Abstract
We have applied a newly developed real-time reverse transcriptase (RT) PCR (RT-PCR) assay for quantification of substance P (SP) mRNA expression (the SP real-time RT-PCR assay) in human blood monocyte-derived macrophages, peripheral blood lymphocytes, and microglia isolated from fetal brain. The SP real-time RT-PCR assay had a sensitivity of 60 mRNA copies, with a dynamic range of detection between 60 and 600,000 copies of the SP gene transcript per reaction mixture. The coefficient of variation of the threshold cycle number between the SP real-time RT-PCR assays was less than 1.16%. This assay with an SP-specific primer pair efficiently recognizes all four isoforms of preprotachykinin A (the SP precursor) gene transcripts. In order to use this assay to measure the levels of SP mRNA in the human immune cells quantitatively, we designed a specific probe (molecular beacon) derived from exon 3 of the SP gene. We demonstrated that the real-time RT-PCR quantitatively detected SP mRNA in the human immune cells, among which the microglia isolated from fetal brain had the highest levels of SP mRNA. The SP real-time PCR assay yielded reproducible data, as the intra-assay variation was less than 1%. Thus, it is feasible to apply the real-time RT-PCR assay for quantification of SP mRNA levels in human immune cells, as well as in other nonneuronal cells. Since SP is a major modulator of neuroimmunoregulation, this assay has the potential for widespread application for basic and clinical investigations.
Substance P (SP), a major modulator in neuroimmunoregulation, is the most studied member of the tachykinin peptide family (2, 22). SP has been described almost exclusively as a peptide of neural origin (24, 27). SP, however, has also been identified in nonneuronal cell types, including murine macrophages (3, 26), human endothelial cells (20, 23), eosinophils (1), and Leydig cells of human and mouse testes (4). We have recently demonstrated that human immune cells (monocytes, T lymphocytes, and microglia) express the SP gene and the SP receptor (12, 15, 18).
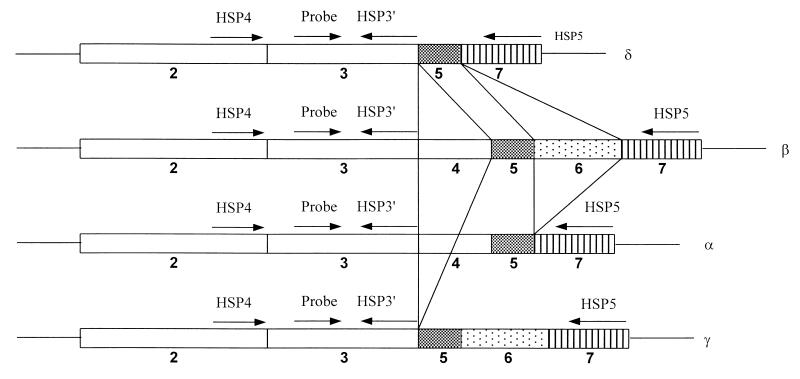
SP is synthesized from four preprotachykinin A (PPT-A) mRNAs (α, β, γ, and δ) (21). These isoforms of mRNA are the result of alternative mRNA splicing of the primary transcript of the PPT-A gene (6). The sequence in exon 3 encodes SP, while the sequence in exon 6 encodes neurokinin A (NKA). The SP sequence is encoded by all four isoforms (α, β, γ, and δ) of PPT-A mRNA, whereas the NKA precursor sequence is present only in β and γ PPT-A mRNAs. These four isoforms of PPT-A mRNA differ in their exon combinations (Fig. 1). β PPT-A mRNA contains all seven exons of the corresponding gene, while α PPT-A mRNA lacks exon 6, γ PPT-A mRNA lacks exon 4 (14, 25), and δ PPT-A mRNA lacks both exon 4 and exon 6 (6, 13).
FIG. 1.
Alternative splicing products of PPT-A mRNA transcripts. Exons 2, 3, 4, 5, 6, and 7 are indicated. Exon 3 encodes SP, and exon 6 encodes NKA. The locations and orientations (arrows) of primers HSP3′, HSP4, HSP5, and the SP MB probe are indicated.
In an attempt to identify SP gene expression in human immune cells, we have recently characterized the splicing products of the PPT-A gene transcript in human blood monocytes, monocyte-derived macrophages (MDMs) (12), lymphocytes (15), and microglia (18) by reverse transcriptase (RT) PCR (RT-PCR). We then developed a strategy of designing a specific primer pair (primer pair human SP4 [HSP4]-HSP3) to amplify SP mRNA-derived cDNA that reflects all four isoforms (α, β, γ, and δ) of PPT-A mRNA transcripts, resulting in a single 121-bp fragment that can be measured quantitatively by the SP mimic-based PCR assay (17). Establishment of the SP mimic-based PCR assay, however, is laborious and needs several dilutions from 1:10 to 10:1 (its dynamic range) for each unknown sample in order to determine a target-to-mimic ratio. Although this assay can determine the number of SP mRNA transcripts in a sample, the differences in SP mRNA copy numbers between samples must be twofold or higher. In addition, the mimic-based PCR quantification involves post-PCR manipulations which are laborious and time-consuming. Recently, a real-time PCR analysis has been developed (10, 31). The real-time PCR method solved these problems with real-time monitoring of the PCR amplification process. The monitoring of the entire reaction rather than just the end product permits the quantification to be based on the early, exponential phase of the reaction. Most importantly, the real-time system for PCR quantification is a probe-based analysis. The molecular beacon [MB] probe has been reported to be used for the construction of probes that are critical for real-time detection of nucleic acid hybridization events (31). Recent studies have successfully used the MB probe for a variety of real-time PCR applications (5, 28, 30–32). The MB probe is a single-stranded nucleic acid molecule that possesses a stem-and-loop structure. In real-time PCR, the MB probe as well as the primers hybridize to the templates during the annealing stage. As the target strands synthesized in a reaction accumulate, the fraction of the MB probe that binds to the targets increases, resulting in a brighter fluorescent signal. Here, we describe a rapid and reproducible RT-PCR method with specific SP primers and a probe that allows the quantification of SP mRNA copies in human immune cells with the ABI Prism 7700 real-time sequence detection system (the SP real-time RT-PCR assay).
MATERIALS AND METHODS
Cells.
Peripheral blood was obtained from healthy adult donors (ages, 24 to 45 years) recruited locally. The blood samples were identified as human immunodeficiency virus type 1 antibody negative by anonymous testing by the enzyme-linked immunosorbent assay method (Coulter Immunology, Hialeah, Fla.). Informed consent was obtained, and the institutional review board of the Children’s Hospital of Philadelphia has approved the present study. Blood was obtained and was used within 4 h of collection. Monocytes were purified by our previously described techniques (7, 9). Freshly isolated monocytes were plated in 24-well culture plates at a density of 106 cells/well in Dulbecco modified Eagle medium containing 10% fetal calf serum. The total length of time in culture for the MDMs was 7 to 10 days. The viabilities of the MDMs were monitored by trypan blue exclusion and cell adherence to the wells. Nonadherent peripheral blood lymphocytes (PBLs) were collected from gelatin-coated flasks, washed three times with phosphate-buffered saline, and cultured in RPMI 1640 medium containing 10% fetal calf serum and 1 μg of phytohemagglutinin P per ml for 72 h. PBL viability was measured by a cell proliferation assay. Human fetal brain microglia were isolated and cultured as described previously (8, 18). NT-2N cells were derived from Ntera2/cl.D1 (NT2) cells, a human teratocarcinoma cell line, and consisted of >95% pure neuronal cells (29). NT-2N cells were used as a positive control for SP mRNA. The Limulus amebocyte lysate assay demonstrated that all media and reagents were free of endotoxin.
RNA extraction.
Total RNA was extracted from MDMs, PBLs (stimulated with phytohemagglutinin P for 72 h), fetal brain microglia, and NT-2N cells (106 cells) with Tri-Reagent (Molecular Research Center, Cincinnati, Ohio), as instructed by the manufacturer. In brief, the total RNA was extracted in a single step by guanidinium thiocyanate-phenol-chloroform extraction. After centrifugation at 13,000 × g for 15 min, the RNA-containing aqueous phase was precipitated in isopropanol. The RNA precipitates were then washed once in 75% ethanol and resuspended in 50 μl of RNase-free water.
Cloning of PPT-A cDNA.
The four isoforms of PPT-A mRNA were cloned and identified with primer pairs HSP4-HSP5 and HSP4-HSP7, as reported earlier (16, 17) (Fig. 1). Briefly, the PCR products amplified with these primers were separated on a 4% agarose gel and then purified with a Wizard PCR Preps DNA purification system (Promega, Madison, Wis.). The purified cDNAs of the PPT-A isoforms (α, β, γ, δ) were then cloned into a plasmid with a Eukaryotic TA cloning kit (Invitrogen Corporation, San Diego, Calif.). The cloned plasmids containing the cDNAs of the PPT-A isoforms were then purified with a Wizard Plus Minipreps DNA purification system (Promega). The presence and orientation of the PPT-A isoform inserts were determined by restriction analysis by using EcoRV digestion and DNA sequencing. The purified plasmids were linearized by EcoRI restriction enzyme digestion and were purified by phenol-chloroform extraction and alcohol precipitation. These cloned PPT-A isoform-containing plasmids were used as a standard control for determination of the accuracy of the quantitative real-time PCR and as a template for the synthesis of mRNA in vitro to evaluate the sensitivity of the real-time RT-PCR and to make a standard curve for the real-time RT-PCR.
In vitro mRNA synthesis.
PPT-A mRNA transcripts were obtained by transcription from the plasmid containing the β-isoform cDNA insert with a MEGAshortscript kit (Ambion, Austin, Tex.). After digestion with RNase-free DNase (Promega), the resulting RNA transcripts were purified by phenol-chloroform extraction and alcohol precipitation as reported previously (16, 17). The purified mRNA was used as a standard to quantitatively measure the SP mRNA transcripts in MDMs, PBLs, and the microglia by the real-time RT-PCR with primer pair HSP4-HSP3′.
Design of molecular beacons and primers.
The PCR primers and the MB probes used were designed with Primer Express software (PE Applied Biosystems) and were synthesized by Integrated DNA Technologies, Inc. (Coralville, Iowa). The HSP4-HSP3′ primer pair (HSP4, 5′-CGACCAGATCAAGGAGGAACTG-3′; HSP3′, 5′-CAGCATCCCGTTTGCCCATT-3′), which is specific for a 121-bp fragment of the SP transcript, was described previously (17), but with modification by the addition of 4 additional nucleotides at the 3′ end of primer HSP3 (underlined) because of the design obtained with the Primer Express software. The sequences of the primers and the MB probe were selected from the sequences of exons 2 and 3 of the PPT-A gene (16) (Fig. 1) so that they amplify a single cDNA fragment that represents the total SP from transcripts of all four isoforms of PPT-A mRNA (17). The primer pair specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is 5′-GGTGGTCTCCTCTGACTTCAACA-3′ (sense) and 5′-GTTGCTGTAGCCAAATTCGTTGT-3′ (antisense). The sequences of the MB probes specific for SP and GAPDH were designed to be perfectly complementary to the target sequence in exon 3 of the SP gene and to be complementary to the sequence between the sequence of each primer in the primer pair for GAPDH, respectively. The following are the sequences of the two MB probes: SP, 5′-FAM-GCGAGCAGA-ATCGCCCGGAGACCCAAGCGCTCGC-DABCYL-3′; GAPDH, 5′-FAM-GCGAGCC-TGGCATTGCCCTCAACGACCACGCTCGC-DABCYL-3′. The underlined sequences are the stem sequences, FAM is 6-carboxyfluorescein, and DABCYL is 4-(4′-dimethylaminophenylaso) benzoic acid. The stem sequences were selected such that they would not complement the sequences within the loop region. The lengths of the MB probes were designed such that the annealing temperature is slightly higher than those for the PCR primers. The MB probes were labeled at the 5′ end with FAM and the quencher DABCYL at the 3′ end. The primers and MB probes were resuspended in TE (Tris-EDTA) buffer and stored at −30°C.
Reverse transcription.
Total RNA (1 μg) was subjected to reverse transcription with a reverse transcription system (Promega) with random primer p(dN)6 (Roche Molecular Biochemicals, Indianapolis, Ind.) and specific primer (antisense) HSP5 (5′-GCATTGCACTCCTTTCATAAGCCA-3′) for 1 h at 42°C. The reaction was terminated by incubation of the reaction mixture at 99°C for 5 min. One-tenth (100 ng) of the resulting cDNA was used as a template for PCR amplification.
Real-time PCR assay.
The Perkin-Elmer ABI Prism 7700 sequence detection system was used for real-time analysis. Thermal cycling conditions were designed as follows: initial denaturation at 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. Fluorescence measurements were recorded during each annealing step. At the end of each PCR run, the data were automatically analyzed by the system and amplification plots were obtained. For each PCR, 2 μl of cDNA template was added to 48 μl of the PCR master mixture (5 μl of 1× PCR buffer II, 5 mM MgCl2, 300 nM deoxynucleoside triphosphates, 400 nM each primer, 1.5 U of AmpliTaq Gold DNA polymerase, 400 nM MB probes, 24.7 μl of water). The PCR buffer contained 5-carboxy-X-rhodamine (ROX) (500 nM) as the reference dye for normalization of the reactions. Any possible fluctuations in the ROX signal are used to correct the sample signal. All amplification reactions were performed in duplicate. To control for the recovery of intact cellular RNA and for the uniform efficiency of each reverse transcription reaction, a GAPDH fragment was amplified by real-time RT-PCR on the same plate as that with SP cDNA. A standard curve was generated with 10-fold dilutions of total RNA quantified with a spectrophotometer. The final results were expressed as the mean number of copies of SP mRNA per microgram of total RNA. In order to prepare a standard curve for SP mRNA, 10-fold serial dilutions of β-isoform PPT-A RNA transcripts ranging from 6 × 101 to 6 × 106 copies/2 μl were amplified.
Amplification efficiency.
The efficiency of real-time amplification was determined by running a standard curve with serial dilutions of a known amount of cloned SP plasmid cDNA. A PCR that amplifies the target sequence with 100% efficiency (E) can double the amount of PCR products in each cycle. The amount of PCR products (Y) from input target molecules (Z) after n cycles can be calculated by the formula s = −1/log(1 + E) (where s is the slope), and after transformation, E is equal to 101/−s − 1 (19).
RESULTS
Sensitivity of real-time RT-PCR.
The analytical sensitivity of the real-time RT-PCR was determined with a series of dilutions of PPT-A β-isoform RNA transcripts containing 0, 6 × 101, 6 × 102, 6 × 103, 6 × 104, 6 × 105, and 6 × 106 molecules. Each assay was performed five times, in duplicate. The real-time RT-PCR could detect as few as 6 SP mRNA copies, although the detection rate was only 60% (6 of 10 replicates) (data not shown). The detection rate, however, was 100% for PPT-A mRNA copy numbers of 60 or higher (10 of 10 replicates). The detection limit, therefore, was 60 RNA molecules per reaction mixture. A representative result is shown in Fig. 2.
FIG. 2.
Sensitivity and linearity of real-time RT-PCR with RNA transcripts derived from plasmid with β-isoform mRNA. A reading of the change in fluorescence (Rn) as a function of the cycle number is demonstrated for a range of known input numbers of copies of the SP RNA transcript. Tenfold serial dilutions of the RNA starting from 60 to 6 × 106 molecules per reaction mixture were amplified by the real-time RT-PCR. (A) Standard curve of serial dilutions of SP RNA (R2 = 0.98). (B) Amplification plot of serial dilutions of SP RNA showing the dynamic detection range of 6 orders of magnitude from 60 to 6 × 106 molecules and the sensitivity of detection of 60 SP mRNA copies per reaction mixture. NC, negative control which lacked PCR-amplified product when RT was omitted from the reverse transcription reaction with 6 × 106 molecules of SP mRNA transcripts.
Linearity, range of quantification, and precision.
Amplification of PPT-A β-isoform RNA transcripts at different concentrations showed linearity over a range of 6 orders of magnitude (Fig. 2), and R2 was 0.98. In order to determine the variations in repetitive measurements of the real-time PCR between different runs, 10-fold serial dilutions of PPT-A β-isoform cDNA-containing plasmids (range, 6 × 101 to 6 × 104 copies per reaction mixture) were examined by real-time PCR in five different experiments. The coefficients of variation (CVs) of the values of the threshold cycle number (CT) between runs were between 0.54 and 1.16% (Table 1 and Fig. 3). The intrarun CV was even lower than the interassay CV (data not shown).
TABLE 1.
Reproducibility of SP real-time PCR assaya
| Run |
CT for the following no. of input copiesb:
|
|||
|---|---|---|---|---|
| 60 | 600 | 6,000 | 60,000 | |
| 1 | 33.38 | 29.94 | 26.37 | 23.05 |
| 2 | 33.44 | 29.72 | 26.56 | 23.46 |
| 3 | 33.97 | 29.68 | 26.27 | 23.58 |
| 4 | 33.45 | 29.58 | 26.60 | 22.70 |
| 5 | 32.88 | 29.36 | 26.56 | 23.42 |
| Mean ± SD | 33.42 ± 0.38 | 29.66 ± 0.21 | 26.47 ± 0.14 | 23.44 ± 0.24 |
| CV(%) | 1.16 | 0.71 | 0.54 | 1.05 |
The data are for five separate assays performed on different days.
Input numbers of copies of plasmid DNA with the β isoform of PPT-A.
FIG. 3.
Standard curves of PPT-A β-isoform plasmid cDNA. The curve was made on the basis of the data from Table 1, and the CT values are the means for five runs. Tenfold serial dilutions of the cDNA ranging from 60 to 6 × 104 copies per reaction mixture were amplified by the real-time PCR. The correlation coefficient was 0.9973.
Amplification efficiency of different isoforms of PPT-A gene.
In order to determine whether SP can be amplified from transcripts of all four isoforms of PPT-A mRNA by the real-time PCR with similar efficiencies, each of the four plasmids that contained the PPT-A α, β, δ, and γ isoform sequences, respectively, was used as a template for the real-time PCR. As shown in Fig. 4 and Table 2, the SP sequence was efficiently amplified by the real-time PCR from plasmids containing all four isoforms of PPT-A. The slopes of the four curves were generated to compare the efficiencies of amplification among the four isoforms of PPT-A cDNA. The efficiencies of the PCR amplifications for these isoforms were between 98 and 110% (Table 2).
FIG. 4.
Real-time PCR amplification plot of the four isoforms of PPT-A cDNA. A reading of a change in fluorescence (Rn) as a function of the cycle numbers is demonstrated for a range of known input copy numbers of plasmids containing the four isoforms of PPT-A. Tenfold serial dilutions of the four plasmids were amplified. The input amounts of targets were 30 to 3 × 106 copies per reaction mixture for plasmids containing the α, β, γ, and δ isoforms of PPT-A, as indicated. NC, negative control, which lacked PCR-amplified product when the template was omitted.
TABLE 2.
Amplification efficiencies and slopes for four isoforms of PPT-A
| PPT-A isoform | % Efficiency | Slope | R2 |
|---|---|---|---|
| α | 103 | 3.25 | 0.9995 |
| β | 98 | 3.37 | 0.9991 |
| γ | 101 | 3.29 | 0.9988 |
| δ | 110 | 3.10 | 0.9983 |
Real-time RT-PCR detection of SP mRNA.
In order to measure the expression of SP mRNA in NT-2N cells, MDMs, PBLs, and microglia, total RNAs isolated from these cells from different healthy donors and a brain specimen were reverse transcribed. The cDNAs were then amplified by the real-time PCR with the specific primers and MB probes. MDMs, PBLs, and the microglia expressed SP mRNA transcripts, as shown in Table 3. As expected, the neuronal (NT-2N) cells express much higher levels of SP mRNA than human immune cells. The cultured fetal brain-derived microglia expressed higher levels of SP mRNA compared with the amount expressed by blood MDMs (Table 3). In order to prove the reproducibility of the SP real-time PCR assay with these immune cells, four cDNA samples from each cell type (PBLs, MDMs, microglia, and NT-2N cells) were run in the assay. The data in Table 4 demonstrate that the SP real-time PCR has excellent reproducibility, with an intra-assay variability of less than 1%.
TABLE 3.
Quantification of SP mRNA in human immune cells
| Expt no. | No. of SP mRNA copies/μg total of RNAa
|
|||
|---|---|---|---|---|
| PBLs | MDMs | Microglia | NT-2N cells | |
| 1 | 1,394 | 7,280 | 198,811 | 10,523,409 |
| 2 | 1,006 | 949 | 310,405 | 13,872,522 |
| 3 | 40,093 | 113,388 | 464,632 | NT |
| 4 | 4,431 | 3,497 | 435,042 | NT |
| 5 | NT | NT | 137,987 | NT |
Human immune cells (PBLs, MDMs, and microglia) were from five different samples. The SP mRNA levels were normalized on the basis of the SP mRNA/GAPDH mRNA ratio obtained by the real-time PCR. NT, not tested.
TABLE 4.
Intra-assay reproducibility of SP real-time PCR assaya
| Repetition |
CT
|
|||
|---|---|---|---|---|
| PBLs | MDMs | Microglia | NT-2N cells | |
| 1 | 39.71 | 39.78 | 30.86 | 26.0 |
| 2 | 39.82 | 39.67 | 30.86 | 25.65 |
| 3 | 39.75 | 39.55 | 30.75 | 25.67 |
| 4 | 39.80 | 39.51 | 30.81 | 25.58 |
| Mean ± SD | 39.77 ± 0.05 | 39.63 ± 0.12 | 30.82 ± 0.05 | 25.73 ± 0.19 |
| CV (%) | 0.12 | 0.31 | 0.17 | 0.73 |
Four cDNA samples from each cell type (PBLs, MDMs, microglia, and NT-2N cells) were amplified by the SP real-time PCR assay.
DISCUSSION
We have recently identified and characterized four isoforms (α, β, γ, and δ) of PPT-A mRNA in human immune cells (12, 15, 16, 18). In order to determine the total SP mRNA transcript levels, we developed a mimic-based PCR to amplify SP mRNA from all four isoforms of PPT-A mRNA transcripts in a single cDNA product with the HSP4-HSP3 primer pair (17). The HSP3 primer sequence is from within the exon 3 sequence of the SP gene (16) (Fig. 1). However, quantification of SP mRNA levels by the mimic-based PCR can be problematic due to the exponential nature of PCR analyses, since small variations in amplification efficiency can lead to dramatic changes in product yields, which obscures differences in the levels of SP mRNA during amplification. This is particularly true for the quantification of low levels of SP mRNA in human immune cells. In the present study, we have successfully used the real-time PCR for the quantification of SP mRNA in human MDMs, PBLs, and microglia (Table 3). The microglia had the highest levels of SP mRNA transcripts among the three types of cells tested, which is in agreement with our previous comparisons with these cell types by mimic-based PCR (18).
The ability to monitor the real-time progress of the amplification completely revolutionizes the PCR-based quantification of DNA and RNA. In real-time PCR, reactions are characterized by the point in time during cycling when the amplification of a PCR product is first detected rather than the amount of PCR product accumulated at the end of the entire PCR process. The higher the numbers of copies of the nucleic acid target at the start of the reaction, the sooner a significant increase in fluorescence is observed (11) and the lower the CT value is. Real-time PCR allows the CT value to be observed when the PCR amplification is still in the exponential phase. Therefore, the CT value is a more reliable measurement of the number of copies of mRNA or DNA at the start of the reaction.
Thus, we have developed an MB probe-based real-time RT-PCR assay for the quantification of SP mRNA copy numbers in human immune cells. We designed an SP-specific MB probe and a pair of SP-specific primers to target the sequence of a single cDNA fragment that reflects total SP mRNA from all four isoforms of PPT-A mRNA. Since it has a wide dynamic detection range (6 × 101 to 6 × 105 copies per reaction), there is no need to dilute or concentrate the samples, which was one of the problems encountered with the mimic-based SP PCR (17). This SP MB-based RT-PCR is much less cumbersome, and the costs associated with the use of this system are the same as those associated with the use of the regular RT-PCR. Because this system allows processing of multiple samples with minimal labor time, the risk of carryover contamination due to post-PCR sample manipulation is minimal. In addition, this assay is highly specific, since the assay uses both specific primers and a probe (the MB probe) to identify SP mRNA transcripts in human immune cells. The SP MB probe-based real-time PCR has an excellent reproducibility (Tables 1 and 4). Furthermore, all four isoforms of PPT-A cDNA in cloned plasmids were efficiently amplified by the SP real-time PCR assay (Fig. 4; Table 2). Although the SP real-time PCR assay that we have developed can amplify cDNA that reflects transcripts of all four isoforms of SP mRNA, it is not feasible to amplify a transcript of a particular SP mRNA isoform. This limitation is due to the PPT-A genome structure and the combination of the seven exons for each isoform of PPT-A mRNA (Fig. 1) (17). However, the assay with different primer pairs has the ability to amplify mRNA of the PPT-A α and δ isoforms or mRNA of the β and γ isoforms (17).
In summary, we have successfully used a newly developed MB probe-based RT-PCR assay to quantitatively measure the total levels of SP mRNA transcripts in both human neural cells and immune cells. Our data demonstrate that this method is precise, sensitive, highly reproducible, and particularly useful for the quantification of SP mRNA levels in nonneuronal cells that have very low levels of expression of the SP gene. This method has the potential for widespread application in basic and clinical studies in neuroimmunology.
Acknowledgments
This work was supported by NIH grant MH 49981 (to S.D.D.) and NIH grant DA12815 (to W.-Z.H.).
REFERENCES
- 1.Aliakbari, J., S. P. Sreedharan, C. W. Turck, and E. J. Goetzl. 1987. Selective localization of vasoactive intestinal peptide and substance P in human eosinophils. Biochem. Biophys. Res. Commun. 148:1440–1445. [DOI] [PubMed] [Google Scholar]
- 2.Black, P. H. 1994. Immune system-central nervous system interactions: effect and immunomodulatory consequences of immune system mediators on the brain. Antimicrob. Agents Chemother. 38:7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bost, K. L., S. A. Breeding, and D. W. Pascual. 1992. Modulation of the mRNAs encoding substance P and its receptor in rat macrophages by LPS. Reg. Immunol. 4:105–112. [PubMed] [Google Scholar]
- 4.Chiwakata, C., B. Brackmann, N. Hunt, M. Davidoff, W. Schulze, and R. Ivell. 1991. Tachykinin (substance-P) gene expression in Leydig cells of the human and mouse testis. Endocrinology 128:2441–2448. [DOI] [PubMed] [Google Scholar]
- 5.Giesendorf, B. A., J. A. Vet, S. Tyagi, E. J. Mensink, F. J. Trijbels, and H. J. Blom. 1998. Molecular beacons: a new approach for semiautomated mutation analysis. Clin. Chem. 44:482–486. [PubMed] [Google Scholar]
- 6.Harmar, A. J., V. Hyde, and K. Chapman. 1990. Identification and cDNA sequence of delta-preprotachykinin, a fourth splicing variant of the rat substance P precursor. FEBS Lett. 275:22–24. [DOI] [PubMed] [Google Scholar]
- 7.Hassan, N. F., D. E. Campbell, and S. D. Douglas. 1986. Purification of human monocytes on gelatin-coated surfaces. J. Immunol. Methods 95:273–276. [DOI] [PubMed] [Google Scholar]
- 8.Hassan, N. F., D. E. Campbell, S. Rifat, and S. D. Douglas. 1991. Isolation and characterization of human fetal brain-derived microglia in in vitro culture. Neuroscience 41:149–158. [DOI] [PubMed] [Google Scholar]
- 9.Hassan, N. F., J. R. Cutilli, and S. D. Douglas. 1990. Isolation of highly purified human blood monocytes for in vitro HIV-1 infection studies of monocyte/macrophages. J. Immunol. Methods 130:283–285. [DOI] [PubMed] [Google Scholar]
- 10.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986–994. [DOI] [PubMed] [Google Scholar]
- 11.Higuchi, R., C. Fockler, G. Dollinger, and R. Watson. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Bio/Technology 11:1026–1030. [DOI] [PubMed] [Google Scholar]
- 12.Ho, W. Z., J. P. Lai, X. H. Zhu, M. Uvaydova, and S. D. Douglas. 1997. Human monocytes and macrophages express substance P and neurokinin-1 receptor. J. Immunol. 159:5654–5660. [PubMed] [Google Scholar]
- 13.Khan, I., and S. M. Collins. 1994. Fourth isoform of preprotachykinin messenger RNA encoding for substance P in the rat intestine. Biochem. Biophys. Res. Commun. 202:796–802. [DOI] [PubMed] [Google Scholar]
- 14.Krause, J. E., J. M. Chirgwin, M. S. Carter, Z. S. Xu, and A. D. Hershey. 1987. Three rat preprotachykinin mRNAs encode the neuropeptides substance P and neurokinin A. Proc. Natl. Acad. Sci. USA 84:881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai, J.-P., S. D. Douglas, and W.-Z. Ho. 1998. Human lymphocytes express substance P and its receptor. J. Neuroimmunol. 86:80–86. [DOI] [PubMed] [Google Scholar]
- 16.Lai, J. P., S. D. Douglas, E. Rappaport, J. M. Wu, and W. Z. Ho. 1998. Identification of a delta isoform of preprotachykinin mRNA in human mononuclear phagocytes and lymphocytes. J. Neuroimmunol. 91:121–128. [DOI] [PubMed] [Google Scholar]
- 17.Lai, J. P., S. D. Douglas, M. Zhao, and W. Z. Ho. 1999. Quantification of substance P mRNA in human mononuclear phagocytes and lymphocytes using a mimic-based RT-PCR. J. Immunol. Methods 230:149–157. [DOI] [PubMed] [Google Scholar]
- 18.Lai, J. P., G. X. Zhan, D. E. Campbell, S. D. Douglas, and W. Z. Ho. 2000. Detection of substance P and its receptor in human fetal microglia. Neuroscience 101:1137–1144. [DOI] [PubMed] [Google Scholar]
- 19.Leutenegger, C. M., J. Higgins, T. B. Matthews, A. F. Tarantal, P. A. Luciw, N. C. Pedersen, and T. W. North. 2001. Real-time TaqMan PCR as a specific and more sensitive alternative to the branched-chain DNA assay for quantitation of simian immunodeficiency virus RNA. AIDS Res. Hum. Retrovir. 17:243–251. [DOI] [PubMed] [Google Scholar]
- 20.Linnik, M. D., and M. A. Moskowitz. 1989. Identification of immunoreactive substance P in human and other mammalian endothelial cells. Peptides 10:957–962. [DOI] [PubMed] [Google Scholar]
- 21.MacDonald, M. R., D. W. McCourt, and J. E. Krause. 1988. Posttranslational processing of alpha-, beta-, and gamma-preprotachykinins. Cell-free translation and early posttranslational processing events. J. Biol. Chem. 263:15176–15183. [PubMed] [Google Scholar]
- 22.McGillis, J. P., M. Mitsuhashi, and D. G. Payan. 1990. Immunomodulation by tachykinin neuropeptides. Ann. N. Y. Acad. Sci. 594:85–94. [DOI] [PubMed] [Google Scholar]
- 23.Milner, P., V. Ralevic, A. M. Hopwood, E. Feher, J. Lincoln, K. A. Kirkpatrick, and G. Burnstock. 1989. Ultrastructural localisation of substance P and choline acetyltransferase in endothelial cells of rat coronary artery and release of substance P and acetylcholine during hypoxia. Experientia 45:121–125. [DOI] [PubMed] [Google Scholar]
- 24.Nakanishi, S. 1987. Substance P precursor and kininogen: their structures, gene organizations, and regulation. Physiol. Rev. 67:1117–1142. [DOI] [PubMed] [Google Scholar]
- 25.Nawa, H., H. Kotani, and S. Nakanishi. 1984. Tissue-specific generation of two preprotachykinin mRNAs from one gene by alternative RNA splicing. Nature 312:729–734. [DOI] [PubMed] [Google Scholar]
- 26.Pascual, D. W., and K. L. Bost. 1990. Substance P production by P388D1 macrophages: a possible autocrine function for this neuropeptide. Immunology 71:52–56. [PMC free article] [PubMed] [Google Scholar]
- 27.Pernow, B. 1983. Substance P. Pharmacol. Rev. 35:85–141. [PubMed] [Google Scholar]
- 28.Piatek, A. S., S. Tyagi, A. C. Pol, A. Telenti, L. P. Miller, F. R. Kramer, and D. Alland. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359–363. [DOI] [PubMed] [Google Scholar]
- 29.Pleasure, S. J., C. Page, and V. M. Lee. 1992. Pure, postmitotic, polarized human neurons derived from NTera 2 cells provide a system for expressing exogenous proteins in terminally differentiated neurons. J. Neurosci. 12:1802–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49–53. [DOI] [PubMed] [Google Scholar]
- 31.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303–308. [DOI] [PubMed] [Google Scholar]
- 32.Vet, J. A., A. R. Majithia, S. A. Marras, S. Tyagi, S. Dube, B. J. Poiesz, and F. R. Kramer. 1999. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl. Acad. Sci. USA 96:6394–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]