 1 mM Na2EGTA, and 10% glycerol with each LOV domain in the presence or absence of white light. The resin was collected by centrifugation and washed with the binding buffer three times under the same light conditions. The sample was separated on SDS/PAGE and stained with Coomassie brilliant blue.
Results
Expression of Atphot2 Polypeptides in E. coli. To characterize the molecular mechanism of photoregulation of the kinase activity, we used three GST-fused N-terminal deletion polypeptides of Atphot2, GST-KD, GST-L2-KD, and GST-L1-L2-KD expressed in E. coli (Fig. 1 A). Unfortunately, preparation of full-length Atphot2 has been unsuccessful, possibly because of in vivo degradation. Each GST-fusion protein, purified by glutathione-Sepharose, showed an approximate molecular mass of 65, 89, or 117 kDa (Fig. 1B), respectively. GST-L1-L2-KD and GST-L2-KD preparations have multiple components with smaller molecular masses. They were degradation products in the C-terminal regions because all were recognizable by GST antibody (data not shown). Judging from their molecular masses, they lacked KD, and their effects on the kinase assay could be ignored.
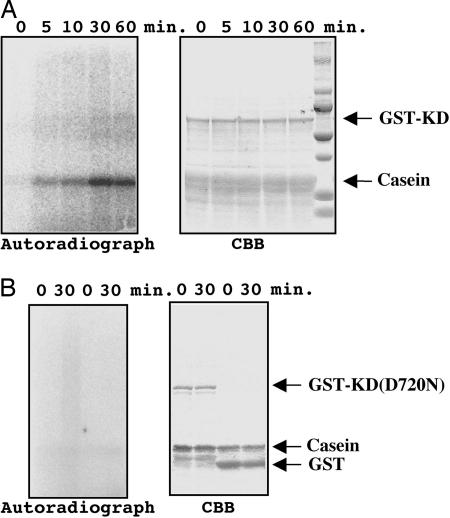
Constitutive Kinase Activity of GST-KD. To see whether phototropin can phosphorylate substrates other than phototropin itself, we tested several common substrates of Ser/Thr kinase, and we found that casein with magnesium ion is the most effective to monitor the GST-KD kinase activity. GST-KD phosphorylated casein in a time-dependent manner, and this phosphorylation was saturated within 30 min (Fig. 2A). Autophosphorylation of the GST-KD was undetectable. Because neither GST itself nor GST-KD(D720), a kinase-inactive mutant, phosphorylated casein (Fig. 2B), the observed kinase activity can be concluded to come from the constitutive kinase activity of KD in phot2.
Fig. 2.
Constitutive protein kinase activity of Atphot2 GST-KD. (A) GST-KD and casein, marked by arrows, were incubated for indicated times in a kinase reaction buffer as described in Materials and Methods. The samples were separated on SDS/PAGE, stained with Coomassie brilliant blue (Right), and then visualized by autoradiography (Left). (B) GST-KD(D720N), a kinase-inactive mutant of GST-KD (two left-most lanes) or GST (two right-most lanes) were incubated with casein as described for A.
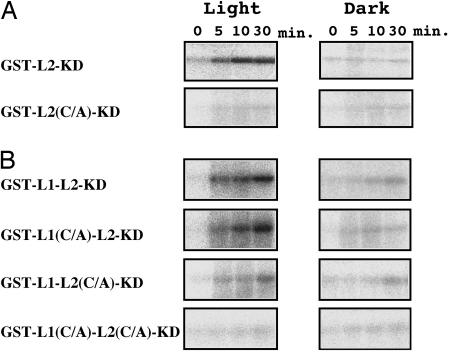
Photoregulation of Kinase Activity in Atphot2 Polypeptides. Using the above in vitro kinase assay system, we studied the roles of each LOV domain in regulation of kinase activity. In contrast to GST-KD, GST-L2-KD did not phosphorylate casein in the dark (Fig. 3A Upper), suggesting the inhibitory role of LOV2 domain. Light irradiation, however, induced phosphorylation of casein that was abolished by introduction of a Cys/Ala mutation into the LOV2 domain (Fig. 3A Lower) indicating the involvement of photoreaction of LOV2 in this photoinduction of the kinase activity. GST-L1-L2-KD also showed a similar photoactivation of the kinase activity that was reduced significantly by introduction of a Cys/Ala mutation into the LOV2 [Fig. 3B, GST-L1-L2-KD and GST-L1-L2(C/A)-KD]. In contrast, when Cys/Ala mutation was introduced into LOV1, there was little effect on this photoactivation even after saturating light irradiation [Fig. 3B, GST-L1(C/A)-L2-KD]. When both LOV domains lost photoreactions in GST-L1(C/A)-L2(C/A)-KD, no kinase activity was detected [Fig. 3B, GST-L1(C/A)-L2(C/A)-KD], suggesting a minor contribution of LOV1 photoreaction in the kinase photoactivation. These results indicate that LOV2 acts as a major light-regulated switch of kinase activity.
Fig. 3.
Light-regulated protein kinase activity of GST-L2-KD (A) and GST-L1-L2-KD (B), including their Cys/Ala (C/A) -mutated homologues. As described in the legend of Fig. 2, kinase activity was detected by phosphorylation of casein by giving either white light (Light) or mock (Dark) irradiations, respectively, for indicated times.
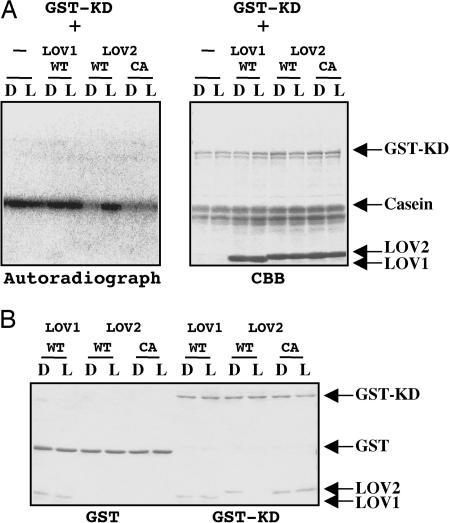
Photoregulation of Kinase Activity by Isolated LOV2. To see the photoregulation mechanism of the kinase activity by LOV2 domain, effects of isolated LOV domains on the kinase activity were studied (Fig. 4A). Presence of LOV1 domain did not reduce the kinase activity of GST-KD either in the dark or under light. LOV2 domain, in contrast, inhibited kinase activity in the dark that was canceled under light in ways similar to those of GST-L2-KD. This light-induced cancellation was not observed in the Cys/Ala mutant of LOV2 domain, indicating that the photoreaction is requisite. Pull-down assay of GST-KD with isolated LOV2 domain clearly showed the association in the dark and dissociation under light of LOV2 domain to GST-KD (Fig. 4B Right, LOV2 WT). This dissociation was not observed in its Cys/Ala mutant (Fig. 4B Right, LOV2 CA), indicating again the involvement of the photoreaction. Furthermore, the assay indicates that the binding site resides in KD but not GST (Fig. 4B Left, LOV2). LOV1 domain, unexpectedly, also binds to GST-KD in a light-independent manner (Fig. 4B Right, LOV1); however, this binding is to GST or possibly the resin because LOV1 was pulled down by GST (Fig. 4B Left, LOV1).
Fig. 4.
Effects of isolated LOV domains on protein kinase activity of GST-KD. (A) Kinase assay of GST-KD was carried out in the absence (-) or presence of purified LOV1 (LOV1) or LOV2 (LOV2) proteins as in Fig. 2. The samples were incubated for 10 min under white light (L) or mock irradiation (D). SDS/PAGE gels were stained with Coomassie brilliant blue (Right) and then visualized by autoradiography (Left). (B) GST pull-down assay on the interaction between GST-KD and each LOV domain. GST or GST-KD was charged to glutathione-Sepharose 4B resin and incubated with either LOV domain under white light (L) or mock irradiation (D). After washing of the resin, the sample was separated on SDS/PAGE gel and stained with Coomassie brilliant blue.
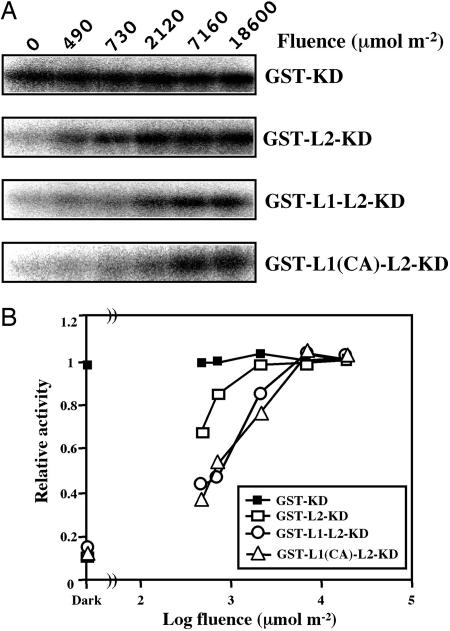
Role of LOV1 Domain. To clarify the role of LOV1 domain in regulation of the kinase activity, we compared fluence response of the photoactivation of the kinase in the Atpho2 polypeptides lacking LOV1 or LOV1 and LOV2 domains (Fig. 5). In GST-L2-KD, kinase activity depended on light intensity, and phosphorylation was saturated at total fluence of 2,120 μmol·m-2. GST-L1-L2-KD also showed a similar fluence response to light irradiation; however, the fluence-response curve is shifted to the higher fluence region. Phosphorylation reached saturation at a total fluence of 7,160 μmol·m-2, which is ≈3.5 times larger than that of GST-L2-KD. Introduction of Cys/Ala mutation to the LOV1 domain did not change the fluence at saturation. However, the shape of the fluence response curve is somewhat different from that of the wild type; it is rather straight compared with that of the wild type with a sigmoidal shape.
Fig. 5.
Fluence response of photoactivation of the kinase in the four N-terminus-deleted polypeptides. GST-KD, GST-L2-KD, GST-L1-L2-KD, or GST-L1(C/A)-L2-KD was incubated with casein in kinase reaction buffer for 10 min at different fluence rates. (A) Phosphorylation of casein was visualized as described in Materials and Methods. Total fluences of irradiated light in the wavelength region from 330 to 500 nm are indicated on the top. (B) Fluence-response curves of the photoactivation of the kinase. Phosphorylation was quantified as described in Materials and Methods and normalized to 1 at the saturated levels. Each symbol represents the average phosphorylation levels of three independent experiments. Symbols on the left indicate the phosphorylation levels in the dark.
Discussion
Construction of in Vitro Assay System of Phototropin Kinase. Phototropin has been shown to be autophosphorylated in response to blue light irradiation in either soluble extracts from phototropin-expressing insect cells (4, 6) or membrane fractions of Arabidopsis (22). There are many reports of expressing LOV domains of phototropin in E. coli for photochemical (11, 13, 27) or structural (17, 18) studies. Its active kinase domain, however, has never been obtained so far. Bacterially expressed oat phot1 kinase domain with a calmodulin-binding protein (CBP) tag was inactive (23). Recently, blue light-dependent autophosphorylation of Vicia faba phototropins was detected in situ in E. coli by far-Western blotting analysis (24), suggesting possibilities to prepare phototropin polypeptides with a kinase activity by an E. coli expression system. In the present study, we succeeded in preparing Atphot2 polypeptides with kinase activity except for the full-length product. They were expressed in E. coli as GST N-terminal fusion proteins. The GST tag (26 kDa) is far bigger than the CBP tag (4 kDa) and known to form a dimer in solution (28) that may contribute to the proper production of kinase-active phototropins in E. coli. GST-KD phosphorylated casein, a common in vitro substrate of Ser/Thr protein kinase (Fig. 2). Phototropin can phosphorylate proteins other than phototropin itself, and this action raises possibilities that target proteins of phototropin kinase may exist in vivo and be involved in the light-signal transduction. Autophosphorylation was undetectable in GST-KD, although the possibility that its autophosphorylation has already been saturated in E. coli cells cannot be excluded. The others two polypeptides, GST-L2-KD and GST-L1-L2-KD, showed light-enhanced autophosphorylation. Light enhancement of GST-L1-L2-KD is much stronger than that of GST-L2-KD (data not shown). Both of the autophosphorylations were inhibited competitively with casein, in which competition in GST-L2-KD is much more obvious than that in GST-L1-L2-KD. These data are consistent with the reported sites of autophosphorylation between LOV1 and LOV2 domains of phototropin (23, 24).
Distinct Roles of LOV Domains in Regulation of Kinase Activity. In GST-L2-KD, LOV2 acted as an inhibitor of phototropin kinase in the dark, and light canceled the inhibition through the cysteine-FMN adduct formation (Fig. 3A). GST-L1-L2-KD also showed a similar photoactivation of the kinase, in which photoreaction of LOV2 is essential (Fig. 3B). Furthermore, kinase of GST-KD was inactivated in the presence of purified LOV2 domain but not LOV1 domain in the dark, and the inactivation by wild-type LOV2 domain is relieved by light, as in GST-L2-KD, though not in a Cys/Ala mutant (Fig. 4A). Thus, all of the results clearly show that LOV2 acts as a light-regulated molecular switch of phototropin kinase in the casein phosphorylation. The observed role of LOV2 in the photoregulation is consistent with the report that LOV2 domain plays a major role in mediating light-dependent autophosphorylation of phototropin and hypocotyl phototropism (25). LOV1, in contrast, shows no inhibition of kinase activity in GST-KD even in the dark (Fig. 4A) and little contribution to the photoactivation of kinase in GST-L1-L2-KD (Fig. 3B). It should be noted, however, that the contribution of LOV1 to the light activation of kinase cannot be totally excluded because the introduction of Cys/Ala mutation into the LOV1 domain brought a minor additive inactivation of the kinase activity (Fig. 3B). Furthermore, introduction of Cys/Ala mutation in the LOV1 domain flattened the fluence response curve, and kinase activity is decreased by ≈18% at the total fluence of 2,120 μmol·m-2. These data suggest a little contribution of LOV1 photoreaction to the light-activation of kinase activity.
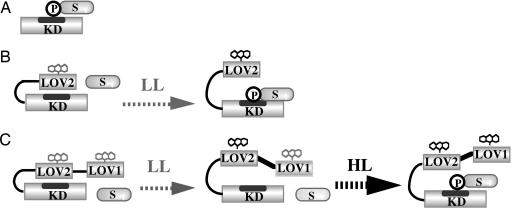
Although bacterially expressed LOV1 domains are reported to show similar photoreactions (27) and the crystal structure (16) is similar to that of LOV2 (17, 18), their roles are still obscure. Recently, LOV1 domain of oat phot1 was reported to be dimeric in solution, whereas LOV2 domain was monomeric (29), suggesting the role of LOV1 as a dimeric site. Present results supplied information regarding the role of LOV1 domain. Addition of LOV1 shifted the fluence-response curve of the kinase photoactivation in GST-L2-KD to the 3.5 times higher fluence region (Fig. 5). This shift suggests that LOV1 acts as an attenuator of the photoactivation, possibly because of a stereochemical blocking of the interactions between LOV2 domain and KD independently of the photoreaction of LOV1. Thus, LOV1 and LOV2 domains showed distinct roles in photoregulation of kinase activity in Atphot2 that are illustrated schematically in Fig. 6. KD expresses kinase activity constitutively (Fig. 6A) and LOV2-KD shows light activation of kinase (Fig. 6B). LOV1-LOV2-KD does not exhibit the activation because of attenuation by LOV1 under low light conditions; however, the attenuation is overcome under high light conditions.
Fig. 6.
Schematic illustration of the roles of LOV1 and LOV2 domains in light regulation of substrate phosphorylation by KD in Atphot2. P, phosphate; S, substrate; LL, low light conditions; HL, high light conditions. (A, B, and C) GST-KD, GST-L2-KD, and GST-L1-L2-KD, respectively, where GST is not indicated.
Interactions Between LOV Domain and Kinase Domain. Present results propose a photoregulation mechanism of phototropin kinase, in which LOV2 domain acts as a molecular inhibitor of the kinase through direct binding between them (Fig. 4), and this inhibition is abolished by light. Among the known Ser/Thr protein kinases, eukaryotic Per-Arnt-Sim kinase (PASK) has the mechanism most similar to that of phototropin (30). PASK has two PAS (PAS-A and PAS-B) domains in the N-terminal region, and the C-terminal half forms a Ser/Thr kinase domain-like phototropin. The kinase activity is inhibited by direct interactions with PAS-A. Furthermore, the activity of substrate phosphorylation is increased by autophosphorylation of PASK, suggesting that blue light-dependent autophosphorylation of phototropin may act in a similar way. On the other hand, kinase domains of phototropin are known to be homologous to those of PKA, which has the same phosphorylation site specificity as that of autophosphorylation in oat phot1 (23). An inactive form of PKA is known to be composed of two regulatory and two catalytic subunits, the former of which inhibit the kinase activity of the latter (31). Binding of cAMP to the regulatory subunit leads to disassembly of the tetramer and release of active catalytic subunits. We, therefore, examined whether PKA is inactivated by Atphot2 LOV2 domain in the dark as GST-KD was. PKA activity, however, was not affected by Atphot2 LOV2 domain (data not shown). Amino acid sequence alignments of KD between phototropin and PKA show that phototropin has additional sequences between subdomains VII and VIII. A docking site of the regulatory and catalytic subunits resides in this region (32). It seems that the interaction site of LOV2 to KD is located in this region of phototropin. The inhibitory effect of LOV2 on casein phosphorylation in darkness is concluded through its direct binding to KD (Fig. 4B) without a linker region between LOV2 and KD. Recently, an intactness of Jα-helix in the linker region (19) was reported to be requisite in photoregulation of Atphot1 autophosphorylation (33). Present results, in contrast, show that a dissected LOV2 domain preserves regulatory activity of the kinase by light. Our purified LOV2 domain has an amino acid sequence additional to the core of LOV2 in its C terminus; however, it did not contain the Jα-helix. The Jα-helix acts as a part of molecular switch that may assist LOV2 domain to bind KD in the dark, and the assistance may be abolished by the photoreaction of LOV2 domain. Our results suggest that the photoreversible binding capacity is in the LOV2 core and does not necessarily require the aid of the hinge region, which may be necessary for the proper interaction and photoregulation of kinase activity in an intact phototropin molecule. There is another possibility that the dispensability of Jα-helix may reflect different molecular mechanisms for the autophosphorylation and the substrate phosphorylation of phototropin.
Concluding Remarks. Atphot2 has been proven to phosphorylate a substrate other than the phototropin itself. This article proposes the presence of novel substrate(s) involved in the signal transduction, e.g., different substrates in each of photoaccumulation and photoavoidance responses of chloroplasts. In Atphot2, two LOV domains function in different ways in the in vitro phosphorylation. Major roles of LOV2 domain in the photoregulation are shown also with the substrate phosphorylation. Atphot2 regulates phototropic response of hypocotyls only at high light intensity (6), which may be explained by the attenuation of the light sensitivity by LOV1 domain. The present in vitro phosphorylation assay system is useful not only to study the photoregulation mechanisms but also to find possible in vivo substrates of Atphot2. Investigations along this line will supply important information to clarify the signal transduction pathways of phot2.
Acknowledgments
We thank Prof. W. R. Briggs of Stanford University for critically reading the manuscript and Prof. M. Iino of Osaka City University for allowing us to use a spectroradiometer. This work was supported by Japanese Ministry of Education, Culture, Sports, Science, and Technology Grant 13139205 (to S.T.).
Author contributions: D.M. and S.T. designed research; D.M. performed research and analyzed data; and S.T. wrote the paper.
Abbreviations: At, Arabidopsis thaliana; KD, kinase domain; LOV, light-oxygen-voltage-sensing; PAS, Per-Arnt-Sim.
References
- 1.Quail, P. H. (2002) Curr. Opin. Cell Biol. 140, 180-188. [DOI] [PubMed] [Google Scholar]
- 2.Briggs, W. R., Beck, C., Cashmore, A. R., Christie, J. M., Hughes, J., Jarillo, J. A., Kagawa, T., Kanegae, H., Liscum, E., Nagatani, A., et al. (2001) Plant Cell 13, 993-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggs, W. R. & Christie, J. M. (2002) Trends Plant Sci. 7, 204-210. [DOI] [PubMed] [Google Scholar]
- 4.Christie, J. M., Reymond, P., Powell, G. K., Bernasconi, P., Raibekas, A., Liscum, E. & Briggs, W. R. (1998) Science 282, 1698-1701. [DOI] [PubMed] [Google Scholar]
- 5.Kagawa, T., Sakai, T., Suetsugu, N., Oikawa, K., Ishiguro, S., Kato, T., Tabata, S., Okada, K. & Wada, M. (2001) Science 291, 2138-2141. [DOI] [PubMed] [Google Scholar]
- 6.Sakai, T., Kagawa, T., Kasahara, M., Swartz, T., Christie, J. M., Briggs, W. R., Wada, M. & Okada, K. (2001) Proc. Natl. Acad. Sci. USA 98, 6969-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarillo, G. A., Gabrys, H., Capel, J., Alonso, J. M., Ecker, J. R. & Cashmore, A. R. (2001) Nature 410, 952-954. [DOI] [PubMed] [Google Scholar]
- 8.Kinoshita, T., Doi, M., Suetsugu, N., Kagawa, T., Wada, M. & Shimazaki, K. (2001) Nature 414, 656-660. [DOI] [PubMed] [Google Scholar]
- 9.Sakamoto, K. & Briggs, W. R. (2002) Plant Cell 14, 1723-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folta, K. M. & Spalding, E. P. (2001) Plant J. 26, 471-478. [DOI] [PubMed] [Google Scholar]
- 11.Christie, J. M., Salomon, M., Nozue, K., Wada, M. & Briggs, W. R. (1999) Proc. Natl. Acad. Sci. USA 96, 8779-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller, S. M., Massy, V., Ballou, D., Williams, C. H., Jr., Distefano, M. D., Moore, M. J. & Walsh, C. T. (1990) Biochemistry 29, 2831-2841. [DOI] [PubMed] [Google Scholar]
- 13.Salomon, M., Christie, J. M., Kneib, E., Lempert, U. & Briggs, W. R. (2000) Biochemistry 39, 9401-9410. [DOI] [PubMed] [Google Scholar]
- 14.Swartz, T. E., Corchnoy, S. B., Christie, J. M., Lewis, J. W., Szundi, I., Briggs, W. R. & Bogomolni, R. A. (2001) J. Biol. Chem. 276, 36493-36500. [DOI] [PubMed] [Google Scholar]
- 15.Iwata, T., Tokutomi, S. & Kandori, H. (2002) J. Am. Chem. Soc. 124, 11840-11841. [DOI] [PubMed] [Google Scholar]
- 16.Fedorov, R., Schlichting, I., Hartmann, E., Domratcheva, T., Fuhrmann, M. & Hegemann, P. (2003) Biophys. J. 84, 2474-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crosson, S. & Moffat, K. (2001) Proc. Natl. Acad. Sci. USA 98, 2995-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crosson, S. & Moffat, K. (2002) Plant Cell 14, 1067-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harper, S. M., Neil, L. C. & Gardner, K. H. (2003) Science 301, 1541-1544. [DOI] [PubMed] [Google Scholar]
- 20.Nakasako, M., Iwata, T., Matsuoka, D. & Tokutomi, S. (2004) Biochemistry 43, 14881-14890. [DOI] [PubMed] [Google Scholar]
- 21.Crosson, S., Rajagopal, S. & Moffat, K. (2003) Biochemistry 42, 2-10. [DOI] [PubMed] [Google Scholar]
- 22.Liscum, E. & Briggs, W. R. (1995) Plant Cell 7, 473-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salomon, M., Kneib, E., von Zeppelin, T. & Rüdiger, W. (2003) Biochemistry 42, 4217-4225. [DOI] [PubMed] [Google Scholar]
- 24.Kinoshita, T., Emi, T., Tominaga, M., Sakamoto, K., Shigenaga, A., Doi, M. & Shimazaki, K. (2003) Plant Physiol. 133, 1453-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christie, J. M., Swartz, T. E., Bogomolni, R. A. & Briggs, W. R. (2002) Plant J. 32, 205-219. [DOI] [PubMed] [Google Scholar]
- 26.Hanks, S. K. & Hunter, T. (1995) FASEB J. 9, 576-596. [PubMed] [Google Scholar]
- 27.Kasahara, M., Swartz, T. E., Olney, M. A., Onodera, A., Mochizuki, N., Fukuzawa, H., Asamizu, E., Tabata, S., Kanegae, H., Takano, M., et al. (2002) Plant Physiol. 129, 762-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McTigue, M. A., Williams, D. R. & Tainer, J. A. (1995) J. Mol. Biol. 246, 21-27. [DOI] [PubMed] [Google Scholar]
- 29.Salomon, M., Lempert, U. & Rüdiger, W. (2004) FEBS Lett. 572, 8-10. [DOI] [PubMed] [Google Scholar]
- 30.Rutter, J., Michnnoff, C. H., Harper, S. M., Gardner, K. H. & McKnight, S. L. (2001) Proc. Natl. Acad. Sci. USA 98, 8991-8996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor, S. S. (1989) J. Biol. Chem. 264, 8443-8446. [PubMed] [Google Scholar]
- 32.Sahara, S., Sato, K., Kaise, H., Mori, K., Sato, A., Aoto, M., Tokmakov, A. A. & Fukami, Y. (1996) FEBS Lett. 384, 138-142. [DOI] [PubMed] [Google Scholar]
- 33.Shannon, M., Harper, J. M., Christie, J. M. & Gardner, K. H. (2004) Biochemistry 43, 16184-16192. [DOI] [PubMed] [Google Scholar]