Abstract
It is likely that human genetic differences mediate susceptibility to viral infection and virus-triggered disorders. OAS genes encoding the antiviral enzyme 2′,5′-oligoadenylate synthetase (2′5′AS) are critical components of the innate immune response to viruses. This enzyme uses adenosine triphosphate in 2′-specific nucleotidyl transfer reactions to synthesize 2′,5′-oligoadenylates, which activate latent ribonuclease, resulting in degradation of viral RNA and inhibition of virus replication. We showed elsewhere that constitutive (basal) activity of 2′5′AS is correlated with virus-stimulated activity. In the present study, we asked whether constitutive activity is genetically determined and, if so, by which variants. Analysis of 83 families containing two parents and two children demonstrated significant correlations between basal activity in parent-child pairs (P<.0001) and sibling pairs (P=.0044), but not spousal pairs, suggesting strong genetic control of basal activity. We next analyzed association between basal activity and 15 markers across the OAS gene cluster. Significant association was detected at multiple markers, the strongest being at an A/G single-nucleotide polymorphism at the exon 7 splice-acceptor site (AG or AA) of the OAS1 gene. At this unusual polymorphism, allele G had a higher gene frequency in persons with high enzyme activity than in those with low enzyme activity (0.44 vs. 0.20; P=3×10-11). Enzyme activity varied in a dose-dependent manner across the GG, GA, and AA genotypes (tested by analysis of variance; P=1×10-14). Allele G generates the previously described p46 enzyme isoform, whereas allele A ablates the splice site and generates a dual-function antiviral/proapoptotic p48 isoform and a novel p52 isoform. This genetic polymorphism makes OAS1 an excellent candidate for a human gene that influences host susceptibility to viral infection.
Introduction
Although viral diseases have undoubtedly been major factors in human evolution, both past and present (e.g., influenza and HIV), there have been relatively few studies aimed at identifying the human genetic differences that influence susceptibility/resistance to viral infection. After exposure to novel viruses, interferon-α is one of the first cytokines released from immune-competent cells, initiating an innate antiviral response via induction of numerous interferon-stimulated genes (Der et al. 1998; Levy and Garcia-Sastre 2001; Sen 2001). Among these are the OAS genes, which encode the critical effector enzyme 2′,5′-oligoadenylate synthetase (2′5′AS); 2′5′AS requires double-stranded (ds) RNA structures, such as viral genomes, to become activated. The activated enzyme then catalyzes the polymerization of ATP into 2′,5′-linked oligoadenylates (2′5′A), and these, in turn, bind to and activate latent ribonuclease (RNaseL), which degrades viral as well as cellular RNA and inhibits protein synthesis. Recently, amino acid sequences of 2′5′AS required for dsRNA binding have been identified through crystallography and structure-guided mutagenesis (Hartmann et al. 2003), and evolutionary conservation analysis has suggested that these sequences may also have nuclease function (Rogozin et al. 2003). The 2′5′AS-RNaseL system may also be involved in growth and apoptosis; for example, several studies have implicated genetic variation at the RNASEL locus (MIM 180435) in susceptibility to prostate cancer (reviewed by Silverman [2003]), possibly through the differential effects of such genetic variants on RNaseL enzyme activity (Casey et al. 2002; Xiang et al. 2003).
2′5′AS is actually a family of enzymes encoded by three closely linked genes on human chromosome 12q24.2, with the following order: OAS1 (MIM 164350), encoding the p42, p44, p46, and p48 isoforms; OAS3 (MIM 603351), encoding p100; and OAS2 (MIM 603350), encoding p69 and p71 (reviewed by Justesen et al. [2000]). In the mouse, the importance of 2′5′AS for clearing viral infections was recently demonstrated by studies showing that host susceptibility to West Nile virus (WNV) and other flaviviruses is controlled by the Oas1b gene, the mouse homologue of OAS1. Mouse strains with an exon 4 point mutation resulting in truncation of the L1 isoform showed 100% mortality after WNV infection, whereas strains lacking this mutation showed restricted virus replication and no mortality (Mashimo et al. 2002; Perelygin et al. 2002). Similarly, in vitro studies demonstrated that WNV replication was less efficient in mouse neuroblastoma cell lines expressing the wild-type Oas1b gene, compared with cells expressing the mutant gene (Lucas et al. 2003). In humans, we have shown that constitutive (basal) activity of 2′5′AS varies among individuals and is associated with susceptibility to type 1 diabetes (Bonnevie-Nielsen et al. 2000), an autoimmune disorder whose etiopathogenesis is thought to involve enterovirus infections (see the studies by Hyöty et al. [1995], Hiltunen et al. [1997], Roivainen et al. [1998], and Lönnrot et al. [2000]). New analysis of data from our earlier study of human 2′5′AS response to yellow fever vaccine (Bonnevie-Nielsen et al. 1989) revealed a highly significant correlation between basal enzyme activity and virus-stimulated activity measured 7 d after vaccination (65 subjects; correlation coefficient 0.65; P<.0001). Thus, any genetic variants that affect 2′5′AS activity could be important determinants of susceptibility/resistance to viral infection and to virus-related diseases. Human genetic variation at another innate immune-system gene, mannose-binding lectin (MBL), has been reported to be associated with basal serum MBL levels and immune response to a variety of pathogens, including HIV and hepatitis viruses (Garred et al. 1997; Yuen et al. 1999; Crosdale et al. 2000; Minchinton et al. 2002). In the present study, we therefore sought to determine, by family studies, whether basal 2′5′AS activity is genetically controlled and, if so, whether the relevant variants reside at the OAS gene cluster.
Material and Methods
Subjects
Danish nuclear families (n=83) with two parents and two children (n = 332 subjects total) were ascertained at the Diabetes Clinic of Odense University Hospital in Odense, Denmark. In each family, one child had type 1 diabetes. Blood was taken from all family members after completion of informed-consent procedures that were approved by Odense University and the University of Calgary (previous appointments of V.B.-N. and L.L.F., respectively), as well as by the University of British Columbia. Peripheral lymphocyte lysates and extracted DNA samples were transported by courier to Canada for analysis.
Marker Genotyping
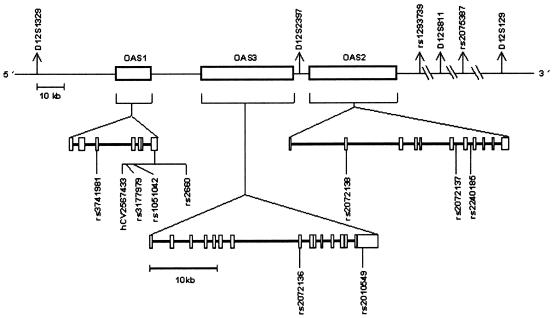
All genotyped markers exist in public databases (National Center for Biotechnology Information, Genome Database, and Applied Biosystems [ABI]/Celera), although, when selected for analysis, several SNPs were unvalidated and lacked allele-frequency information. Microsatellites were genotyped on LI-COR 4200S DNA sequencers after PCR amplification by use of publicly available primer sequences. SNPs were genotyped on an ABI 7000 real-time thermocycler with the use of TaqMan probes and primers purchased from ABI. SNP probes and primers were designed using the ABI Assays-by-Design service (or, in the case of the Celera SNP hCV2567433, using the ABI Assays-on-Demand service, in which probe and primer information is not released to the public). The genomic locations of the typed markers are shown in figure 1, and the sequences of PCR primers and probes used for genotyping these markers (or, for hCV2567433, the genomic sequence including the SNP) are given in table A1 (online only).
Figure 1.
Schematic map showing the genomic positions of the genetic markers typed
DNA Sequencing and mRNA Analysis
Genomic DNA from 10 unrelated children (5 with high enzyme activity and 5 with low enzyme activity) was sequenced for all seven exons (including UTRs), flanking intronic sequences, and regulatory regions of OAS1 (primer sequences are provided in table A2 [online only]). Sequencing was performed by the Core Sequencing Laboratory of the Canadian Genetic Diseases Network of Centres of Excellence in Vancouver, by use of direct sequencing methods, on an ABI 3100 Genetic Analyzer (BigDye Terminator Cycle Sequencing Kit, version 3.1). Multiple sequences were aligned with ClustalW software (see the ClustalW Web site) to identify sequence variants. The results of sequencing exon 7 and flanking regions in the 10 unrelated persons are given in table A3 (online only).
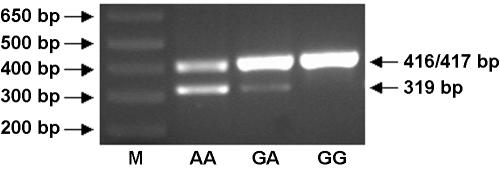
Fresh lymphocytes from healthy individuals (two with genotype AA and one with genotype GA) and lymphoid Daudi cells (with genotype GG) were used for OAS1 mRNA analysis. Primers for RT-PCR amplification of cDNA from p46 and p48 mRNA were positioned in exon 5 and in exon 7, downstream of the p48 splice-acceptor site (primer sequences are given in appendix A [online only]). These primers were designed to yield 417-bp and 319-bp fragments for the p46 and p48 splice variants, respectively, which were separated on 2% agarose gel, stained with ethidium bromide, and detected under UV light (fig. 2). Both nonstimulated and interferon-α–stimulated cells were employed, with qualitatively similar results. The existence of mRNA for the novel p52 variant was determined by excision and sequencing of the upper band in the two persons with the AA genotype. The identities of the p46 and p48 bands were confirmed by sequencing cDNA from the excised 417-bp band in the person with the GG genotype and from the 319-bp band in the two persons with the AA genotype, respectively.
Figure 2.
Expression of OAS1 p46, p48, and p52 in interferon-α–stimulated human cells. M = molecular-weight standard. Lane 1, AA genotype; lane 2, GA genotype; and lane 3, GG genotype. RT-PCR primers were designed to amplify cDNA segments of sizes 417 bp and 319 bp, corresponding to the p46 and p48 splice variants, respectively. Unexpectedly, the primers also amplified a 416-bp segment, corresponding to a novel p52 variant (see text). Results produced by use of nonstimulated cells were qualitatively similar, but bands were fainter.
Measurement of 2′5′AS Enzyme Activity
Basal (constitutive) 2′5′AS activity was assayed in lysates prepared from peripheral blood lymphocytes by measurement of the rate of conversion of radiolabeled ATP to 2′5′A, as described elsewhere (Bonnevie-Nielsen et al. 2000). Radioactive 32P-ATP was added to a reaction mixture containing an ATP-generating system (creatine kinase plus creatine phosphate) to ensure constant molarity of ATP during incubation. Sample protein was adjusted to a concentration optimal for the enzyme’s conversion of ATP. Samples containing generated 2′5′A were spotted on polyethylene-imine (PEI) plates, separated overnight by ascending thin-layer chromatography (TLC), and autoradiographed for detection. The radioactive ATP and generated 2′5′A spots were cut from the TLC plate and quantitated in a β-scintillation counter. Enzyme activity is expressed in units per mg protein, with “1 unit” defined as nanomoles of ATP converted to 2′5′A per minute. Samples were assayed in duplicate. The analytical variance (SD) of the assay was calculated to be 36% from internal-control samples of baculovirus-expressed 2′5′AS (p46).
Statistical Analyses
The enzyme activities among the 332 subjects ranged from 1.30 U/mg to 21.78 U/mg, with a mean ± SD of 3.79±2.19. Prior to data analysis, four outliers with enzyme activities >3 SD above the mean (a father and three children from four different families, with enzyme activities of 19.82, 11.14, 17.26, and 21.78) were excluded from all analyses involving enzyme activity, since such extreme activity measures might reflect an ongoing response to virus exposure rather than constitutive levels. (The 2′5′AS activity observed after viral infection or stimulation with polyI:C, a synthetic dsRNA, is typically 5–10 times higher than the basal activity.) Among the remaining 328 subjects, enzyme activities ranged from 1.30 U/mg to 9.90 U/mg, with a mean ± SD of 3.62±1.52. One additional child who was assayed for enzyme activity was included in the activity correlation analyses but was excluded from the activity by marker association analyses because no DNA sample was available for marker genotyping. Diabetes status in the children was ignored in all analyses.
The Statistical Analysis System (SAS) software package was used to calculate correlations of enzyme activity between relative pairs, P values associated with the correlation coefficients, analysis of variance (ANOVA) of enzyme activity versus OAS1 splice-site genotypes, and the P value associated with the ANOVA statistic. To analyze association between genetic marker-allele frequencies and enzyme activity, all 327 subjects were categorized as belonging to either “high-activity” or “low-activity” groups, which were defined by the median cutoff value of 3.33, resulting in 164 persons assigned to the high-activity group and 163 persons assigned to the low-activity group. The significance of differences in genetic marker-allele frequencies between these two groups was then analyzed by χ2 testing with a 2×2 contingency table. For microsatellite markers, the most common (major) allele was compared with the sum of all other alleles. Similar procedures were used to analyze the effects of markers while controlling for genetic variation at the splice-site SNP hCV2567433. For example, to test for association between enzyme activity and rs3741981 in the subset of subjects who had splice-site genotype AA, the median enzyme activity value in that subset was used to create high- and low-activity groups, and then rs371981 allele frequencies in the two groups were compared.
Results and Discussion
Analysis of basal 2′5′AS activity in members of 83 Danish families showed highly significant correlations between genetically related individuals, but not between spouses. Correlation coefficients were 0.33 for mother-child pairs (P<.0001), 0.36 for father-child pairs (P<.0001), and 0.32 for sibling pairs (P=.0044) but were 0.14 for parental pairs (P>.05 [not significant]). A completely genetic quantitative trait would show correlation coefficients of 0.5 for relative pairs who share 50% of their genes (i.e., parent-child or sibling pairs) and no correlation between genetically unrelated spouses. Thus, the results suggest that basal 2′5′AS activity is under strong genetic control.
To determine whether the genetic variants controlling 2′5′AS activity were located in the OAS gene cluster, we genotyped the SNP rs3741981 in OAS1, the microsatellite D12S2397 located between OAS3 and OAS2, and the SNP rs2240185 in OAS2. Association with 2′5′AS activity was analyzed by classifying all subjects as having either high or low enzyme activity and then comparing marker-allele frequencies in these two groups (for more details, see the “Material and Methods” section). Results showed highly significant associations between enzyme activity and all three OAS-region markers. We therefore genotyped an additional 12 markers across and flanking the OAS gene cluster and found that the strongest associations were with markers in OAS1 (see table 1). To identify all other polymorphic OAS1 variants, we sequenced all seven OAS1 exons, including UTRs, flanking intronic sequences, and the 5′ regulatory region in 10 unrelated individuals (5 with high enzyme activity and 5 with low enzyme activity). Sequencing identified only one additional (previously known) SNP in exon 7, which was then typed in all subjects. Thus, no new OAS1 polymorphisms were found by sequencing. Figure 1 shows the locations of genotyped markers, and table A3 (online only) gives the exon 7 genomic sequences of the 10 persons.
Table 1.
Allele Frequencies for OAS-Region Genetic Markers in Subjects with Low Versus High 2′5′AS Enzyme Activity[Note]
|
Minor-Allele Frequencyin Subjects from |
||||||
| Marker (12cen→qter) | Locationa | Variant (Major/Minor) | Low-Activity Group | High-Activity Group | χ2 (1 df) | P |
| D12S1329b | Upstream of OAS1 | Microsatellitec 158 bp/other | .423 | .460 | .9 | NSd |
| rs3741981 | OAS1 exon 3 (exon C) | A/G | .301 | .494 | 25.5 | 4 × 10−7 |
| hCV2567433e |
OAS1 intron 5/exon 7 splice-acceptor site (AA→AG) |
A/G | .199 | .442 | 44.2 | 3 × 10−11 |
| rs3177979 | OAS1 exon 7 | A/G | .196 | .436 | 43.4 | 4 × 10−11 |
| rs1051042 | OAS1 exon 7 | C/G | .196 | .436 | 43.4 | 4 × 10−11 |
| rs2660 | OAS1 exon 7 | A/G | .196 | .436 | 43.4 | 4 × 10−11 |
| rs2072136 | OAS3 exon 8 | G/A | .298 | .238 | 3.0 | NSd |
| rs2010549 | OAS3 3′ UTR | G/C | .230 | .177 | 2.9 | NSd |
| D12S2397 | Between OAS3 and OAS2 | Microsatellitec 236 bp/other | .619 | .432 | 22.5 | 2 × 10−6 |
| rs2072138 | OAS2 exon 2 | C/G | .402 | .244 | 18.7 | 2 × 10−5 |
| rs2072137 | OAS2 intron 6 | T/C | .540 | .393 | 14.1 | 2 × 10−4 |
| rs2240185 | OAS2 intron 7 | G/C | .414 | .561 | 14.1 | 2 × 10−4 |
| rs1293739 | Downstream of OAS2 | G/A | .261 | .195 | 4.0 | .045 |
| D12S811 | Downstream of OAS2 | Microsatellitec 350 bp/other | .790 | .834 | 2.0 | NSd |
| rs2075387 | Downstream of OAS2 (KIAA0682 exon 3) | C/T | .298 | .325 | .6 | NSd |
| D12S129 | Downstream of OAS2 | Microsatellitec 214 bp/other | .696 | .643 | 2.1 | NSd |
Note.— Findings for haplotype block markers are shown in bold italics.
Nomenclature for OAS1 follows Justesen et al. (2000). There are seven exons: A–E (or 1–5), 6, and 7. Exons 5 and 6 have no intervening intron, so intron 5 precedes exon 7. Exon 5 is included in all OAS1 isoforms. Exon 6 is included in p42 but is excluded from p46, p48, and p52. Exon 7 is excluded from p42, but it is included in p46, p48, and p52. (An EST with predicted molecular weight p44 excludes both exon 6 and exon 7 and includes an additional exon 8.)
Also called “D12S1340” and “D12S2396.”
For microsatellite markers, the most common allele is defined as the “major allele,” and the sum of all other alleles is defined as the “minor allele.”
NS = not significant (P>.05).
Celera ID.
The results presented in table 1, which suggested that genetic markers in OAS1 were the most strongly associated with enzyme activity, were derived by analyzing gene frequencies in two artificial “high enzyme activity” and “low enzyme activity” groups. To attempt to confirm these results by use of a different method, we analyzed the variance in enzyme activity across groups defined by marker genotype. Table 2 shows the ANOVA results for all markers that produced significant differences between the high- and low-activity groups in table 1. These ANOVA results also suggested that the markers most strongly associated with enzyme activity were OAS1 markers.
Table 2.
ANOVA for Enzyme Activity in Subjects Categorized by Genetic Marker Genotype[Note]
|
Mean Enzyme Activity ± SD (Genotype) |
|||||
| Marker (12cen→qter) | Lower-Activity Genotype | Intermediate-Activity Genotype | Higher-Activity Genotype | F | P |
| rs3741981 | 2.97 ± 1.05 (AA) | 3.88 ± 1.55 (GA) | 4.28 ± 1.85 (GG) | 19.4 | 1 × 10−8 |
| hCV2567433a | 2.96 ± 1.00 (AA) | 4.02 ± 1.56 (GA) | 4.87 ± 1.98 (GG) | 35.5 | 1 × 10−14 |
| rs3177979 | 2.97 ± 1.00 (AA) | 4.03 ± 1.56 (GA) | 4.85 ± 2.02 (GG) | 34.5 | 3 × 10−14 |
| rs1051042 | 2.97 ± 1.00 (CC) | 4.03 ± 1.56 (GC) | 4.85 ± 2.02 (GG) | 34.5 | 3 × 10−14 |
| rs2660 | 2.97 ± 1.00 (AA) | 4.03 ± 1.56 (GA) | 4.85 ± 2.02 (GG) | 34.5 | 3 × 10−14 |
| D12S2397 | 3.04 ± 1.09 (X/X) | 3.71 ± 1.50 (X/236) | 4.11 ± 1.73 (236/236) | 11.9 | 1 × 10−5 |
| rs2072138 | 3.03 ± .88 (GG) | 3.50 ± 1.56 (CG) | 3.88 ± 1.58 (CC) | 6.0 | 3 × 10−3 |
| rs2072137 | 3.26 ± 1.23 (CC) | 3.42 ± 1.37 (TC) | 4.25 ± 1.79 (TT) | 12.2 | 8 × 10−6 |
| rs2240185 | 3.19 ± 1.22 (GG) | 3.50 ± 1.35 (GC) | 4.34 ± 1.88 (CC) | 14.0 | 1 × 10−6 |
| rs1293739 | 3.09 ± .99 (AA) | 3.51 ± 1.49 (GA) | 3.74 ± 1.57 (GG) | 2.0 | .14 |
Note.— Findings for haplotype block markers are in bold italics. “X” represents any allele other than 236 bp.
Celera ID.
The strongest genetic association with 2′5′AS activity, both by analysis of gene frequencies in enzyme activity groups (table 1) and by analysis of enzyme activity in genotype groups (table 2), occurred at SNP marker hCV2567433, whose G and A alleles govern splicing to the last OAS1 exon (exon 7), and at a block of three other markers downstream within exon 7 (rs3177979, rs1051042, and rs2660) (see bold italics in tables 1 and 2). These four SNPs were in virtually complete linkage disequilibrium, such that (with one exception) only two haplotypes were found among 332 parental chromosomes: G-G-G-G, with an overall frequency of 0.32, and A-A-C-A, with a frequency of 0.68. The one exception was a single G-A-C-A haplotype found in one parent and transmitted to both children (thus, this rare haplotype has an estimated frequency of 0.3%).
The sequence AG is required for normal splicing at splice-acceptor sites. The G allele at SNP hCV2567433 (AG at acceptor site) retains the splice site and is associated with high enzyme activity, whereas the more common A allele (AA at acceptor site) ablates the splice site and is associated with low enzyme activity. The frequency of the G allele was 0.199 in the low-activity group, compared with 0.442 in the high-activity group (P=3×10-11) (table 1). The G-allele frequency difference was even larger between the lowest and highest 25% of the enzyme activity distribution: 0.152 versus 0.512 (P=4×10-12). Analysis of enzyme activities for the three splice-site genotypes revealed a dose-dependent effect (table 2). Among 147 individuals with genotype AA, 150 individuals with genotype GA, and 30 individuals with genotype GG, the mean (U/mg) ± SD of the enzyme activity was 2.96±1.00, 4.02±1.56, and 4.87±1.98, respectively, a highly significant progression (ANOVA P=1×10-14) (table 2). In summary, the more G alleles that a person has at the splice-acceptor site, the higher the person's basal 2′5′AS activity.
In the above analyses, we included all 327 subjects to maximize sample size. The use of related subjects is not expected to increase the chance of false-positive results, since all families are of identical structure (two parents and two children) and therefore receive equal weight. To confirm that this is the case, we tested for association between enzyme activity and splice-site genotype with the use of only the 165 unrelated parents. Results for both analysis methods showed a highly significant association: (1) in analysis of allele frequencies, the G-allele frequencies in the low- and high-activity groups were essentially identical to those observed with the use of all subjects (0.201 in the low-activity group, compared with 0.434 in the high-activity group; χ2=20.56; P=6×10-6), and, (2) in analysis of enzyme activity variance across splice-site genotypes, a highly significant relationship was also demonstrated (ANOVA F=14.6; P=2×10-6).
The significant associations between enzyme activity and other OAS markers could be independent effects or could be merely secondary to linkage disequilibrium between the splice-site variants and the other markers. To discriminate between these two possibilities, we tested for association with the other markers (except the three markers in the exon 7 haplotype block discussed above) while fixing genetic variation at the splice site. When analysis was restricted to individuals with splice-site genotype AA, no association was detected between enzyme activity and any of the other markers that showed significant results in table 1 (i.e., rs3741981, D12S2397, rs2072138, rs2072137, rs2240185, and rs1293739). Similarly, no significant associations were found with other markers when analysis was restricted to individuals with splice-site genotype GA. For example, table 3 presents the results of testing for association with rs3741981 (the second most strongly associated marker in the total data set) while fixing variation at the splice-site SNP. These results indicate no relation between rs3741981 and enzyme activity when the effects of variation at the splice site were removed. However, table 4 shows that, when the reverse procedure was performed by testing for association with the splice-site SNP while fixing variation at rs3741981, significant effects of the splice-site variants were still detectable (with an increased frequency of the G allele in the high-activity group). Similar results were obtained for other markers. For example, there was no significant association between D12S2397 and enzyme activity when variation at the splice-site SNP was fixed, but, when the reverse procedure was performed by considering only persons with the most common genotype 236/236 at D12S2397, a significant increase of the splice-site G allele was seen in persons with high enzyme activity versus those with low enzyme activity (0.65 vs. 0.47; P=.024). Finally, it is theoretically possible that the association of the splice-site polymorphism with enzyme activity could be secondary to one of the three SNPs in almost complete linkage disequilibrium with the splice-site SNP (indicated by the bold italics that designate the haplotype block markers in tables 1 and 2). It is impossible to test this by use of the type of restricted analysis performed above for other associated markers. However, as will be shown below, the magnitude of the protein structural change due to the splice-site polymorphism is much greater than that due to the remaining three SNPs. Together, these results strongly suggest that the primary association of enzyme activity is with the splice-site polymorphism (i.e., the splice-site SNP is the causal variant) and that the associations observed with other OAS markers are secondary to linkage disequilibrium between the splice-site variants and those other markers (although the possibility of minor effects due to other variants cannot be ruled out).
Table 3.
Association of rs3741981 with Enzyme Activity (with Variation Fixed at the Splice-Site SNP)
|
rs3741981 Allele Count (Frequency) for |
||||||
| Individuals with Splice-Site Genotype AAa |
Individuals with Splice-Site Genotype GAa |
Individuals with Splice-Site Genotype GGb |
||||
| rs3741981Allele | Low-ActivityGroup | High-ActivityGroup | Low-ActivityGroup | High-ActivityGroup | Low-ActivityGroup | High-ActivityGroup |
| A | 127 | 132 | 66 | 69 | 0 | 0 |
| G | 19 (.13) | 16 (.11) | 84 (.56) | 81 (.54) | 30 (1.0) | 30 (1.0) |
| Total | 146 | 148 | 150 | 166 | 30 | 30 |
P value for the low-activity group versus the high-activity group was not significant.
No variation between the low-activity group and the high-activity group.
Table 4.
Association of Splice-Site SNP with Enzyme Activity (with Variation Fixed at rs3741981)
|
Splice-Site Allele Count (Frequency) for |
||||||
| Individuals with rs3741981 Genotype AAa |
Individuals with rs3741981 Genotype GAb |
Individuals with rs3741981 Genotype GGc |
||||
| Splice-SiteAllele | Low-ActivityGroup | High-ActivityGroup | Low-ActivityGroup | High-ActivityGroup | Low-ActivityGroup | High-ActivityGroup |
| A | 114 | 116 | 104 | 89 | 16 | 5 |
| G | 0 (.0) | 0 (.0) | 58 (.36) | 77 (.46) | 32 (.67) | 43 (.90) |
| Total | 114 | 116 | 162 | 166 | 48 | 48 |
No variation between the low-activity group and the high-activity group.
P=.052 for the low-activity group versus the high-activity group.
P=.007 for the low-activity group versus the high-activity group.
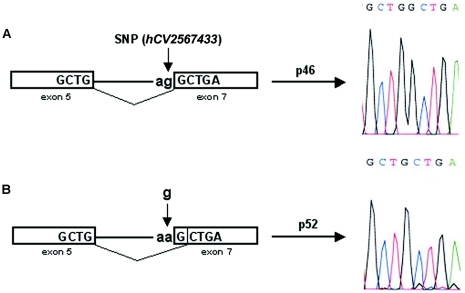
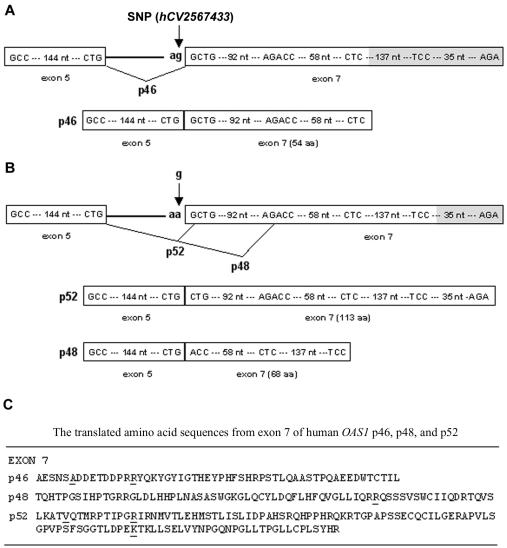
Since the A allele ablates the splice-acceptor site, we next asked whether splicing to exon 7 occurs in mRNA transcripts derived from A-allele sequences and, if so, which splice site is used. Ghosh et al. (2001) reported elsewhere that exon 7 splicing can occur both at an upstream site, producing the p46 isoform, and—apparently by classical alternative splicing of the same transcript—further downstream, producing the p48 isoform. (Note that lack of exon 7 splicing results in the smaller p42 isoform [see footnote a in table 1].) Because splicing at the more downstream site causes a reading-frame change, the p46 and p48 isoforms have different amino acid sequences at their carboxyl-terminal tails. In the present study, we have shown that, since the upstream site is genetically variable, only the G allele can generate p46. To determine whether A-allele transcripts use the downstream p48 splice site, we amplified mRNA from persons with AA, GA, and GG genotypes by use of primers designed to amplify 417-bp and 319-bp segments from p46 and p48 mRNAs, respectively. Figure 2 shows that genotypes AA and GA, but not GG, have the lower band corresponding to p48 splicing, suggesting that only the A allele generates p48 (although more individuals with the GG genotype need to be tested to verify this). Unexpectedly, all three genotypes showed the upper band corresponding to p46 (G allele) splicing. However, DNA sequencing of this band from the two individuals with the AA genotype revealed that the actual size was 416 bp, rather than 417 bp—splicing had occurred at the next AG sequence, located only one base downstream of the splice-site polymorphism (see fig. 3). Splicing at this single-base–shifted position alters the reading frame, resulting in a large, novel, previously undescribed 2′5′AS isoform with predicted molecular weight p52. Figure 2 implies that this shifted A-allele splice site is not optimal, since alternative splicing using the downstream p48 site also occurs in persons with the AA and GA genotypes. In summary, only persons possessing at least one G allele can produce the p46 isoform; persons having only A alleles cannot make p46 but can produce other isoforms, such as p48 and p52. Figure 4 summarizes the splicing events leading to the p46, p48, and p52 isoforms, as well as the amino acid sequences translated from each of these exon 7 splice variants, which result in very diverse protein sequences for the isoform C-terminal tails.
Figure 3.
Schematic diagram of splicing events generating the p46 and novel p52 cDNA sequences. Exons 5 and 7 (boxes) are spliced together following the “ag” at the splice-acceptor site. A, Sequence containing a “g” (G allele) at the splice-site SNP (hCV2567433) (vertical arrow). B, Sequence containing an “a” (A allele) at the SNP, which shifts the splice-acceptor site one base downstream. The resulting splice variants were determined to have the cDNA sequences shown to the right of the horizontal arrows, with the p46 sequence having an extra G, compared with the p52 sequence.
Figure 4.
Schematic diagram of the DNA sequences, splicing events, and translated proteins for the p46, p48, and p52 isoforms. A, p46, generated from the G allele at the splice-acceptor site SNP (hCV2567433). B, p52 and p48, generated from the A allele at splice-site SNP. C, Exon 7 translated amino acid sequences for p46, p48, and p52. SNPs rs3177979, rs1051042, and rs2660 are within codons of the underlined amino acids. For p46, rs2660 is untranslated; the splice-site G allele is in linkage disequilibrium with allele G at rs3177979 and allele G at rs1051042, producing amino acids A (GCA) and R (AGG), respectively. For p48, rs3177979 and rs1051042 are untranslated; splice-site allele A is in disequilibrium with allele A at rs2660, producing amino acid R (AGA). For p52, splice-site allele A is in disequilibrium with alleles A, C, and A at the three SNPs, producing amino acids V (GTA), R (CGT), and K (AAG), respectively.
Our findings suggest that the high-enzyme-activity G allele produces the p46 isoform, whereas the low-activity A allele produces the novel p52 isoform as well as the p48 isoform (by alternative splicing). We surmised that the HT1080 cell line, which was reported by Ghosh et al. (2001) to produce mRNAs for both p46 and p48, came from a person with the GA genotype, and we have confirmed this by genotyping HT1080 cells. Most interestingly, these investigators showed that the p48 isoform has proapoptotic activity, which the investigators localized to a BH3 (Bcl-2 homology-3) domain encoded by exon 7. This proapoptotic activity of p48 was independent of its synthetase and RNaseL-activating capabilities. It remains to be determined why basal 2′5′AS activity is higher in persons carrying the G (p46) allele than in those having only the A (p48/p52) allele. Perhaps p46 is a more efficient synthetase than p48 and/or p52; however, this is highly speculative, and there is no evidence that the C-terminal tail is involved in functional aspects of the 2′5′AS molecule, such as ATP binding, dsRNA binding (necessary for enzyme activation), or tetramer formation (required for p48 activation [Ghosh et al. 1997]). It should be noted that other 2′5′AS isoforms (e.g., OAS1 p42 isoform and OAS2 isoforms) most likely also contribute to total basal 2′5′AS activity; however, the present study suggests that interindividual differences in total enzyme activity result primarily from genetically determined splice variants of OAS1.
Examples exist of rare splice-site mutations causing genetic disease as a consequence of impaired or absent functioning of the resultant protein. However, to our knowledge, this is the first example of a common splice-site polymorphism (both alleles abundant in the general population) that generates different functional proteins that may have differential effects on predisposition to common diseases. The three genotypes AA, GA, and GG show systematic variation in basal 2′5′AS antiviral enzyme activity. A recent study reported that European patients with persistent hepatitis C virus infection had a significantly higher frequency of genotype GG at OAS1 SNP rs2660, which we have shown above to be almost perfectly correlated with genotype GG at the splice-site polymorphism, compared with patients with self-limiting hepatitis C infection (Knapp et al. 2003). By inference, patients with persistent infection also had increased frequencies of genotype GG at the splice-site polymorphism that (paradoxically) is associated with high basal antiviral enzyme activity. Similarly, we have evidence that patients with type 1 diabetes have higher basal 2′5′AS activity (Bonnevie-Nielsen et al. 2000) and significantly higher frequencies of splice-site genotypes GG and GA than control subjects (Field et al., in press). To reconcile this apparent paradox, we hypothesize that persons with GG and GA genotypes, despite having higher basal 2′5′AS activity than persons with AA, are unable or less able to mount an important antiviral response through the (non-2′5′AS–RNaseL) p48 proapoptotic pathway. Clearly, further studies are needed to determine the precise relationship between p46/p48/p52 isozymes, 2′5′AS antiviral/apoptotic activities, and the degree to which the OAS1 splice-site genotypes predict variation in immune response to important viral pathogens and variation in cellular apoptotic parameters.
In this Danish sample, the G-allele frequency was 0.32, similar to that found in 99 unrelated Danes (0.34) (L.L.F., F.P., and V.B.-N., unpublished data) and to that reported in the ABI/Celera database for European-originating people (0.35). In contrast, the G-allele frequency for 45 African Americans in the ABI/Celera database is higher (0.47) and for 288 healthy Hong Kong Chinese is lower (0.24) (L.L.F., Y.-L.L., and V.B.-N., unpublished data). Hong Kong Chinese subjects also demonstrate very strong disequilibrium between the splice-site polymorphism and rs2660, with the same two common haplotypes (G-G and A-A) as were found in Danes in the present study. The G variant at the splice site is ancient and possibly “ancestral,” since the recently released sequence of our closest relative, the chimpanzee Pan troglodytes, displays a G allele (both at the splice site and at rs2660) (see the UCSC Genome Browser). Thus, the frequency of the common A allele at the splice site could vary significantly across human populations, perhaps as a result of differences in their virus-exposure history and concomitant natural selection.
Acknowledgments
Research funding was provided by grants from the Alberta Children’s Hospital Research Foundation and the Juvenile Diabetes Research Foundation (to L.L.F. and V.B.-N.), the British Columbia Children’s Hospital Foundation (BCCHF) (to V.B.-N.), and the Canadian Genetic Diseases Network of Centres of Excellence program and the Canadian Institutes of Health Research (to L.L.F.). V.B.-N. and L.L.F. hold Senior Investigatorships from the BCCHF. We thank E. Swiergala, for invaluable technical expertise; Z.-W. Wang, for assistance in data analysis; and Dr. T. Bech-Hansen, for insightful comments.
Appendix A: Supplemental Material
Table A1.
Sequences of Primers for Amplifying Genetic Markers, of TaqMan Probes, and of hCV2567433-Flanking Regions
|
Primer Sequencesa |
TaqMan Assay Probe Sequences |
|||
| Marker | Forward | Reverse | VIC | FAM |
| D12S1329 | 5′-CCTATCCCACCCAGGC-3′ | 5′-AGTCTGCCCCAGGCAC-3′ | … | … |
| rs3741981 | 5′-TGCCCGAACAGGTCAGTTG-3′ | 5′-TCGATGAGCTTGACATAGATTTGG-3′ | 5′-ACTGGCAGCTATAAA-3′ | 5′-ACTGGCGGCTATAA-3′ |
| hCV2567433b | Ordered from ABIc | Ordered from ABIc | Ordered from ABIc | Ordered from ABIc |
| rs3177979 | 5′-TGTGATCATGTGTCTCACCCTTTC-3′ | 5′-GAGGGTACTCATGTGTTCCAATGTA-3′ | 5′-AGCAACAGTGCAGACG-3′ | 5′-AAAGCAACAGTACAGACG-3′ |
| rs1051042 | 5′-GCAACAGTGCAGACGATGAGA-3′ | 5′-GAGGGTACTCATGTGTTCCAATGTA-3′ | 5′-CCAGGAGGTATCAGAA-3′ | 5′-CCAGGACGTATCAGA-3′ |
| rs2660 | 5′-CATTTTCAGGTGGGACTCTTGATC-3′ | 5′-CTTGGATTATACACCAGCTCACTGA-3′ | 5′-AGAGAAGACAAAGCT-3′ | 5′-AGAGAGGACAAAGC-3′ |
| rs2072136 | 5′-TCTGGCACCAAACCAAATCC-3′ | 5′-CTCCTGGCAGCCACTGGT-3′ | 5′-CTACTCGAGGCTCC-3′ | 5′-TCTACTCAAGGCTCCT-3′ |
| rs2010549 | 5′-AAAGGCTGGAGGAGCAGAAG-3′ | 5′-TCTGATTCGGCTACAGTGGTCTAA-3′ | 5′-ACTGGACTATTGGTTTCA-3′ | 5′-ACTGGACTATTCGTTTCA-3′ |
| D12S2397 | 5′-GCTTTATGGTATTGACTTGTATC-3′ | 5′-AATTGAAAGCTAAGTGAGAGAG-3′ | … | … |
| rs2072138 | 5′-TCCTATGGACGGAAAACAGTCTTAA-3′ | 5′-CTTCTCTTCTGATCCTGGAATTGTT-3′ | 5′-ACAAGGGTACCATCG-3′ | 5′-AGGACAAGCGTACCAT-3′ |
| rs2072137 | 5′-CAACAGCCAAGATCCAGATTGTC-3′ | 5′-GCCCAATATTACATCATTCCAACAA-3′ | 5′-CTGGCCTTTCTCA-3′ | 5′-TGGCCCTTCTCATG-3′ |
| rs2240185 | 5′-TGCTCCCTGTGTCTTAGACATCAG-3′ | 5′-ACAGATTCCAAAACATTTTTGACAAA-3′ | 5′-ATTCACAGTAATTTC-3′ | 5′-TATTCAGAGTAATTTCC-3′ |
| rs1293739 | 5′-CCTAACCACAAAAGGGAAACGA-3′ | 5′-GGTCTTGACCTTGAACTCTGCTTAA-3′ | 5′-ACCTAAGAAGAATCCA-3′ | 5′-AGAACCTAAAAAGAATC-3′ |
| D12S811 | 5′-GCACCTGTAATCCCAGATA-3′ | 5′-CAATGCAGGACACTCTTCTA-3′ | … | … |
| rs2075387 | 5′-CCGCTCCTACCGTGAAAGTG-3′ | 5′-CTCCGAGGTGACCCTGCA-3′ | 5′-CGTCTTCCGCCGCA-3′ | 5′-TCTTCTGCCGCAGGG-3′ |
| D12S129 | 5′-CAGCATCTTCATGCCACG-3′ | 5′-ATGTGTGTGTTTTTCCCAAGG-3′ | … | … |
PCR primers for microsatellite markers have M13 tail in forward or reverse primer (M13 sequence not shown).
The flanking sequence of hCV2567433 is TGTGATCATGTGTCTCACCCTTTCA[A/G]GCTGAAAGCAACAGT.
ABI Assay ID:_2567433_10.
Table A2.
Sequences of Primers Used for Genomic DNA Sequencing of OAS1 Exons and Flanking Regions in 10 Unrelated Persons
|
Primer Sequences |
||
| Exon | Forward | Reverse |
| Exon 1 (Exon A) | 5′-AATAAGATTTATGTTGGCTGGAGGTT-3′ | 5′-AGGGTCACTTGTTAATAATAGGGATGTAG-3′ |
| Exon 2 (Exon B) | 5′-GAGCATCCATTTTCCCATCTG-3′ | 5′-TGTCCCCACATTATGCCAATTT-3′ |
| Exon-3 (Exon C) | 5′-GCAGCATTCTCTAGGTGCCAG-3′ | 5′-TTCTGTTGCAGGCTCCTCTC-3′ |
| Exon 4 (Exon D) | 5′-ATCCAGCGCTCTTAACAAGGG-3′ | 5′-TAGTGCCTGCCTCACAGGGT-3′ |
| Exon 5 (Exon E) and Exon 6 | 5′-GAGCCCTTCCTCATGTTCTG-3′ | 5′-CACTATTTGGGCGACAGGAT-3′ |
| Exon 7 | 5′-GCTTTGAGCAAAAGGCTCTC-3′ | 5′-CAGTGCCCAGAGCTATGCTT-3′ |
Table A3.
Multiple Alignment of OAS1 Exon 7 DNA Sequencing Results from 10 Unrelated Persons (5 with Low Enzyme Activity and 5 with High Enzyme Activity)[Note]
| Individual ID | DNA Sequence |
| BN-00-0004-202-LOW | ggctgacggctgaagtctgtctgctgaccactgtcccagcagctggggcttgttagtccttcctcaaagggggatccagatggcatgtcacagtgtctaccgtaaatgctcactgaatccagctgcaatgcaggaagactccctgatgtgatcatgtgtctcaccctttcaaGCTGAAAGCAACAGTACAGACGATGAGACCGACGATCCCAGGACGTATCAGAAATATGGTTACATTGGAACACATGAGTACCCTCATTTCTCTCATAGACCCAGCACACTCCAGGCAGCATCCACCCCACAGGCAGAAGAGGACTGGACCTGCACCATCCTCTGAATGCCAGTGCATCTTGGGGGAAAGGGCTCCAGTGTTATCTGGACCAGTTCCTTCATTTTCAGGTGGGACTCTTGATCCAGAGAAGACAAAGCTCCTCAGTGAGCTGGTGTATAATCCAGGACAGAACCCAGGTCTCCTGACTCCTGGCCTTCTATGCCCTCTATCCTATCATAGATAACATTCTCCACAGCCTCACTTCATTCCACCTATTCTCTGAAAATATTCCCTGAGAGAGAACAGAGAGATTTAGATAAGAGAATGAAATTCCAGCCTTGACTTTCTTCTGTGCACCTGATGGGAGGGTAATGTCTAATGTATTATCAATAACAATAAAAATAAAGCAAATACCAtttattgggtgtttattaacttcaaggcacaga |
| BN-00-0020-202-LOW | ggctgacggctgaagtctgtctgctgaccactgtcccagcagctggggcttgttagtccttcctcaaagggggatccagatggcatgtcacagtgtctaccgtaaatgctcactgaatccagctgcaatgcaggaagactccctgatgtgatcatgtgtctcaccctttcarGCTGAAAGCAACAGTRCAGACGATGAGACCGACGATCCCAGGASGTATCAGAAATATGGTTACATTGGAACACATGAGTACCCTCATTTCTCTCATAGACCCAGCACACTCCAGGCAGCATCCACCCCACAGGCAGAAGAGGACTGGACCTGCACCATCCTCTGAATGCCAGTGCATCTTGGGGGAAAGGGCTCCAGTGTTATCTGGACCAGTTCCTTCATTTTCAGGTGGGACTCTTGATCCAGAGARGACAAAGCTCCTCAGTGAGCTGGTGTATAATCCAGGACAGAACCCAGGTCTCCTGACTCCTGGCCTTCTATGCCCTCTATCCTATCATAGATAACATTCTCCACAGCCTCACTTCATTCCACCTATTCTCTGAAAATATTCCCTGAGAGAGAACAGAGAGATTTAGATAAGAGAATGAAATTCCAGCCTTGACTTTCTTCTGTGCACCTGATGGGAGGGTAATGTCTAATGTATTATCAATAACAATAAAAATAAAGCAAATACCAtttattgggtgtttattaacttcaaggcacaga |
| BN-00-0023-202-LOW | ggctgacggctgaagtctgtctgctgaccactgtcccagcagctggggcttgttagtccttcctcaaagggggatccagatggcatgtcacagtgtctaccgtaaatgctcactgaatccagctgcaatgcaggaagactccctgatgtgatcatgtgtctcaccctttcaaGCTGAAAGCAACAGTACAGACGATGAGACCGACGATCCCAGGACGTATCAGAAATATGGTTACATTGGAACACATGAGTACCCTCATTTCTCTCATAGACCCAGCACACTCCAGGCAGCATCCACCCCACAGGCAGAAGAGGACTGGACCTGCACCATCCTCTGAATGCCAGTGCATCTTGGGGGAAAGGGCTCCAGTGTTATCTGGACCAGTTCCTTCATTTTCAGGTGGGACTCTTGATCCAGAGAAGACAAAGCTCCTCAGTGAGCTGGTGTATAATCCAGGACAGAACCCAGGTCTCCTGACTCCTGGCCTTCTATGCCCTCTATCCTATCATAGATAACATTCTCCACAGCCTCACTTCATTCCACCTATTCTCTGAAAATATTCCCTGAGAGAGAACAGAGAGATTTAGATAAGAGAATGAAATTCCAGCCTTGACTTTCTTCTGTGCACCTGATGGGAGGGTAATGTCTAATGTATTATCAATAACAATAAAAATAAAGCAAATACCAtttattgggtgtttattaacttcaaggcacaga |
| BN-00-0033-202-LOW | ggctgacggctgaagtctgtctgctgaccactgtcccagcagctggggcttgttagtccttcctcaaagggggatccagatggcatgtcacagtgtctaccgtaaatgctcactgaatccagctgcaatgcaggaagactccctgatgtgatcatgtgtctcaccctttcaaGCTGAAAGCAACAGTACAGACGATGAGACCGACGATCCCAGGACGTATCAGAAATATGGTTACATTGGAACACATGAGTACCCTCATTTCTCTCATAGACCCAGCACACTCCAGGCAGCATCCACCCCACAGGCAGAAGAGGACTGGACCTGCACCATCCTCTGAATGCCAGTGCATCTTGGGGGAAAGGGCTCCAGTGTTATCTGGACCAGTTCCTTCATTTTCAGGTGGGACTCTTGATCCAGAGAAGACAAAGCTCCTCAGTGAGCTGGTGTATAATCCAGGACAGAACCCAGGTCTCCTGACTCCTGGCCTTCTATGCCCTCTATCCTATCATAGATAACATTCTCCACAGCCTCACTTCATTCCACCTATTCTCTGAAAATATTCCCTGAGAGAGAACAGAGAGATTTAGATAAGAGAATGAAATTCCAGCCTTGACTTTCTTCTGTGCACCTGATGGGAGGGTAATGTCTAATGTATTATCAATAACAATAAAAATAAAGCAAATACCAtttattgggtgtttattaacttcaaggcacaga |
| BN-00-0072-202-LOW | ggctgacggctgaagtctgtctgctgaccactgtcccagcagctggggcttgttagtccttcctcaaagggggatccagatggcatgtcacagtgtctaccgtaaatgctcactgaatccagctgcaatgcaggaagactccctgatgtgatcatgtgtctcaccctttcaaGCTGAAAGCAACAGTACAGACGATGAGACCGACGATCCCAGGACGTATCAGAAATATGGTTACATTGGAACACATGAGTACCCTCATTTCTCTCATAGACCCAGCACACTCCAGGCAGCATCCACCCCACAGGCAGAAGAGGACTGGACCTGCACCATCCTCTGAATGCCAGTGCATCTTGGGGGAAAGGGCTCCAGTGTTATCTGGACCAGTTCCTTCATTTTCAGGTGGGACTCTTGATCCAGAGAAGACAAAGCTCCTCAGTGAGCTGGTGTATAATCCAGGACAGAACCCAGGTCTCCTGACTCCTGGCCTTCTATGCCCTCTATCCTATCATAGATAACATTCTCCACAGCCTCACTTCATTCCACCTATTCTCTGAAAATATTCCCTGAGAGAGAACAGAGAGATTTAGATAAGAGAATGAAATTCCAGCCTTGACTTTCTTCTGTGCACCTGATGGGAGGGTAATGTCTAATGTATTATCAATAACAATAAAAATAAAGCAAATACCAtttattgggtgtttattaacttcaaggcacaga |
| BN-00-0017-202-HIGH | ggctgacggctgaagtctgtctgctgaccactgtcccagcagctggggcttgttagtccttcctcaaagggggatccagatggcatgtcacagtgtctaccgtaaatgctcactgaatccagctgcaatgcaggaagactccctgatgtgatcatgtgtctcaccctttcagGCTGAAAGCAACAGTGCAGACGATGAGACCGACGATCCCAGGAGGTATCAGAAATATGGTTACATTGGAACACATGAGTACCCTCATTTCTCTCATAGACCCAGCACACTCCAGGCAGCATCCACCCCACAGGCAGAAGAGGACTGGACCTGCACCATCCTCTGAATGCCAGTGCATCTTGGGGGAAAGGGCTCCAGTGTTATCTGGACCAGTTCCTTCATTTTCAGGTGGGACTCTTGATCCAGAGAGGACAAAGCTCCTCAGTGAGCTGGTGTATAATCCAGGACAGAACCCAGGTCTCCTGACTCCTGGCCTTCTATGCCCTCTATCCTATCATAGATAACATTCTCCACAGCCTCACTTCATTCCACCTATTCTCTGAAAATATTCCCTGAGAGAGAACAGAGAGATTTAGATAAGAGAATGAAATTCCAGCCTTGACTTTCTTCTGTGCACCTGATGGGAGGGTAATGTCTAATGTATTATCAATAACAATAAAAATAAAGCAAATACCAtttattgggtgtttattaacttcaaggcacaga |
| BN-00-0043-202-HIGH | ggctgacggctgaagtctgtctgctgaccactgtcccagcagctggggcttgttagtccttcctcaaagggggatccagatggcatgtcacagtgtctaccgtaaatgctcactgaatccagctgcaatgcaggaagactccctgatgtgatcatgtgtctcaccctttcarGCTGAAAGCAACAGTRCAGACGATGAGACCGACGATCCCAGGASGTATCAGAAATATGGTTACATTGGAACACATGAGTACCCTCATTTCTCTCATAGACCCAGCACACTCCAGGCAGCATCCACCCCACAGGCAGAAGAGGACTGGACCTGCACCATCCTCTGAATGCCAGTGCATCTTGGGGGAAAGGGCTCCAGTGTTATCTGGACCAGTTCCTTCATTTTCAGGTGGGACTCTTGATCCAGAGARGACAAAGCTCCTCAGTGAGCTGGTGTATAATCCAGGACAGAACCCAGGTCTCCTGACTCCTGGCCTTCTATGCCCTCTATCCTATCATAGATAACATTCTCCACAGCCTCACTTCATTCCACCTATTCTCTGAAAATATTCCCTGAGAGAGAACAGAGAGATTTAGATAAGAGAATGAAATTCCAGCCTTGACTTTCTTCTGTGCACCTGATGGGAGGGTAATGTCTAATGTATTATCAATAACAATAAAAATAAAGCAAATACCAtttattgggtgtttattaacttcaaggcacaga |
| BN-00-0058-202-HIGH | ggctgacggctgaagtctgtctgctgaccactgtcccagcagctggggcttgttagtccttcctcaaagggggatccagatggcatgtcacagtgtctaccgtaaatgctcactgaatccagctgcaatgcaggaagactccctgatgtgatcatgtgtctcaccctttcarGCTGAAAGCAACAGTRCAGACGATGAGACCGACGATCCCAGGASGTATCAGAAATATGGTTACATTGGAACACATGAGTACCCTCATTTCTCTCATAGACCCAGCACACTCCAGGCAGCATCCACCCCACAGGCAGAAGAGGACTGGACCTGCACCATCCTCTGAATGCCAGTGCATCTTGGGGGAAAGGGCTCCAGTGTTATCTGGACCAGTTCCTTCATTTTCAGGTGGGACTCTTGATCCAGAGARGACAAAGCTCCTCAGTGAGCTGGTGTATAATCCAGGACAGAACCCAGGTCTCCTGACTCCTGGCCTTCTATGCCCTCTATCCTATCATAGATAACATTCTCCACAGCCTCACTTCATTCCACCTATTCTCTGAAAATATTCCCTGAGAGAGAACAGAGAGATTTAGATAAGAGAATGAAATTCCAGCCTTGACTTTCTTCTGTGCACCTGATGGGAGGGTAATGTCTAATGTATTATCAATAACAATAAAAATAAAGCAAATACCAtttattgggtgtttattaacttcaaggcacaga |
| BN-00-0062-202-HIGH | ggctgacggctgaagtctgtctgctgaccactgtcccagcagctggggcttgttagtccttcctcaaagggggatccagatggcatgtcacagtgtctaccgtaaatgctcactgaatccagctgcaatgcaggaagactccctgatgtgatcatgtgtctcaccctttcarGCTGAAAGCAACAGTRCAGACGATGAGACCGACGATCCCAGGASGTATCAGAAATATGGTTACATTGGAACACATGAGTACCCTCATTTCTCTCATAGACCCAGCACACTCCAGGCAGCATCCACCCCACAGGCAGAAGAGGACTGGACCTGCACCATCCTCTGAATGCCAGTGCATCTTGGGGGAAAGGGCTCCAGTGTTATCTGGACCAGTTCCTTCATTTTCAGGTGGGACTCTTGATCCAGAGARGACAAAGCTCCTCAGTGAGCTGGTGTATAATCCAGGACAGAACCCAGGTCTCCTGACTCCTGGCCTTCTATGCCCTCTATCCTATCATAGATAACATTCTCCACAGCCTCACTTCATTCCACCTATTCTCTGAAAATATTCCCTGAGAGAGAACAGAGAGATTTAGATAAGAGAATGAAATTCCAGCCTTGACTTTCTTCTGTGCACCTGATGGGAGGGTAATGTCTAATGTATTATCAATAACAATAAAAATAAAGCAAATACCAtttattgggtgtttattaacttcaaggcacaga |
| BN-00-0063-202-HIGH | ggctgacggctgaagtctgtctgctgaccactgtcccagcagctggggcttgttagtccttcctcaaagggggatccagatggcatgtcacagtgtctaccgtaaatgctcactgaatccagctgcaatgcaggaagactccctgatgtgatcatgtgtctcaccctttcagGCTGAAAGCAACAGTGCAGACGATGAGACCGACGATCCCAGGAGGTATCAGAAATATGGTTACATTGGAACACATGAGTACCCTCATTTCTCTCATAGACCCAGCACACTCCAGGCAGCATCCACCCCACAGGCAGAAGAGGACTGGACCTGCACCATCCTCTGAATGCCAGTGCATCTTGGGGGAAAGGGCTCCAGTGTTATCTGGACCAGTTCCTTCATTTTCAGGTGGGACTCTTGATCCAGAGAGGACAAAGCTCCTCAGTGAGCTGGTGTATAATCCAGGACAGAACCCAGGTCTCCTGACTCCTGGCCTTCTATGCCCTCTATCCTATCATAGATAACATTCTCCACAGCCTCACTTCATTCCACCTATTCTCTGAAAATATTCCCTGAGAGAGAACAGAGAGATTTAGATAAGAGAATGAAATTCCAGCCTTGACTTTCTTCTGTGCACCTGATGGGAGGGTAATGTCTAATGTATTATCAATAACAATAAAAATAAAGCAAATACCAtttattgggtgtttattaacttcaaggcacaga |
Note.— Exon 7 is shown in uppercase letters, and flanking introns are shown in lowercase letters. Bold, underlined letters indicate SNPs hCV2567433, rs3177979, rs1051042, and rs2660, respectively. r = a/g; R = A/G; and S = C/G. Therefore, r, R, or S indicate heterozygosity at the respective SNPs.
Sequences of RT-PCR Primers Used to Amplify cDNA from p46 and p48 mRNA
The sequences of primers for amplification of cDNA from p46 and p48 mRNA were derived from E18 cDNA (NCBI GenBank accession number X02875). The forward primer was positioned in exon 5 (exon E), and the reverse primer was positioned in exon 7, downstream of the p48 splice-acceptor site. These primers were designed to produce 417-bp and 319-bp fragments for the p46 and p48 splice variants, respectively. They also inadvertently detected the novel p52 splice variant, which has a fragment size of 416 bp.
Forward primer: 5′-GGCGGACCCTACAGGAAACT-3′.
Reverse primer: 5′-ACACCAGCTCACTGAGGAGC-3′.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Applied Biosystems (ABI)/Celera, http://www.appliedbiosystems.com/ (for SNP variants, locations, and frequency data)
- ClustalW, http://www.ebi.ac.uk/clustalw/ (for DNA sequence alignment software)
- Genome Database, http://www.gdb.org/ (for microsatellite primer sequences)
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/ (for marker locations and frequency data)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for locus designations of RNASEL, OAS1, OAS3, and OAS2)
- UCSC Genome Browser, http://www.genome.ucsc.edu/ (for human and chimpanzee OAS sequence data from the Genome Project sequence draft)
References
- Bonnevie-Nielsen V, Larsen ML, Frifelt JJ, Michelsen B, Lernmark Å (1989) Association of IDDM and attenuated response of 2′,5′-oligoadenylate synthetase to yellow fever vaccine. Diabetes 38:1636–1642 [DOI] [PubMed] [Google Scholar]
- Bonnevie-Nielsen V, Martensen PM, Justesen J, Levin K, Beck-Nielsen H, Worsaa A, Dyrberg T (2000) The antiviral defense system is persistently activated in human type 1 diabetes. Clinical Immunology 96:11–18 [DOI] [PubMed] [Google Scholar]
- Casey G, Neville PJ, Plummer SJ, Xiang Y, Krumroy LM, Klein EA, Catalona WJ, Nupponen N, Carpten JD, Trent JM, Silverman RH, Witte JS (2002) RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat Genet 32:581–583 [DOI] [PubMed] [Google Scholar]
- Crosdale DJ, Ollier WE, Thomson W, Dyer PA, Jensenious J, Johnson RW, Poulton KV (2000) Mannose binding lectin (MBL) genotype distributions with relation to serum levels in UK Caucasoids. Eur J Immunogenet 27:111–117 [DOI] [PubMed] [Google Scholar]
- Der SD, Zhou A, Williams BRG, Silverman RH (1998) Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc Natl Acad Sci USA 95:15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field LL, Bonnevie-Nielsen V, Pociot F, Lu S, Nielsen TB, Beck-Nielsen H. OAS1 splice site polymorphism controlling antiviral enzyme activity influences susceptibility to type 1 diabetes. Diabetes (in press) [DOI] [PubMed] [Google Scholar]
- Garred P, Madsen HO, Balslev U, Hofmann B, Pedersen C, Gerstoft J, Svejgaard A (1997) Susceptibility to HIV infection and progression of AIDS in relation to variant alleles of mannose-binding lectin. Lancet 349:236–240 [DOI] [PubMed] [Google Scholar]
- Ghosh A, Sarkar SN, Guo W, Bandyopadhyay S, Sen GC (1997) Enzymatic activity of 2′-5′-oligoadenylate synthetase is impaired by specific mutations that affect oligomerization of the protein. J Biol Chem 272:33220–33226 [DOI] [PubMed] [Google Scholar]
- Ghosh A, Sarkar SN, Rowe TM, Sen GC (2001) A specific isozyme of 2′-5′ oligoadenylate synthetase is a dual function proapoptotic protein of the Bcl-2 family. J Biol Chem 276:25447–25455 [DOI] [PubMed] [Google Scholar]
- Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC (2003) Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol Cell 12:1173–1185 [DOI] [PubMed] [Google Scholar]
- Hiltunen M, Hyöty H, Knip M, Ilonen J, Reijonen H, Vähäsalo P, Roivainen M, Lönnrot M, Leinikki P, Hovi T, Åkerblom HK (1997) Islet cell antibody seroconversion in children is temporally associated with enterovirus infections. J Infect Dis 175:554–560 [DOI] [PubMed] [Google Scholar]
- Hyöty H, Hiltunen M, Knip M, Laakkonen M, Vähäsalo P, Karjalainen J, Koskela P, Roivainen M, Leinikki P, Hovi T, Åkerblom HK, and the Childhod Diabetes in Finland (DiMe) Study Group (1995) A prospective study of the role of coxsackie B and other enterovirus infections in the pathogenesis of IDDM. Diabetes 44:652–657 [DOI] [PubMed] [Google Scholar]
- Knapp S, Yee LJ, Frodsham AJ, Hennig BJ, Hellier S, Zhang L, Wright M, Chiaramonte M, Graves M, Thomas HC, Hill AV, Thursz MR (2003) Polymorphisms in interferon-induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS-1 and PKR. Genes Immun 4:411–419 [DOI] [PubMed] [Google Scholar]
- Justesen J, Hartmann R, Kjeldgaard NO (2000) Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell Mol Life Sci 57:1593–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DE, Garcia-Sastre A (2001) The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev 12:143–156 [DOI] [PubMed] [Google Scholar]
- Lönnrot M, Korpela K, Knip M, Ilonen J, Simell O, Korhonen S, Savola K, Muona P, Simell T, Koskela P, Hyöty H (2000) Enterovirus infection as a risk factor for beta-cell autoimmunity in a prospectively observed birth cohort: the Finnish diabetes prediction and prevention study. Diabetes 49:1314–1318 [DOI] [PubMed] [Google Scholar]
- Lucas M, Mashimo T, Frenkiel MP, Simon-Chazottes D, Montagutelli X, Ceccaldi PE, Guénet JL, Desprès P (2003) Infection of mouse neurones by West Nile virus is modulated by the interferon-inducible 2′-5′-oligoadenylate synthetase 1b protein. Immunol Cell Biol 81:230–236 [DOI] [PubMed] [Google Scholar]
- Mashimo T, Lucas M, Simon-Chazottes D, Frenkiel MP, Montagutelli X, Ceccaldi PE, Deubel V, Guénet JL, Desprès P (2002) A nonsense mutation in the gene encoding 2′-5′-oligoadenylate synthetase/L1 isoform is associated with West Nile virus susceptibility in laboratory mice. Proc Natl Acad Sci USA 99:11311–11316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchinton RM, Dean MM, Clark TR, Heatley S, Mullighan CG (2002) Analysis of the relationship between mannose-binding lectin (MBL) genotype, MBL levels and function in an Australian blood donor population. Scand J Immunol 56:630–641 [DOI] [PubMed] [Google Scholar]
- Perelygin AA, Scherbik SV, Zhulin IB, Stockman BM, Li Y, Brinton MA (2002) Positional cloning of the murine flavivirus resistance gene. Proc Natl Acad Sci USA 99:9322–9327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Aravind L, Koonin EV (2003) Differential action of natural selection on the N and C-terminal domains of 2′-5′ oligoadenylate synthetases and the potential nuclease function of the C-terminal domain. J Mol Biol 326:1449–1461 [DOI] [PubMed] [Google Scholar]
- Roivainen M, Knip M, Hyöty H, Kulmala P, Hiltunen M, Vähäsalo P, Hovi T, Åkerblom HK, and the Childhood Diabetes in Finland (DiMe) Study Group (1998) Several different enterovirus serotypes can be associated with prediabetic autoimmune episodes and onset of overt IDDM. J Med Virol 56:74–78 [DOI] [PubMed] [Google Scholar]
- Sen GC (2001) Viruses and interferons. Annu Rev Microbiol 55:255–281 [DOI] [PubMed] [Google Scholar]
- Silverman RH (2003) Implications for RNase L in prostate cancer biology. Biochemistry 42:1805–1812 [DOI] [PubMed] [Google Scholar]
- Xiang Y, Wang Z, Murakami J, Plummer S, Klein EA, Carpten JD, Trent JM, Isaacs WB, Casey G, Silverman RH (2003) Effects of RNase L mutations associated with prostate cancer on apoptosis induced by 2′,5′-oligoadenylates. Cancer Res 63:6795–6801 [PubMed] [Google Scholar]
- Yuen MF, Lau CS, Lau YL, Wong WM, Cheng CC, Lai CL (1999) Mannose binding lectin gene mutations are associated with progression of liver disease in chronic hepatitis B infection. Hepatology 29:1248–1251 [DOI] [PubMed] [Google Scholar]