Abstract
Autoimmune disorders constitute a diverse group of phenotypes with overlapping features and a tendency toward familial aggregation. It is likely that common underlying genes are involved in these disorders. Until very recently, no specific alleles—aside from a few common human leukocyte antigen class II genes—had been identified that clearly associate with multiple different autoimmune diseases. In this study, we describe a unique collection of 265 multiplex families assembled by the Multiple Autoimmune Disease Genetics Consortium (MADGC). At least two of nine “core” autoimmune diseases are present in each of these families. These core diseases include rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), type 1 diabetes (T1D), multiple sclerosis (MS), autoimmune thyroid disease (Hashimoto thyroiditis or Graves disease), juvenile RA, inflammatory bowel disease (Crohn disease or ulcerative colitis), psoriasis, and primary Sjögren syndrome. We report that a recently described functional single-nucleotide polymorphism (rs2476601, encoding R620W) in the intracellular tyrosine phosphatase (PTPN22) confers risk of four separate autoimmune phenotypes in these families: T1D, RA, SLE, and Hashimoto thyroiditis. MS did not show association with the PTPN22 risk allele. These findings suggest a common underlying etiologic pathway for some, but not all, autoimmune disorders, and they suggest that MS may have a pathogenesis that is distinct from RA, SLE, and T1D. DNA and clinical data for the MADGC families are available to the scientific community; these data will provide a valuable resource for the dissection of the complex genetic factors that underlie the various autoimmune phenotypes.
Introduction
Autoimmune diseases share a number of characteristics that suggest common etiologic pathways or mechanisms, including reactivity to self-antigens by the humoral and/or cellular immune system, as well as genetic associations with human leukocyte antigen (HLA). Although familial clustering of autoimmunity has long been recognized, the patterns of aggregation across different autoimmune diseases have not been clearly delineated. Certain rare familial clusters of autoimmune phenotypes, such as the polyglandular autoimmune endocrinopathies, are known to have a common genetic basis (Eisenbarth and Gottlieb 2004). Among the more common autoimmune disorders, probably the best evidence for familial clustering among different autoimmune diseases involves rheumatoid arthritis (RA [MIM 180300]), autoimmune thyroid disease (AITD) (especially Hashimoto thyroiditis [MIM 140300]), and type 1 diabetes (T1D [MIM 222100]) (Torfs et al. 1986; Lin et al. 1998). However, the degree and significance of familial clustering among the larger group of autoimmune disorders is uncertain; only a few large or well-controlled epidemiological studies of this issue have been reported (Torfs et al. 1986; Ginn et al. 1998; Lin et al. 1998; Broadley et al. 2000), in part, because many of these diseases are relatively uncommon and have heterogeneous phenotypes that are difficult to confirm in population surveys.
Clustering of diseases within families may be explained by shared environmental exposures, shared genes, or interactions between genetic and environmental factors. A growing body of evidence suggests that exposure to cigarette smoke, for example, may increase the risk of multiple autoimmune diseases, including AITD (Vestergaard 2002), RA (Heliovaara et al. 1993; Symmons et al. 1997; Uhlig et al. 1999), systemic lupus erythematosus (SLE [MIM 152700]) (Hardy et al. 1998; Bengtsson et al. 2002), and multiple sclerosis (MS [MIM 126200]) (Riise et al. 2003). Exposure to crystalline silica (Steenland and Goldsmith 1995) and to Epstein-Barr virus infection (Harley and James 1999; James et al. 2001) also appears to influence the risk of multiple autoimmune diseases. In addition to these environmental factors, there is now increasing evidence that common genes may underlie autoimmunity. Becker and colleagues (1998) have described a pooled analysis of genome-screening results and have identified 18 distinct clusters with evidence of genetic linkage to two or more autoimmune or immune disorders. However, evidence that specific risk alleles are associated with multiple autoimmune diseases is relatively sparse. Even within the HLA system, different alleles are associated with different autoimmune phenotypes (Todd et al. 1988; Hall et al. 1996; Tan and Arnett 1998; Barcellos et al. 2003), and, indeed, it is not always clear that the HLA alleles are the only relevant risk factor within the major histocompatibility complex (MHC) (Jawaheer et al. 2002). Until very recently, CTLA4 was the only gene that had been convincingly associated with two different autoimmune diseases—AITD and T1D—but the strength of these associations is rather modest, with odds ratios (ORs) of ∼1.45 and ∼1.15, respectively (Ueda et al. 2003).
In this study, we describe a unique registry of 265 multiplex families that have been specifically identified as having two or more different autoimmune diseases in the same family. Detailed clinical data for each family member—along with a blood sample—have been collected by the Multiple Autoimmune Disease Genetics Consortium (MADGC), under a contract with the National Institute of Allergy and Infectious Diseases (NIAID). Genetic analysis of these families reveals that PTPN22 is associated with several different autoimmune disorders, confirming and extending several recent reports (Begovich et al. 2004; Bottini et al. 2004; Kyogoku et al. 2004). For the first time, we demonstrate an association with Hashimoto thyroiditis, a common form of AITD. This work establishes PTPN22 as the first major genetic variant that clearly confers substantial risk of multiple different autoimmune phenotypes, and it points the way to understanding common disease mechanisms. The MADGC collection of multiplex families is likely to provide an important scientific resource for further studies in this area.
Material and Methods
Multiplex Families: Entry Criteria and Approach to Recruitment
At the request of NIAID, investigators with expertise in the genetics of a wide range of autoimmune diseases were asked to serve as consultants for this effort and met with the study investigators at the National Institutes of Health on November 1, 1999 (see the “Acknowledgments” section for the names of participants). The primary goal of that meeting was to determine which autoimmune diseases should be included in the collection, on the basis of a variety of considerations, including strength of evidence for genetic susceptibility, clustering with other autoimmune diseases, population prevalence, impact on public health, and feasibility of collection. Eight “core” autoimmune diseases were targeted initially: adult RA, SLE, MS, AITD (Graves disease [MIM 275000] or Hashimoto thyroiditis), T1D, psoriasis (MIM 177900), inflammatory bowel disease (IBD [MIM 266600]) (Crohn disease or ulcerative colitis), and juvenile rheumatoid arthritis (JRA [MIM 604302]). Subsequently, primary Sjögren syndrome (SS [MIM 270150]) was added to this core list of diseases. To be eligible for inclusion in the registry/repository, the multiplex families were required to have at least two members affected with at least two of these nine core diseases. Furthermore, at least two of the affected members had to be informative for linkage (e.g., a single affected parent/offspring pair would not meet enrollment criteria).
Families were recruited from a variety of sources, including patient advocacy groups and publications (e.g., the American Autoimmune Related Diseases Association [AARDA] and the Multiple Sclerosis Quarterly Report), as well as other forms of publicity and direct mailing of flyers and posters to physicians’ offices. Once a family was deemed eligible for inclusion, all individuals affected with any autoimmune disease (see the MADGC Web site) were invited to participate, as were all unaffected individuals who were likely to be useful for genetic linkage or association studies. Informed consent was obtained from all enrolled subjects, and the study was approved by the institutional review boards at each of the three participating centers: North Shore University Hospital, University of Minnesota, and University of California San Francisco (UCSF).
Demographic and Clinical Information
Basic demographic information was collected from all enrolled subjects via telephone or mailed questionnaire. All subjects were also asked about the presence of any of the nine core diseases or other autoimmune diseases. Subjects who reported at least one of these conditions provided additional clinical information over the telephone, including date of diagnosis; name, address, and specialty of treating physician(s); and a history of the symptoms and treatment received for the disease(s). Specific lists of symptoms and treatments were developed for each of the nine core diseases with the assistance of the aforementioned consultants; these lists are available either by request from the authors or on the MADGC Web site. For other autoimmune diseases (i.e., noncore diseases), similar information was collected using a standardized form that was designed to be more general.
Confirmation of Autoimmune Disease Diagnoses
Autoimmune disease diagnoses were confirmed either by the treating physician or on the basis of a review of medical records by one of the study investigators or consultants. Criteria for diagnostic confirmation were those established either by the appropriate professional body (e.g., American College of Rheumatology) or on the advice of consultants. Standardized diagnostic confirmation forms were developed for each of the nine core diseases and were completed by the confirming physician/investigator. For use in documenting diagnosis of other autoimmune diseases, a more general form was developed for completion by the study physician/investigator. All subjects who reported an autoimmune disease that could not be confirmed were considered “unknown” for that disease, unless it was clear from the available information that the disease was not present. Subjects could be confirmed for more than one autoimmune disease, or they could be confirmed for one or more diseases and considered unknown for other autoimmune diseases.
Collection of Biological Material
All enrollees were asked to provide blood for processing and storage at a centralized biorepository located at the North Shore Long Island Jewish Research Institute. Samples were processed and were stored as plasma and as frozen viable peripheral blood mononuclear cells (PBMCs). After shipment of frozen PBMCs in batches, immortalized lymphoblastoid cell lines were successfully established from the majority of samples at a second site (University of Minnesota). As a security precaution, all samples are stored in at least two geographically separate sites (Manhasset and Minneapolis).
White control subjects (n=2,064) were derived from several sample sets. The PTPN22 genotyping data for the majority of these control subjects have all been reported elsewhere (Begovich et al. 2004; Kyogoku et al. 2004). Additional PTPN22 typing for this study was performed on 103 white subjects derived from the New York Cancer Project collection (Mitchell et al. 2004).
Genotyping
To confirm family relationships, all samples were genotyped, as described elsewhere (Baechler et al. 2003), for a set of ∼100 polymorphic markers (ABI, version 2.5; marker sets 1–7) and were analyzed using the programs Relative (Göring and Ott 1997) and PedCheck (O’Connell and Weeks 1998). HLA-DR typing was performed at Roche Molecular Systems, as described elsewhere (Erlich et al. 1991). Genotyping of the PTPN22 SNP (rs2476601, 1858C→T, R620W) was performed using a PSQ HS 96A Pyrosequencer. A quantity of 2 ng of DNA was amplified by PCR in a 10-μl reaction by use of the following primers: 5′-GTTGCGCAGGCTAGTCTTGA-3′ (forward), 5′-GCTGCTCCGGTTCATAGATTGGATAGCAACTGCTCCAAGG-3′ (reverse), and 5′-Biotin-GCTGCTCCGGTTCATAGATT-3′ (Univ1_B). The addition of specific sequences to the 5′ end of the reverse primer (shown in bold italics) allowed the use of a biotinylated universal primer, Univ1_B. These primers were used at a ratio of 1:9 (reverse primer:universal primer). PCR conditions were as follows: 95°C for 12 min; 50 cycles at 95°C for 45 s, at 56.4°C for 45 s, and at 72°C for 45 s; and 72°C for 10 min. The amplicon was denatured with NaOH, separated, washed, and neutralized. To detect the polymorphism, the sequencing primer 5′-AAATGATTCAGGTGTCC-3′ was used in combination with appropriate Pyrosequencing substrates and enzymes, in accordance with manufacturer instructions. Genotyping of PTPN22 R620W on control subjects was performed as described elsewhere (Begovich et al. 2004).
Data Management and Statistical Analysis
All demographic and clinical information was entered into a Microsoft Access database. Pedigrees were created for each family by use of the program Cyrillic, version 2.1 (Cherwell Scientific), and the pedigree data were imported into the Access database. These core data are also available upon request as downloadable Microsoft Excel files (see the MADGC Web site). Information about biological material—such as sample status, sample location, and genotyping results—is managed with a custom-designed relational database in SQL at the North Shore Long Island Jewish Research Institute.
Basic summary statistics were generated using the program SAS, version 8.02 (SAS Institute). ORs and 95% CIs were computed to describe associations between specific autoimmune diseases (and groups of autoimmune diseases) and the presence of the minor allele of PTPN22, on the basis of a comparison with control subjects. All of the affected individuals in MADGC families were used in these case-control association analyses. Since affected siblings obtained from the same parental mating type exhibit the same genotype and allele frequencies (W.L. and P.K.G., unpublished results), the inclusion of more than one affected sibling per family will not bias the estimates of ORs, as long as no particular parental mating type is over- or underrepresented in the sample. The inclusion of affected relatives other than siblings has an even smaller effect on an association analysis, because the genotype correlation among distant relatives is much lower.
Results
Demographic and Disease Characteristics of the MADGC Family Collection
The demographic and disease characteristics of the MADGC family collection reflect, in part, the recommendations of an expert advisory panel that was convened in late 1999 (see the “Material and Methods” section). The entry criteria for these families include a requirement for at least two affected individuals per family and at least two of the nine core diseases in these affected individuals.
Demographic characteristics of the currently enrolled 265 families with multiplex autoimmune diseases are summarized in table 1. The majority of families were white, with only nine families from other ethnic backgrounds. Families consisted of an average of 3.2 affected individuals, but 11 families each had between 6 and 9 affected individuals. Nearly half of the families (48%) were recruited through patient advocacy groups, primarily the AARDA and the National Multiple Sclerosis Society. Approximately 20% of the families were ascertained by one of the three major recruitment sites for the MADGC: North Shore University Hospital, the University of Minnesota, and UCSF. Since the emphasis of research at these sites has been primarily on population-genetics studies of SLE and/or RA, there is a relative enrichment of probands with these rheumatic diseases. The remaining families were recruited using a variety of other approaches, including articles in the lay press. Thus, by virtue of the criteria for entry and the modes of recruitment, these families are not—and were not intended to be—a representative sample of families with multiple autoimmune diseases in the general population.
Table 1.
Demographic Characteristics of 265 Multiplex Autoimmune Families
| Characteristic | Finding |
| Mean no. of affected individuals per family (range) | 3.2 (2–9) |
| Mean total no. of individualsa enrolled per family (range) | 5.7 (2–19) |
| Ethnicityb (% white) | 96.6 |
| No. from recruitment source (%): | |
| Patient advocacy group/publication | 128 (48) |
| Other | 80 (30) |
| Local recruitmentc | 57 (22) |
Affected plus unaffected.
Other ethnicities included Hispanic (1.1%), mixed ethnicity (1.9%), and unknown ethnicity (0.4%).
Enrollment at these sites included recruitment onsite (e.g., clinics) and offsite (e.g., patients known to investigators through other research activities).
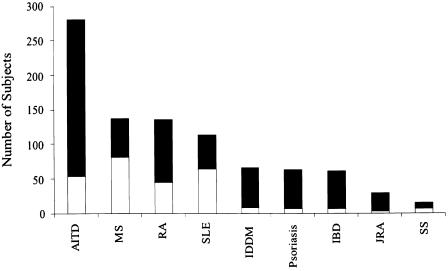
The distribution of the nine core diseases among all 806 affected individuals, as well as among the 265 probands in these families, is shown in figure 1. Note that the total number of core diseases tallied in figure 1 (907) exceeds the number of affected individuals (806) because 97 subjects were affected with two or more different core diseases. Overall, AITD is the most common disorder among all affected family members. This is expected, given that it is the disorder with the highest population prevalence of the nine core diseases. MS, RA, and SLE were the next most common diseases present in these families. The high prevalence of RA in the families is not unexpected, given the overall ∼1% estimated prevalence of this disease in the general population (Gabriel 2000). However, both SLE and MS are substantially overrepresented—both among all affected individuals and among the probands—compared with their low population prevalence (Broadley et al. 2000; Gabriel 2000). This undoubtedly reflects the ascertainment bias inherent in the modes of recruitment discussed above—namely, that existing collections of patients with RA, SLE, and MS were used as a source of recruitment for ∼20% of the MADGC families.
Figure 1.
Distribution of the core autoimmune diseases among 806 affected members of 265 multiplex autoimmune families. Probands are shown in the lower (unblackened) portion of bars; other affected individuals are shown in the upper (blackened) portion of bars. A total of 907 instances of these core autoimmune diseases are tabulated, since 97 individuals had two or more such diseases (most frequently involving AITD [see text]). IDDM = insulin-dependent diabetes mellitus.
Despite the ascertainment bias in this collection, we also examined the patterns of disease aggregation in the MADGC families. In an attempt to reduce the effects of ascertainment bias—as well as for simplicity of interpretation and data presentation—we stratified the families by the seven most common diseases present in the probands, and then we examined the distribution of these seven diseases in the siblings of these probands. We also calculated ORs to determine whether there was any significant enrichment of particular autoimmune diseases among siblings in families selected by proband diagnosis, compared with the distribution of these diseases among all the affected siblings in the entire set of 265 families. The results are shown in table 2. For example, the data show that AITD is present in 69 (30%) of the 232 siblings in the 265 families (first row). Among the families in whom the proband has AITD, 19 (48%) of the 40 siblings also have AITD (second row). This represents a nominally significant enrichment of AITD in these siblings, compared with siblings overall in these families (OR 2.13; 95% CI 1.08–4.2; P=.043 [P value uncorrected]).
Table 2.
Comparison of Frequency of Autoimmune Diseases in Siblings in Family Groups, Stratified by Diagnosis of Proband[Note]
|
No. (%) of Siblings Affected with |
||||||||
| Family Groupa(No. of Probands) | No. of Affected Siblings | AITD | RA | MS | SLE | T1D | IBD | Psoriasis |
| All probands (265) | 232 | 69 (30) | 36 (16) | 16 (7) | 19 (8) | 23 (10) | 15 (6) | 25 (11) |
| Probands with: | ||||||||
| AITD (43) | 40 | 19 (48)b | 9 (23) | 2 (5) | 3 (8) | 6 (15) | 3 (8) | 0 (0) |
| RA (46) | 51 | 15 (29) | 20 (39)c | 2 (4) | 2 (4) | 3 (6) | 5 (10) | 6 (12) |
| MS (82) | 64 | 17 (27) | 5 (8) | 3 (5) | 6 (9) | 9 (14) | 7 (11) | 12 (19)d |
| SLE (65) | 53 | 23 (43)b | 9 (17) | 4 (8) | 6 (11) | 4 (8) | 1 (2) | 4 (8) |
| T1D (10) | 9 | 2 (22) | 0 (0) | 0 (0) | 1 (11) | 3 (33)e | 0 (0) | 1 (11) |
| IBD (7) | 8 | 2 (25) | 1 (13) | 3 (38)f | 0 (0) | 2 (25) | 0 (0) | 0 (0) |
| Psoriasis (8) | 8 | 1 (13) | 2 (25) | 1 (13) | 0 (0) | 0 (0) | 0 (0) | 3 (38) |
Note.— Bold italics indicate P<.1 (two-sided Fisher’s exact test).
Stratified by proband diagnosis.
AITD in siblings of probands with AITD: OR 2.13, 95% CI 1.08–4.2, P=.043; AITD in siblings of probands with SLE: OR 1.81, 95% CI 0.98–3.34, P=.073.
RA in siblings of probands with RA: OR 3.5, 95% CI 1.8–6.8, P=.0003.
Psoriasis in siblings of probands with MS: OR 1.9, 95% CI 0.90–4.0, P=.09; psoriasis in siblings of probands with psoriasis: OR 4.96, 95% CI 11.1–22.0, P=.053.
T1D in siblings of probands with T1D: OR 4.54, 95% CI 1.06–19.40, P=.06.
MS in siblings of probands with IBD: OR 8.1, 95% CI 1.77–37.0, P=.018.
Similar trends toward disease aggregation among siblings can be seen in table 2. However, with the exception of families with multiple siblings with RA, none of these associations remains significant after correcting for multiple comparisons. The reason for the enrichment of sibling pairs with RA in these families is that a significant number of these families were originally recruited from pre-existing collections of sibling pairs with RA (Jawaheer et al. 2004). Therefore, although this collection offers a rich resource of families with multiple autoimmune diseases, it does not provide anything more than suggestive information concerning the normal patterns of familial aggregation of autoimmune diseases in the general population.
High Degree of Female Predominance in RA and MS
As expected for autoimmune diseases in general, there was a predominance of females among the affected members of these families. Overall, 82% of affected individuals were female, compared with 54% of unaffected individuals who were contacted directly for the study. When all unaffected individuals in the family are considered, regardless of direct contact, 43% are female. This suggests an overall increase in females among participants versus nonparticipants in the study. Nevertheless, the percentage of females varied by autoimmune disease, ranging from 57% of individuals with T1D to 94% of individuals with SLE and individuals with SS (see table 3). The degree of enrichment for females was unexpectedly high for diseases such as RA and MS, which typically exhibit 3:1 and 2:1 female-to-male ratios, respectively, in sporadic cases. The enrichment of females among all diseases is even more dramatic if only probands are considered (table 3). This may reflect a tendency for female subjects to be more knowledgeable about family history and therefore more likely to participate in family research studies. However, it is possible that the female predominance in our families may also have biological explanations, perhaps reflecting enhanced penetrance in females when multiple risk genes are present, as may be the case in these families.
Table 3.
Sex Distribution among 806 Affected Members of 265 Multiplex Autoimmune Families, by Core Autoimmune Disease
|
% Female |
||
| Disease (No. of Affected Subjectsa) | All Affected Individuals | Probands Only |
| AITD (282) | 88 | 100 |
| MS (138) | 85 | 93 |
| RA (137) | 89 | 93 |
| SLE (113) | 94 | 94 |
| T1D (67) | 57 | 100 |
| Psoriasis (63) | 68 | 100 |
| IBD (61) | 66 | 86 |
| JRA (30) | 83 | 100 |
| SS (16) | 94 | 100 |
The total number of individuals exceeds 806 because of the presence of more than one autoimmune disease in some individuals.
HLA Typing Reveals Typical Patterns of Disease Association
First-level DNA-based HLA-DR typing was performed for all members of these multiplex families. Table 4 summarizes the results for the five core diseases that have widely recognized HLA-DR associations in whites—namely, DR4 for RA, DR2 and DR3 for SLE, DR3 for AITD, DR2 for MS, and DR3 and DR4 for T1D. The frequencies of these HLA-DR allelic groups in a white control population (n=926) from the New York Cancer Project cohort are shown for comparison, with calculated ORs. The observed HLA associations are consistent with previous observations in white subjects with these disorders (Hall et al. 1996; Tan and Arnett 1998; Todd et al. 1988; Barcellos et al. 2003). Of course, recent studies have indicated that additional loci, other than HLA-DRB1, within the MHC also contribute to risk of several autoimmune diseases (Graham et al. 2002; Jawaheer et al. 2002; Harbo et al. 2004; Vandiedonck et al. 2004).
Table 4.
HLA-DR Associations with Selected Autoimmune Diseases among 256 White Multiplex Autoimmune Families[Note]
|
DR-Allele Frequency (%a) in |
|||
| Disease (No. of Individuals) and DR Allele | Cases | Controls | OR (95% CI)b |
| RA (129): | |||
| DR4 | 58 | 28 | 2.02 (1.62–2.42) |
| SLE (107): | |||
| DR2 | 35 | 29 | 1.2 (.91–1.59) |
| DR3 | 45 | 20 | 2.24 (1.75–2.87) |
| MS (135): | |||
| DR2 | 53 | 29 | 1.84 (1.54–2.24) |
| AITD (270): | |||
| DR3 | 38 | 20 | 1.89 (1.58–2.30) |
| T1D (65): | |||
| DR3 | 57 | 20 | 2.89 (2.24–3.65) |
| DR4 | 69 | 28 | 2.44 (2.0–2.96) |
Note.— DR typing results are shown by use of serologic equivalence. DR2 contains both DRB1*15 and DRB1*16 alleles, and DR4 contains all DRB1*04 subtypes.
Frequency (%) is rounded to the nearest integer.
Compared with a group of 926 white controls from the New York metropolitan area.
Other Autoimmune Diseases
One or more other (noncore) autoimmune diseases were confirmed among 80 individuals (67 families). The most frequent of these diseases were pernicious or hemolytic anemia (MIM 170900 and MIM 205700, respectively) (confirmed in 19 members of 14 families), idiopathic thrombocytopenic purpura (MIM 188030) (12 individuals), antiphospholipid syndrome (MIM 107320) (7 individuals), mixed or undifferentiated connective tissue disease (6 individuals), scleroderma (MIM 181750) (6 individuals), and vitiligo (MIM 193200) (7 individuals).
Patterns of Disease Aggregation in Single Individuals
In the families we studied, 128 affected individuals had more than one autoimmune disease. This includes the 97 individuals with two or more core diseases, as summarized in figure 1. The co-occurrence of AITD with another disease was by far the most common pattern and was present in 84 (65.6%) of the 128 subjects. These included 13 subjects with MS/AITD, 23 subjects with RA/AITD, 12 subjects with T1D/AITD, and 16 subjects with SLE/AITD.
Association of the PTPN22 R620W Polymorphism with Multiple Autoimmune Diseases
To begin to use this family resource to study genetic polymorphisms that may underlie multiple autoimmune diseases, we genotyped 249 white families in this initial collection for a case-control study of the PTPN22 R620W polymorphism. Recent studies have indicated that PTPN22 is associated with T1D, RA, SLE, Graves disease, and autoimmune Addison disease. No information is currently available for other autoimmune disorders, although unpublished data indicate that there is no evidence of association with MS (Begovich et al. 2005). Using a Pyrosequencing assay, we determined the prevalence of the minor (T) allele (rs2476601, encoding tryptophan at amino acid position 620 of the full-length protein) in 746 affected members of these families, compared with 2,064 healthy white controls (the majority of whom had been genotyped previously) (Begovich et al. 2004; Kyogoku et al. 2004).
The results, given in table 5, show striking variation in genetic association for different diseases, with ORs ranging from 0.35 to 2.49. When all 746 affected individuals were compared with controls, the combined OR was 1.35 (P=.002). Notably, in addition to T1D and SLE, Hashimoto thyroiditis was also significantly associated with the 620W risk allele (OR 1.63; 95% CI 1.24–2.17). The association with RA was positive but was of borderline significance. The OR for Graves disease was >1 but was not statistically significant. When all individuals in disease groups with an OR of >1 (T1D, SLE, AITD, and RA) were combined (n=483), the cumulative OR was 1.59 (P<.0001). The very low sample sizes for some disease subgoups make it impossible to draw definitive conclusions for all diseases. Nevertheless, the absence of any evidence of association of PTPN22 with MS is striking, given that 120 patients with MS were available for study, and this confirms recent data (Begovich et al. 2005). Positive associations with T1D, RA, SLE, Graves disease, and autoimmune Addison disease have been reported elsewhere (Begovich et al. 2004; Bottini et al. 2004; Kyogoku et al. 2004; Smyth et al. 2004; Velaga et al. 2004). We also examined PTPN22 associations in individuals with more than one autoimmune disorder (data not shown). Since most of these individuals had AITD as one of these diseases (see above), it is not surprising that the OR for this group (OR 1.73; 95% CI 1.17–2.55) was very similar to that seen for Hashimoto thyroiditis alone.
Table 5.
Association of the PTPN22 Missense SNP with Four Autoimmune Diseases in the White MADGC Families (249 Families, 746 Affected Individuals)
| AutoimmuneDisease(s) | No. of Cases | T-AlleleFrequency (%) | OR (95% CI)a |
| T1D | 61 | 18.9 | 2.49 (1.56–3.97) |
| Hashimoto thyroiditis | 194 | 14.2 | 1.77 (1.31–2.40) |
| SLEb | 101 | 12.9 | 1.58 (1.04–2.43) |
| RAc | 122 | 11.9 | 1.45 (.97–2.16) |
| Graves disease | 58 | 10.3 | 1.24 (.67–2.27) |
| MS | 120 | 8.3 | .98 (.61–1.56) |
| Psoriasis | 51 | 7.8 | .91 (.44–1.89) |
| JRA | 27 | 7.4 | .86 (.31–2.39) |
| IBD | 41 | 7.3 | .85 (.37–1.96) |
| SS | 16 | 3.1 | .35 (.05–2.54) |
| T1D, AITD, SLE, and RA | 483 | 12.9 | 1.59 (1.28–1.98) |
The OR was calculated on the basis of a T-allele frequency of 8.5% in 2,064 white control subjects; 95% CIs are not corrected for the complex patterns of relatedness of cases.
Thirty-two cases from affected sib pair families reported elsewhere (Kyogoku et al. 2004).
Forty-three cases from affected sib pair families reported elsewhere (Begovich et al. 2004).
The significant association with Hashimoto thyroiditis is a novel observation. To address any concern about including multiple affected members from the same family in a case-control analysis, we performed an association analysis by restricting the cases with Hashimoto thyroiditis to one affected sibling per sibship (165 individuals). A particular random sampling of one affected sibling per sibship led to a T-allele frequency of 13.9%, an OR of 1.70, a 95% CI of 1.20–2.42, and a P value of .003. We repeated this random sampling 100 times; the averaged T-allele frequency is 13.91%, with an SD of 0.37%, which is consistent with the result given in table 5.
Discussion
Autoimmune diseases constitute a diverse set of related phenotypes that are presumed to have common underlying mechanisms and, thus, some degree of shared genetic predisposition (Becker 2001). The diversity of phenotypes among autoimmune diseases makes them difficult to study as a unified group, and their relative rarity as individual diseases poses a challenge for accumulating sufficient sample sizes that contain specific disease subgroups or subphenotypes. In general, with the exception of the HLA region, attempts to identify genes that confer genetic predisposition across the different autoimmune diseases have been unsuccessful or, in the case of CTLA4, have provided only modest and incomplete evidence that a common allele underlies different forms of autoimmunity (Ueda et al. 2003). Several recent reports suggest that another member of the CTLA4 family of T-cell regulatory receptors, PD-1, may also predispose to several autoimmune diseases (Prokunina et al. 2002, 2004; Nielsen et al. 2003; Lin et al. 2004).
The MADGC collection of families was originally conceived to be a resource for identifying the common genetic elements involved in autoimmunity and was based on the idea that certain alleles may act to predispose to multiple autoimmune diseases. We posited that genes acting across autoimmune phenotypes would be enriched in families in whom multiple different autoimmune phenotypes are present. These might be common genes with low penetrance, and, thus, they would probably also be found in sporadic disease or in families with only one autoimmune phenotype. Alternatively, more rare and highly penetrant genes for autoimmunity may also be present in the population, and these will likely only be detected by focusing on larger multiplex families. The MADGC collection provides a valuable resource for identifying both types of autoimmunity genes.
In this study, we show that a relatively common variant of PTPN22, 620W, confers susceptibility for four different autoimmune disorders: T1D, RA, SLE, and Hashimoto thyroiditis. There have been initial reports of separate associations of PTPN22 with three of these diseases (Begovich et al. 2004; Bottini et al. 2004; Kyogoku et al. 2004). This is the first reported association with Hashimoto thyroiditis. The OR for Graves disease was >1, but our sample size was inadequate to definitively address this question. However, two recent reports have shown clear evidence of association with Graves disease (Smyth et al. 2004; Velaga et al. 2004). In contrast, the lack of association with MS is of interest, since the sample size of MS cases in this data set (n=120) provides a reasonable likelihood of detecting such an association. For this analysis, a case sample size of 120 has 62% power to detect an OR of 1.8, under the assumption of a control risk-allele frequency of 0.085. Furthermore, this lack of association has recently been confirmed in a larger MS data set (Begovich et al. 2005).
These results have implications for the diversity of pathogenic mechanisms that are likely to underlie autoimmune diseases. It is clear that PTPN22 is directly involved in setting thresholds for T-cell receptor (TCR) signaling and, in this context, is acting, in part, through binding to intracellular kinases such as Csk (Gregorieff et al. 1998; Cloutier and Veillette 1999). These interactions control the phosphorylation state of regulatory tyrosines on key signaling molecules in the TCR complex, such as Lck and ZAP70 (Mustelin et al. 2004). PTPN22-knockout animals display rather subtle changes in a number of immune parameters (Hasegawa et al. 2004)—most prominently, in the enhanced T-cell proliferation in the memory-effector compartment. In the older animals (aged 6 mo), there was an expansion in the number of T cells and an enlargement of the spleen and the lymph nodes. This was accompanied by the spontaneous formation of germinal centers and higher levels of several Ig isotypes. Thus, there was an up-regulation of the humoral immune response in these animals that appeared to be largely secondary to lowered thresholds for signaling in T cells (Hasegawa et al. 2004).
The existence of humoral abnormalities in the PTPN22-knockout mice is consistent with the fact that autoantibody production is a prominent feature of all the human diseases that are associated with PTPN22. Indeed, SLE, RA, T1D, and Hashimoto thyroiditis are all characterized by the frequent development of autoantibodies, often before the onset of clinical disease (Arbuckle et al. 2003; Strieder et al. 2003; Hoppu et al. 2004; Nielen et al. 2004). We also have recently shown that the PTPN22 association is almost exclusively with seropositive RA (Begovich et al. 2004; Lee et al., in press). In addition, the formation of ectopic germinal centers can be seen in both RA and AITD (Weyand et al. 2001), and increased germinal center formation is observed in autoimmune mouse models for SLE and T1D (Luzina et al. 2001). In contrast, human MS does not appear to have a prominent humoral component (Haffler 2004). These combined observations suggest that the PTPN22 620W risk allele may act primarily as a predisposing factor for the development of autoantibodies in these disorders. It will therefore be of great interest to examine (1) whether the PTPN22 risk allele is also associated with the presence of autoantibodies in individuals who do not display a clinical phenotype and (2) whether PTPN22 confers additional risk of the eventual development of clinical autoimmunity in these individuals. The MADGC families should be useful for such an investigation, since we have recruited many unaffected individuals in these families and plasma is available for additional antibody testing of all subjects.
The absence of an association with MS appears to imply that lowered thresholds for TCR signaling may not have an important role in the predisposition to MS, at least at stages of T-cell differentiation in which PTPN22 levels are critical for regulation. This is somewhat surprising, given the wealth of data implicating T cells in the pathogenesis of this disease (Haffler 2004). However, knowledge is still very incomplete concerning exactly which T-cell subsets depend on PTPN22 for regulation. Overall, phosphatases have a negative regulatory function in T cells, and there may be other phosphatases, such as PTP-PEST (another Csk-binding phosphatase), that might serve a function that is redundant to PTPN22 in certain cell types (Mustelin et al. 2004). It should also be pointed out that PTPN22 is broadly expressed in hematopoietic cells, and, thus, the mechanism for disease susceptibility may extend beyond T cells (Begovich et al. 2004). Knowledge of the regulatory circuits controlled by phosphatases in T lymphocytes (or other cell types), combined with population-genetics data such as we describe here, may eventually make it possible for researchers to identify the critical lymphocyte subsets and pathogenic mechanisms that are involved in the different autoimmune diseases.
The overall goal of the MADGC is to collect family data that will facilitate the identification of genes that predispose to multiple different autoimmune diseases. Data from additional families are currently being collected to increase the statistical power for future studies of putative disease-susceptibility alleles, as they are discovered. We intend to pursue a more detailed analysis of intermediate phenotypes in this family collection, including a comprehensive analysis of autoantibodies in both affected and unaffected individuals. Data from these families can also be used to examine other biomarkers that may be relevant to autoimmunity, including patterns of gene expression (Baechler et al. 2003). Numerous other questions need to be addressed, including the significance of female predominance among the affected individuals in these families and the possible role of environmental factors in accounting for familial aggregation. The very large extended pedigrees in this collection may be useful in linkage analyses for the identification of rare, high-penetrance genes. Finally, the data from these families may prove to be particularly useful for discovering gene-gene interactions, under the assumption that the final autoimmune phenotype depends on an interaction of common autoimmunity genes with more disease-specific susceptibility alleles segregating in these families. It is likely that very large sample sizes will be required to successfully perform such studies, since stochastic and environmental factors will complicate the elucidation of these relationships.
Acknowledgments
A large number of individuals contributed to these studies. We thank the families who generously contributed their time, and we are grateful to the numerous physicians who provided clinical information and confirmation of diagnosis for the affected individuals in these families. The recruitment of these families would not have been possible without the devoted efforts of the following individuals: Jennifer Pearce, Molly Mollin, Peggy Rasmussen, Sarah Kupfer, Christine Melanson, Karina DiLuzio, Gila Klein, Mary McFeely, and Amber Leiran. We are also grateful to the large number of consultants who offered advice and guidance at the planning meeting that took place in Bethesda on November 1, 1999, and at various times thereafter. These individuals include Brian Apatoff, Theodore Bayliss, Steven Brant, Carol Brownscheidle, Patrick Concannon, Bruce Cree, Terry Davies, J. T. Elder, Lawrence Jacobs, Daniel Kastner, Henry MacFarland, Frederick Miller, Steven Rich, Yaron Tomer, and Ronald Wilder. This work was supported by NIAID contract NO1-AI95386.
Electronic-Database Information
The URLs for data presented herein are as follows:
- MADGC, http://www.madgc.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for RA, Hashimoto thyroiditis, T1D, SLE, MS, Graves disease, psoriasis, IBD, JRA, SS, pernicious or hemolytic anemia, idiopathic thrombocytopenic purpura, antiphospholipid syndrome, scleroderma, and vitiligo)
References
- Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, James JA, Harley JB (2003) Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med 349:1526–1533 10.1056/NEJMoa021933 [DOI] [PubMed] [Google Scholar]
- Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW (2003) Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA 100:2610–2615 10.1073/pnas.0337679100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos LF, Oksenberg JR, Begovich AB, Martin ER, Schmidt S, Vittinghoff E, Goodin DS, Pelletier D, Lincoln RR, Bucher P, Swerdlin A, Pericak-Vance MA, Haines JL, Hauser SL, for the Multiple Sclerosis Genetics Group (2003) HLA-DR2 dose effect on susceptibility to multiple sclerosis and influence on disease course. Am J Hum Genet 72:710–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KG (2001) The common genetic hypothesis of autoimmune/inflammatory disease. Curr Opin Allergy Clin Immunol 1:399–405 10.1097/01.all.0000011052.77127.a6 [DOI] [PubMed] [Google Scholar]
- Becker KG, Simon RM, Bailey-Wilson JE, Freidlin B, Biddison WE, McFarland HF, Trent JM (1998) Clustering of non-major histocompatibility complex susceptibility candidate loci in human autoimmune diseases. Proc Natl Acad Sci USA 95:9979–9984 10.1073/pnas.95.17.9979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begovich AB, Caillier SJ, Alexander HC, Penko JM, Hauser SL, Barcellos LF, Oksenberg JR (2005) The R620W polymorphism of the protein tyrosine phosphatase PTPN22 is not associated with multiple sclerosis. Am J Hum Genet 76:184–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, et al (2004) A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet 75:330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson AA, Rylander L, Hagmar L, Nived O, Sturfelt G (2002) Risk factors for developing systemic lupus erythematosus: a case-control study in southern Sweden. Rheumatology (Oxford) 41:563–571 [DOI] [PubMed] [Google Scholar]
- Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, Mustelin T (2004) A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 36:337–338 10.1038/ng1323 [DOI] [PubMed] [Google Scholar]
- Broadley SA, Deans J, Sawcer SJ, Clayton D, Compston DA (2000) Autoimmune disease in first-degree relatives of patients with multiple sclerosis: a UK survey. Brain 123:1102–1111 10.1093/brain/123.6.1102 [DOI] [PubMed] [Google Scholar]
- Cloutier JF, Veillette A (1999) Cooperative inhibition of T-cell antigen receptor signaling by a complex between a kinase and a phosphatase. J Exp Med 189:111–121 10.1084/jem.189.1.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth G, Gottlieb PA (2004) Autoimmune polyendocrine syndromes. N Engl J Med 350:2068–2079 10.1056/NEJMra030158 [DOI] [PubMed] [Google Scholar]
- Erlich H, Bugawan T, Begovich AB, Scharf S, Griffith R, Saiki R, Higuchi R, Walsh PS (1991) HLA-DR, DQ and DP typing using PCR amplification and immobilized probes. Eur J Immunogenet 18:33–55 [DOI] [PubMed] [Google Scholar]
- Gabriel S (2000) The epidemiology of the rheumatic diseases. In: Ruddy S, Harris EA, Sledge CB (eds) Kelly’s textbook of rheumatology, 6th ed. W. B. Saunders, Philadelphia [Google Scholar]
- Ginn LR, Lin JP, Plotz PH, Bale SJ, Wilder RL, Mbauya A, Miller FW (1998) Familial autoimmunity in pedigrees of idiopathic inflammatory myopathy patients suggests common genetic risk factors for many autoimmune diseases. Arthritis Rheum 41:400–405 [DOI] [PubMed] [Google Scholar]
- Göring HH, Ott J (1997) Relationship estimation in affected sib pair analysis of late-onset diseases. Eur J Hum Genet 5:69–77 [PubMed] [Google Scholar]
- Graham RR, Ortmann WA, Langefeld CD, Jawaheer D, Selby SA, Rodine PR, Baechler EC, Rohlf KE, Shark KB, Espe KJ, Green LE, Nair RP, Stuart PE, Elder JT, King RA, Moser KL, Gaffney PM, Bugawan TL, Erlich HA, Rich SS, Gregersen PK, Behrens TW (2002) Visualizing human leukocyte antigen class II risk haplotypes in human systemic lupus erythematosus. Am J Hum Genet 71:543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A, Cloutier JF, Veillette A (1998) Sequence requirements for association of protein-tyrosine phosphatase PEP with the Src homology 3 domain of inhibitory tyrosine protein kinase p50csk. J Biol Chem 273:13217–13222 10.1074/jbc.273.21.13217 [DOI] [PubMed] [Google Scholar]
- Haffler D (2004) Science in medicine: multiple sclerosis. J Clin Invest 113:788–794 10.1172/JCI200421357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall FC, Weeks DE, Camilleri JP, Williams LA, Amos N, Darke C, Gibson K, Pile K, Wordsworth BP, Jessop JD (1996) Influence of the HLA-DRB1 locus on susceptibility and severity in rheumatoid arthritis. QJM 89:821–829 [DOI] [PubMed] [Google Scholar]
- Harbo HF, Lie BA, Sawcer S, Celius EG, Dai KZ, Oturai A, Hillert J, Lorentzen AR, Laaksonen M, Myhr KM, Ryder LP, Fredrikson S, Nyland H, Sorensen PS, Sandberg-Wollheim M, Andersen O, Svejgaard A, Edland A, Mellgren SI, Compston A, Vartdal F, Spurkland A (2004) Genes in the HLA class I region may contribute to the HLA class II-associated genetic susceptibility to multiple sclerosis. Tissue Antigens 63:237–247 10.1111/j.0001-2815.2004.00173.x [DOI] [PubMed] [Google Scholar]
- Hardy CJ, Palmer BP, Muir KR, Sutton AJ, Powell RJ (1998) Smoking history, alcohol consumption, and systemic lupus erythematosus: a case-control study. Ann Rheum Dis 57:451–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley JB, James JA (1999) Epstein-Barr virus infection may be an environmental risk factor for systemic lupus erythematosus in children and teenagers. Arthritis Rheum 42:1782–1783 [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Martin F, Huang G, Tumas D, Diehl L, Chan AC (2004) PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science 303:685–689 10.1126/science.1092138 [DOI] [PubMed] [Google Scholar]
- Heliovaara M, Aho K, Aromaa A, Knekt P, Reunanen A (1993) Smoking and risk of rheumatoid arthritis. J Rheumatol 20:1830–1835 [PubMed] [Google Scholar]
- Hoppu S, Ronkainen MS, Kulmala P, Akerblom HK, Knip M, and the Childhood Diabetes in Finland Study Group (2004) GAD65 antibody isotypes and epitope recognition during the prediabetic process in siblings of children with type I diabetes. Clin Exp Immunol 136:120–128 10.1111/j.1365-2249.2004.02416.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JA, Neas BR, Moser KL, Hall T, Bruner GR, Sestak AL, Harley JB (2001) Systemic lupus erythematosus in adults is associated with previous Epstein-Barr virus exposure. Arthritis Rheum 44:1122–1126 [DOI] [PubMed] [Google Scholar]
- Jawaheer D, Li W, Graham RR, Chen W, Damle A, Xiao X, Monteiro J, Khalili H, Lee A, Lundsten R, Begovich A, Bugawan T, Erlich H, Elder JT, Criswell LA, Seldin MF, Amos CI, Behrens TW, Gregersen PW (2002) Dissecting the genetic complexity of the association between human leukocyte antigens and rheumatoid arthritis. Am J Hum Genet 71:585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawaheer D, Lum RF, Amos CI, Gregersen PK, Criswell LA (2004) Clustering of disease features within 512 multicase rheumatoid arthritis families. Arthritis Rheum 50:736–741 10.1002/art.20066 [DOI] [PubMed] [Google Scholar]
- Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, Carlton VE, Chang M, Ramos P, Baechler EC, Batliwalla FM, Novitzke J, Williams AH, Gillett C, Rodine P, Graham RR, Ardlie KG, Gaffney PM, Moser KL, Petri M, Begovich AB, Gregersen PK, Behrens TW (2004) Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet 75:504–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AT, Li W, Liew A, Bombardier C, Weisman M, Massarotti EM, Kent J, Wolfe F, Begovich A, Gregersen PK. The PTPN22 R620W polymorphism associates with RF positive rheumatoid arthritis in a dose-dependent manner but not with HLA shared epitope status. Genes Immun (in press) [DOI] [PubMed] [Google Scholar]
- Lin JP, Cash JM, Doyle SZ, Peden S, Kanik K, Amos CI, Bale SJ, Wilder RL (1998) Familial clustering of rheumatoid arthritis with other autoimmune diseases. Hum Genet 103: 475–482 10.1007/s004390050853 [DOI] [PubMed] [Google Scholar]
- Lin SC, Yen JH, Tsai JJ, Tsai WC, Ou TT, Liu HW, Chen CJ (2004) Association of a programmed death 1 gene polymorphism with the development of rheumatoid arthritis, but not systemic lupus erythematosus. Arthritis Rheum 50:770–775 10.1002/art.20040 [DOI] [PubMed] [Google Scholar]
- Luzina IG, Atamas SP, Storrer CE, daSilva LC, Kelsoe G, Papadimitriou JC, Handwerger BS (2001) Spontaneous formation of germinal centers in autoimmune mice. J Leukoc Biol 70:578–584 [PubMed] [Google Scholar]
- Mitchell MK, Gregersen PK, Johnson S, Parsons R, Vlahov D (2004) The New York Cancer Project: rationale, organization, design, and baseline characteristics. J Urban Health 81:301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustelin T, Alonso A, Bottini N, Huynh H, Rahmouni S, Nika K, Louis-dit-Sully C, Tautz L, Togo SH, Bruckner S, Mena-Duran AV, al-Khouri AM (2004) Protein tyrosine phosphatases in T cell physiology. Mol Immunol 41:687–700 10.1016/j.molimm.2004.04.015 [DOI] [PubMed] [Google Scholar]
- Nielen MM, van Schaardenburg D, Reesink HW, van de Stadt RJ, van der Horst-Bruinsma IE, de Koning MH, Habibuw MR, Vandenbroucke JP, Dijkmans BA (2004) Specific autoantibodies precede the symptoms of rheumatoid arthritis: a study of serial measurements in blood donors. Arthritis Rheum 50:380–386 10.1002/art.20018 [DOI] [PubMed] [Google Scholar]
- Nielsen C, Hansen D, Husby S, Jacobsen BB, Lillevang ST (2003) Association of a putative regulatory polymorphism in the PD-1 gene with susceptibility to type 1 diabetes. Tissue Antigens 62:492–497 10.1046/j.1399-0039.2003.00136.x [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identifying genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokunina L, Castillejo-Lopez C, Oberg F, Gunnarsson I, Berg L, Magnusson V, Brookes AJ, Tentler D, Kristjansdottir H, Grondal G, Bolstad AI, Svenungsson E, Lundberg I, Sturfelt G, Jonssen A, Truedsson L, Lima G, Alcocer-Varela J, Jonsson R, Gyllensten UB, Harley JB, Alarcon-Segovia D, Steinsson K, Alarcon-Riquelme ME (2002) A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet 32:666–669 10.1038/ng1020 [DOI] [PubMed] [Google Scholar]
- Prokunina L, Padyukov L, Bennet A, de Faire U, Wiman B, Prince J, Alfredsson L, Klareskog L, Alarcon-Riquelme M (2004) Association of the PD-1.3A allele of the PDCD1 gene in patients with rheumatoid arthritis negative for rheumatoid factor and the shared epitope. Arthritis Rheum 50:1770–1773 10.1002/art.20280 [DOI] [PubMed] [Google Scholar]
- Riise T, Nortvedt MW, Ascherio A (2003) Smoking is a risk factor for multiple sclerosis. Neurology 61:1122–1124 [DOI] [PubMed] [Google Scholar]
- Smyth D, Cooper JD, Collins JE, Heward JM, Franklyn JA, Howson JM, Vella A, Nutland S, Rance HE, Maier L, Barratt BJ, Guja C, Ionescu-Tirgoviste C, Savage DA, Dunger DB, Widmer B, Strachan DP, Ring SM, Walker N, Clayton DG, Twells RC, Gough SC, Todd JA (2004) Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes 53:3020–3023 [DOI] [PubMed] [Google Scholar]
- Steenland K, Goldsmith DF (1995) Silica exposure and autoimmune diseases. Am J Ind Med 28:603–608 [DOI] [PubMed] [Google Scholar]
- Strieder TG, Prummel MF, Tijssen JG, Endert E, Wiersinga WM (2003) Risk factors for and prevalence of thyroid disorders in a cross-sectional study among healthy female relatives of patients with autoimmune thyroid disease. Clin Endocrinol (Oxf) 59:396–401 [DOI] [PubMed] [Google Scholar]
- Symmons DP, Bankhead CR, Harrison BJ, Brennan P, Barrett EM, Scott DG, Silman AJ (1997) Blood transfusion, smoking, and obesity as risk factors for the development of rheumatoid arthritis. Arthritis Rheum 40:1955–1961 [DOI] [PubMed] [Google Scholar]
- Tan FK, Arnett FC (1998) The genetics of lupus. Curr Opin Rheumatol 10:399–408 [DOI] [PubMed] [Google Scholar]
- Todd JA, Bell JI, McDevitt HO (1988) A molecular basis for genetic susceptibility to insulin-dependent diabetes mellitus. Trends Genet 4:129–134 10.1016/0168-9525(88)90135-7 [DOI] [PubMed] [Google Scholar]
- Torfs CP, King MC, Huey B, Malmgren J, Grumet FC (1986) Genetic interrelationship between insulin-dependent diabetes mellitus, the autoimmune thyroid diseases, and rheumatoid arthritis. Am J Hum Genet 38:170–187 [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, et al (2003) Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 423:506–511 10.1038/nature01621 [DOI] [PubMed] [Google Scholar]
- Uhlig T, Hagen KB, Kvien TK (1999) Current tobacco smoking, formal education, and the risk of rheumatoid arthritis. J Rheumatol 26:47–54 [PubMed] [Google Scholar]
- Vandiedonck C, Beaurain G, Giraud M, Hue-Beauvais C, Eymard B, Tranchant C, Gajdos P, Dausset J, Garchon HJ (2004) Pleiotropic effects of the 8.1 HLA haplotype in patients with autoimmune myasthenia gravis and thymus hyperplasia. Proc Natl Acad Sci USA 101:15464–15469 10.1073/pnas.0406756101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velaga MR, Wilson V, Jennings CE, Owen CJ, Herington S, Donaldson PT, Ball SG, James RA, Quinton R, Perros P, Pearce SH (2004) The codon 620 tryptophan allele of the lymphoid tyrosine phosphatase (LYP) gene is a major determinant of Graves’ disease. J Clin Endocrinol Metab 89:5862–5865 10.1210/jc.2004-1108 [DOI] [PubMed] [Google Scholar]
- Vestergaard P (2002) Smoking and thyroid disorders—a meta-analysis. Eur J Endocrinol 146:153–161 10.1530/eje.0.1460153 [DOI] [PubMed] [Google Scholar]
- Weyand CM, Kurtin PJ, Goronzy JJ (2001) Ectopic lymphoid organogenesis: a fast track for autoimmunity. Am J Pathol 159:787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]