Abstract
Autoimmune, inflammatory, and infectious diseases present a major burden to human health and are frequently associated with loci in the human major histocompatibility complex (MHC). Here, we report a high-resolution (1.9 kb) linkage-disequilibrium (LD) map of a 4.46-Mb fragment containing the MHC in U.S. pedigrees with northern and western European ancestry collected by the Centre d'Etude du Polymorphisme Humain (CEPH) and the first generation of haplotype tag single-nucleotide polymorphisms (tagSNPs) that provide up to a fivefold increase in genotyping efficiency for all future MHC-linked disease-association studies. The data confirm previously identified recombination hotspots in the class II region and allow the prediction of numerous novel hotspots in the class I and class III regions. The region of longest LD maps outside the classic MHC to the extended class I region spanning the MHC-linked olfactory-receptor gene cluster. The extended haplotype homozygosity analysis for recent positive selection shows that all 14 outlying haplotype variants map to a single extended haplotype, which most commonly bears HLA-DRB1*1501. The SNP data, haplotype blocks, and tagSNPs analysis reported here have been entered into a multidimensional Web-based database (GLOVAR), where they can be accessed and viewed in the context of relevant genome annotation. This LD map allowed us to give coordinates for the extremely variable LD structure underlying the MHC.
Introduction
Characterization of patterns of linkage disequilibrium (LD) at a fine scale across the genome is central to the optimal design and execution of association studies of common variants that influence susceptibility of common diseases, as well as the understanding of processes such as recombination, mutation, and selection. LD maps report directly on marker correlation in the population and can thus guide marker selection for association studies. It is well established that regions of high LD display low haplotype diversity; thus, common haplotypes can be efficiently tagged with only a subset of all common variants, known as “haplotype tag SNPs” (tagSNPs) (Daly et al. 2001; Johnson et al. 2001; Chapman et al. 2003; Goldstein et al. 2003; Clayton et al. 2004). However, the use of tagSNPs carries some loss of power for detection of all other common variants, with the exception of completely correlated markers. Many methods exist to identify an optimal set of tagSNPs that are based on observed correlations among an initial set of markers, to avoid the collection of redundant information (see Goldstein et al. [2003] for review). These methods take advantage of high-resolution LD maps that show increasing efficiency at higher marker density (Ke et al. 2004a).
Several linkage scans and association-mapping studies of most, if not all, autoimmune diseases—such as type 1 diabetes, multiple sclerosis, systemic lupus erythematosus (Davies et al. 1994; Marchini et al. 2003; Harbo et al. 2004; Marrosu et al. 2004; Oksenberg et al. 2004)—and infectious diseases like malaria and AIDS (Hill et al. 1991; Carrington and O’Brien 2003) have given strong signals in the major histocompatibility complex (MHC), a biologically and medically important region on human chromosome 6p that harbors many immune genes. Partly because of these disease studies, this region attracted a lot of attention, particularly with respect to the structure of LD and recombination. Whereas, historically, only the classical HLA loci were studied by genotyping, recent systematic efforts to characterize LD patterns across the whole of the MHC have been reported at increasing marker densities by use of SNPs (Walsh et al. 2003; Stenzel et al. 2004). These complement a number of studies of the recombinational rates in the region. In three different studies, Cullen and colleagues (1995, 1997, 2002) have mapped recombination rates across the MHC class II region and have demonstrated the presence of recombination hotspots near HLA-DNA, BRD2 (formerly RING3), and HLA-DQB1 and within the TAP2 gene. But, perhaps most importantly, examining the relationship between LD and recombination with a combination of SNP genotyping and sperm recombination mapping, Jeffreys et al. (2001) demonstrated directly the correlation between regions of low LD and the presence of recombination hot spots in a discrete 200-kb segment of the MHC. However, all the studies named above have been limited by the unavailability of a dense SNP map of the entire MHC. Recently, the total number of known SNPs across the MHC has risen to >36,000 (dbSNP121), mainly through efforts of the HapMap Project (The International HapMap Consortium 2003) and the complete resequencing of the MHC region in homozygous cell lines, which contributed >50% of the available SNPs (Stewart et al. 2004). This vast resource allowed us to construct a 1.9-kb–resolution LD map across the MHC in U.S. pedigrees with northern and western European ancestry collected by CEPH and to suggest a first-generation set of haplotype tagSNPs for use in association studies. The map shows a high degree of variability in average LD between MHC subregions and reveals that the olfactory-receptor gene cluster in the extended class I region falls within the largest regions of high LD currently known in the human genome. Using Phase 2.1 (Li and Stephens 2003; Crawford et al. 2004) and the method described by McVean et al. (2004), we obtain estimates of recombination rates across the entire MHC region and show that 90% of predicted peaks (hotspots) correlate with LD breaks.
Material and Methods
DNA Samples and Genotyping
We selected a total of 3,892 SNPs, across a 4.46-Mb region spanning the MHC (human chromosome 6, 28,918,812–33,377,873 bp [National Center for Biotechnology Information build 34]), from dbSNP121 and the International HapMap Project. Selection criteria required that SNPs have spacing >500 bp and have an assay design for Illumina’s Golden Gate genotyping platform (Oliphant et al. 2002). Markers were typed across a panel of 190 DNA samples from CEPH families (Utah residents with ancestry from northern and western Europe); DNA was obtained from the Coriell Cell Repository. In total, 180 founder chromosomes were used for the haplotype construction. Note that this panel includes all the samples used both by the International HapMap Project and in the map by Walsh et al. (2003) and hence allowed full integration of data on the ∼820 markers genotyped by this effort (see sample list at the Human Chromosome 6 Project Overview Web site). Genotyping was performed at a multiplex level of 1,536 SNPs per well, and data quality was assessed by duplicate DNAs (n=8) and inheritance tests among parent-child trios. SNPs with more than two discrepant calls were removed. PEDCHECK (O’Connell and Weeks 1998) and PEDSTATS (Center for Statistical Genetics) were used to identify and remove genotypes that lead to Mendelian inconsistencies. The genotype confidence score for keeping data was set to 0.25. Finally, we removed loci with <80% of all possible genotypes, out of Hardy-Weinberg equilibrium (χ2⩾10) and zero heterozygosity.
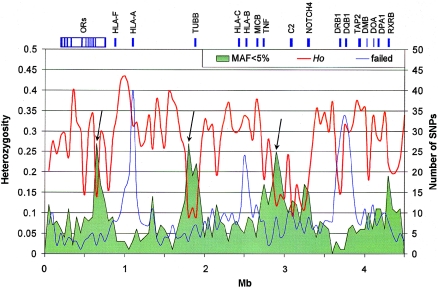
In total, we retained 3,122 SNPs for further analysis. Of those, 787 were either nonpolymorphic or had a minor-allele frequency (MAF) <5% in our sample (5% is the MAF cutoff used by International HapMap Project). The distribution of these 787 markers is not random, which suggests variation deserts in three regions in which the density of such markers is clearly increased (fig. 1, black arrows). Thus, we compared the heterozygosity distribution obtained here with that from a genotyping data set of 396 SNPs assayed with the MassEXTEND assay and mass spectrometry (Sequenom platform) in the same sample (Walsh et al. 2003). Data sets from both platforms showed the same trend, which supports the presence of low-heterozygosity regions rather than genotyping problems. We also observed a higher failure rate (fig. 1, blue line) around the highly polymorphic genes HLA-A, HLA-B, HLA-C, HLA-DQB, and HLA-DRB1, which suggests that alternative typing assays may be needed in these regions. The 2,335 loci with MAF ⩾5%—an average spacing of 1 SNP per 1.9 kb—were used in all further analyses (marker and raw genotypes are available at the Human Chromosome 6 Project Overview Web site and have been submitted to dbSNP).
Figure 1.
Low-heterozygosity regions in the MHC. The observed heterozygosity (“Ho,” red line) averaged in 50-kb windows is plotted with the number of loci with MAF <5% (green area). SNPs showing MAF <5% are not randomly distributed across the analyzed 4.459-Mb region. Specifically, three regions (black arrows) presented loss of heterozygosity; most SNPs are monomorphic in this population. These fragments contain OR2H1 and MAS1L; DHX16, NRM, MDC1 TUBB, and FLOT1; BAT5, LY6GD, C6orf25, DDAH2, C6orf26, and VARS2, respectively. The uneven distribution of the 770 SNPs that failed to generate genotyping data is represented by the blue line (“failed,” number of SNPs in 50-kb windows). Typing failures are concentrated mainly in three genomic regions (blue peaks), including HLA-A, HLA-B, HLA-C, HLA-DQB1, and HLA-DRB1, which suggests that the highly polymorphic nature of these genes might be responsible for the failure.
Estimation of LD
To capture the strength of LD between markers, we estimated two LD measurements on the basis of the pairwise disequilibrium coefficient—D′ (Lewontin 1964) and r2—by using the HaploXT program in the GOLD package (Abecasis and Cookson 2000). Nonrecombinant haplotypes were derived by employing Merlin (Abecasis et al. 2002) and were then used to estimate haplotype frequencies in founder chromosomes. To compare decay rates among MHC regions, the decay of LD was plotted as a function of physical distance. The decay rates represent the average r2 and D′ values for all markers separated by distance S (for S=10 kb, 20 kb, 30 kb,…500 kb). Moving average (sliding window [SW]) of pairwise LD coefficients (r2) was performed at 500-kb windows across the MHC to display general trends of LD. Pairwise r2 values from all markers spaced between 25 kb and 250 kb were averaged and plotted in 500-kb successive windows. At each step, the 500-kb window moved forward 50 kb. A short increment between windows—or large overlapping—contributes to smoothing the curve and avoids distinct peaks. LD patterns, particularly LD breaks and high-LD long-range regions, were visualized using the GOLDsurfer program (Pettersson et al. 2004) independent of block assessment.
To define haplotype blocks, a set of consecutive sites between which there is little or no evidence of historical recombination, we adopted the D′-based criteria of Gabriel et al. (2002). Haplotype-block–structure assessment, as implemented in Haploview 2.05 (Haploview Web site), was performed for the whole data set without segmentations (3,122 SNPs), and the “block-like” features—number, distribution, and average size—were evaluated.
Exact map positions of LD breaks—determined by SNP coordinates at block boundaries—were identified by analyzing the haplotype-block distribution across the region, with consideration of multiallelic D′ values between consecutive blocks. Pairwise disequilibrium estimates, regardless of the haplotype-block definition (Haplo.XT), were interpreted using GOLDsurfer, and the resultant LD-break distribution was compared with that from the haplotype-block analysis.
tagSNP Selection
Application of a stringent r2 threshold (r2>0.8) between SNPs would allow the selected tagSNPs to resolve >80% of all existing haplotypes (Chapman et al. 2003). The LDselect algorithm (Carlson et al. 2004) groups tagSNPs in bins in which all pairwise r2 values exceed the threshold, so that only one tagSNP would need to be genotyped per bin. The binning process takes place, regardless of the strength of the underlying LD (low or high), for a given region, and behaves independent of any block requirements. Tagging efficiency was defined as n/nh, where nh is the number of tagSNPs selected to cover the region and n the total number of genotyped SNPs in the region (Ke et al. 2004a). The first SNP from each bin SNP list was selected for the final list, which was loaded into the GLOVAR database (Human Genome Server).
Variation of Recombination Rate across the MHC Region
Phase 2.1 was used for estimation of recombination rate and hotspot evidence (Li and Stephens 2003; Crawford et al. 2004). The region was separated into windows of 50 SNPs, with 5 SNPs overlapped between neighboring windows. Phased founder haplotypes, which were obtained by running Merlin, were fed into the program (Abecasis et al. 2002). To rescale population-genetic estimates, Ne was estimated from a comparison of the genetic map distance across the MHC region (Cullen et al. 2002) and the estimated population-recombination rate for the same region (McVean et al. 2004).
Coordinates of recombination hotspots were mapped to the current genome assembly to determine the context in which hotspots take place (intra- or intergenic), and consistency was verified by comparison with the haplotype-block (boundaries) distribution
Extended-Haplotype Homozygosity Analysis
Initial SNP chromosomal phasing was done using the Genehunter program (Kruglyak et al. 1996). Further haplotype inference of SNP genotyping data was performed using a refinement of the model used in PHASE (Stephens et al. 2001), with allowance for recombination (Fearnhead and Donnelly 2001). Both unphased and missing SNP data were inferred in this manner. Since we have a dense set of markers and since most markers are in strong LD with several other markers, we do not believe that the phasing has introduced serious bias into our results. Only independent grandparental chromosomes from all families were analyzed.
Extended haplotype homozygosity (EHH) analysis (Sabeti et al. 2002) was performed for each haplotype-block allele (Gabriel et al. 2002), by use of Finch software (P. C. Sabeti, P. Varilly, B. Fry, E. Lander, unpublished material) and with centimorgan estimations as distance. Genetic distances were estimated using the empirical sperm recombination map of Cullen et al. (2002). Since the mapping efforts of Cullen et al. (2002) covered only a subset of the region analyzed in our study, genetic distances from marker rs422331 to marker rs1233384 (749 kb) were estimated by examination of ancestral recombination by use of methods of McVean et al. (2004). Outlying variants were chosen on the basis of two criteria designed to pick alleles with high EHH values for their frequency class. First, as a simple approximation of the distribution, we ranked scores by EHH value × allele frequency. Values >4.5 SD above the mean were considered outliers. Second, all variants were sorted by frequency into 5% bins. Outliers had EHH values ⩾4.5 SD above the mean for the remaining values in that bin.
Results
High-Resolution LD Map of the MHC
We considered 2,335 loci with an MAF ⩾5% for construction of an LD map across a 4.46-Mb region of chromosome 6 that contains the MHC. The average marker density is one SNP per 1.9 kb (median 1.2 kb), with only two gaps >25 kb; the largest was 63 kb. Of all gaps, 85% were <3 kb.
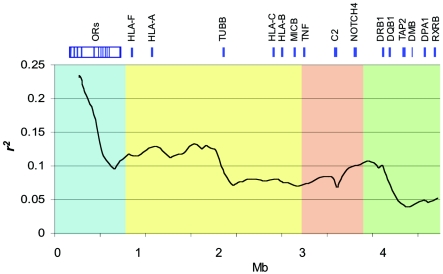
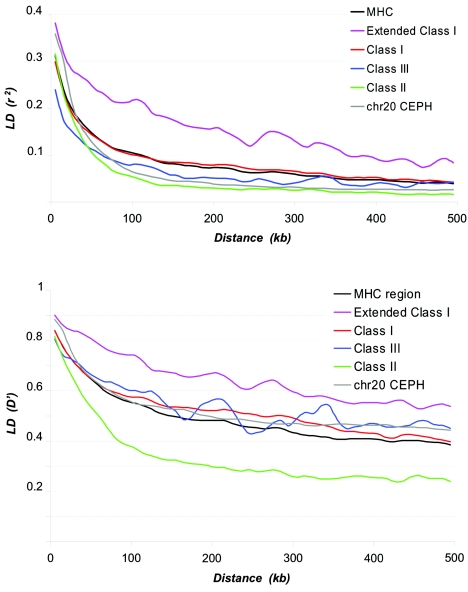
We first examined LD at a broad level across the region using an SW plot of average r2. Figure 2 shows that LD fluctuates greatly along the MHC, consistent with the findings of extreme LD variability throughout the human genome. It is interesting, though, to note the striking differences in long-range LD trends between MHC subregions. For example, the extended class I region, which harbors an olfactory-receptor gene cluster (Younger et al. 2001), is a contiguous block of high LD that extends ∼540 kb. The class I region shows lower overall LD than does the extended class I region, with the telomeric half having higher LD than the centromeric half. LD range is lowest within the MHC class II and extended class II regions, consistent with the presence of multiple known recombination hotspots (Cullen et al. 1995, 1997, 2002; Jeffreys et al. 2001; Jeffreys and May 2004). We also examined the decay of LD as a function of physical distance for each MHC domain separately. The decay plots show fairly distinctive slopes (fig. 3), consistent with the patterns emerging from the SW analysis (fig. 2). The LD-decay pattern of the extended class I and class II regions are likely to fall within the extremes of genome distribution. For example, we compared LD decay between the MHC and a 10-Mb region of chromosome 20 (Ke et al. 2004b), which represents the longest genomic fragment with available typing information close to the SNP density reported here. LD decay between the two regions is comparable only when values are averaged across the MHC (fig. 3).
Figure 2.
SW plot of average r2 across the MHC. Average r2 was calculated from 25 kb to 250 kb in 500-kb SWs, with 50-kb increments between windows. The SW plot captures trends of LD (r2) by averaging r2 values between a given marker and all the SNPs up to 250 kb. Avoiding pairwise comparisons with surrounding markers (25 kb each side) excludes the raising effect of closely linked loci on LD. MHC extended class I, class I, class III, and class II regions (blue, yellow, orange, and green, respectively) present comparatively distinct variation patterns of long-range LD, which is reflected in the haplotype-block analysis and interferes in the SNP-tagging process.
Figure 3.
Decay of LD as a function of distance. The decay rates represented by average r2 (A) and D′ (B) for all marker pairs separated by distance S (S=10 kb, 20 kb, 30 kb,…500 kb). Line colors represent different MHC subregions and chromosome 20 genotyping data from CEPH families (Ke et al. 2004b). Whereas LD decay values averaged across the whole MHC region are comparable with the chromosome 20 LD decay, the extended class I and classical class II regions show distinct slopes consistent with the underlying LD structure observed in fig. 2.
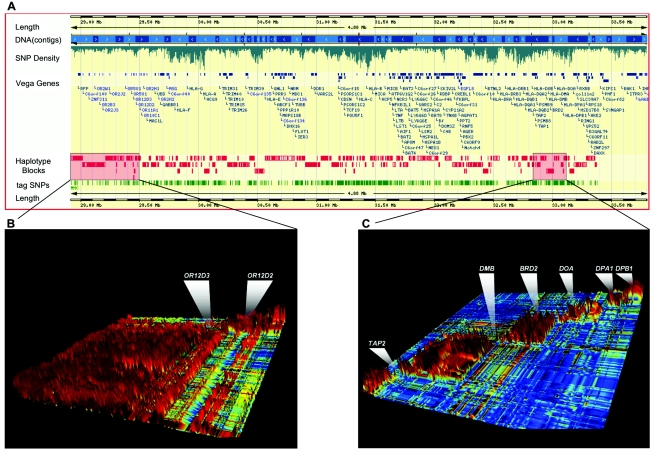
To examine the microstructure of LD obtained with the SW analysis, we used the method described by Gabriel et al. (2002) to define haplotype blocks. This method was previously employed for another data set from this region by Walsh et al. (2003), who found 17 haplotype blocks covering only 25% of the MHC sequence; additional block-like regions have been reported by Stenzel et al. (2004). Using the same criteria (Gabriel et al. 2002), we obtained 202 blocks covering 81.95% of the MHC region as a direct result of the much higher marker density employed in the present study. Haplotype blocks have an average size of 18 kb and 6.4 haplotypes per block. To visualize the distribution of haplotype blocks across the MHC in the context of other genomic features such as gene structures and known SNPs, we used the GLOVAR genome browser maintained at the Sanger Institute. Figure 4A provides an overview of the whole region and shows the SNP density, manually annotated protein-coding genes (Mungall et al. 2003), and haplotype blocks.
Figure 4.
LD structure across the MHC. A, Distribution of haplotype blocks across the MHC region, as viewed in the GLOVAR genome browser. Haplotype blocks, according to criteria of Gabriel et al. (2002) implemented by Haploview 2.05 (Haploview Web site), are represented by red bars. Each bar corresponds to an individual haplotype block comprising a number of SNPs (red marks), which are located according their map position. This enables an accurate interpretation of the LD-block distribution, size, and gaps in the context of additional genomic features, such as gene annotation, SNP density, and physical distance. The distribution of tagSNPs selected in this work is generated in the GLOVAR genome browser and is indicated by a green track under the haplotype blocks. B, High-resolution view of 720 kb of the extended MHC class I region, as represented by GOLDsurfer 3D view of D′ values (Pettersson et al. 2004). This region contains a large cluster of olfactory-receptor genes in high LD (540 kb), interrupted by a single recombination hotspot between OR12D3 and OR12D2. This long-range LD region includes 13 contiguous haplotype blocks, according to the criteria of Gabriel et al. (2002) (see corresponding inset in panel A). C, View of the LD structure (D′ values) within the MHC class II region in which experimental evidence for recombination hotspots have been described elsewhere (Cullen et al. 1997; Jeffreys et al. 2001). High-LD areas (red blocks) are separated by recombination hotspots. The first three LD breaks correspond to recombination hotspots mapped at TAP2 and HLA-DMB and between BRD2 and HLA-DOA (Cullen et al. 1997; Jeffreys et al. 2001). Another LD break is visualized between HLA-DOA and HLA-DPA1.
As expected from the SW analysis, a large portion of the extended class I region (540 kb) that extends over the olfactory-receptor cluster is represented in just two LD blocks interrupted by a single recombination hotspot spanning ∼750 bp (fig. 4B). In general, large blocks cover most of the classical class I region, except for areas surrounding HLA-A, HLA-B, HLA-C, and MICA, in which the LD pattern is disrupted and blocks are shorter. Several studies have shown experimentally that most recombination takes place in narrow hotspots, delimiting regions of high LD (Cullen et al. 1997, 2002; Jeffreys et al. 2001; Jeffreys and May 2004). There are at least four prominent LD breaks intermingled among relatively long blocks in the telomeric portion of the class I region. On its proximal side, recombination appears to take place more uniformly, which results in a number of short stretches of high LD. Three additional hotspots are apparent in the class III region; the most centromeric one resides after NOTCH4 before the class II border, 70 kb 5′ of the hotspot mapped by Stenzel et al. (2004). Within the class II region, those recombination hotspots described elsewhere (Cullen et al. 1997; Jeffrey et al. 2001) at TAP2, HLA-DMB, and between BRD2 and HLA-DOA were clearly identified (fig. 4C). Two additional hotspots are evident within the centromeric portion of the class II region, the stronger one between HLA-DOA and HLA-DPA1 and the other between HLA-DPB1 and HLA-DPB2.
For each MHC subregion and for the region as a whole, table 1 summarizes the number of blocks and block characteristics, average size, sequence coverage, and number of markers per block. At an average marker density of one SNP per 1.9 kb, sequence coverage in blocks is >80% across the MHC, with a slight drop in the class II region. The corresponding figures at one SNP per 5 kb (table 1), which are indicative of the first phase of the International HapMap Project, show a net gain of ∼15% in coverage at the higher marker density. The corresponding figures from the chromosome 20q study (Ke et al. 2004b) show the same trend and suggest that, because of the extreme variability of LD across the genome, sequence coverage will differ between genomic regions. The average size of blocks in CEPH individuals is 18 kb in both data sets, suggesting that this figure may represent the genome average.
Table 1.
LD-Structure Features and tagSNPs in the MHC, at Different Marker Densities[Note]
|
Haplotype Block Structure |
SNP Tagging |
|||||||||||||
|
No. of SNPs |
No. of Blocks |
Block Size(kb) |
Sequence Coverage(%) |
SNPs per Block |
Total No. of tagSNPs |
Tagging Efficiency |
||||||||
| Region | Full | 5 | Full | 5 | Full | 5 | Full | 5 | Full | 5 | Full | 5 | Full | 5 |
| Extended class I | 408 | 161 | 22 | 18 | 34 | 35 | 90 | 77 | 17.9 | 8.3 | 75 | 52 | 5.44 | 3.10 |
| Class I | 1106 | 368 | 99 | 46 | 15 | 18 | 82 | 71 | 10.1 | 7 | 356 | 180 | 3.11 | 2.04 |
| Class III | 280 | 142 | 27 | 24 | 23 | 19 | 81 | 60 | 9.6 | 5 | 110 | 82 | 2.54 | 1.73 |
| Class II | 466 | 215 | 48 | 35 | 12 | 19 | 73 | 64 | 8.8 | 5.3 | 204 | 109 | 2.28 | 1.97 |
| MHCa | 2,260 | 896 | 202 | 116 | 18 | 26 | 82 | 67 | 10.7 | 6.7 | 745 | 416 | 3.03 | 2.15 |
| Chromosome 20b | 3,372c | 2,020 | 367c | 278 | 18c | 20 | 64c | 55 | 7.8c | 5.9 | 1,338c | 1013 | 2.52c | 1.99 |
Note.— “Full” density indicates that all genotyped markers were included, whereas 5-kb density means there was, on average, one marker per 5 kb. Haplotype-block analysis was performed using Haploview. tagSNPs were selected using LDSelect (Carlson et al. 2004), with r2threshold=.80. Tagging efficiency was defined as n/nh, where nh is the number of tagSNPs selected to cover the region, and n is the total number of genotyped SNPs in the region (Ke et al. 2004a).
Includes data averaged across the 4.459-kb region covering the complete MHC.
Data from chromosome 20 include a 10-Mb region (Ke et al. 2004a)
Chromosome 20 data at density of one SNP per 3 kb.
tagSNP Selection
To exploit the dense LD map described above, we proceeded to select a list of maximally informative SNPs for each MHC region that can be used for disease-association studies. We chose to define tagSNPs through a process that disregards the strength of the underlying LD (low or high) in a given region and hence behaves independent of any block requirements. The caveat in this analysis is that there is an abundance of methods for selecting tagSNPs and that different methods result in different lists of tagSNPs. The results in table 1 show that, at the same marker density (one SNP per 5 kb), the tagging efficiency of the whole MHC region (2.15) is slightly higher than that of chromosome 20 (1.99). There is an overall gain in tagging efficiency (40.9%) when the whole MHC data set is used (one SNP per 1.9 kb), probably as a consequence of the extensive LD in the extended class I region. We observed very good correlation between tagging efficiency and the strength of LD in MHC subregions. Specifically, the extended class I and class III regions appear to approach saturation with only five and three additional tagSNPs necessary to capture the information provided by the extra 113 and 45 assayed SNPs, respectively, in the progression from 1 SNP per 3 kb (70 and 107 tagSNPs, respectively [data not shown]) to full density (table 1). As suggested elsewhere (Ke et al. 2004a), selection of tagSNPs in high-LD regions is largely robust to variability in marker density. Given the high LD underlying the extended class I region, it appears that many features—number of tagSNPs, block size, sequence coverage, and number of blocks—come close to steady state at one SNP per 3 kb (data not shown) where increasing SNP density does not result in further efficiency. A different trend is seen in the class III region, in which sequence coverage is 60% at one SNP per 5 kb, increases to 73% at one SNP per 3 kb, and reaches 81% at full density.
As marker density increases, tagging efficiency also increases in the class I region, but the overall number of tagSNPs is much higher at maximum density here. This latter trend is depicted in figure 4A (“tag SNPs”) and is particularly evident in regions in which LD is low and sequence coverage needs to be improved. In contrast, sequence coverage at full density (90%) in the extended class I region experienced only a slight increment compared with that at one SNP per 3 kb (89% [data not shown]). Since class II has the shortest LD range and hence the lowest sequence coverage in blocks, the number of tagSNPs increases substantially at maximum marker density (table 1). The relationship between marker density and tagSNPs remain constant, which indicates that LD is very diffuse across the class II region and that more markers will need to be typed. The significantly larger number of blocks (shorter in average) at maximum marker density is consistent with low LD patterns in class II, in which approximately half of the haplotype blocks contain >4 SNPs. Differences in the extended class I and class III regions can be explained by dissimilarities in the underlying LD and LD-block sequence coverage in each region (table 1). The complete list of tagSNPs generated for each MHC region, as well as their distribution, is available at GLOVAR.
Ke et al. (2004a) showed that tagSNPs selected in western Europeans (to explain 100% of haplotype diversity) can explain 96% of haplotype variation in CEPH founders, who are of western and northern European ancestry, and that ∼90% of the tagSNPs selected in one sample would also be selected as tagSNPs in the other. Thus, the MHC list of tagSNPs presented here should be useful for studies involving individuals of western and northern European ancestry.
Ancestral Recombination Estimation across the MHC Region
Previous studies have shown that the rate of recombination varies substantially across certain regions of the MHC (Cullen et al. 1997, 2002; Jeffreys and May 2004). Sperm typing within a 200-kb class II fragment (Jeffreys et al. 2001) has determined the location of recombination hot spots and has shown that they correlate very strongly with LD breaks. The LD map described above has the resolution required to accurately map such events as shown in figure 4C. Thus, we employed the recently described method of McVean et al. (2004) to estimate fine-scale recombination-rate variations based on population-genetic data and to predict the location and intensity of hotspots across the whole MHC. Also, we wanted to assess whether the haplotype-block pattern reflects genuine recombination-rate variation and whether block boundaries are actually delimited by recombination hotspots.
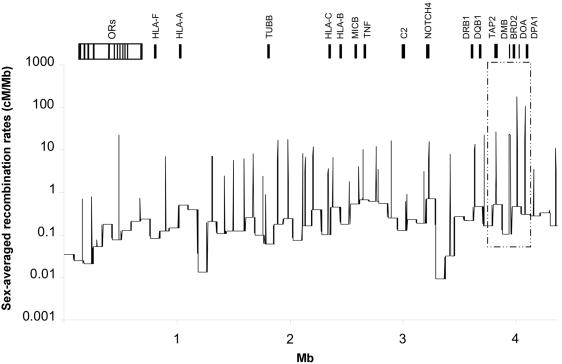
The nature of recombination rates inferred from patterns of genetic variation varied immensely across the region (fig. 5) and showed patterns mirroring the LD patterns described above. The average recombination rate for the 4.459-Mb region is 0.66714 cM/Mb, with the extended class I region showing a very low rate, 0.195 cM/Mb. The classic MHC has a rate of 0.7852 cM/Mb. More specifically, class I and III recombination rates are somewhat similar to each other (0.443 cM/Mb and 0.469 cM/Mb, respectively), whereas class II has the highest rate (1.712 cM/Mb). It is worth highlighting the extreme variation in recombination rates—a difference of up to 4 orders of magnitude. The data suggest the presence of 29 hotspots—and show >10 times the background recombination rates—corresponding to an average density of one per 150 kb (fig. 5). An additional recombination site falling short of the hotspot threshold, sevenfold higher, was observed between HLA-C and HLA-B. In total, hotspots cover only 1.6% of the genomic sequence and have an average size of 2.43 kb. Slightly more hotspots were observed between genes than within genes (n=19 vs. n=11, respectively).
Figure 5.
Recombination-rate variation across the MHC. Recombination rates estimated from population-genetic data are far from being uniform; their distribution fluctuates considerably in both, by scale (cM/Mb) and by map position. Recombination hotspots are represented by peaks 10 times higher than the local background level of recombination. Peaks enclosed in the inset correspond to hotspots identified by sperm typing (Jeffreys et al. 2001) located at or near TAP2, HLA-DMB, BRD2, and HLA-DOA, observable as LD breaks in figure 4C. The recombination hotspot between OR12D3 and OR12D2 in the olfactory-receptor gene cluster (“ORs”), inferred from population genetic data, correlates perfectly with the LD break visible in figure 4B. Note the presence of two coldspots showing recombination rates 10 times lower than the local background level of recombination.
All but two of the inferred hotspots coincide with the haplotype-block boundaries we calculated using the method of Gabriel et al. (2002). Interestingly, we find recombination hotspots in TAP2 and HLA-DMB and between BRD2 and HLA-DOA at the same locations as reported by sperm-typing analysis (Jeffreys et al. 2001; Cullen et al. 2002), (fig. 5, inset). We observed two recombination coldspots (0.013 and 0.009, 10 times lower than the background recombination), both of which overlap with high-LD fragments (fig. 5). In one case, the lack of recombination perfectly corresponds to one large haplotype block (109 kb) lying in the class III/II border, which contains the C6orf10 gene and is surrounded by two hotspots and a diffuse pattern of a number of short LD blocks.
Extended-Haplotype Analysis
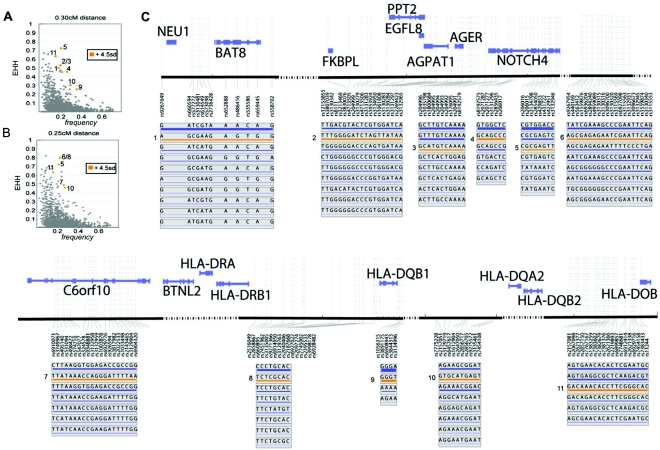
Positive selection brings rare alleles to higher frequency in relatively few generations, thus affording fewer opportunities for recombination events to separate an allele from its original chromosomal context. Therefore, haplotype-block alleles showing a high EHH score and high frequency in the population may have experienced recent positive selection and could represent functionally important alleles. To identify potential signals of positive selection in the MHC, we used EHH analysis, which determines the length of the chromosomal haplotypes extending from a specific allele at a particular locus (Sabeti et al. 2002). To control for the high positional variation in recombination rates in the MHC that would artificially affect the length of haplotypes, we used the sperm-typing recombination map of the region described by Cullen et al. (2002). Given this map, we scanned the MHC, using each haplotype block as an independent locus from which to determine EHH values, assessing each of the 1,966 alleles examined. Since no similarly large region of the genome has a direct sperm-based estimate of recombination rate, we compared the EHH values of haplotype-block alleles within the MHC data set with each other and identified allelic variants that are outliers, on the basis of statistical rank of the EHH value relative to allele frequency.
We observed eight alleles at 0.3 cM and six alleles at 0.25 cM that deviate by >4.5 SD from the mean of the EHH×frequency statistic and also from the mean of the 5% frequency bins (table 2 and fig. 6). All 14 of these alleles map to a single extended haplotype. An example of one of these alleles is shown in figure 7. A mutation on this haplotype may have experienced recent directional positive selection. Interestingly, this haplotype bears allele HLA-DRB1*1501 94% of the time (in one case, the allele was HLA-DRB1*0301). Although HLA-DRB1*1501 is an exciting candidate, we cannot rule out other possibly functional variation on the haplotype.
Table 2.
EHH Outliers at .3cM and .25cM
|
SNP |
Base |
Haplotype | |||||||
| EHH Analysis and Panela | Start | End | Start | End | Length(bp) | Sequence | Frequency | EHH LengthcM | Haplotype Frequency × EHH |
| .30cM: | |||||||||
| C1 | rs9267649 | rs558702 | 31929262 | 31974756 | 45,494 | AGCGAAGAGTGG | .19355 | .49638 | .09607 |
| C2 | rs12153855 | rs3132965 | 32146274 | 32218440 | 72,166 | TTTGGGGATCTAGTTATAA | .20968 | .52923 | .11097 |
| C3 | rs204996 | rs2071278 | 32221326 | 32236884 | 15,558 | GCATGTCAAAA | .20968 | .52923 | .11097 |
| C4 | rs8192575 | rs206018 | 32237824 | 32249325 | 11,501 | GCAGCCC | .30645 | .29872 | .09154 |
| C5 | rs206016 | rs3132946 | 32252628 | 32261496 | 8,868 | CGCGAGTT | .21774 | .72080 | .15695 |
| C9 | rs3891175 | rs3134996 | 32680881 | 32683289 | 2,408 | GGGT | .37097 | .25411 | .09427 |
| C10 | rs2157051 | rs7454108 | 32705045 | 32727862 | 22,817 | GTGCATGAGT | .27419 | .45276 | .12414 |
| C11 | rs2157081 | rs11244 | 32808752 | 32827699 | 18,947 | GACAGACACCTTCGGGCAC | .16129 | .63158 | .10187 |
| .25cM: | |||||||||
| C5 | rs206016 | rs3132946 | 32252628 | 32261496 | 8,868 | CGCGAGTT | .21774 | .72080 | .15695 |
| C6 | rs9267954 | rs3115553 | 32284504 | 32317371 | 32,867 | AGCGAGAGAATCGAATTCAG | .22581 | .79365 | .17921 |
| C7 | rs6910071 | rs6904320 | 32390832 | 32420060 | 29,228 | TTATAAACCAGGGATTTTAA | .26613 | .48295 | .12853 |
| C8 | rs660895 | rs2187668 | 32628857 | 32657048 | 28,191 | TCTCGCAC | .22581 | .79365 | .17921 |
| C10 | rs2157051 | rs7454108 | 32705045 | 32727862 | 22,817 | GTGCATGAGT | .27419 | .45276 | .12414 |
| C11 | rs2157081 | rs11244 | 32808752 | 32827699 | 18,947 | GACAGACACCTTCGGGCAC | .16129 | .63158 | .10187 |
Panels are from figure 6.
Figure 6.
EHH outliers at 0.3 cM and 0.25 cM. A, EHH by frequency plot at 0.3cM, indicating the eight outliers at that distance. B, EHH by frequency plot at 0.25cM, indicating six outliers at that distance. C, Physical mapping of outlying variants. A subset of genes in the region is shown for reference. Haplotype variants with extended LD are indicated in orange and with numbers that correlate to their position in the EHH by frequency plots in panels A and B.
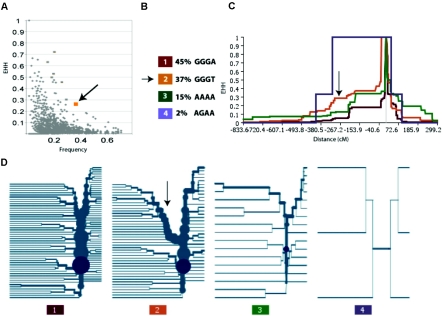
Figure 7.
Representative EHH outlier. A, EHH by frequency plot at 0.3 cM distance, indicating one of the eight variants at that an outlier with distance that is >4.5 SD in its frequency bin and per its frequency×EHH statistic. This variant is indicated in figure 6 (panel 9) and is part of a block slightly centromeric to the DQB1 gene. B, Haplotype structure of the block containing this haplotype variant. The outlier is the 37% allele (“GGGT”) indicated in orange in all remaining plots (arrow). C, EHH by distance (cM) plot of all variants in the block. The arrow indicates the region of extended LD for the orange haplotype. D, Haplotype bifurcation plots of all variants in the block. The arrow indicates the region of interest for the orange haplotype.
Discussion
We genotyped 2,335 loci across the MHC in CEPH pedigrees. The constructed high-resolution map, one marker per 1.9 kb, provides a comprehensive guide to fine-scale patterns of LD and recombination, as well as the means to select optimal marker sets for disease-association studies. To that end, we suggested one such set of tagSNPs, although we realize that many investigators will use their preferred method.
High and long-range LD has long been interpreted as one of the hallmarks of the MHC (The MHC Sequencing Consortium 1999). The present study revealed that the olfactory-receptor gene cluster falls within a 540-kb region of uninterrupted high LD. Ehlers et al. (2000) showed that these polymorphic olfactory loci contribute to extended HLA/OR haplotypes, and a recent study by Füst et al. (2004) indicates that a specific haplotype associated with smoking behavior may even extend to the central region (class III) of the MHC. The HapMap data (International HapMap Project) suggest that this region of strong LD extends farther toward the telomere for 350 kb. It will be interesting to determine the full extent of what appears to be one of the longest regions of high LD in the genome. It also remains to be investigated whether this LD block—which seems to extend over ∼900 kb, according to the HapMap data (International HapMap Project)—is also present in other populations.
EHH analysis compares an allele’s frequency “age” to its extended haplotype’s recombinational “age.” Under neutral evolution, new variants require a long time to reach high frequency; during that time, LD adjacent to the new variant will be decreased because of recombination. Consequently, common alleles will be surrounded by short-range LD. High-frequency variants within a long-range LD context may represent variants under positive selection. Our EHH analysis identified a handful of variants in which haplotypes showed comparatively extended homozygosity, given their population frequencies. These variants map to a single long-range haplotype that bears HLA-DRB1*1501 94% of the time. This haplotype also bears other variants from the so-called ancestral DR2 haplotype; however, the correlation is best with the HLA-DRB1*1501 allele, which suggests that it or another nearby variant may have been responsible for the hypothesized selective event. Variation on the DR2 haplotype has been associated with susceptibility to multiple sclerosis and systemic lupus erythematosus (Barcellos et al. 2003; Heggarty et al. 2003; Larsen and Alper 2004), whereas it has been associated with a protective effect for type I diabetes (Cucca et al. 2001)
Population-based recombination-rate estimates (coalescent model) provide an average transmission of recombination rates (female-male) over thousands of generations, whereas pedigree analyses are based on only a few meioses and sperm-typing measures of recombination in few extant males. We find that pedigree analysis, sperm typing, and coalescent approaches identify identical recombination hotspots for the 200-kb MHC class II fragment studied by Jeffreys et al. (2001) (figs. 4C and fig. 5, inset) This is in agreement with other studies (McVean et al. 2004; Crawford et al. 2004), and it provides additional weight to the new recombination hotspots identified in the present study. As a consequence of the number of recombination hotspots predicted for the MHC region, we would expect to find a fairly disrupted LD pattern. However, extended haplotypes exist at relatively high frequency in whites. In addition, according to population frequencies, the MHC can be divided into only a few blocks that contain nonrandomly associated alleles at different loci (Yunis et al. 2003). We also observed regions that showed excessively long haplotypes, given their population frequencies. Selective sweeps and population history (genetic drift) can explain the disproportionately longrange LD in some haplotypes, but there is also evidence indicating a strong influence of recombination activity shaping the LD landscape within the MHC. Specifically, a considerable proportion of recombination hotspots can be resolved as gene-conversion events (Jeffreys and May 2004). Additionally, differential intensity in crossover activity and haplotype-specific recombination patterns (Ahmad et al. 2003) could also affect the extent of LD. However, the relationship between haplotype-specific hotspots and population-specific haplotypes observed in a 75-kb fragment of the MHC class II region (Kauppi et al. 2003) remains to be studied.
Our analysis shows that the number of tagSNPs required to explain variation across large genomic regions fluctuates and depends on the extent of LD. High marker density is required in regions of low LD to allow comprehensive coverage with tagSNPs. Extrapolating to the rest of the genome, the International HapMap Project database will provide efficient tagging in regions of extended LD already in phase I (one SNP per 5 kb) but will require the higher density of phase II to achieve comprehensive coverage of the whole genome.
We identified a subset (tagSNPs) of all the typed SNPs with MAF >5% that captures haplotype variation in this sample. The tags are displayed in the context of annotated genomic sequence through GLOVAR. As expected, variation in the underlying LD greatly affects the SNP-tagging process and efficiency in each MHC subregion. The highest SNP tagging efficiency (5.44) could be achieved for the extended class I region, which indicates attainable gains in typing cost for subsequent studies. Higher efficiency can be obtained by using haplotype-based methods (Ke et al. 2004a), although haplotypes and their frequencies must be carefully estimated in regions of complex local LD and recombination profile (e.g., the class II region). The list of tagSNPs generated within the context of LD profiles and distribution of recombination hotspot will allow MHC association studies to be conducted more efficiently. On the basis of data presented here, given a sufficiently large population, it will also be possible to identify rare haplotypes, thus increasing sensitivity for association with MHC-linked disease. In addition, a reduction in alloreactivity can be envisioned on the basis of the complementary information provided by SNP typing. At least for the extended haplotypes, which represent 25%–30% of all haplotypes in European-derived populations, patient-donor matching can be improved by the identification of haplotype-specific and block-specific variation (Yunis et al. 2003).
Acknowledgments
M.M.M. was supported by a Wellcome Trust postdoctoral fellowship. E.W. was supported by a Cancer Research Institute fellowship. J.D.R. was supported by National Institute of Diabetes and Digestive and Kidney Diseases grant DK64869. M.D., M.G., S.H., J.M., P.W., D.R.B., S.B., and P.D. were supported by The Wellcome Trust. We thank all members of the MHC Haplotype Project Consortium, in particular S. Sawcer, J. Trowsdale, and J. Todd. We thank P. Sabeti for allowing the use of the Finch software prior to publication.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Center for Statistical Genetics, http://www.sph.umich.edu/csg/abecasis/PedStats/ (for PEDSTATS)
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/index.html
- GLOVAR Genome Browser, http://www.glovar.org/Homo_sapiens/
- Haploview, http://www.broad.mit.edu/mpg/haploview/index.php
- Human Chromosome 6 Project Overview, http://www.sanger.ac.uk/HGP/Chr6/
- International HapMap Project, http://www.hapmap.org/
- MHC Haplotype Project, http://www.sanger.ac.uk/HGP/Chr6/MHC/
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) MERLIN—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cookson WOC (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 [DOI] [PubMed] [Google Scholar]
- Ahmad T, Neville M, Marshall SE, Armuzzi A, Mulcahy-Hawes K, Crawshaw J, Sato H, Ling K-L, Barnardo M, Goldthorpe S, Walton R, Bunce M, Jewell DP, Welsh KI (2003) Haplotype-specific linkage disequilibrium patterns define the genetic topography of the human MHC. Hum Mol Genet 12:647–656 [PubMed] [Google Scholar]
- Barcellos LF, Oksenberg JR, Begovich AB, Martin ER, Schmidt S, Vittinghoff E, Goodin DS, Pelletier D, Lincoln RR, Bucher P, Swerdlin A, Perick-Vance MA, Haines JL, Hauser SL, for the Multiple Sclerosis Genetics Group (2003) HLA-DR2 dose effect on susceptibility to multiple sclerosis and influence on disease course. Am J Hum Genet 72:710–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA (2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74:106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington M, O’Brien SJ (2003) The influence of HLA genotype on AIDS. Annu Rev Med 54:535–551 [DOI] [PubMed] [Google Scholar]
- Chapman JM, Cooper JD, Todd JA, Clayton DG (2003) Detecting disease associations due to linkage disequilibrium using haplotype tags: a class of tests and the determinants of statistical power. Hum Hered 56:18–31 [DOI] [PubMed] [Google Scholar]
- Clayton D, Chapman J, Cooper J (2004) Use of unphased multilocus genotype data in indirect association studies. Genet Epidemiol 27:415–428 [DOI] [PubMed] [Google Scholar]
- Crawford DC, Bhangale T, Li N, Hellenthal G, Rieder MJ, Nickerson DA, Stephens M (2004) Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat Genet 36:700–706 [DOI] [PubMed] [Google Scholar]
- Cucca F, Lampis R, Congia M, Angius E, Nutland S, Bain SC, Barnett AH, Todd JA (2001) A correlation between the relative predisposition of MHC class II alleles to type 1 diabetes and the structure of their proteins. Hum Mol Genet 10:2025–2037 [DOI] [PubMed] [Google Scholar]
- Cullen M, Erlich H, Klitz W, Carrington M (1995) Molecular mapping of a recombination hotspot located in the second intron of the human TAP2 locus. Am J Hum Genet 56:1350–1358 [PMC free article] [PubMed] [Google Scholar]
- Cullen M, Noble J, Erlich H, Thorpe K, Beck S, Klitz W, Trowsdale J, Carrington M (1997) Characterization of recombination in the HLA class II region. Am J Hum Genet 60:397–407 [PMC free article] [PubMed] [Google Scholar]
- Cullen M, Perfetto SP, Klitz W, Nelson G, Carrington M (2002) High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am J Hum Genet 71:759–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SE, Hudson TJ, Lander ES (2001) High-resolution haplotype structure in the human genome. Nat Genet 29:229–232 [DOI] [PubMed] [Google Scholar]
- Davies JL, Kawaguchi Y, Bennett ST, Copeman JB, Cordell HJ, Pritchard LE, Reed PW, Gough SCL, Jenkins SC, Palmer SM, Balfour KM, Rowe BR, Farrall M, Barnett AH, Bain SC, Todd JA (1994) A genome-wide search for human type-1 diabetes susceptibility genes. Nature 371:130–136 [DOI] [PubMed] [Google Scholar]
- Ehlers A, Beck S, Forbes SA, Trowsdale J, Volz A, Younger R, Ziegler A (2000) MHC-linked olfactory receptor loci exhibit polymorphism and contribute to extended HLA/OR-haplotypes. Genome Res 10:1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnhead P, Donnelly P (2001) Estimating recombination rates from population genetic data. Genetics 159:1299–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füst G, Arason GJ, Kramer J, Szalai C, Duba J, Yang Y, Chung EK, Zhou B, Blanchong CA, Lokki ML, Bodvarsson S, Prohaszka Z, Karadi I, Vatay A, Kovacs M, Romics L, Thorgeirsson G, Yu CY (2004) Genetic basis of tobacco smoking: strong association of a specific major histocompatibility complex haplotype on chromosome 6 with smoking behavior. Int Immunol 16:1507–1514 [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Ahmadi KR, Weale ME, Wood NW (2003) Genome scans and candidate gene approaches in the study of common diseases and variable drug responses. Trends Genet 19:615–622 [DOI] [PubMed] [Google Scholar]
- Harbo HF, Lie BA, Sawcer S, Celius EG, Dai KZ, Oturai A, Hillert J, Lorentzen AR, Laaksonen M, Myhr KM, Ryder LP, Fredrikson S, Nyland H, Sorensen PS, Sandberg-Wollheim M, Andersen O, Svejgaard A, Edland A, Mellgren SI, Compston A, Vartdal F, Spurkland A (2004) Genes in the HLA class I region may contribute to the HLA class II-associated genetic susceptibility to multiple sclerosis. Tissue Antigens 63:237–247 [DOI] [PubMed] [Google Scholar]
- Heggarty S, Sawcer S, Hawkins S, McDonnell G, Droogan A, Vandenbroeck K, Hutchinson M, Setakis E, Compston A, Graham C (2003) A genome wide scan for association with multiple sclerosis in a N. Irish case control population. J Neuroimmunol 143:93–96 [DOI] [PubMed] [Google Scholar]
- Hill AVS, Allsopp CEM, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA, Bennett S, Brewster D, McMichael AJ, Greenwood BM (1991) Common West African HLA antigens are associated with protection from severe malaria. Nature 352:595–600 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R (2001) Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet 29:217–222 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, May CA (2004) Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet 36:151–156 [DOI] [PubMed] [Google Scholar]
- Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RC, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SC, Clayton DG, Todd JA (2001) Haplotype tagging for the identification of common disease genes. Nat Genet 29:233–237 [DOI] [PubMed] [Google Scholar]
- Kauppi L, Sajantila A, Jeffreys AJ (2003) Recombination hotspots rather than population history dominate linkage disequilibrium in the MHC class II region. Hum Mol Genet 12:33–40 [DOI] [PubMed] [Google Scholar]
- Ke X, Durrant C, Morris AP, Hunt S, Bentley DR, Deloukas P, Cardon LR (2004a) Efficiency and consistency of haplotype tagging of dense SNP maps in multiple samples. Hum Mol Genet 13:2557–2565 [DOI] [PubMed] [Google Scholar]
- Ke XY, Hunt S, Tapper W, Lawrence R, Stavrides G, Ghori J, Whittaker P, Collins A, Morris AP, Bentley D, Cardon LR, Deloukas P (2004b) The impact of SNP density on fine-scale patterns of linkage disequilibrium. Hum Mol Genet 13:577–588 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Larsen CE, Alper CA (2004) The genetics of HLA-associated disease. Curr Opin Immunol 16:660–667 [DOI] [PubMed] [Google Scholar]
- Lewontin RC (1964) The interaction of selection and linkage. I. General considerations: heterotic models. Genetics 49:49–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Stephens M (2003) Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165:2213–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini M, Antonioli R, Lleo A, Barili M, Caronni M, Origgi L, Vanoli M, Scorza R (2003) HLA class II antigens associated with lupus nephritis in Italian SLE patients. Hum Immunol 64:462–468 [DOI] [PubMed] [Google Scholar]
- Marrosu MG, Sardu C, Cocco E, Costa G, Murru MR, Mancosu C, Murru R, Lai M, Contu P (2004) Bias in parental transmission of the HLA-DR3 allele in Sardinian multiple sclerosis. Neurology 63:1084–1086 [DOI] [PubMed] [Google Scholar]
- McVean GAT, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P (2004) The fine-scale structure of recombination rate variation in the human genome. Science 304:581–584 [DOI] [PubMed] [Google Scholar]
- Mungall AJ, Palmer SA, Sims SK, Edwards CA, Ashurst JL, Wilming L, Jones MC, et al (2003) The DNA sequence and analysis of human chromosome 6. Nature 425:805–811 [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksenberg JR, Barcellos LF, Cree BAC, Baranzini SE, Bugawan TL, Khan O, Lincoln RR, Swerdlin A, Mignot E, Lin L, Goodin D, Erlich HA, Schmidt S, Thomson G, Reich DE, Pericak-Vance MA, Haines JL, Hauser SL (2004) Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am J Hum Genet 74:160–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant A, Barker DL, Stuelpnagel JR, Chee MS (2002) BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques Suppl 56–61 [PubMed] [Google Scholar]
- Pettersson F, Jonsson O, Cardon LR (2004) GOLDsurfer: three dimensional display of linkage disequilibrium. Bioinformatics 20:3241–3243 [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZP, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, Ackerman HC, Campbell SJ, Altshuler D, Cooper R, Kwiatkowski D, Ward R, Lander ES (2002) Detecting recent positive selection in the human genome from haplotype structure. Nature 419:832–837 [DOI] [PubMed] [Google Scholar]
- Stenzel A, Lu T, Koch WA, Hampe J, Guenther SM, De La Vega FM, Krawczak M, Schreiber S (2004) Patterns of linkage disequilibrium in the MHC region on human chromosome 6p. Hum Genet 114:377–385 [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CA, Horton R, Allcock RJN, Ashurst JL, Atrazhev AM, Coggill P, Dunham I, et al (2004) Complete MHC haplotype disease gene mapping. Genome Res 14:1176–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium (2003) The International HapMap Project. Nature 426:789–796 [DOI] [PubMed] [Google Scholar]
- The MHC Sequencing Consortium (1999) Complete sequence and gene map of a human major histocompatibility complex. Nature 401:921–923 [DOI] [PubMed] [Google Scholar]
- Walsh EC, Mather KA, Schaffner SF, Farwell L, Daly MJ, Patterson N, Cullen M, Carrington M, Bugawan TL, Erlich H, Campbell J, Barrett J, Miller K, Thomson G, Lander ES, Rioux JD (2003) An integrated haplotype map of the human major histocompatibility complex. Am J Hum Genet 73:580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger RM, Amadou C, Bethel G, Ehlers A, Lindahl KF, Forbes S, Horton R, Milne S, Mungall AJ, Trowsdale J, Volz A, Ziegler A, Beck S (2001) Characterization of clustered MHC-linked olfactory receptor genes in human and mouse. Genome Res 11:519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis EJ, Larsen CE, Fernandez-Vina M, Awdeh ZL, Romero T, Hansen JA, Alper CA (2003) Inheritable variable sizes of DNA stretches in the human MHC: conserved extended haplotypes and their fragments or blocks. Tissue Antigens 62:1–20 [DOI] [PubMed] [Google Scholar]