Abstract
We characterized the T-cell receptor (TCR) repertoire in freshly harvested tumor lesions, in short-term-expanded CD4+ tumor infiltrating lymphocytes (TIL) as well as in CD4+ and CD8+ peripheral blood lymphocytes (PBL) from three patients with cervical cancer. Skewing of the T-cell repertoire as defined by measuring the length of the complementarity-determining region 3 (CDR3) of the TCR VA and VB chains was observed in CD8+ PBL, in freshly harvested tumor tissue, as well as in CD4+ TIL. Comparative analysis of the TCR repertoire revealed unique monoclonal TCR transcripts within the tumor lesion which were not present in PBL, suggesting selection of TCR clonotypes due to antigenic stimulation. TCR repertoire analysis of the short-term (7-day) CD4+ TIL lines revealed that the TCR composition is markedly different from that in CD4+ PBL or in the freshly harvested tumor tissue. Only one-third of CD4+ TIL lines showed HLA-DR-restricted recognition of autologous tumor cells as defined by cytolysis. These data provide support for the antigen-driven selection of T cells within cervical cancer lesions and suggest that analysis of the TCR repertoire may aid in obtaining an objective description of the immune response in patients with cervical cancer who are undergoing epitope-based immunotherapy.
Several methods have been used to analyze the cellular immune response in patients with cervical cancer, including enzyme-linked immunospot analysis of T cells obtained from peripheral blood (32) and characterization of tumor-infiltrating lymphocytes (TIL) directed against human papillomavirus type 16 (HPV-16) (6) or HPV-derived peptides which bind to different major histocompatibility complex (MHC) class II alleles, including DR3, DR4, DR15 or DQ2 (12, 13, 32). CD8+ cytotoxic T cells directed against HPV-16 E6 and E7 antigens have also been detected in peripheral blood lymphocytes (PBL) from women with cervical dysplasia (1, 29). Recruitment and maintenance of HPV-specific and MHC class II-restricted CD4+ T cells may be beneficial for patients at high risk of ultimately developing cervical cancer since the majority of early cervical cancer lesions show allelic loss of MHC class I expression (16) and defects in the MHC class I antigen-processing and presentation machinery (3).
One of the crucial experiments in evaluating anticancer immune responses is to show that the antipeptide-specific cellular immune responses elicited by in vitro stimulation with HPV oncoproteins (9, 31) or with peptides (15, 28) are also directed against the autologous tumor cells. This has not yet been demonstrated, since cervical cancer cell lines are not easily established. Thus, HLA-matched cancer cells served as targets for evaluating the anti-HPV-directed immune responses (15, 31).
In cancers of different histologies, the molecular definition of an antitumor response was initiated by demonstrating anticancer CD4+ or CD8+ responses. These tumor-specific T cells have been used to identify a number of antigens, which may serve as tumor rejection antigens (for a review, see reference 35).
A different scenario is true for cervical cancer, since the majority of cervical cancers express the HPV E6 and E7 oncoproteins that may provide targets for an antitumor cellular immune response (35). Thus, most studies used HPV-derived targets as a readout and surrogate marker of anti-cervical cancer-directed immunity. To gauge the cellular immune response in patients with cervical cancer, we evaluated the T-cell receptor (TCR) repertoire by using measurement of the TCR CDR3 region in PBL, tumor lesions, and TIL from three patients with cervical cancer, from which autologous tumor cell lines could be established. This method allows us to objectively describe the TCR repertoire within different anatomic compartments. Limited TCR diversity suggests selection of T cells by antigenic stimulation (10, 22). In addition, TIL were tested for recognition of autologous tumor cells.
MATERIALS AND METHODS
Immunomagnetic cell sorting.
Heparinized blood was drawn, and peripheral blood mononuclear cells (PBMCs) were obtained by separation over a Ficoll gradient and stored in liquid nitrogen at 1 × 107 to 5 × 107 cells/vial in 90% fetal calf serum-10% dimethyl sulfoxide. CD4+ or CD8+ T cells were separated from PBMCs using anti-CD4- or anti-CD8-coated immunomagnetic beads (Miltenyi, Bergisch Gladbach, Germany). Briefly, PBMCs or TIL were incubated for 15 min at 4°C with immunomagnetic beads (10 μl of beads/105 cells) and run by gravity through a separation column with a magnet. The cells were washed twice with 2 ml of phosphate-buffered saline-0.01% bovine serum albumin and eluted with 1 ml of phosphate-buffered saline-0.01% bovine serum albumin after removal of the magnet. This procedure yields >96% pure CD4+ or CD8+ T cells (data not shown). Tumor sections were obtained from freshly isolated cancer tissue from patients undergoing curative surgery for cervical cancer (Table 1). The tumor specimens were split into two aliquots. One aliquot was cut into small pieces and seeded for 7 days in 48-well plates containing 50% AIM-V medium (Invitrogen, Groningen, The Netherlands) and 50% Dulbecco modified Eagle medium (high glucose) obtained from Gibco (Eggenstein, Germany) supplemented with 10% fetal calf serum and 50 ng of human recombinant interleukin-7 (IL-7) per ml generously provided by Adrian Minty, Sanofi, France. T cells grew out from the tumor tissue and were expanded in the presence of autologous tumor cells. The other aliquot was snap-frozen in liquid nitrogen.
TABLE 1.
MHC alleles and HPV typea
| Patient | HLA-A | HLA-B | HLA-Cw | DR | DQ | HPV type |
|---|---|---|---|---|---|---|
| MZ-CC01-1 | 1,11 | 8,56 | 1,7 | 3,4 | 2,8 | 16 |
| MZ-CC01-2 | 1,2 | 35,44 | 4,5 | 7,5 | 3,0 | 16 |
| MZ-CC01-3 | 1,32 | 7,62 | 4,7 | 8,15 | 4,6 | 16 |
Three patients from whom we could establish an autologous tumor cell line were characterized for MHC class I and II alleles. The resected tumor tested positive for HPV16 DNA in all individuals.
TCR-CDR3 spectratyping.
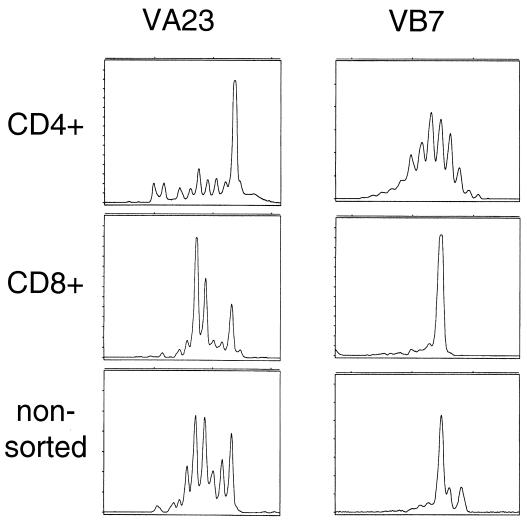
Qualitative analysis of either CD8+- or CD4+-sorted T cells (using immunomagnetic beads) was carried out as described in detail previously (14). Briefly, RNA was extracted, reverse transcribed into cDNA, and amplified by individual 29 TCR VA- and 24 VB-specific primer pairs, and a runoff reaction using a fluorophore-labeled TCR-CA- or TCR-CB-specific primer was performed. Labeled amplicons were analyzed by DNA fragment analysis using appropriate size standards and a model 310 sequencer and Genescan software (ABI, Weiterstadt, Germany). To identify monoclonal and oligoclonal TCR transcripts, amplicons were subcloned into the TA sequencing vector (Invitrogen, Groningen, The Netherlands). TCR VA and VB were reported as monoclonal only if direct sequencing of the PCR amplicon or all subcloned PCR transcripts yielded the identical TCR sequence. To condense the information from a single sample analysis, the individual TCR VA or VB families may be grouped into a single figure with VA1 to VA29 or VB1 to VB24 along with the CDR3 length expressed as the number of amino acids. This TCR-CDR3 landscape provides the structural anatomy as defined by the TCR-CDR3 length for each TCR family in a T-cell subpopulation, which can be compared to the TCR-landscape in a different compartment (e.g., CD4+ TIL versus CD4+ PBL) (23). The CDR3 analysis was carried out with separated CD4+ and CD8+ T cells, since potential monoclonal or oligoclonal TCR VA or VB families in one T-cell population could be potentially “masked” if the same TCR VA or VB family shows a polyclonal composition (Fig. 1). TCR-CDR3 spectratyping of clinical specimens has been described in detail in the accompanying paper (23).
FIG. 1.
CD4+ and CD8+ T cells must be segregated for TCR CDR3 analysis. CD4+ sorted T cells (top), CD8+ sorted T cells (middle), or unseparated T cells (bottom) were subjected to TCR CDR3 analysis. As a paradigm, the TCR VA23 and the VCR VB7 analyses from TIL are depicted. Note that the rather monoclonal TCR VA23 peak in the CD4+ T-cell pool would be missed if nonsorted T cells had served as the source to generate cDNA for TCR CDR3 analysis.
Cytotoxicity and cytokine release assays.
The NK/LAK-sensitive target cell line Daudi or the NK-sensitive target cell line K562 served as target cells in cytotoxicity and cytokine release assays. Autologous fibroblasts were isolated and used as negative control targets for testing TIL obtained from individual patients. Autologous tumor cell lines were obtained using Dulbecco modified Eagle medium supplemented with 20% fetal calf serum and keratinocyte growth factor (1 μg/ml) obtained from R&D (Wiesbaden, Germany) and short-term exposure (3 days) to 200 μg of G418 per ml. which reduced the viability of fibroblasts but not of tumor cells. We could establish only 3 of 25 tumor samples in vitro. A different approach involved the transplantation of small pieces of tumor tissue under the kidney capsule of scid/beige mice, which yielded a success rate of 1 of 6 tumor samples. A standard 4-h chromium release assay was used to assess cytolytic recognition of target cells. Unless otherwise indicated, an effector-to-target ratio of 10:1 was used. 51Cr-labeled target cell suspensions were adjusted to 105 cells/ml, and 100 μl of this cell suspension was added to individual assay wells in triplicate determinations. Suspensions (100 μl) of effector cells were added to experimental wells, and the plates were incubated for 4 h at 37°C. Spontaneous-release wells received 100 μl of RPMI medium supplemented with fetal calf serum, and maximum-release wells received 100 μl of Triton X-100 (10% [vol/vol] in water). Aliquots of 100 μl were harvested from each well and counted in a gamma counter (Pharmacia, Stockholm, Sweden). Cytokine production was measured using the same effector-to-target ratio in a standard 24-h gamma interferon (IFN-γ) release assay as specified by the manufacturer (Diaclone, Besançon, France).
RESULTS
TCR composition in PBL, TIL, and tumor tissue: evidence for antigen-driven TCR selection.
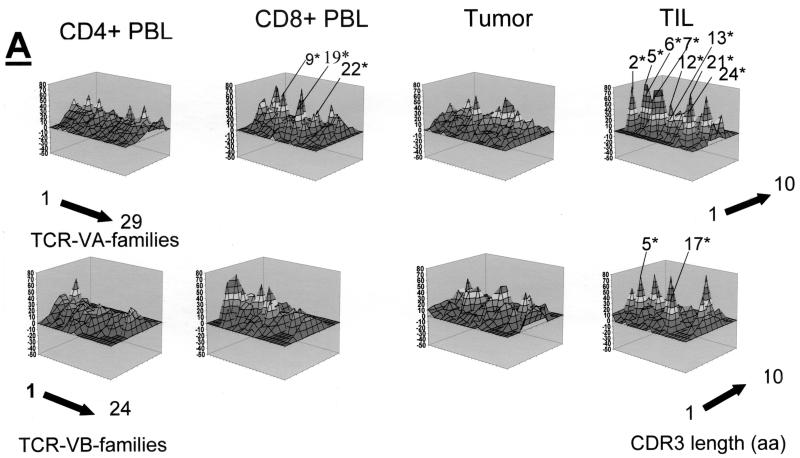
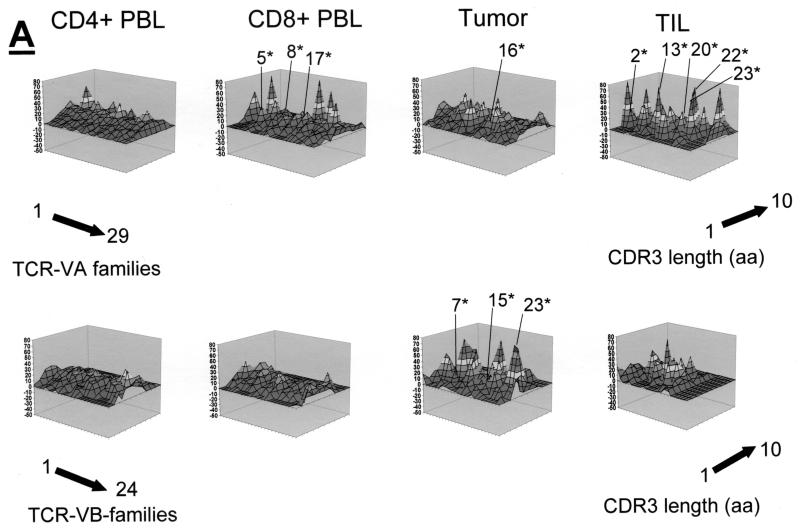
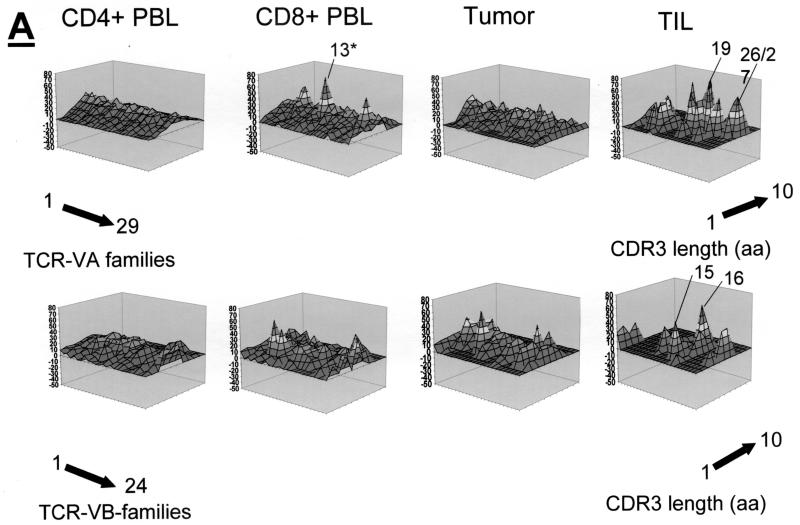
Qualitative TCR VA and VB analysis was carried out in CD4+, CD8+ PBL, and CD4+ TIL, as well as in fresh frozen tumor tissue sections from three patients who underwent surgery for cervical cancer. The qualitative TCR VA and VB CDR3 profiles from CD4+ PBL showed a normal Gaussian distribution in all three patients. TCR CDR3 VB analysis in CD8+ PBL showed an oligoclonal CDR3 pattern in the TCR VB as and TCR VA families; two of three patients exhibited at least three monoclonal TCR VA transcripts in the CD8+ population: patient MZ-CC01-1 had VA9, VA19, and VA22 (Fig. 2A), and patient MZ-CC01-2 had VA5, VA8S2, and VA17 (Fig. 3A). Patient MZ-CC01-3 exhibited a single TCR VA13 transcript in the CD8+ T-cell population (Fig. 4A).
FIG. 2.
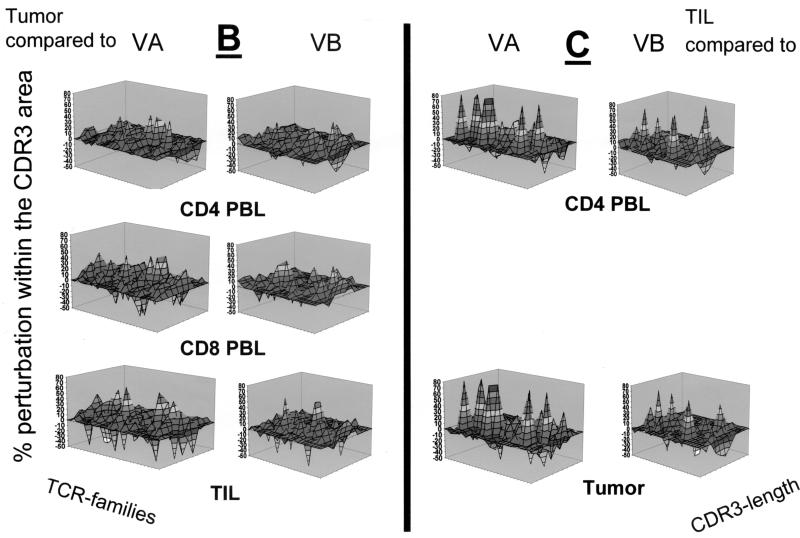
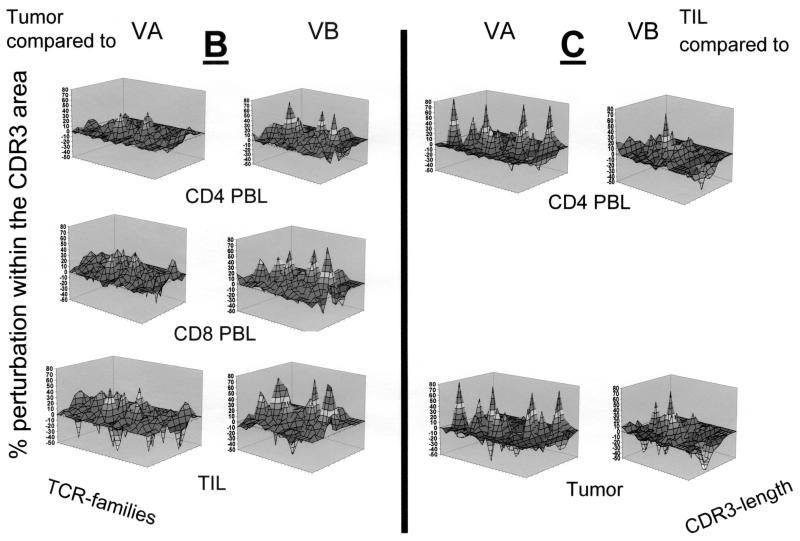
Qualitative assessment of the TCR repertoire in patient MZ-CC01-1. (A) TCR VA and VB landscape in PBL, freshly harvested tumor, and TIL. Monoclonal TCRs (listed in Table 2) are indicated by asterisks. Each shade represents 10% of the area under the curve in each individual VA (n = 29) or VB (n = 24) family. (B) Perturbation analysis of the TCR VA and VB repertoire in the tumor specimen compared to CD4+ and CD8+ PBL or CD4+ TIL. Each shade represents a 10% difference in the area under the curve in individual CDR3 peaks. An unperturbed (i.e., identical) repertoire would represent a smooth surface. Differences are shown as 10% over- or underrepresentation of VA and VB family peaks. (C) Perturbation analysis in CD4+ TIL compared to CD4+ PBL or tumor tissue. aa, amino acids.
FIG. 3.
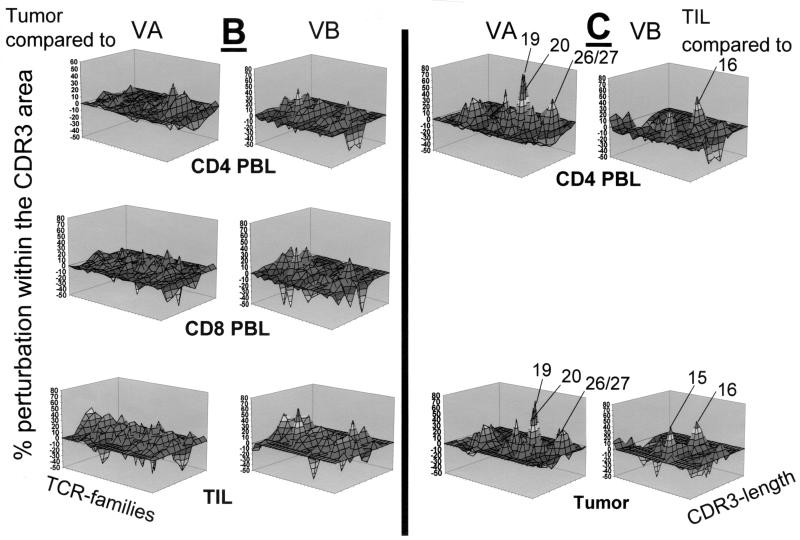
TCR repertoire in patient MZ-CC01-2. (A) TCR VA and VB analysis in CD4+ and CD8+ PBL, tumor, and TIL. (B) Comparative TCR analysis (perturbation) in tumor tissue compared to PBL or TIL. (C) TCR perturbation analysis in CD4+ TIL compared to CD4+ PBL or tumor tissue. Note that a single TCR VB family is overrepresented in TIL compared to CD4+ PBL. In contrast, a large number of TCR VA chains are overrepresented and show monoclonality as defined by DNA sequence analysis (Table 2). Asterisks indicate monoclonal TCRs. aa, amino acids.
FIG. 4.
TCR repertoire in patient MZ-CC01-3. (A) TCR VA and VB analysis in PBL, tumor, and TIL. Note that only a limited number of TCR VB chains are present in CD4+ TIL. (B) TCR perturbation analysis in tumor tissue compared to PBL or TIL. There are no major differences in regard to the TCR VA or VB composition in tumor tissue compared to PBL. (C) TCR perturbation analysis in CD4+ TIL compared to CD4+ PBL or tumor tissue. Note the overrepresentation of a selected number of TCR VA or VB chains in TIL. This CD4+ TIL line showed recognition of autologous tumor cells (Table 3). Asterisks indicate monoclonal TCRs. aa, amino acids.
In contrast to PBL, short-term-established TIL (>90% CD4+ [data not shown]) exhibited an oligoclonal TCR pattern. In patient MZ-CC01-1, nine individual TCR VA transcripts were monoclonal as determined by DNA sequence analysis. TIL from patient MZ-CC01-2 showed four TCR VA transcripts (Table 2). No monoclonal TCR mRNA transcripts were found in TIL from patient MZ-CC01-3. In contrast to TIL, two of three freshly harvested tumor samples did not show monoclonal TCR transcripts; the exception was the tumor sample from patient MZ-CC01-3. Of note, these monoclonal TCR transcripts could not be identified in TIL (Fig. 2A to 4A)
TABLE 2.
Monoclonal TCR transcriptsa
| Patient | Source | V family | V region | CDR3 region | Joining region | J family |
|---|---|---|---|---|---|---|
| MZ-CC01-1 | CD8+ PBL | VA 9 | SAMYYCA | PASRS | NTGKLIFGQG | AJ 37 |
| VA 19 | SAVYICA | VDSR | AAGNKLTFGGG | AJ 17 | ||
| VA 22 | SAVYFCA | LSPI | SSGSARQLTFGSG | AJ 22 | ||
| CD4+ TIL | VA 2 | SATYLCA | VNTY | SGNTPLVFGKG | AJ 29 | |
| VA 5 | SATYLCA | LDQG | DYKLSFGAG | AJ 20 | ||
| VA 6 | SAMYFCA | MREGY | SGNTPLVFGKG | AJ 29 | ||
| VA 7S2 | SASYLCA | VREQ | AAGNKLTFGGG | AJ 17 | ||
| VA 12 | SAVYFCA | PL | TNAGKSTFGDG | AJ 27 | ||
| VA 13 | SGVYFCA | V | SGYSTLTFGKG | AJ 11 | ||
| VA 21 | SAVYFCA | AKD | QAGTALIFGKG | AJ 15 | ||
| VA 24 | SASYICV | VP | NTDKLIFGTG | AJ 34 | ||
| VB 5S8 | SALYLCA | SSFRIGY | GYTFGSG | BJ 1-2 | ||
| VB 17 | TAFYLCA | SREWGD | TEAFFGQG | BJ 1.1 | ||
| MZ-CC01-2 | CD8+ PBL | VA 5 | SATYLCA | LDSR | AAGNKLTFGGG | AJ 17 |
| VA 8S2 | SAVYFCA | ENTF | NQGGKLIFGQG | AJ 23 | ||
| VA 17 | SATYFCA | ATYV | NSGGGADGLTFGKG | AJ 45 | ||
| CD4+ TIL | VA 2 | SATYLCV | VPDGI | NDMRFGAG | AJ 43 | |
| VA 13 | SGVYFCA | VERW | ARLMFGDG | AJ 31 | ||
| VA 20 | TAVYYCL | VGPLM | NSGGYQKVTFGIG | AJ 13 | ||
| VA 22 | SAVYFCA | LKGR | NNNDMRFGAG | AJ 43 | ||
| VA 23 | SATYLCA | VR | SGAGSYQLTFGKG | AJ 28 | ||
| Tumor | VA 16 | SALYFCA | VRDHS | SGGYNKLIFGAG | AJ 4 | |
| VB 7S1 | SALYLCA | SSQEGDL | YEQYFGPG | BJ 2-7 | ||
| VB 15 | TALYFCA | TSDSDSGD | TDTQYFGPG | BJ 2-3 | ||
| VB 23 | SALYFCA | SSQDN | SYEQYFGPG | BJ 2-7 | ||
| MZ-CC01-3 | CD8+ PBL | VA 13 | SGVYFCA | VDAGY | GNEKLTFGTG | AJ 48 |
cDNA was generated from CD4+ and CD8+ PBL, freshly harvested tumor tissue, and CD4+ TIL and tested for monoclonal TCR VA or VB transcripts as described in the text. TCR VA or VB families were characterized for monoclonality by direct sequencing of the amplified product from each TCR family. Note the large number of monoclonal TCR VA mRNA transcripts in CD4+ TIL from patient MZ-CC01-1. Monoclonal TCR transcripts detected in freshly harvested tumor tissue could not be detected in TIL or in PBL. Thus, individual TCR clonotypes could not be expanded in vitro. Short-term (7 days) in vitro culture of T cells (TIL) in the presence of autologous tumor and IL-7 resulted in an oligoclonal T-cell population.
TCR composition in freshly harvested tumor tissue.
TCR VA and VB transcripts revealed that the TCR VA chain landscape exhibits a Gaussian distribution with no major alterations. In contrast, the TCR VB repertoire showed an oligoclonal TCR composition. Of note, the definition of a Gaussian distribution of TCR VA transcripts within the tumor tissue suggests only that a broad repertoire of TCR VA chains may be available within the lesion. It does not imply that the Gaussian distribution identified within the tumor tissue is identical to a (potential) Gaussian distribution of the TCR VA or VB chains in PBL or TIL, since the TCR landscape describes the length of the TCR CDR3 region, which may have a different length distribution in PBL or TIL (Fig. 2A to 4A).
Comparative TCR analysis in tumor tissue and TIL.
Recent studies suggested that monoclonal or oligoclonal TCR transcripts are indicative of ongoing cellular immune responses. Therefore, for each individual we compared the CDR3 TCR repertoire of the tumor lesion to the TCR repertoire obtained from CD4+ PBL, CD8+ PBL, or CD4+ TIL (Fig. 2B to 4B). In general, differences were more visible in the TCR VA landscape than in the VB repertoire if TCRs present in tumor tissue were compared to those in CD4+ or CD8+ PBL. Substantial differences were identified in the composition of both the TCR VA and VB repertoires in tumor lesions with respect to the short-term-established TIL lines defined by over- or underrepresentation of individual TCR VA or VB families. As expected, comparison of the CD4+ TIL lines to TCR VA and VB transcripts in PBL and tumor tissue (Fig. 2C to 4C) exhibited substantial differences pertaining to TCR VA and VB mRNA transcripts. Of note, CD4+ TIL from patient MZ-CC01-3 showed overrepresentation of the TCR VB16 family compared to that of CD4+ PBL or snap-frozen tumor tissue (Fig. 4C).
Recognition of autologous tumor cells.
CD4+ TIL lines were established from three patients (Table 1) and tested for recognition of autologous tumor cells using a standard 51Cr release and a 48-h IFN-γ release assay. Only one of three TIL lines (i.e., the line from patient MZ-CC01-3) showed MHC class II (DR)-restricted recognition of the autologous tumor cells (Table 3). TIL did not lyse MHC class I-matched target cells of different histology or HLA-DR,-DQ-matched cervical cancer cell lines harboring the identical HPV type (e.g., the cell line Caski, HPV-16+). Exclusively autologous tumor cells, but not autologous fibroblasts, were lysed. Lysis could be blocked using an anti-HLA-DR but not an anti-MHC class I monoclonal antibody (MAb). No difference was observed between cytolysis and IFN production; only TIL from patient MZ-CCO1-3 reacted in a HLA-DR restricted fashion.
TABLE 3.
Recognition of autologous tumor cellsa
| Histology | Target cell lines | HPV type | Effector TIL % spe- cific lysis | Shared MHC alleles |
|---|---|---|---|---|
| Hematopoietic | Daudi | 0 | ||
| K562 | 0 | |||
| Melanoma | Mel 397 | 15 | A1 | |
| Mel 642 | 2 | |||
| Renal cell cancer | RCC1940 | 0 | A1 | |
| RCC1897 | 0 | A1, C1 | ||
| Colorectal cancer | SW480 | 0 | B7 | |
| HT29 | 0 | A1 | ||
| Cervical cancer | HT3 | 0 | Nil | |
| C33-A | 0 | B7 | ||
| Caski | 16 | 0 | B7, DR8, 15, DQ4, 6 | |
| Siha | 16 | 0 | DR15, DQ6 | |
| C41 | 18 | 0 | A1, DR8, DQ4 | |
| MS751 | 18 | 0 | ||
| Me180 | 68 | 0 | A1, C7 | |
| CC-H1 | 16 | 40 | All alleles (autol- ogous tumor) | |
| CC-H1+ anti- class I MAb | 16 | 38 | ||
| CC-H1+ anti- class II MAb | 16 | 15 | ||
| Fibroblasts | CC-H1− fibroblasts | 0 |
Tumor cell lines of different histology were used to evaluate MHC class II-restricted recognition of the CD4+ TIL line from patient MZ-CC01-3 in a standard 51Cr-release assay. Note that TIL recognize exclusively autologous tumor cells, which could be blocked using an anti-MHC class II (DR)-specific MAb (clone 243). TIL do not recognize the cervical cancer cell line Caski, which has both HLA-DR and HLA-DQ alleles and also harbors HPV-16. All target cell lines could potentially be lysed, since they represent targets for 7-day LAK cells cultured in 1,000 IU of IL-2/ml. TIL lines from patients MZ-CC01-1 or MZ-CC01-2 did not lyse autologous tumor cells (data not shown). We noted no difference between lysis and cytokine (IFN-γ) production regarding the response to the targets listed in the table (data not shown).
DISCUSSION
The three salient and novel findings of this report are as follows. (i) Monoclonal TCR transcripts were detected predominantly in CD8+ PBL but not in CD4+ T lymphocytes and monoclonal TCR (VA) transcripts in the freshly harvested tumor from patients with cervical cancer. (ii) A large number of monoclonal TCR mRNA (VA) transcripts were found in short-term-established CD4+ TIL from two of three patients. These monoclonal TCRs are not present in PBL or in tumor tissue. (iii) Evidence was found for a skewed TCR repertoire in in vitro-expanded TIL compared to the cancer lesions. Thus, T-cell reactivity may not describe the in situ TCR reactivity. (iv) The autologous tumor cells defined by cytolysis in an HLA-DR restricted fashion were recognized in one of three in TIL lines. The last observation has not been reported previously. Of note, TIL lines have not been restimulated in vitro with peptides or recombinant proteins.
The TCR VA and VB CDR3 analysis has been used to gauge the cellular immune response in patients with cancer, including renal cell cancer (17), melanoma (19), non-small-cell lung cancer (5), and lymphoma (27). In general, the identification of monoclonal (or oligoclonal) TCR mRNA transcripts has been interpreted as a selected TCR repertoire due to antigenic expansion of single T-cell clones (10). Caution should be exercised if only PBL are analyzed: multiple monoclonal TCR mRNA transcripts can be detected in PBL from apparently healthy individuals and may reflect the remnants of previous encounters with any antigen (11). However, only a few monoclonal TCR transcripts can be detected in PBL from healthy individuals (10, 11, 22, 24). The identification of an oligoclonal TCR repertoire or more than two or three monoclonal TCRs is indicative of an ongoing cellular immune response resulting in the (limited) selection of a few TCR clonotypes (10, 22). Of interest, the detection of monoclonal TCR VA chains is more frequent than that of TCR VB chains (Table 2). Skewing of the TCR repertoire associated with a narrow TCR VA chain repertoire has been attributed to structural constraints by MHC class I-restricted T cells (30); the role of MHC class II-restricting molecules has not yet been defined. A similar bias to a narrowed TCR VA repertoire has been observed in the cellular immune response to Epstein-Barr virus (18). It is not clear if such a bias is indicative of any active MHC class I- or II-restricted immune response or if the selection of certain VA families may be associated with viral infections.
The notion that freshly harvested tumor tissue obtained from patients with cervical cancer showed oligo- or monoclonal expansion of some TCR VB families (Table 2) suggests an ongoing cellular immune response in situ. Oligo- or monoclonal TCR transcripts may also not be accessible to this reverse transcription-PCR-based detection, since the tumor tissue has not been dissected for CD4 and CD8 T cells. Monoclonal selection in either of these T cell subsets may be masked by a polyclonal TCR pattern in the CD4 or the CD8 T-cell population (Fig. 1). The tumor lesions showed CD4 and CD8 T-cell infiltrates (data not shown). This is not the case for the sorted CD4+ and CD8+ T cells obtained from PBL or for the TIL lines, which showed >90% CD4+ T cells (data not shown). Three of three short-term-cultured TIL lines exhibited a very oligoclonal TCR pattern, and more than 30% of the entire TCR VA repertoire (9 of 29 VA families in patient MZ-CC01-1) was composed of single clones. It is also evident from these data that the TIL lines used in this study did not reflect the original T-cell composition present within the tumor lesions. Thus, T-cell reactivity to autologous tumor cells (in TIL) may not reflect the in situ composition of the T-cell infiltrate present within the cervical cancer lesion. These TIL were cultured in 50 IU of IL-2 and 50 pg of IL-7 per ml in the presence of autologous tumor. Recent results suggested that IL-7 does not result in nonspecific alterations of the T-cell population but, rather, exerts a homeostatic effect by increasing the number of antigen-specific T cells (8). Therefore, differences in the TCR composition of TIL observed in this report may also reflect the antigen-driven expansion of certain TCR VA and VB subsets which may have already been selected for IL-7-dependent growth in vivo (20). In addition, gain of TIL effector function may not be correlated with expansion of certain TCR families (8). However, the ultimate proof that ex vivo-cultured TIL or PBL are different from the “tumor compartment” pertaining to the T-cell repertoire can be obtained only if the tumor sample can be subjected to microdissection, allowing the separation of CD4+ and CD8+ T cells.
In contrast to the freshly harvested tumor tissue, CD4+ TIL lines could be compared to CD4+ PBL harvested at the time of tumor resection. TCR CDR3 analysis may represent a useful tool to describe the T-cell infiltration in situ, particularly if cancer lesions are easily accessible, e.g., in patients with melanoma or with carcinoma in situ of the cervix. Thus, the in situ T-cell infiltrate can be measured as a surrogate marker for (antigen-driven) inflammation in the context of a therapeutic or prophylactic HPV vaccine, which is currently being tested in patients with cervical cancer or in individuals at elevated risk of ultimately developing cervical cancer (2, 26).
The hypothesis that TCR CDR3 analysis represents a surrogate marker for antigen-driven selection is supported by the observation that one of three TIL lines showed recognition of the autologous tumor cells as defined by cytolysis. The observation that only one of three TIL recognized the autologous tumor may be because (i) tumor-reactive T-cells may have undergone activation-induced cell death (21) during in vitro culture or (ii) immunosuppressive factors of the tumor microenvironment, potentially mediated by IL-10 or transforming growth factor β (33), may be counterproductive in expansion or maintenance of tumor-specific T cells (4, 7). A number of cytotoxic CD4+ T cells have been identified in tumors of different histology. A CD4+ T-cell population which is able to lyse autologous tumor cells is particularly interesting as a tool to define the antitumor CD4+ T-cell response at the molecular level. However, cytotoxic CD4+ T cells isolated from cervical cancer lesions appear not to be common in our experience.
It may be argued that the detection of MHC class II-restricted CD4+ T cells targeting the autologous tumor may not be of biological significance, since T-helper cells may instead aid either B cells or CD8+ cytotoxic T-cells in exerting and sustaining their function. However, recent studies suggested that MHC class II-positive tumor cells may themselves act as antigen-presenting cells and present a different set of epitopes from those presented by professional antigen-presenting cells (i.e., dendritic cells) (25). Here we show that HLA-DR-restricted and tumor-specific CD4+ T cells are indeed capable of lysing autologous cervical cancer cells. The fact that an HLA-DR-matched cervical cancer cell line with the identical HPV type (Table 3) is not lysed indicates that the antitumor response in cervical cancer may not always represent an anti-HPV response. Alternatively, defects in the MHC class II antigen-processing and presentation machinery may also account for differences in target cell recognition (34). Indeed, “private antigens,” unique for individual patients, have been identified (for a review, see reference 35) and may also be present in patients with cervical cancer.
Acknowledgments
This work was supported by grant SFB 432 (A9) from the Deutsche Forschungsgemeinschaft to M.M. and a core grant from the Deutsche Krebshilfe (Tumor Vaccination Center, Mainz, Germany).
REFERENCES
- 1.Bontkes, H. J., T. D. de Gruijl, A. J. van den Muysenberg, R. H. Verheijen, M. J. Stukart, C. J. Meijer, R. J. Scheper, S. N. Stacey, M. F. Duggan-Keen, P. L. Stern, S. Man, L. K. Borysiewicz, and J. M. Walboomers. 2000. Human papillomavirus type 16 E6/E7-specific cytotoxic T lymphocytes in women with cervical neoplasia. Int. J. Cancer 88:92-98. [PubMed] [Google Scholar]
- 2.Connett, H. 2001. HPV vaccine moves into late stage trials. Nat. Med. 7:388.. [DOI] [PubMed] [Google Scholar]
- 3.Cromme, F. V., J. Airey, M. T. Heemels, H. L. Ploegh, P. J. Keating, P. L. Stern, C. J. Meijer, and J. M. Walboomers. 1994. Loss of transporter protein, encoded by the TAP-1 gene, is highly correlated with loss of HLA expression in cervical carcinomas. J. Exp. Med. 179:335-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dworacki, G., N. Meidenbauer, I. Kuss, T. K. Hoffmann, W. Gooding, M. Lotze, and T. L. Whiteside. 2001. Decreased zeta chain expression and apoptosis in CD3+ peripheral blood T lymphocytes of patients with melanoma. Clin. Cancer Res. 7:947S-957S. [PubMed]
- 5.Echchakir, H., C. Asselin-Paturel, G. Dorothee, I. Vergnon, D. Grunenwald, S. Chouaib, and F. Mami-Chouaib. 1999. Analysis of T-cell-receptor beta-chain-gene usage in peripheral-blood and tumor-infiltrating lymphocytes from human non-small-cell lung carcinomas. Int. J. Cancer 81:205-213. [DOI] [PubMed] [Google Scholar]
- 6.Evans, E. M., S. Man, A. S. Evans, and L. K. Borysiewicz. 1997. Infiltration of cervical cancer tissue with human papillomavirus-specific cytotoxic T-lymphocytes. Cancer Res. 57:2943-2950. [PubMed] [Google Scholar]
- 7.Gastman, B. R., D. E. Johnson, T. L. Whiteside, and H. Rabinowich. 1999. Caspase-mediated degradation of T-cell receptor zeta-chain. Cancer Res. 59:1422-1427. [PubMed] [Google Scholar]
- 8.Geiselhart, L. A., C. A. Humphries, T. A. Gregorio, S. Mou, J. Subleski, and K. L. Komschlies. 2001. IL-7 administration alters the CD4:CD8 ratio, increases T cell numbers, and increases T cell function in the absence of activation. J. Immunol. 166:3019-3027. [DOI] [PubMed] [Google Scholar]
- 9.Gerard, C. M., N. Baudson, K. Kraemer, C. Ledent, D. Pardoll, and C. Bruck. 2001. Recombinant human papillomavirus type 16 E7 protein as a model antigen to study the vaccine potential in control and E7 transgenic mice. Clin. Cancer Res. 7:838S-847S. [PubMed]
- 10.Gorochov, G., A. U. Neumann, A. Kereveur, C. Parizot, T. Li, C. Katlama, M. Karmochkine, G. Raguin, B. Autran, and P. Debre. 1998. Perturbation of CD4+ and CD8+ T-cell repertoires during progression to AIDS and regulation of the CD4+ repertoire during antiviral therapy. Nat. Med. 4:215-221. [DOI] [PubMed] [Google Scholar]
- 11.Hingorani, R., I. H. Choi, P. Akolkar, B. Gulwani-Akolkar, R. Pergolizzi, J. Silver, and P. K. Gregersen. 1993. Clonal predominance of T cell receptors within the CD8+ CD45RO+ subset in normal human subjects. J. Immunol. 151:5762-5769. [PubMed] [Google Scholar]
- 12.Hohn, H., H. Pilch, S. Gunzel, C. Neukirch, K. Freitag, A. Necker, and M. J. Maeurer. 2000. Human papillomavirus type 33 E7 peptides presented by HLA-DR*0402 to tumor-infiltrating T cells in cervical cancer. J. Virol. 74:6632-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hohn, H., H. Pilch, S. Gunzel, C. Neukirch, C. Hilmes, A. Kaufmann, B. Seliger, and M. J. Maeurer. 1999. CD4+ tumor-infiltrating lymphocytes in cervical cancer recognize HLA-DR-restricted peptides provided by human papillomavirus-E7. J. Immunol. 163:5715-5722. [PubMed] [Google Scholar]
- 14.Hohn, H., T. Reichert, C. Neukirch, H. Pilch, and M. J. Maeurer. 1999. Monoclonal TCR mRNA transcripts are preferentially detected in the TCR variable alpha chain in CD8(+) T-lymphocytes: implications for immunomonitoring. Int. J. Mol. Med. 3:139-144. [DOI] [PubMed] [Google Scholar]
- 15.Jochmus, I., W. Osen, A. Altmann, G. Buck, B. Hofmann, A. Schneider, L. Gissmann, and H. G. Rammensee. 1997. Specificity of human cytotoxic T lymphocytes induced by a human papillomavirus type 16 E7-derived peptide. J. Gen. Virol. 78:1689-1695. [DOI] [PubMed] [Google Scholar]
- 16.Koopman, L. A., W. E. Corver, A. R. van der Slik, M. J. Giphart, and G. J. Fleuren. 2000. Multiple genetic alterations cause frequent and heterogeneous human histocompatibility leukocyte antigen class I loss in cervical cancer. J. Exp. Med. 191:961-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurokawa, T., M. Oelke, and A. Mackensen. 2001. Induction and clonal expansion of tumor-specific cytotoxic T lymphocytes from renal cell carcinoma patients after stimulation with autologous dendritic cells loaded with tumor cells. Int. J. Cancer 91:749-756. [DOI] [PubMed] [Google Scholar]
- 18.Lim, A., L. Trautmann, M. A. Peyrat, C. Couedel, F. Davodeau, F. Romagne, P. Kourilsky, and M. Bonneville. 2000. Frequent contribution of T cell clonotypes with public TCR features to the chronic response against a dominant EBV-derived epitope: application to direct detection of their molecular imprint on the human peripheral T cell repertoire. J. Immunol. 165:2001-2011. [DOI] [PubMed] [Google Scholar]
- 19.Maccalli, C., C. Farina, M. Sensi, G. Parmiani, and A. Anichini. 1997. TCR beta-chain variable region-driven selection and massive expansion of HLA-class I-restricted antitumor CTL lines from HLA-A*0201+ melanoma patients. J. Immunol. 158:5902-5913. [PubMed] [Google Scholar]
- 20.Maeurer, M. J., W. Walter, D. Martin, L. Zitvogel, E. Elder, W. Storkus, and M. T. Lotze. 1997. Interleukin-7 (IL-7) in colorectal cancer: IL-7 is produced by tissues from colorectal cancer and promotes preferential expansion of tumour infiltrating lymphocytes. Scand. J. Immunol. 45:182-192. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen, T., and J. Russell. 2001. The regulation of FasL expression during activation-induced cell death (AICD). Immunology 103:426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pannetier, C., J. Even, and P. Kourilsky. 1995. T-cell repertoire diversity and clonal expansions in normal and clinical samples. Immunol. Today 16:176-181. [DOI] [PubMed] [Google Scholar]
- 23.Pilch, H., H. Höhn, K. Freitag, C. Neukirch, A. Necker, P. Haddad, P. G. Knapstein, and M. J. Maeurer. 2002. Improved assessment of the T-cell receptor (TCR) VB repertoire in clinical specimens: combination of TCR-CDR3 spectratyping with flow cytometry-based TCR VB frequency analysis. Clin. Diagn. Lab. Immunol. 9:257-266. [DOI] [PMC free article] [PubMed]
- 24.Posnett, D. N., R. Sinha, S. Kabak, and C. Russo. 1994. Clonal populations of T cells in normal elderly humans: the T cell equivalent to “benign monoclonal gammapathy.” J. Exp. Med. 179:609-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi, L., J. M. Rojas, and S. Ostrand-Rosenberg. 2000. Tumor cells present MHC class II-restricted nuclear and mitochondrial antigens and are the predominant antigen presenting cells in vivo. J. Immunol. 165:5451-5461. [DOI] [PubMed] [Google Scholar]
- 26.Ressing, M. E., W. J. van Driel, R. M. Brandt, G. G. Kenter, J. H. de Jong, T. Bauknecht, G. J. Fleuren, P. Hoogerhout, R. Offringa, A. Sette, E. Celis, H. Grey, B. J. Trimbos, W. M. Kast, and C. J. Melief. 2000. Detection of T helper responses, but not of human papillomavirus-specific cytotoxic T lymphocyte responses, after peptide vaccination of patients with cervical carcinoma. J. Immunother. 23:255-266. [DOI] [PubMed] [Google Scholar]
- 27.Rezvany, M. R., M. Jeddi-Tehrani, A. Osterborg, E. Kimby, H. Wigzell, and H. Mellstedt. 1999. Oligoclonal TCRBV gene usage in B-cell chronic lymphocytic leukemia: major perturbations are preferentially seen within the CD4 T-cell subset. Blood 94:1063-1069. [PubMed] [Google Scholar]
- 28.Rudolf, M. P., S. Man, C. J. Melief, A. Sette, and W. M. Kast. 2001. Human T-cell responses to HLA-A-restricted high binding affinity peptides of human papillomavirus type 18 proteins E6 and E7. Clin. Cancer Res. 7:788S-795S. [PubMed]
- 29.Santin, A. D., P. L. Hermonat, A. Ravaggi, S. Bellone, J. J. Roman, S. Jayaprabhu, S. Pecorelli, G. P. Parham, and M. J. Cannon. 2001. Expression of CD56 by human papillomavirus E7-specific CD8+ cytotoxic T lymphocytes correlates with increased intracellular perforin expression and enhanced cytotoxicity against HLA-A2-matched cervical tumor cells. Clin. Cancer Res. 7:804S-810S. [PubMed]
- 30.Sim, B. C., L. Zerva, M. I. Greene, and N. R. Gascoigne. 1996. Control of MHC restriction by TCR Valpha CDR1 and CDR2. Science 273:963-966. [DOI] [PubMed] [Google Scholar]
- 31.Thornburg, C., D. Boczkowski, E. Gilboa, and S. K. Nair. 2000. Induction of cytotoxic T lymphocytes with dendritic cells transfected with human papillomavirus E6 and E7 RNA: implications for cervical cancer immunotherapy. J. Immunother. 23:412-418. [DOI] [PubMed] [Google Scholar]
- 32.van der Burg, S. H., M. E. Ressing, K. M. Kwappenberg, A. de Jong, K. Straathof, J. de Jong, A. Geluk, K. E. van Meijgaarden, K. L. Franken, T. H. Ottenhoff, G. J. Fleuren, G. Kenter, C. J. Melief, and R. Offringa. 2001. Natural T-helper immunity against human papillomavirus type 16 (HPV16) E7-derived peptide epitopes in patients with HPV16-positive cervical lesions: identification of 3 human leukocyte antigen class II-restricted epitopes. Int. J. Cancer 91:612-618. [DOI] [PubMed] [Google Scholar]
- 33.von Bernstorff, W., M. Voss, S. Freichel, A. Schmid, I. Vogel, C. Johnk, D. Henne-Bruns, B. Kremer, and H. Kalthoff. 2001. Systemic and local immunosuppression in pancreatic cancer patients. Clin. Cancer Res. 7:925S-932S. [PubMed]
- 34.Walter, W., K. Lingnau, E. Schmitt, M. Loos, and M. J. Maeurer. 2000. MHC class II antigen presentation pathway in murine tumors: tumour evasion from immunosurveillance? Br. J. Cancer 83:1192-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, R. F., and S. A. Rosenberg. 1999. Human tumor antigens for cancer vaccine development. Immunol. Rev. 170:85-100. [DOI] [PubMed] [Google Scholar]