Abstract
Campomelic dysplasia (CD) is a semilethal skeletal malformation syndrome with or without XY sex reversal. In addition to the multiple mutations found within the sex-determining region Y–related high-mobility group box gene (SOX9) on 17q24.3, several chromosome anomalies (translocations, inversions, and deletions) with breakpoints scattered over 1 Mb upstream of SOX9 have been described. Here, we present a balanced translocation, t(4;17)(q28.3;q24.3), segregating in a family with a mild acampomelic CD with Robin sequence. Both chromosome breakpoints have been identified by fluorescence in situ hybridization and have been sequenced using a somatic cell hybrid. The 17q24.3 breakpoint maps ∼900 kb upstream of SOX9, which is within the same bacterial artificial chromosome clone as the breakpoints of two other reported patients with mild CD. We also report a prenatal identification of acampomelic CD with male-to-female sex reversal in a fetus with a de novo balanced complex karyotype, 46,XY,t(4;7;8;17)(4qter→4p15.1::17q25.1→17qter;7qter→7p15.3::4p15.1→4pter;8pter→8q12.1::7p15.3→7pter;17pter→17q25.1::8q12.1→8qter). Surprisingly, the 17q breakpoint maps ∼1.3 Mb downstream of SOX9, making this the longest-range position effect found in the field of human genetics and the first report of a patient with CD with the chromosome breakpoint mapping 3′ of SOX9. By using the Regulatory Potential score in conjunction with analysis of the rearrangement breakpoints, we identified a candidate upstream cis-regulatory element, SOX9cre1. We provide evidence that this 1.1-kb evolutionarily conserved element and the downstream breakpoint region colocalize with SOX9 in the interphase nucleus, despite being located 1.1 Mb upstream and 1.3 Mb downstream of it, respectively. The potential molecular mechanism responsible for the position effect is discussed.
Mammalian gene expression is regulated at many levels. Despite the near completion of the human genome sequence, little is known about the genomic aspects of gene regulation in humans. A useful model for studying such regulation is provided by genomic rearrangements that result in diseases and in which chromosome breakpoints do not disrupt the causative gene but instead map outside the intact gene. Kleinjan and van Heyningen (1998) reviewed position effects of chromosomal rearrangements and proposed a few possible mechanisms that may cause such effects: (1) separation of the gene from its enhancer or promoter region, (2) juxtaposition with an enhancer element from another gene, (3) removal of the long-range insulator or boundary element, (4) competition with another enhancer, and (5) position-effect variegation—that is, the insertion of the gene in a new heterochromatin environment. Recently, Tufarelli et al. (2003) described yet another mechanism of position-effect–related gene regulation, “antisense-mediated cis-acting methylation utilizing non-coding RNA,” which is similar to XIST/TSIX–mediated X-chromosome inactivation (Kleinjan and van Heyningen 2003). Cis-acting regulatory elements in humans as distant as 1 Mb 5′ from the target gene have been described (de Kok et al. 1996; Davies et al. 1999; Pfeifer et al. 1999; Jamieson et al. 2002; Lettice et al. 2002; Nobrega et al. 2003). In addition, it has been proposed that abnormal chromosome structure and/or chromatin remodeling affect gene expression (Bickmore and Maarel 2003; Cho et al. 2004).
Haploinsufficiency of the sex-determining region Y (SRY)–related high-mobility group box gene (SOX9) is known to cause campomelic dysplasia (CD [MIM 114290]), a clinically distinct syndrome characterized by skeletal anomalies, such as bowed femurs and tibiae, hypoplastic scapulae, 11 pairs of ribs, pelvic malformations, Robin sequence, and clubbed feet (Foster et al. 1994; Wagner et al. 1994). In two-thirds of individuals with CD with a 46,XY karyotype, male-to-female sex reversal has been described (Houston et al. 1983; Mansour et al. 1995). The two mechanisms responsible for SOX9 haploinsufficiency are intragenic mutations (such as point mutations, insertions, and deletions) and chromosome rearrangements (Maraia et al. 1991; Young et al. 1992; Tommerup et al. 1993; Foster et al. 1994; Wagner et al. 1994; Kwok et al. 1995; Cameron et al. 1996; Meyer et al. 1997; Goji et al. 1998; Hageman et al. 1998; Pfeifer et al. 1999). Although no consistent phenotype-genotype correlations have been established on the basis of intragenic mutations, the patients with chromosome rearrangements tend to have milder phenotypes (Pfeifer et al. 1999). The analysis of 12 patients with CD who had apparently balanced chromosome rearrangements showed the breakpoints to be scattered ∼140–950 kb upstream of the SOX9 gene, whereas the gene itself was intact (Wunderle et al. 1998; Pfeifer et al. 1999; Erdel et al. 2004). Recently, Pop et al. (2004) reported an ∼1.5-Mb microdeletion located ∼380 kb upstream of SOX9 in a patient with CD. It is interesting that Huang et al. (1999) described a chromosome duplication, dup(17)(q24.1q24.3), associated with XX female-to-male sex reversal and that Bishop et al. (2000) reported XX female-to-male sex reversal in a transgenic mouse with a 134-kb insertional deletion resulting from a recombinant construct insertion 0.98 Mb upstream of SOX9. Qin et al. (2004) proposed that the transgenic insertion of a promoter from the recombinant construct in these mice interacts with gonad-specific enhancer elements leading to sex reversal (Qin et al. 2004). Thus, both the chondrogenic and gonadal functions of SOX9 appear to be precisely regulated by elements located as far as 1 Mb from the gene itself. Detailed DNA analysis of the genomic region extending up to 1 Mb proximal to SOX9 failed to uncover any protein-coding genes, suggesting that the chromosomal rearrangements remove one or more cis-regulatory elements from an extended SOX9 region (Pfeifer et al. 1999; Bagheri-Fam et al. 2001; Pop et al. 2004; Qin et al. 2004). Here, we report two new patients with acampomelic CD with chromosomal breakpoints mapping ∼900 kb upstream and ∼1.3 Mb downstream of SOX9.
Patient 1: The proband AA is a 6-year-old white girl, the second child born to a 31-year-old mother and a 32-year-old father. Her prenatal history is uneventful, with the mother undergoing amniocentesis because of a reciprocal chromosome translocation t(4;17) segregating in the family. Amniotic fluid chromosome analysis showed that the proband is a carrier of the same balanced t(4;17). The proband was delivered vaginally at term without complications. She was diagnosed with Robin sequence; the cleft palate was repaired at age 3 years. She also had tracheostenosis. A tracheostomy was performed at 10 wk, and she received a G-tube and underwent fundoplication. She had a history of frequent croup as a child and also required speech therapy. On physical examination at age 6 years, her height was 103 cm and her weight was 15 kg (both <5th percentile). She had flat malar surfaces, a depressed nasal bridge, prominent eyes, and 11 pairs of ribs. Her left ear was posteriorly rotated. Skeletal survey revealed hypoplastic scapulae and iliac wings, as well as an irregularity of the end plates of the thoracic vertebrae. There was an anomaly of the upper cervical spine, consisting of underdevelopment of the neural arch of C2, a congenital defect of C2, and probable fibrous occipitalization of C1. The dens was slightly hypoplastic, but there was no instability of the cervical spine on flexion.
To characterize whether the balanced t(4;17) was involved in the pathophysiology of these conditions, repeat chromosome studies were requested for all family members. G-banded chromosome analysis from phytohemagglutinin-stimulated peripheral blood lymphocytes showed that the proband is a carrier of a balanced reciprocal translocation with the karyotype 46,XX,t(4;17)(q28.3;q24.3). Chromosome analysis of the parents showed that the proband’s father was a carrier of the same translocation, whereas the karyotype of the mother was normal. The proband’s older brother was also found to be a carrier of this translocation. The brother and father have many of the same clinical features as the proband (fig. 1). In addition, both the brother and the father have myopia, and hearing impairment was present in the father. An initial differential diagnosis included acampomelic CD and Stickler syndrome; however, a thorough physical examination and skeletal survey diagnosed mild acampomelic CD with Robin sequence.
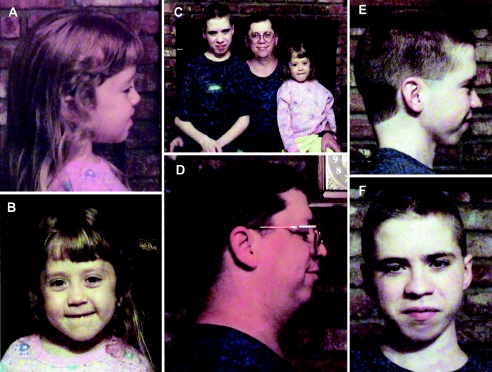
Figure 1.
Photographs of the proband (A, B, and C), her father (C and D), and her brother (C, E, and F). Note the Robin sequence.
FISH with a set of several BAC probes mapped the 17q breakpoint within BAC clone RP11-879D6 (fig. 2A). The der(4) breakpoint was mapped between BAC clones RP11-79M4 and RP11-11I23. To narrow the chromosome 17q breakpoint within BAC clone RP11-879D6, we used a 6,234-bp long-range PCR product, amplified using forward primer 5′-GTAGCTATCTTAGCCCTGGCTGACAGTCACTT-3′ and reverse primer 5′-GGACACTTGCCAGATAAGAACTGGGTAGAC-3′ (Takara Bio), as a probe in FISH mapping. The breakpoint was localized to the distal one-third portion of the BAC clone and was further narrowed by a standard PCR walking method with the use of DNA from a somatic cell hybrid. DNA sequencing of the 1.2-kb PCR product obtained with forward primer 5′-AATCATGAAGATGGCCTTGC-3′ (for chromosome 4) and reverse primer 5′-TCCAGCCAAAGGGAAGAGTA-3′ (for chromosome 17) identified the der(4) breakpoint at nucleotide position 69815055 on chromosome 17 (899,164 bp upstream of SOX9) and at 136261648 on chromosome 4 (UCSC Genome Browser, build 34, July 2003) (fig. 2B). The der(17) breakpoint was identified by PCR in genomic DNA by use of forward primer 5′-TTGATGTATGGCCTGAACCA-3′ and reverse primer 5′-CAGCAAGATGGGGTCTATCAA-3′. An associated deletion of 13 bp on chromosome 4 was found (fig. 2). The breakpoint on chromosome 4 maps within a mariner transposon-like element HSMAR2, suggesting its potential causative role in the formation of the translocation.
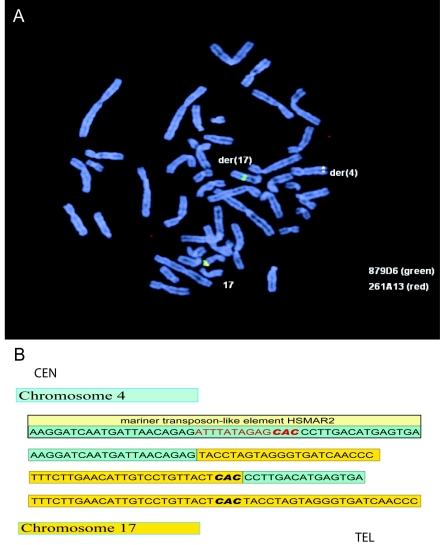
Figure 2.
Mapping of chromosome breakpoints in patient 1. A, Metaphase chromosomes after FISH with BAC clones RP11-879D6 (green) and RP11-261A13 (red) showed that RP11-879D6 spans the breakpoint. B, DNA sequencing of both breakpoints of the derivative chromosomes 4 (green) and 17 (yellow) identified a 13-bp deletion on 4q28.3 generated by the translocation breakpoint. The breakpoint on chromosome 4 maps within a mariner transposon-like element HSMAR2. The trinucleotide CAC in bold represents the site of the translocation where there is identical sequence on both chromosomes 4 and 17 and may designate the origin of the translocation.
In an effort to identify possible transcripts that may be responsible for the CD phenotype, we used several gene-prediction programs and identified seven hypothetical transcripts in the region that spans 100 kb in either direction from the breakpoint on chromosome 17—Ecgenes H17C12306.1 and H17C12308.1, SGP genes Chr17_1538.1 and Ch17_1539.1, Fgenesh++ gene C17001650, and Genscan genes NT_010641.44 and NT_010641.45. Expression of these genes was explored by PCR analyses of human gonadal and fetal brain tissue cDNAs, in which SOX9 transcription is known to occur. Only exons from transcripts H17C12306.1, H17C12308.1, and Chr17_1538.1 were expressed, but none of these transcripts overlapped the breakpoint, making it unlikely that any genes in this region contribute to the CD phenotype. However, it is also possible that the translocation breakpoint may have disrupted the regulation of one of these more proximally located transcripts. Mutation screening of SOX9 revealed only one heterozygous common polymorphism, PM 879 in exon 2, which is known to be not associated with a CD phenotype.
Patient 2: A 15-year-old, G1P0 white woman was referred at 12 wk of gestation because of a family history of Down syndrome. Ultrasound examination showed multiple congenital anomalies, including increased nuchal fold, prominent cisterna magna, micrognathia, deviation of the cardiac axis, and hypoplasia of the middle phalanx of the fifth digit. After receiving genetic counseling, she elected to undergo amniocentesis.
GTG-banding analysis of amniotic fluid chromosomes from in situ cultures showed a complex karyotype: 46,XY,t(4;7;8;17)(4qter→4p15.2::17q25→17qter;7qter→7p15::4p15.2→4pter;8pter→8q12.2::7p15→7pter;17pter→17q25::8q12.2→8qter). At the chromosome level, the complex translocation appeared to be balanced. Parental chromosomes were normal. Ultrasound examination showed a female fetus. On the basis of the presence of the 17q25 breakpoint, the XY sex reversal was inferred to be the result of SOX9 gene malfunction. At 38 wk of gestation, the fetus showed decreased movements. The amniotic fluid index was decreased, vaginal delivery was induced, and a baby girl was delivered. The Apgar scores were 5 and 8 at 1 min and 5 min, respectively. External examination revealed a weight of 2,720 g, with a head circumference of 33.4 cm. The infant was noted to have several congenital anomalies, including a cleft palate, micrognathia, small mouth, posteriorly rotated ears, and nail and digital abnormalities. Three days after birth, the baby was extubated but developed respiratory distress, immediately requiring reintubation. After 3 wk, because of the poor prognosis, the infant was extubated, after which she expired. At autopsy, additional multiple congenital malformations of the skeletal system were observed, including abnormally developed cartilage and bone, 11 pairs of ribs, and normal female genitalia with abnormally formed ovaries with no oocytes (fig. 3). Acampomelic CD with male-to-female sex reversal was diagnosed.
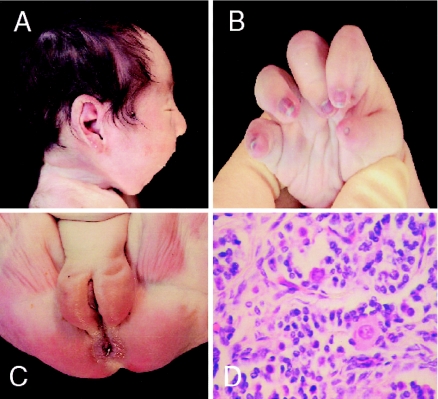
Figure 3.
Autopsy study of patient 2. A, Micrognathia as part of Robin sequence. B, Cone-shaped fingers and hypoplastic thumb, with poorly formed distal interphalangeal creases and hypoplastic nails. C, Normal female external genitalia. D, Absence of oocytes.
The identified chromosome aberration was confirmed by whole-chromosome painting and SKY (Vysis) (data not shown). Each of the four chromosome breakpoints was then mapped at a BAC clone resolution: chromosome 4 within BAC clone RP11-93M12 (4p15.1), chromosome 7 between BAC clones RP11-764N24 and RP11-233O19 (7p15.3), chromosome 8 within BAC clone RP11-17A4 (8q12.1), and chromosome 17 within two overlapping BAC clones, RP11-661C3 and RP11-449L23 (17q25.1). Using two-color interphase FISH with BAC clones RP11-1116I6 (SOX9 specific), RP11-879D6 (spanning the 17q upstream breakpoint), and RP11-661C3 (adjacent to the 17q downstream breakpoint on the centromeric side), we found no evidence of additional chromosome 17 rearrangements, such as paracentric inversion (data not shown). The patient's final karyotype was designated as 46,XY,t(4;7;8;17)(4qter→4p15.1::17q25.1→17qter;7qter→7p15.3::4p15.1→4pter;8pter→8q12.1::7p15.3→7pter;17pter→17q25.1::8q12.1→8qter). Genomic analysis of the breakpoint regions identified the disruption of the PCDH7 gene on chromosome 4. PCDH7 encodes a type I membrane protein and is predominantly expressed in the heart and brain. The chromosome 7 breakpoint is in the vicinity of RAPGEF5, a gene expressed in the brain that encodes a guanine nucleotide exchange factor. The breakpoint located on 17q25.1 likely interrupts the SDK2 gene (which encodes a protein involved in laminar-specific synaptic connectivity in the retina) ∼1.3 Mb downstream of SOX9. None of these genes are thought to contribute to the CD phenotype. Although the clinical features may be complicated by the disruption of genes in the other breakpoints, the classic CD features suggest that the chromosome 17 translocation is most likely responsible for the phenotype of this patient.
It has been hypothesized that distal enhancers interact with promoters through a set of proteins that “bend” DNA to bring the enhancers within physical proximity of their respective target genes, thus regulating their function (Carter et al. 2002; Tolhuis et al. 2002; Palstra et al. 2003). To investigate the possibility of spatial proximity of cis-acting regulatory elements to SOX9, we performed a two-color interphase FISH assay with the use of the SOX9-specific BAC clone RP11-1116I6, the 17q upstream breakpoint–specific clone RP11-879D6 (∼1 Mb upstream from SOX9), and the BAC clone RP11-661C3, which is adjacent to the 17q downstream breakpoint on the proximal side (∼1.2 Mb downstream from SOX9) (fig. 4A). Probes that span the RAI1 gene on 17p11.2 (RP11-525O11) and a region ∼1 Mb proximal to RAI1 (RP11-28B23) that is not known to harbor enhancers for that gene were used as negative controls. As positive controls, BAC clones specific to the POU3F4 gene and its predicted enhancer localized ∼900 kb upstream (de Kok et al. 1996) were used (RP11-246G22 and RP11-54M14, respectively). The distances between SOX9 and its potential enhancer regions (∼1.1 Mb upstream and ∼1.3 Mb downstream) and that between POU3F4 and its predicted enhancer (∼900 kb upstream), the positive control, was measured in 50 cells. There were no statistically different distance measurements in any of these three intervals. Remarkably, these three distances were each significantly smaller (P value of the F test = 1.56 × 10−8) than the distance between RAI1 and a region ∼1 Mb upstream (fig. 4). These results suggest the presence of three-dimensional proximity between both upstream and downstream chromosome breakpoint regions and SOX9 that, when disrupted, could lead to separation of the potential cis-regulatory element(s). Our FISH data suggest that the architectural component may be necessary but not sufficient for proper expression of SOX9. In support of this notion, Gilbert et al. (2004) recently demonstrated that open chromatin fibers correlate with regions of the highest gene density but not with gene expression. Moreover, Poirier et al. (2004) proposed changes of chromatin conformation around Sox9 during its upregulation by the SRY protein.
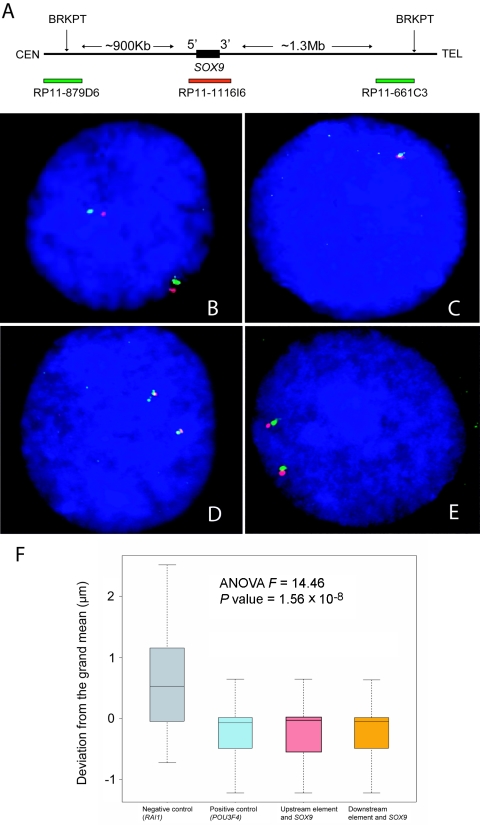
Figure 4.
Interphase FISH colocalization experiment. A, Physical map of the region surrounding SOX9. BAC clones are depicted as horizontal bars, with the colors corresponding to the probes in D and E. The upstream and downstream breakpoints are shown as vertical arrows. B, Interphase FISH with two probes, RAI1-specific BAC clone RP11-525O11 (red) and RP11-28B23 (green), located ∼1 Mb upstream from RAI1, in a male control individual without CD. The distance between the red and green signals was measured in 50 cells. The average distance between these two probes was 1.35 μm. There was no significant correlation (r=0.55) between the signal distance and the cell diameter. These measurements served as the negative control for the colocalization experiment. C, FISH with BAC clones that encompass the gene POU3F4 (RP11-246G22 [red]) and its suspected enhancer (RP11-54M14 [green]). The average distance between these markers was 0.67 μm. This constituted the positive control for the experiment. D, FISH with SOX9-specific BAC clone RP11-1116I6 (red) and the ∼1 Mb upstream breakpoint–spanning clone RP11-879D6 (green). The average distance between these probes was 0.62 μm. E, FISH with SOX9-specific BAC clone RP11-1116I6 (red) and the BAC clone RP11-661C3 (green), which is adjacent to the downstream breakpoint on the proximal side. The average distance between these probes was 0.61 μm. F, Graphic representation of the results of the experiment. The average distances between markers were analyzed using a one-way ANOVA test. The graph is centered around the grand mean (0.81 μm). The boxes contain 75% of the data for each group. The bars contain 95% of the data, and the line in the box is the mean. The distances from SOX9 to SOX9cre1 and from SOX9 to the downstream breakpoint are significantly shorter than the negative control (P=1.56×10-8) and are highly similar to the positive control. All distances were measured with the aid of a stage micrometer for accuracy (0.01 mm).
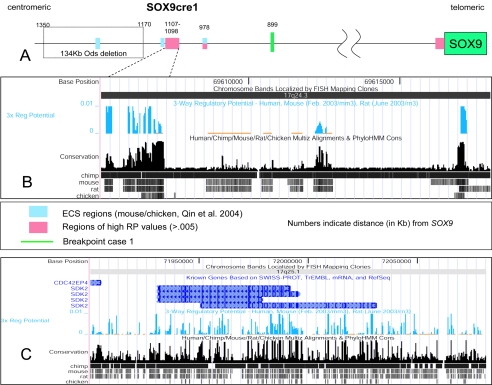
To search for potential cis-acting regulatory elements, we conducted in silico analysis of both chromosome breakpoint regions. PipMaker analysis of the regions surrounding the breakpoints with those from the syntenic mouse chromosome 11 region demonstrated intervals of significant homology in noncoding regions, several of which contain >80% sequence identity for >100 bp (data not shown). A Regulatory Potential (RP) score has been described recently (Kolbe et al. 2004). By comparing human, mouse (two-way RP), and rat (three-way RP) syntenic DNA sequences, the RP score predicts DNA elements with significantly reduced background, as compared with the results of other similar programs. The three-way RP values revealed several distinct sequences both upstream and downstream of SOX9. Most of these sequences fall outside of coding regions and may be potential regulators of SOX9. It is interesting that an ∼1.1-kb DNA element was identified (nucleotide positions 69605974–69607150) between the Ods deletion region (∼50 kb proximal to this element) and the upstream breakpoint in patient 1 (∼200 kb distal to this element) (Bishop et al. 2000; Qin et al. 2004) (fig. 5A and 5B). A secondary 226-bp DNA sequence with a high RP score was also found, 12 kb distal to the first element. The RP program also predicted several DNA segments with high scores throughout the downstream breakpoint area, signifying potential regulatory elements of SOX9 (fig. 5C). We suggest that the upstream, highly conserved genomic DNA region, which we term “SOX9cre1” (SOX9 conserved regulatory element 1), could contain the SOX9 cis-regulatory element that was modified by the Dct promoter in Odd Sex mice (Qin et al. 2004) and is dissociated from SOX9 by translocation breakpoints. In addition, this element may be responsible for the isolated Robin sequence phenotype reported recently by Jamshidi et al. (2004). It is interesting that the 17q breakpoint in the family reported in that article maps very close and proximal to the breakpoint in patient 1 of the present study. SOX9 regulates cartilage formation by activating the expression of chondrocyte-specific genes, such as COL2A1, COL9A1, and COL11A2 (Bi et al. 1999; Zhang et al. 2003). It is also possible that some patients with isolated Robin sequence have mutations involving this or other regulatory elements of SOX9. Thus, the mild CD phenotype found in our patient and others (Stalker and Zori 1997; Stalker et al. 2001; Hill-Harfe et al. 2005 [in this issue]) may be the result of slightly modified expression of chondrocyte-specific genes, secondary to altered SOX9 regulation as a result of spatial dissociation from the distal SOX9cre1. Mutations in COL2A1 and COL11A2 were shown to cause Stickler syndrome type I (STLI) and Stickler syndrome type II, respectively (Maumenee 1979; Ahmad et al. 1991; Vikkula et al. 1995; Williams et al. 1996; Donoso et al. 2002). A differential diagnosis of STLI was considered for patient 1 of the present study as well as for the two other patients with 17q24.3 translocation breakpoints (Vintiner et al. 1991; Stalker et al. 2001).
Figure 5.
Computer analysis of the regulatory elements around SOX9. A, Graphical representation of the region ∼1.35 Mb upstream of SOX9. The evolutionary conserved sequences (ECS) regions described by Qin et al. (2004) and the nearby region of high RP values are shown as blue and pink boxes, respectively. B, A more detailed view of the region of high RP values, with the UCSC Genome Browser map of three-way (human, mouse, and rat) RP analysis of the candidate cis-regulatory element SOX9cre1, ∼1.1 Mb upstream of SOX9. The presence of such an element has been hypothesized elsewhere (Pfeifer et al. 1999; Bagheri-Fam et al. 2001; Pop et al. 2004; Qin et al. 2004). C, UCSC map of the region surrounding the downstream breakpoint within the SDK2 gene in 17q25.1. There are several regions of high RP scores throughout the breakpoint area, signifying areas of potential regulatory elements of SOX9. Most RP peaks fall outside coding regions. BLAST results of SOX9cre1 suggest that this may be a pseudogene of the PAI-1 mRNA binding protein from chromosome 1. PhyloHMM Cons = phylogenetic hidden Markov model. URLs for SWISS-PROT, TrEMBL, and RefSeq can be found in the Electronic-Database Information section. For information on Multiz, see Blanchette et al. (2004).
Lettice et al. (2003) demonstrated that point mutations in a regulatory element located ∼1 Mb from the target gene Shh (localized in the intron of the LMBR1 gene) are capable of causing congenital abnormalities and possess the capacity to modify gene activity. Such abnormalities (reviewed by Kleinjan and van Heyningen [2005]) may yet help to describe a new type of genomic regulation, whereby the proper positions of key distal regulators are necessary for adequate tissue and temporal specificity of gene expression. This type of regulation may be similar to the active chromatin hub, in which the upstream and downstream regulatory elements must be adequately positioned for the formation of the correct three-dimensional structure necessary for the proper regulation of a tissue-specific and temporally specific gene (Tolhuis et al. 2002). Supporting this notion, Horike et al. (2005) recently demonstrated that formation of a silent-chromatin loop is a new mechanism underlying gene regulation.
In summary, we provide evidence for position effects on SOX9 associated with chromosome breakpoints mapping ∼1.3 Mb downstream and ∼0.9 Mb upstream of SOX9 and show FISH data consistent with physical interaction between these genomic regions. A potential upstream SOX9 cis-acting regulatory element (SOX9cre1) was delineated by comparative genomics in combination with breakpoint mapping. Finally, our studies suggest that, in some individuals with isolated Robin sequence, their phenotype may result from dysregulation of SOX9 by mutations or genomic rearrangements that affect the cis-acting regulatory elements of this gene.
Acknowledgments
We thank the patients and their families for their participation in this study. We appreciate the critical reviews of Drs. Veronica van Heyningen, Dirk-Jan Kleinjan, Brendan Lee, and Gerd Scherer. We thank Dr. Weimin Bi, for helpful discussion; Dr. Chad Shaw, for assistance with the statistical analyses; Dr. Marjorie R. Grafe, for the autopsy notes and pictures; and Dr. Svetlana A. Yatsenko and Charles Zaremba, for technical assistance. This work was generously supported by grants from the National Institute of Child Health and Human Development (PO1 HD39420) and the Baylor College of Medicine Mental Retardation Research Center (HD24064).
Electronic-Database Information
The URLs for data presented herein are as follows:
- ECgene, http://genome.ewha.ac.kr/ECgene/
- Fgenesh++, http://www.softberry.com
- Genscan, http://genes.mit.edu/GENSCAN.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CD) [PubMed]
- PipMaker, http://pipmaker.bx.psu.edu/pipmaker/
- RefSeq, http://www.ncbi.nlm.nih.gov/RefSeq/
- SGP, http://nemo.imim.es/grib/
- Swiss-Prot, TrEMBL, http://us.expasy.org/sprot/
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- Ahmad NN, Ala-Kokko L, Knowlton RG, Jimenez SA, Weaver EJ, Maguire JI, Tasman W, Prockop DJ (1991) Stop codon in the procollagen II gene (COL2A1) in a family with the Stickler syndrome (arthro-opthalmopathy). Proc Natl Acad Sci USA 88:6624–6627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri-Fam S, Ferraz C, Demaille J, Scherer G, Pfeifer D (2001) Comparative genomics of the SOX9 region in human and Fugu rubripes: conservation of short regulatory sequence elements within large intergenic regions. Genomics 78:73–82 [DOI] [PubMed] [Google Scholar]
- Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B (1999) Sox9 is required for cartilage formation. Nat Genet 22:85–89 [DOI] [PubMed] [Google Scholar]
- Bickmore WA, van der Maarel SM (2003) Perturbations of chromatin structure in human genetic disease: recent advances. Hum Mol Genet 12:R207–R213 [DOI] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, Haussler D, Miller W (2004) Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res 14:708–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop CE, Whitworth DJ, Qin Y, Agoulnik AI, Agoulnik IU, Harrison WR, Behringer RR, Overbeek PA (2000) A transgenic insertion upstream of Sox9 is associated with dominant XX sex reversal in the mouse. Nat Genet 26:490–494 [DOI] [PubMed] [Google Scholar]
- Cameron FJ, Hageman RM, Cooke-Yarborough C, Kwok C, Goodwin LL, Sillence DO, Sinclair AH (1996) A novel germ line mutation in SOX9 causes familial campomelic dysplasia and sex reversal. Hum Mol Genet 5:1625–1630 [DOI] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P (2002) Long-range chromatin regulatory interactions in vivo. Nat Genet 32:623–626 [DOI] [PubMed] [Google Scholar]
- Cho KS, Elizondo LI, Boerkoel CF (2004) Advances in chromatin remodeling and human disease. Curr Opin Genet Dev 14:308–315 [DOI] [PubMed] [Google Scholar]
- Davies AF, Mirza G, Flinter F, Ragoussis J (1999) An intersititial deletion of 6p24-p25 proximal to the FKHL7 locus and including AP-2α that affects anterior eye chamber development. J Med Genet 36:708–710 [PMC free article] [PubMed] [Google Scholar]
- de Kok YJM, Vossenaar ER, Cremers CWRJ, Dahl N, Laporte J, Hu LJ, Lacombe D, Fischel-Ghodsian N, Friedman RA, Parnes LS, Thorpe P, Bitner-Glindzicz M, Pander H-J, Heilbronner H, Graveline J, den Dunnen JT, Brunner HG, Ropers H-H, Cremers FPM (1996) Identification of a hot spot for microdeletions in patients with X-linked deafness type 3 (DFN3) 900 kb proximal to the DFN3 gene POU3F4. Hum Mol Genet 5:1229–1235 [DOI] [PubMed] [Google Scholar]
- Donoso LA, Edward AO, Frost AT, Ritter R III, Ahmad NN, Vrabec T, Rogers J, Meyer D (2002) Identification of a stop codon mutation in exon 2 of the collagen 2A1 gene in a large Stickler syndrome family. Am J Ophthalmol 134:720–727 [DOI] [PubMed] [Google Scholar]
- Erdel M, Lane AH, Fresser F, Probst P, Utermann G, Scherer G (2004) A new campomelic dysplasia translocation breakpoint maps 400 kb from SOX9 [abstract P0249]. European Society of Human Genetics Munich. Eur J Hum Genet Suppl 12:136 [Google Scholar]
- Foster JW, Dominguez-Steglich MA, Guioli S, Kwok C, Weller PA, Stevanovic M, Weissenbach J, Mansour S, Young ID, Goodfellow PN, Brook JD, Schafer AJ (1994) Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372:525–530 [DOI] [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA (2004). Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 118:555–566 [DOI] [PubMed] [Google Scholar]
- Goji K, Nishijima E, Tsugawa C, Nishio H, Pokharel RK, Matsuo M (1998) Novel missense mutation in the HMG box of SOX9 gene in a Japanese XY male resulted in campomelic dysplasia and severe defect in masculinization. Hum Mutat Suppl 1:S114–S116 [DOI] [PubMed] [Google Scholar]
- Hageman RM, Cameron FJ, Sinclair AH (1998) Mutation analysis of the SOX9 gene in a patient with campomelic dysplasia. Hum Mutat Suppl 1:S112–S113 [DOI] [PubMed] [Google Scholar]
- Hill-Harfe KL, Kaplan L, Stalker HJ, Zori RT, Pop R, Scherer G, Wallace MR (2005) Fine mapping of chromosome 17 translocation breakpoints ⩾900 kb upstream of SOX9 in acampomelic campomelic dysplasia and a mild, familial skeletal dysplasia. Am J Hum Genet 76:663–671 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horike S, Cai S, Miyano M, Cheng J-F, Kohwi-Shigematsu T (2005) Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet 37:31–40 [DOI] [PubMed] [Google Scholar]
- Houston CS, Opitz JM, Spranger JW, Macpherson RI, Reed MH, Gilbert EF, Herrmann J, Schinzel A (1983) The campomelic syndrome: review, report of 17 cases, and follow-up on the currently 17-year-old boy first reported by Maroteaux et al. in 1971. Am J Med Genet 15:3–28 [DOI] [PubMed] [Google Scholar]
- Huang B, Wang S, Ning Y, Lamb AN, Bartley J (1999) Autosomal XX sex reversal caused by duplication of SOX9. Am J Med Genet 87:349–353 [DOI] [PubMed] [Google Scholar]
- Jamieson RV, Perveen R, Kerr B, Carette M, Yardley J, Heon E, Wirth MG, van Heyningen V, Donnai D, Munier F, Black GCM (2002) Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum Mol Genet 11:33–42 [DOI] [PubMed] [Google Scholar]
- Jamshidi N, Macciocca I, Dargaville PA, Thomas P, Kilpatrick N, McKinlay Gardner RJ, Farlie PG (2004) Isolated Robin sequence associated with a balanced t(2;17) chromosomal translocation. J Med Genet 41:e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V (2003) Turned off by RNA. Nat Genet 34:125–126 [DOI] [PubMed] [Google Scholar]
- ——— (2005) Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet 76:8–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan D-J, van Heyningen V (1998) Position effect in human genetic disease. Hum Mol Genet 7:1611–1618 [DOI] [PubMed] [Google Scholar]
- Kolbe D, Taylor J, Elnitski L, Eswara P, Li J, Miller W, Hardison R, Chiaromonte F (2004) Regulatory potential scores from genome-wide three-way alignments of human, mouse, and rat. Genome Res 14:700–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok C, Weller PA, Guioli S, Foster JW, Mansour S, Zuffardi O, Punnett HH, Dominguez-Steglich MA, Brook JD, Young ID, Goodfellow PN, Schafer AJ (1995) Mutations in SOX9, the gene responsible for campomelic dysplasia and autosomal sex reversal. Am J Hum Genet 57:1028–1036 [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E (2003) A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12:1725–1735 [DOI] [PubMed] [Google Scholar]
- Lettice LA, Horikoshi T, Heaney SJH, van Baren MJ, van der Linde HC, Breedveld GJ, Joosse M, Akarsu N, Oostra BA, Endo N, Shibata M, Suzuki M, Takahashi E, Shinka T, Nakahori Y, Ayusawa D, Nakabayashi K, Scherer SW, Heutink P, Hill RE, Noji S (2002) Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci USA 99:7548–7553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S, Hall CM, Pembrey ME, Young ID (1995) A clinical and genetic study of campomelic dysplasia. J Med Genet 32:415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia R, Saal HM, Wangsa D (1991) A chromosome 17q de novo paracentric inversion in a patient with campomelic dysplasia: case report and etiologic hypothesis. Clin Genet 39:401–408 [DOI] [PubMed] [Google Scholar]
- Maumenee IH (1979) Vitreoretinal degeneration as a sign of generalized connective tissue disease. Am J Ophthalmol 88:432–449 [DOI] [PubMed] [Google Scholar]
- Meyer J, Südbeck P, Held M, Wagner T, Schmitz ML, Bricarelli FD, Eggermont E, Friedrich U, Haas OA, Kobelt A, Leroy JG, Van Maldergem L, Michel E, Mitulla B, Pfeiffer RA, Schinzel A, Schmidt H, Scherer G (1997) Mutational analysis of the SOX9 gene in campomelic dysplasia and autosomal sex reversal: lack of genotype/phenotype correlations. Hum Mol Genet 6:91–98 [DOI] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM (2003) Scanning human gene deserts for long-range enhancers. Science 302:413 [DOI] [PubMed] [Google Scholar]
- Palstra R-J, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W (2003) The β-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35:190–194 [DOI] [PubMed] [Google Scholar]
- Pfeifer D, Kist R, Dewar K, Devon K, Lander ES, Birren B, Korniszewski L, Back E, Scherer G (1999) Campomelic dysplasia translocation breakpoints are scattered over 1 Mb proximal to SOX9: evidence for an extended control region. Am J Hum Genet 65:111–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier C, Qin Y, Adams CP, Anaya Y, Singer JB, Hill AE, Lander ES, Nadeau JH, Bishop CE (2004) A complex interaction of imprinted and maternal-effect genes modifies sex determination in Odd Sex (Ods) mice. Genetics 168:1557–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop R, Conz C, Lindenberg KS, Blesson S, Schmalenberger B, Briault S, Pfeifer D, Scherer G (2004) Screening of the 1 Mb SOX9 5′ control region by array CGH identifies a large deletion in a case of campomelic dysplasia with XY sex reversal. J Med Genet 41:e47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Kong Lk, Poirier C, Truong C, Overbeek PA, Bishop CE (2004) Long-range activation of Sox9 in Odd Sex (Ods) mice. Hum Mol Genet 13:1213–1218 [DOI] [PubMed] [Google Scholar]
- Stalker HJ, Gray BA, Zori RT (2001) Dominant transmission of a previously unidentified 13/17 translocation in a five-generation family with Robin cleft and other skeletal defects. Am J Med Genet 103:339–341 [DOI] [PubMed] [Google Scholar]
- Stalker HJ, Zori RT (1997) Variable expression of rib, pectus, and scapular anomalies with Robin-type cleft palate in a 5-generation family: a new syndrome? Am J Med Genet 73:247–250 [PubMed] [Google Scholar]
- Tolhuis B, Palstra R-J, Splinter E, Grosveld F, de Laat W (2002) Looping interaction between hypersensitive sites in the active β-globin locus. Mol Cell 10:1453–1465 [DOI] [PubMed] [Google Scholar]
- Tommerup N, Schempp W, Meinecke P, Pedersen S, Bolund L, Brandt C, Goodpasture C, Guldberg P, Held K, Reinwein H, Saugstad OD, Scherer G, Skjeldal O, Toder R, Westvik J, van der Hagen CB, Wolf U (1993) Assignment of an autosomal sex reversal locus (SRA1) and campomelic dysplasia (CMPD1) to 17q24.3-q25.1. Nat Genet 4:170–174 [DOI] [PubMed] [Google Scholar]
- Tufarelli C, Stanley JAS, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR (2003) Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet 34:157–165 [DOI] [PubMed] [Google Scholar]
- Vikkula M, Mariman ECM, Lui VCH, Zhidkova NI, Tiller GE, Goldring MB, van Beersum SE, de Wall Malefijt MC, van den Hoogen FH, Ropers HH, Mayne R, Cheah KSE, Olsen BR, Warman ML, Brunner HG (1995) Autosomal dominant and recessive osteochondrodysplasias associated with the COL11A2 locus. Cell 80:431–437 [DOI] [PubMed] [Google Scholar]
- Vintiner GM, Temple IK, Middleton-Price HR, Baraitser M, Malcolm S (1991) Genetic and clinical heterogeneity of Stickler syndrome. Am J Med Genet 41:44–48 [DOI] [PubMed] [Google Scholar]
- Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E, Wolf U, Tommerup N, Schempp W, Scherer G (1994) Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79:1111–1120 [DOI] [PubMed] [Google Scholar]
- Williams CJ, Ganguly A, Considine E, McCarron S, Prockop DJ, Walsh-Vockley C, Michels VV (1996) A to G transition at the 3′ acceptor splice site of IVS17 characterizes the COL2A1 gene mutation in the original Stickler syndrome kindred. Am J Med Genet 63:461–467 [DOI] [PubMed] [Google Scholar]
- Wunderle VM, Critcher R, Hastie N, Goodfellow PN, Schedl A (1998) Deletion of long-range regulatory elements upstream of SOX9 causes campomelic dysplasia. Proc Natl Acad Sci USA 95:10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ID, Zuccollo JM, Maltby EL, Broderick NJ (1992) Campomelic dysplasia associated with a de novo 2q;17q reciprocal translocation. J Med Genet 29:251–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Jimenez SA, Stokes DG (2003) Regulation of human COL9A1 gene expression: activation of the proximal promoter region by SOX9. J Biol Chem 278:117–123 [DOI] [PubMed] [Google Scholar]