Abstract
Autosomal recessive primary microcephaly (MCPH) is a neurodevelopmental disorder. It is characterized by two principal features, microcephaly present at birth and nonprogressive mental retardation. The microcephaly is the consequence of a small but architecturally normal brain, and it is the cerebral cortex that shows the greatest size reduction. There are at least seven MCPH loci, and four of the genes have been identified: MCPH1, encoding Microcephalin; MCPH3, encoding CDK5RAP2; MCPH5, encoding ASPM; and MCPH6, encoding CENPJ. These findings are starting to have an impact on the clinical management of families affected with MCPH. Present data suggest that MCPH is the consequence of deficient neurogenesis within the neurogenic epithelium. Evolutionary interest in MCPH has been sparked by the suggestion that changes in the MCPH genes might also be responsible for the increase in brain size during human evolution. Indeed, evolutionary analyses of Microcephalin and ASPM reveal evidence for positive selection during human and great ape evolution. So an understanding of this rare genetic disorder may offer us significant insights into neurogenic mitosis and the evolution of the most striking differences between us and our closest living relatives: brain size and cognitive ability.
Introduction
The aim of this review is to summarize the recent findings in autosomal recessive primary microcephaly (MCPH). First, the clinical features of the disorder are discussed, especially that “microcephaly” is the finding of a reduced head circumference (HC) and that MCPH is but one of many conditions in which the principal features are “microcephaly and mental retardation. Second, we review the molecular genetic findings for the six known recessive loci, for which four of the genes have been discovered. Thirdly, we present the evidence that MCPH is the consequence of deficient mitosis in neural precursors. And finally, we discuss the role that MCPH genes might have played during the evolution of the human brain, which is approximately three times larger than that expected for a primate of our body size.
Clinical Features of MCPH
Microcephaly and HC
The term “microcephaly” refers to a clinical finding: an HC significantly less than expected for an individual’s age and sex (Warkany 1981; Aicardi 1998). HC is used as a surrogate measurement of brain size; however, it is only imperfectly correlated with brain volume. Other methods have been used (e.g., volumetric nuclear magnetic resonance [NMR] scanning), but HC remains the common, simple method for evaluating gross brain size (Aicardi 1998), although it needs to be accurately measured and charted relative to age and sex (Nellhaus 1968; Sher and Brown 1975; Babson and Benda 1976; Sells 1977; Gross et al. 1983; Roche et al. 1987). An HC of three standard deviations below the mean (−3 SD) is usually the cut-off for defining microcephaly (Ross and Frias 1977; Baraitser 1990). Importantly, microcephaly is strongly correlated with mental retardation (Warkany 1981; Gross et al. 1983; Dolk 1991). Individuals with an HC of −3 SD and normal intelligence are occasionally reported, but individuals with an HC of −4 SD or less and normal intelligence are very rare (Teebi et al. 1987).
Microcephaly is divided into primary microcephaly, which is present at birth, and secondary microcephaly, which develops postnatally (Woods 2004). The crucial difference between these groupings is that primary microcephaly is usually a static developmental anomaly, whereas secondary microcephaly indicates a progressive neurodegenerative condition (Qazi and Reed 1973; Opitz and Holt 1992; Dobyns 2002; Rosenberg et al. 2002). There are many nongenetic and genetic causes of “primary microcephaly with mental retardation,” such as congenital infection with toxoplasma, maternal alcohol overconsumption during pregnancy, and Rubenstein Taybi syndrome, all of which must be excluded before the diagnosis of MCPH is considered (Cowie 1960; Winter and Baraitser 2003).
Definition of MCPH as a Phenotype
“True microcephaly,” “microcephaly vera,” and “autosomal recessive primary microcephaly” are terms in the literature that were probably all intended to describe the same phenotype. In “true microcephaly,” the sloping forehead was a defining feature; however, it is not seen in all cases of MCPH (Bundey 1997; Roberts at al. 2002). Over time, “microcephaly vera” has come to include the condition of any child with congenital microcephaly accompanied by a range of neurological features, and hence it has lost specificity as a diagnosis. This led to the introduction of a new diagnostic label, “autosomal recessive primary microcephaly,” shortened to “MCPH” (MIM #251200) (Jackson et al. 1998).
The initial defining clinical features of MCPH in the studies by Jackson et al. (2002) and Roberts et al. (2002) were:
-
1.
Congenital microcephaly at least 4 SD below age and sex means.
-
2.
Mental retardation but no other neurological finding, such as spasticity, seizures, or progressive cognitive decline.
-
3.
Normal height and weight, appearance, and results on chromosome analysis and brain scan.
MCPH is a disorder of fetal brain growth. The timing of the reduction in growth has been elucidated by ultrasound of affected pregnancies. Normal head measurements are found up to 20 wk of gestation, whereas a decreased HC is seen by 32 wk (C.G.W., personal observation). After birth, HC lies between −4 and −12 SD (Roberts et al. 2002). The relative degree of microcephaly does not vary throughout life, and within a family HC usually does not vary by >2 SD between affected individuals (Roberts et al. 2002). NMR and CAT scans of the brain show that MCPH causes a central nervous system of reduced size, with the greatest effect on the cerebral cortex (Bond et al. 2002). MCPH is associated with a simplification of the cerebral cortical gyral pattern and a slight reduction in the volume of the white matter, consistent with the small size of the brain (Barkovich et al. 2001). In general, the architecture of the CNS is normal, with no evidence of a neuronal migration defect (Bond et al. 2002).
Individuals with MCPH are all mentally retarded, but the retardation is usually only of mild to moderate severity (Roberts et al. 2002). There is no evidence of progressive cognitive decline or of any motor deficit. Early milestones such as alertness, smiling to parents, and head control are often normal. Later motor and social milestones are mildly delayed, but speech development is consistently delayed. As teenagers, children with MCPH have good fine motor control, balance, and grace in movements. They achieve well in sports, compared with other mentally retarded children, and often have a “happy affect.” Individuals with MCPH tend to be well behaved and are “biddable”—that is, they are compliant with instruction and can be taught skills of daily living, common-sense safety, and some reading and writing (Ross and Frias 1977; Roberts et al. 2002).
Subsequent to MCPH gene discovery, genotype/phenotype studies showed that the original MCPH diagnostic criteria required revision. Whereas the original definition excluded seizures, height reduction, and abnormal cytogenetic findings, these features have now been reported in some cases of MCPH, and the diagnosis of MCPH is no longer excluded by their presence. Specific genotype-phenotype correlations have been made for individuals with MCPH1 mutations. These individuals may have a reduction in height, although it is not as marked as the degree of microcephaly (Neitzel et al. 2002; Trimborn et al. 2004). In one MCPH1 family, a small number of periventricular neuronal heterotopias (groups of neurons in an abnormal position within the brain) were found—which is indicative of abnormal neuronal migration (Trimborn et al. 2004). Periventricular neuronal heterotopias were not reported in the original families used to link and identify the MCPH1 gene, so their relative incidence is unknown (Jackson et al. 2002). MCPH1 mutations are also consistently associated with an increased proportion of cells with premature chromosome condensation leading to a prophaselike appearance seen on cytogenetic analysis (see below).
The current clinical definition of MCPH is as follows:
-
1.
Congenital microcephaly, with HC at least 4 SD below age and sex means.
-
2.
Mental retardation but no other neurological finding, such as spasticity, or progressive cognitive decline. Fits are unusual but do not exclude the diagnosis.
-
3.
For the majority of people with MCPH, normal height, weight, appearance, chromosome analysis, and brain scan are reported. For people with MCPH1 mutations, a reduction in height may be found, but the HC will always be significantly more reduced than height; on NMR scan, some MCPH1 patients show evidence of periventricular neuronal heterotopias suggestive of neuronal migration defects; and cytogenetic analysis may indicate an increased proportion of prophaselike cells.
Incidence of MCPH
Prior to the discovery of MCPH genes, autosomal recessive microcephaly was reported to have an incidence of 1/30,000 in Japan, 1/250,000 in Holland, and 1/2,000,000 in Scotland (Komai et al. 1955; Van den Bosch 1959; Tolmie et al. 1987). More-recent studies have not been performed; however, MCPH seems rarer in whites than in Asian and Arab populations where consanguineous marriage is practiced. Our anecdotal experience in the Yorkshire region of Britain is a white MCPH incidence of 1 per million compared to 1/10,000 in those originating from northern Pakistan. An increased incidence of autosomal recessive diseases in consanguineous populations is expected on theoretical grounds and has been observed (Freire-Maia 1990; Bundey and Alam 1993; Tuncbilek 2001). Given the number of MCPH genes and mutations found in the Pakistani population (see below), the high incidence of MCPH probably reflects the effects of consanguinity and not a founder effect.
Inheritance of Primary Microcephaly
Prior to the discovery of MCPH genes, “true microcephaly” or “microcephaly vera” had been considered to be inherited as an autosomal recessive disorder (Kloepfer et al. 1964; Bundey 1997). This hypothesis was based on the reports of pedigrees with multiple affected siblings but unaffected parents, frequent parental consanguinity, and segregation ratios that suggested a large proportion of primary microcephaly, particularly in consanguineous populations, is caused by recessive genes (Böök et al. 1953; Komai et al. 1955; Van den Bosch 1959; Sujatha et al. 1989). Although the expected recurrence risk for autosomal recessive traits is 1 in 4, the empiric recurrence risk for a nonconsanguineous couple who have had one child with a diagnosis of MCPH has been calculated as 1 in 5 if detailed chromosome studies and neuroimaging yield normal results (Tolmie et al. 1987). It has been suggested that there may be a phenotype in heterozygous carriers of MCPH mutations, because of an increased frequency of intellectual impairment in parents of individuals with recessive microcephaly (Kloepfer et al. 1964; Qazi and Reed 1975). However, this has not been a consistent finding, and this matter awaits reassessment (Van den Bosch 1959; Roberts et al. 2002).
Molecular Genetics of MCPH
Linkage and Heterogeneity
MCPH was expected to exhibit genetic heterogeneity, because of its broad clinical phenotype (Cowie 1960). Six loci have been identified to date (MCPH1–MCPH6; see table 1) (Jackson et al. 1998; Jamieson et al. 1999, 2000; Roberts et al. 1999, 2002; Moynihan et al. 2000; Pattison et al. 2000; Leal et al. 2003). Each recessive locus was initially mapped in a large consanguineous family by autozygosity mapping techniques, using panels of ∼400 polymorphic autosomal microsatellite markers spread throughout the human genome at a distance of ∼12 cM (Lander and Botstein 1987; Mueller and Bishop 1993).
Table 1.
MCPH Loci[Note]
| Locus | AutosomalLocalization | Linkage Reference | Gene | Ethnicity |
| MCPH1 | 8p22-pter | Jackson et al. 1998 | Microcephalin | Northern Pakistani |
| MCPH2 | 19q13.1-13.2 | Roberts et al. 1999 | Unknown | Northern Pakistani and Indian |
| MCPH3 | 9q34 | Moynihan et al. 2000 | CDK5RAP2 | Northern Pakistani |
| MCPH4 | 15q15-q21 | Jamieson et al. 1999 | Unknown | Moroccan |
| MCPH5 | 1q31 | Pattison et al. 2000; Jamieson et al. 2000 | ASPM | Northern Pakistani, Turkish, Jordanian, Dutch, Saudi Arabian, Yemeni, and Indian |
| MCPH6 | 13q12.2 | Leal et al. 2003 | CENPJ | Northern Pakistani and Brazilian |
Note.— The known MCPH loci are listed in chronological order of discovery. For each locus, the autosomal location is given, with the appropriate reference, the name of the gene (if known), and the ethnicities of the individuals in whom a mutation has been reported.
Four of the six loci (MCPH1–MCPH3 and MCPH5) were discovered in families of northern Pakistani origin, with two loci (MCPH1 and MCPH3) remaining exclusive to this population.
MCPH heterogeneity studies have been performed in a northern Pakistani population and MCPH5 was identified as the most common locus, accounting for linkage of nearly half of the amassed families (24/56 families) (Roberts et al. 2002). MCPH2 accounted for 10 families (14%); MCPH1 and MCPH3 were each associated with two families (4%), whereas MCPH4 linkage has not been found in the Pakistani population. MCPH6 had not been identified at the time of the heterogeneity investigation; however, subsequently this locus has been linked to families of northern Pakistani origin (Bond et al. 2005). MCPH2, MCPH5, and MCPH6 are represented in multiple populations (Jamieson et al. 2000; Pattison et al. 2000; Bond et al. 2002, 2003; Leal et al. 2003, Kumar et al. 2004). Further MCPH loci await discovery, as 18/56 northern Pakistani families and 5/9 Indian families show no evidence of linkage to the known loci (Roberts et al. 2002; Kumar et al. 2004).
Genes
Positional cloning strategies have been employed to identify causative genes at four loci (fig. 1 and table 1): MCPH1 (Jackson et al. 2002), MCPH5 (Bond et al. 2002), MCPH3 (Bond et al. 2005), and MCPH6 (Bond et al. 2005).
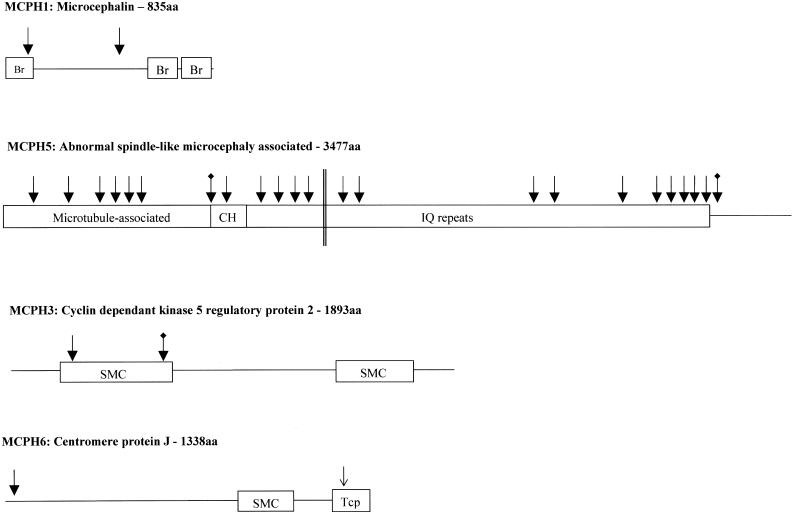
Figure 1.
The MCPH proteins, probable functional domains, and sites of mutations. The MCPH loci and protein names are given above a cartoon of the protein. The known or well-predicted domains are shown. Reported mutations are shown at their positions within the gene: nonsense mutations as filled arrows, splicing mutations that lead to premature termination codons as blackened arrows with marked tail, and a missense mutation as an unfilled arrow. Parallel lines indicate the translocation breakpoint in the ASPM gene described by Pichon et al. (2004). Each MCPH protein is marked with the known or hypothesized functional domains. Br is a BRCT Breast Cancer Suppressor Protein (BRCA1) carboxy-terminal domain, pfam00533. SMC is a chromosome segregation ATPase domain, COG1196, but all are only possible predictions. CH is calponin homology domain, pfam00307. IQ repeats is an isoleucine-glutamine calmodulin-binding domain, pfam00612. Tcp is a T-complex protein 10 C-terminus domain, pfam07202.
MCPH1/Microcephalin
Using ancestral haplotyping of novel microsatellite markers, Jackson and colleagues further refined the MCPH1 region from 13 cM to 2.1 Mb before commencing candidate gene sequencing. In two consanguineous families of northern Pakistani origin, a single homozygous 74C→G (S25X) nonsense mutation was identified in the second exon of a 2,505-nucleotide, 14-exon uncharacterized gene named “Microcephalin” (Jackson et al. 2002) The mutation segregated correctly within the families and was not present in >200 population-matched control alleles. Microcephalin is expressed in fetal brain, fetal liver, and fetal kidney at similar levels and is expressed at low levels in other fetal and adult tissues. Tissue in situ hybridization studies of Microcephalin mRNA in mouse fetal sections identified regions of Microcephalin expression in the developing forebrain during the period of neurogenesis.
MCPH5/ASPM
The most common cause of MCPH is homozygous mutations in the MCPH5 gene, Abnormal Spindle–like, Microcephaly associated (Drosophila) (ASPM) (Bond et al. 2002, 2003; Kumar et al. 2004; Pichon et al. 2004; Shen et al., in press). ASPM is a large gene with 28 exons and a 10.4-kb ORF, and its name is derived from the Drosophila gene abnormal spindles. The 24 published ASPM mutations comprise nonsense mutations, deletions of 1–7 base pairs, a breakpoint translocation, and intronic splice-donor point mutations (Bond et al. 2002, 2003; Kumar et al. 2004; Pichon et al. 2004). The mutations are distributed throughout the gene and are all predicted to produce truncated protein products ranging in size from 116 amino acids (R117X) to 3,357 amino acids (K3328fsX29). There is no correlation between the position of the mutation in the gene and the degree of microcephaly or mental retardation (Bond et al. 2002, 2003). This may indicate that the common mechanism of action of ASPM mutation is nonsense-mediated decay (Cartegni et al. 2002). Investigation of the fetal expression pattern of Aspm mRNA in mouse brain revealed expression at sites of active neurogenesis in the neuroepithelium (Bond et al. 2002). Postnatally, Aspm expression decreases with the completion of neurogenesis and upregulation of gliogenesis in the cortex. This expression pattern indicates that Aspm is involved in neuron rather than glia production. Thus, the neuronal reduction associated with MCPH due to ASPM mutations is compatible with the expression pattern of Aspm in mouse brain.
It has been suggested that a second gene causing MCPH exists at the MCPH5 locus, as mapping studies in five Jordanian and Dutch families linked the families to the MCPH5 locus but excluded the genetic region containing ASPM (Wallerman et al. 2003). If this is the case, it must be a rare cause of MCPH, since all MCPH5 linked Pakistani families have mutations in ASPM and since ASPM mutations in the Dutch and Jordanian populations have been identified (Bond et al. 2002, 2003)
MCPH3/CDK5RAP2
The MCPH3 locus was refined from 12 cM to 2.2 Mb by the identification and subsequent microsatellite screening of a second family of northern Pakistani origin that shows linkage and with multiple affected members (Moynihan et al. 2000; Bond et al. 2005). A 34-exon candidate gene, Cyclin Dependent Kinase 5 Regulatory Associated protein 2 (CDK5RAP2) was sequenced in the families. A homozygous 243T→A (S81X) nonsense mutation and an intronic mutation 15 bases prior to the normal splice-acceptor sequence IVS26-15A→G (E385fsX4) were identified. Both mutations segregated with the disease within the families and were not present in >380 control alleles. The intronic mutation creates a new, superior splice-acceptor site for exon 27, leading to the translation of 4 amino acids before a termination codon is reached. Gene expression of Cdk5rap2 in murine sagittal sections was highest in the brain and the spinal cord, particularly in the neuroepithelium of the frontal cortex (Bond et al. 2005).
MCPH6/CENPJ
The 6-Mb MCPH6 region was refined by the use of novel polymorphic microsatellite markers that were genotyped in the original Brazilian family used to map the locus and in two additional northern Pakistani multiply-affected consanguineous families (Leal et al. 2003; Bond et al. 2005). This facilitated the reduction of the locus to 3.1 Mb and enabled the selection of a candidate gene, Centromere associated Protein J (CENPJ, also known as “CPAP,” for centrosomal protein 4.1- associated protein), in which two homozygous mutations were identified: the frameshift mutation 17delC in exon 2 (T6fsX3), in two pedigrees, and the missense mutation 3704A→T in exon 16 (E1235V), in one pedigree. 3704A→T is the only MCPH mutation described to date that is not predicted to result in a protein truncation. The E1235V mutation occurs in a highly conserved Tcp10 binding domain, which is involved in binding to the protein 4.1R-135, but how it leads to a functional null allele is unknown (fig. 1d) (Hung et al. 2004).
Mitosis and MCPH
Microcephalin, Cell Cycle, and DNA Repair
The 835-aa Microcephalin gene product is predicted to contain three BRCA1 C-terminal domains (BRCT domains), with the S25X protein-truncating mutation occurring in the first of these domains (fig. 1a). Other proteins containing BRCT domains have roles in cell-cycle control and DNA repair (Hyton et al. 2000), and a loss of DNA repair genes can lead to apoptosis during neurogenesis (Mochida and Walsh 2001). This led Jackson and colleagues to hypothesize that Microcephalin may have a function in DNA repair or cell-cycle control and that the phenotype of MCPH might result from perturbation of normal cell-cycle regulation in neuronal progenitors (Jackson et al. 2002).
Two potential roles of Microcephalin have emerged, one on the control of cell-cycle timing and the other of DNA repair following ionizing radiation damage (Trimborn et al. 2004; Xu et al. 2004). A novel autosomal recessive disorder was described that was characterized by premature chromosome condensation (PCC) in siblings from a consanguineous family exhibiting microcephaly, growth retardation, and mental retardation (Neitzel et al. 2002). Patient cell preparations exhibited a high proportion of prophaselike cells (7%–17%), signifying early onset of chromosome condensation. The disorder was found to result from a 427insA mutation in Microcephalin (Trimborn et al. 2004). MCPH1 primary microcephaly shares this cellular phenotype of PCC, and it can also be recapitulated by small-interfering-RNA–mediated depletion of Microcephalin (Trimborn et al. 2004). Experimental depletion of Microcephalin also impairs radiation-induced intra-S-phase and G2/M cell-cycle checkpoints by decreasing the levels of BRCA1 and Checkpoint kinase 1 (Xu et al. 2004). MCPH may therefore result from a perturbation of the cell cycle in neurogenesis (Trimborn et al. 2004).
ASPM and the Spindle Pole Body
Bioinformatics analyses suggest that the 3,477-aa human ASPM protein contains a putative amino terminal microtubule binding domain (Saunders et al. 1997); at least one calponin homology domain; 74 isoleucine-glutamine (IQ) domains, which potentially bind calmodulin (Craig and Norbury 1998; Bond et al. 2002); and a carboxy terminal region of no known function (fig. 1b).
Asp, the probable ortholog of ASPM in Drosophila, is essential for organization and bundling of microtubules at the spindle poles and has been shown to be necessary for the proper formation of a central mitotic spindle in mitosis and meiosis (Ripoll et al. 1985; Gonzalez et al. 1990; Avides and Glover 1999; Avides et al. 2001; Wakefield et al. 2001; Riparbelli et al. 2001). This role is supported by results from immunofluorescence studies, which have shown wild-type Asp to be distributed throughout the cytoplasm in interphase but to localize to the spindle poles during mitosis when the bipolar spindle is formed (Saunders et al. 1997; Wakefield et al. 2001; Riparbelli et al. 2002). During late anaphase and telophase, when the spindle begins to disassemble, Asp relocates to the minus ends of the central spindle microtubules, where it remains until the central spindle has broken down (Saunders et al. 1997; Avides and Glover 1999; Wakefield et al. 2001).
CDK5RAP2, CENPJ, and the Centrosome
Human CDK5RAP2 and CENPJ are also centrosomal proteins, being localized to the centrosomes in interphase and to the spindle poles during mitosis (Hung et al. 2000; Bond et al. 2005). The probable Drosophila ortholog of CDK5RAP2, centrosomin (cnn), interacts with the gamma tubulin ring complexes within the centrosome, which are responsible for the production of the microtubules forming the mitotic spindle (Li and Kaufman 1996; Megraw et al. 1999; Terada et al. 2003). Drosophila cnn mutants exhibit a gross reduction in cell number in the central and peripheral nervous systems (Li and Kaufman 1996). CENPJ is known to interact via a Tcp10 domain with nonerythrocyte 4.1 protein 135 splice variant (4.1R-135) (Hung et al. 2000). CENPJ is associated with the γ-tubulin ring complex (Hung et al. 2004). In vitro evidence suggests that CENPJ might be able to inhibit microtubule nucleation and to depolymerize microtubules (Hung et al. 2004). Therefore, CDK5RAP2 and CENPJ may be involved in the control of the centrosomal production of microtubules during neurogenic mitosis (Bond et al. 2005).
Neurodevelopmental Etiology of MCPH
All data to date suggest that MCPH is a primary disorder of neurogenic mitosis and not one of neural migration, neural apoptosis, or neural function. This is because all four known MCPH genes are expressed in the neuroepithelium, and phenotypic features and brain scans suggest that MCPH patients have a small brain that functions normally for its size.
The majority of neurons in the brain are thought to originate from neuronal progenitors that are the predominant cells in the neuroepithelium, which line the ventricles of the brain (Brand and Rakic 1979). Neural progenitor cells have a specific pattern of mitotic activity that was first delineated in fly neuroblasts: symmetric cell divisions, which have mitotic spindles in the plane of the neuroepithelium that yield two neural progenitor cells, or asymmetric divisions, which occur when the mitotic spindle is oriented perpendicular to the neuroepithelium, giving rise to one postmitotic neuron and one progenitor cell (Doe and Bowerman 2001; Haydar et al. 2003; Albertson and Doe 2003). Since the timing of these two types of divisions during development is likely to be crucial to the final number of neurons, it has been proposed that changes in its regulation might be responsible for some cortical disorders, like microcephaly (Rakic 1988), and also for differences in brain size among different species (Rakic 1995). In mammals, it has also been shown experimentally that larger brains can result from increasing the number of neural progenitor symmetric cell divisions (Kornack and Rakic 1998; Chen and Walsh 2003).
The function of MCPH genes during neurogenic mitosis is unknown but is likely to be either (1) controlling the expansion of the neural progenitor pool or (2) involvement in the decision to switch from symmetric to asymmetric cell division. The latter hypothesis is supported by Drosophila studies showing that homozygous asp mutations result in metaphase arrest during asymmetric cell division in larval brain and that cnn mutants result in deficient asymmetric cell division in the male germ line (Wakefield et al. 2001; Yamashita et al. 2003). The cellular localization of three of the MCPH proteins—CDK5RAP2, ASPM, and CENPJ—is known to be centrosomal during mitosis. This suggests that the centrosome is an organelle of great importance during neurogenic mitosis.
MCPH, Brain Size, and Evolution
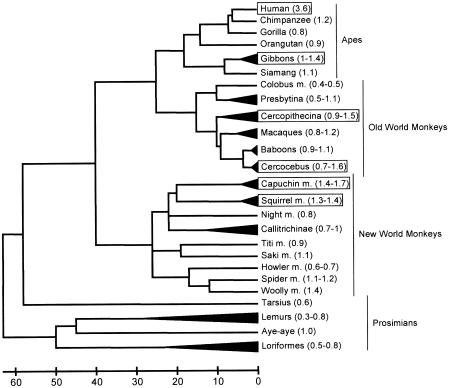
Brain size differs among animals mainly because animals come in different sizes. In mammals, >90 % of variation in brain weight can be explained by variation in body weight (Jerison 1973). Outliers from this relationship are of interest and if certain species or groups of species have brain sizes that are larger than expected from their body size (i.e., that are more “encephalized”), this is usually interpreted as an adaptation of these species (Harvey and Krebs 1990). No other species is as encephalized as humans, which have an ∼5-fold larger brain than expected for a mammal of their body size and an ∼3-fold larger brain than expected for an ape of their body size. But increases in relative brain size have also occurred in many other lineages among primates (see fig. 2 and, e.g., Armstrong 1985; Stephan et al. 1988; Marino 1998). Also notable are dolphins and related whales, which are more encephalized than any primate other than humans (Marino 1998). Fossil evidence suggests that encephalization increased several times during human evolution, starting around 2 million years ago and reaching its current state probably before the lineage leading to modern humans split from that leading to Neanderthals, ∼500,000 years ago (McHenry 1994). Interestingly, at least one reduction of relative brain size occurred during the evolution of human species, as well: a recently discovered species, Homo florensis, which lived until 18,000 years ago on the Indonesian island Flores, evolved probably from an ancestral Homo erectus population but had a relative brain size substantially lower than that in Homo erectus (Brown et al. 2004; Morwood et al. 2004).
Figure 2.
Encephalization in primates. The relative encephalization of various primate species was mapped on a phylogeny of primates (Goodman 1999). Data on brain weight and body weight was taken from Marino (1998) for apes, Old World monkeys, and New World monkeys and from Stephan et al. (1981), as given by Clark et al. (2001), for prosimians. The encephalization quotient (i.e., the ratio of observed brain weight to expected brain weight) is given in parentheses. The range is given when the data from several species are collapsed in the tree. The expected brain weight was calculated from a linear regression on log transformed brain and body weights for the Marino data, excluding humans (brainweight=10exp(0.6765×bodyweight+2.3406)). Species showing at least a 1.4-fold–heavier brain than expected for their body weights are marked by rectangles.
MCPH is an intriguing disease from an evolutionary standpoint, since it affects specifically the ratio of brain size to body size and since the brain of affected individuals can be of comparable size to that of a chimpanzee or gorilla (Bond et al. 2002; Jackson et al. 2002). Maybe even more intriguing is that MCPH genes—as discussed above—seem to play a role in the mitosis of neuronal precursors and that this is the process that has been hypothesized to be affected during the evolution of larger brains (Rakic 1995). So, from a genetic point of view, MCPH genes are excellent candidate genes for gain-of-function mutations, because they can carry loss-of-function mutations that affect the phenotype of interest. With the discovery of the first two MCPH genes, Microcephalin and ASPM, it has been possible to study whether there is evidence for adaptive changes in these genes and whether such changes correlate with changes in relative brain size during evolution.
Studying Protein Evolution
When one compares the coding regions of a gene in two species, one can find two types of differences: synonymous differences, which do not change the encoded protein sequence, and nonsynonymous differences, which do. The ratio of synonymous differences to synonymous sites (Ks) and the ratio of nonsynonymous differences to nonsynonymous sites (Ka) can then be calculated and compared (for reviews, see Yang and Bielawski 2000; Hurst 2002; Fay and Wu 2003). If one assumes that differences at synonymous sites do not have any functional consequences (i.e., do not affect the fitness of an organism), Ks can be regarded as an estimate of the neutral mutation rate. So, if mutations at all nonsynonymous sites also have no functional consequence—as is the case in a pseudogene—then Ka should be not different from Ks, and the expected Ka/Ks ratio would be 1. Since often mutations that change the protein sequence have negative consequences, these are not observed as differences between two species and the Ka/Ks ratio is <1. Sometimes, however, mutations at nonsynonymous sites can have advantageous consequences and these are then more likely to be observed between two species. Therefore, a Ka/Ks ratio (significantly) larger than one is interpreted as evidence for positive selection in that gene. Of course, both of these effects can act on a given gene, so that for example Ka/Ks can be smaller than one if only a few nonsynonymous mutations have been positively selected and many nonsynonymous sites are under constraint. Consequently, if Ka/Ks ratios <1 differ (e.g., between two evolutionary lineages), this can be due to a different amount of positive selection or to a different amount of constraint in these two lineages. One possible way to estimate the amount of constraint is to analyze common polymorphisms within a species. If this estimate of Ka/Ks is (significantly) smaller than the Ka/Ks estimate of fixed differences along the species lineage, positive selection can be inferred (McDonald 1991).
Unfortunately, other genetic changes in a gene (such as insertions and deletions, changes in splicing pattern, regulatory changes), can not currently be tested in a similar way, mainly because a neutral mutation rate like Ks is missing for these kinds of changes so that expected values can not rigorously be estimated. Although single adaptive genetic changes can in principle be identified by their signature of selection at closely linked sites within a population, this signature vanishes quickly after the selected site has been fixed in a population, so that for example in humans such a signature is not expected if the fixation of the adaptive mutation is more than 200,000 years ago (Przeworski 2002). The relative brain size of modern humans was probably attained around 500,000 years ago (see above). Therefore, seeking the signature of selection in this way does not seem a very promising method by which to find human-specific adaptive changes related to brain size increase.
Evolutionary Studies of Microcephalin
Two studies have examined the evolution of MCPH1, and both find some evidence for positive selection (Evans et al. 2004a; Wang and Su 2004). In the study by Wang and Su, a signature of positive selection on the lineage leading from the ancestor of all apes to the ancestor of great apes was found, as well as some signature of positive selection during recent human evolution (i.e., very likely less than half a million years ago). In the study by Evans et al., a signature of positive selection on the lineages leading to the ancestor of Old World Monkeys, apes, great apes, African apes, and finally humans was found. Given the relatively low power to detect positive selection at all, these findings are remarkable. However, their results need to be viewed with caution, as the strongest signatures of selection are detected on lineages, which show no particular evidence for an increase in encephalization, and the human lineage, for which the biggest change in brain size has occurred, does not show a tendency for higher Ka/Ks compared with other lineages. As discussed above, this does not exclude the possibility that some changes in Microcephalin might have been positively selected which lead to the human brain increase, but independent evidence for this (e.g., by functional studies) is needed.
Evolutionary Studies of ASPM
The evolution of the ASPM gene in primates and mammals has been examined in three studies (Zhang 2003; Evans et al. 2004b; Kouprina et al. 2004). An initial and tempting speculation that the number of IQ repeats (74 in man, 62 in mouse, and 24 in Drosophila) could correlate with brain size in animals could not be confirmed, since many mammals have as many IQ repeats as humans and since the difference in IQ length between mouse and human is specific for the lineage leading to mouse (Bond et al. 2002; Zhang 2003). Interestingly, however, the highest Ka/Ks ratio is found on the human lineage, and analyses of polymorphisms among humans suggest that ASPM is currently under a high evolutionary constraint, indicating that human-specific amino acid changes might have been fixed by positive selection. As for Microcephalin, these findings are remarkable in the light of the relatively low power of these tests, and this time the biggest increase in Ka/Ks correlates with the biggest increase in brain size. However, high Ka/Ks values have also been found on other ape lineages, and polymorphisms in gorillas suggest that amino acid changes in ASPM might already have been fixed by positive selection in the ancestors of humans, chimpanzees, and gorillas (Kouprina et al. 2004). Additionally, the high Ka/Ks ratio on the human lineage seems to be caused, to a large extent, by a lower number of fixed synonymous substitutions. This would imply a lower mutation rate on the human lineage, in addition to positive selection. So some aspects of these evolutionary findings do not have a straightforward interpretation for human brain expansion.
Implications of Evolutionary Findings
Although the signatures of positive selection for ASPM and for Microcephalin are not so strong that they clearly implicate a role for these genes in human brain expansion, they have an encouraging tendency. Further evolutionary analyses of other primate or mammalian lineages in which increases in relative brain size occurred (see above) might be fruitful, to clarify how well evolutionary changes in MCPH genes correlate with brain size increase. In addition, the identification of genes that interact with MCPH genes, especially in neuronal progenitor cell mitosis, will lead to further candidates that might be involved in the evolution of brain size increase in humans and other species. It is worth noting that an increase in relative brain size is not the only phenotypic change that occurred during the evolution of the human brain (Preuss 2004) and that recent studies suggest that positive selection might have acted on a considerable number of genes important for brain function and development during human evolution (Dorus et al. 2004). It will be important (and difficult) to find functional model systems in which these kinds of evolutionary hypotheses can be further tested. Transgenic mice could be one such possibility, and, although it is unclear to what extent mice would work as a model system in this respect, it is encouraging that artificial genetic changes can cause a brain size increase in mice (Chenn and Walsh 2002).
Conclusions
Given the many possible causes of microcephaly, the diagnosis of MCPH should only be made after differential diagnoses have been sought and eliminated. With identification of MCPH genes, the following are becoming increasingly available for patients: prenatal diagnosis (to detect a recurrence of the disorder), postnatal diagnosis (to distinguish the disorder from the many differential diagnoses), and carrier testing (in consanguineous families in which the disease is known to occur). There are at least seven genes that can cause this neurodevelopmental disorder; four have been identified, and current evidence suggests that MCPH is probably a primary disorder of neurogenic mitosis. As such, it may have much to teach us about the control of neuron production by neural stem cells and about how this has been modulated by evolution to control the brain sizes of different species.
Acknowledgments
We thank Professor Arthur Boylston, for helpful discussion, and Jeffrey Keen, for first setting our minds thinking about evolution.
Electronic-Database Information
URLs for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MCPH) [PubMed]
- pfam, http://pfam.wustl.edu/, and COG, http://www.ncbi.nlm.nih.gov/COG/ (for protein domain identification)
References
- Aicardi J (1998) Malformations of the central nervous system. In: Aicardi J (ed) Diseases of the nervous system in childhood, 3rd ed. McKeith Press, London, pp 90–91 [Google Scholar]
- Albertson R, Doe CQ (2003) Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat Cell Biol 5:166–170 [DOI] [PubMed] [Google Scholar]
- Armstrong E (1985) Relative brain size in monkeys and prosimians. Am J Phys Anthropol 66:263–273 [DOI] [PubMed] [Google Scholar]
- Avides MC, Glover DM (1999) Abnormal spindle protein, Asp, and the integrity of mitotic centrosomal microtubule organizing centres. Science 283:1733–1735 [DOI] [PubMed] [Google Scholar]
- Avides MC, Tavares A, Glover DM (2001) Polo kinase and Asp are needed to promote the mitotic organizing activity of centrosomes. Nat Cell Biol 3:421–424 [DOI] [PubMed] [Google Scholar]
- Babson SG, Benda GI (1976) Growth graphs for the clinical assessment of infants of varying gestational age. J Pediatr 89:814–820 [DOI] [PubMed] [Google Scholar]
- Baraitser M (1990) Microcephaly. In: The genetics of neurological disorders, Vol. 18, 2nd ed. Oxford Medical Publications, Oxford, pp 26–33 [Google Scholar]
- Barkovich AJ, Kuzniecky RI, Dobyns WB (2001) Radiologic classification of malformations of cortical development. Curr Opin Neurol 14:145–149 [DOI] [PubMed] [Google Scholar]
- Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, Springell K, Mahadevan M, Crow YJ, Markham AF, Walsh CA, Woods CG (2002) ASPM is a major determinant of cerebral cortical size. Nat Genet 32:316–320 [DOI] [PubMed] [Google Scholar]
- Bond J, Scott S, Hampshire DJ, Springell K, Corry P, Abramowicz MJ, Mochida G, Hennekam RCM, Maher ER, Alswaid A, Jafri H, Rashid Y, Mubaidin A, Walsh C, Roberts E, Woods CG (2003) Protein truncating mutations in ASPM cause variable reduction in brain size. Am J Hum Genet 73:1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böök JA, Schut JW, Reed SC (1953) A clinical and genetical study of microcephaly. Am J Ment Defic 57:637–660 [PubMed] [Google Scholar]
- Brand S, Rakic P (1979) Genesis of the primate neostriatum: [3H]thymidine autoradiographic analysis of the time of neuron origin in the rhesus monkey. Neuroscience 4:767–778 [DOI] [PubMed] [Google Scholar]
- Brown P, Sutikna T, Morwood MJ, Soejono RP, Jatmiko, Saptomo EW, Due RA (2004) A new small-bodied hominin from the Late Pleistocene of Flores, Indonesia. Nature 431:1055–1061 [DOI] [PubMed] [Google Scholar]
- Bundey S (1997) Abnormal mental development. In: Rimoin DL, Connor JM, Pyeritz RE (eds) Emery and Rimoin’s principles and practice of medical genetics, 3rd ed. Churchill Livingstone. New York, p 730 [Google Scholar]
- Bundey S, Alam H (1993) A five-year prospective study of the health of children in different ethnic groups, with particular reference to the effect of inbreeding. Eur J Hum Genet 1:206–219 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3:285–298 [DOI] [PubMed] [Google Scholar]
- Chenn A, Walsh CA (2002) Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297:365–369 [DOI] [PubMed] [Google Scholar]
- ——— (2003) Increased neuronal production, enlarged forebrains and cytoarchitectural distortions in β-catenin overexpressing transgenic mice. Cereb Cortex 13:599–606 [DOI] [PubMed] [Google Scholar]
- Clark DA, Mitra PP, Wang SS (2001) Scalable architecture in mammalian brains. Nature 411:189–193 [DOI] [PubMed] [Google Scholar]
- Cowie V (1960) The genetics and sub-classification of microcephaly. J Ment Defic Res 4:42–47 [DOI] [PubMed] [Google Scholar]
- Craig R, Norbury C (1998) The novel murine calmodulin-binding protein Sha1 disrupts mitotic spindle and replication checkpoint functions in fission yeast. J Cell Sci 111:3609–3619 [DOI] [PubMed] [Google Scholar]
- Dobyns WB (2002) Primary microcephaly: new approaches for an old disorder. Am J Med Genet 112:315–317 [DOI] [PubMed] [Google Scholar]
- Doe CQ, Bowerman B (2001) Asymmetric cell division: fly neuroblast meets worm zygote. Curr Opin Cell Biol 13:68–75 [DOI] [PubMed] [Google Scholar]
- Dolk H (1991) The predictive value of microcephaly during the first year of life for mental retardation at seven years. Dev Med Child Neurol 33:974–983 [DOI] [PubMed] [Google Scholar]
- Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, Mahowald M, Wyckoff GJ, Malcom CM Lahn BT (2004) Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell 119:1027–1040 [DOI] [PubMed] [Google Scholar]
- Evans PD, Anderson JR, Vallender EJ, Choi SS, Lahn BT (2004a) Reconstructing the evolutionary history of microcephalin, a gene controlling human brain size. Hum Mol Genet 13:1139–1145 [DOI] [PubMed] [Google Scholar]
- Evans PD, Anderson JR, Vallender EJ, Gilbert SL, Malcom CM, Dorus S, Lahn BT (2004b) Adaptive evolution of ASPM, a major determinant of cerebral cortical size in humans. Hum Mol Genet 13:489–494 [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu CI (2003) Sequence divergence, functional constraint, and selection in protein evolution. Annu Rev Genomics Hum Genet 4:213–235 [DOI] [PubMed] [Google Scholar]
- Freire-Maia N (1990) Consanguinity marriages in Brazil. Rev Bras Biol 50:863–866 [PubMed] [Google Scholar]
- Gonzalez C, Saunders RD, Casal J, Molina I, Carmena M, Ripoll P, Glover DM (1990) Mutations at the asp locus of Drosophila lead to multiple free centrosomes in syncytial embryos, but restrict centrosome duplication in larval neuroblasts. J Cell Sci 96:605–616 [DOI] [PubMed] [Google Scholar]
- Goodman M (1999) The genomic record of humankind’s evolutionary roots. Am J Hum Genet 64:31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross SJ, Oehler JM, Eckerman CO (1983) Head growth and developmental outcome in very low birth weight infants. Pediatrics 71:70–75 [PubMed] [Google Scholar]
- Harvey PH, Krebs JR (1990) Comparing brains. Science 249:140–146 [DOI] [PubMed] [Google Scholar]
- Haydar TF, Ang E Jr, Rakic P (2003) Mitotic spindle rotation and mode of cell division in the developing telencephalon. Proc Natl Acad Sci USA 100:2890–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung LY, Chen H-L, Dhang C-W, Li B-R, Tang TK (2004) Identification of a novel microtubule-destabilizing motif in CPAP that binds to tubulin heterodimers and inhibits microtubule assembly. Mol Biol Cell 15:2697–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung LY, Tang CJ, Tang TK (2000) Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the γ-tubulin complex. Mol Cell Biol 20:7813–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD (2002) The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet 18:486–487 [DOI] [PubMed] [Google Scholar]
- Huyton T, Bates PA, Zhang X, Sternberg MJ, Freemont PS (2000) The BRCA1 C-terminal domain: structure and function. Mutat Res 460:319–332 [DOI] [PubMed] [Google Scholar]
- Jackson AP, Eastwood H, Bell SM, Adu J, Toomes C, Carr IM, Roberts E, Hampshire DJ, Crow YJ, Mighell AJ, Karbani G, Jafri H, Rashid Y, Mueller RF, Markham AF, Woods CG (2002) Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet 71:136–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AP, McHale DP, Campbell DA, Jafri H, Rashid Y, Mannan J, Karbani G, Corry P, Levene MI, Mueller RF, Markham AF, Lench NJ, Woods CG (1998) Primary autosomal recessive microcephaly (MCPH1) maps to chromosome 8p22-pter. Am J Hum Genet 63:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CR, Fryns JP, Jacobs J, Matthijs G, Abramowicz MJ (2000) Primary autosomal recessive microcephaly: MCPH5 maps to 1q25-q32. Am J Hum Genet 67:1575–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CR, Govaerts C, Abramowicz MJ (1999) Primary autosomal recessive microcephaly: homozygosity mapping of MCPH4 to chromosome 15. Am J Hum Genet 65:1465–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerison HJ (1973) Evolution of the brain and intelligence. Academic Press, New York [Google Scholar]
- Kloepfer HW, Platou RV, Hansche WJ (1964) Manifestations of a recessive gene for microcephaly in a population isolate. J Genet Hum 13:52–59 [PubMed] [Google Scholar]
- Komai T, Kishimoto K, Ozaki Y (1955) Genetic study of microcephaly based on Japanese material. Am J Hum Genet 47:51–65 [PMC free article] [PubMed] [Google Scholar]
- Kornack DR, Rakic P (1998) Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc Natl Acad Sci USA 95:1242–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Pavlicek A, Mochida GH, Solomon G, Gersch W, Yoon YH, Collura R, Ruvolo M, Barrett JC, Woods CG, Walsh CA, Jurka J, Larionov V (2004) Accelerated evolution of the ASPM gene controlling brain size begins prior to human brain expansion. PLoS Biol 2:E126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Blanton SH, Babu M, Markandaya M, Girimaji SC (2004) Genetic analysis of primary microcephaly in Indian families: novel ASPM mutations. Clin Genet 66:341–348 [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D (1987) Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science 236:1567–1570 [DOI] [PubMed] [Google Scholar]
- Leal GF, Roberts E, Silva EO, Costa SMR, Hampshire DJ, Woods CG (2003) A Brazilian locus for autosomal recessive primary microcephaly maps to 13q12.2. J Med Genet 40:540–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Kaufman TC (1996) The homeotic target gene centrosomin encodes an essential centrosomal component. Cell 88:585–596 [DOI] [PubMed] [Google Scholar]
- Marino L (1998) A comparison of encephalization between odontocete cetaceans and anthropoid primates. Brain Behav Evol 51:230–238 [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654 [DOI] [PubMed] [Google Scholar]
- McHenry HM (1994) Tempo and mode in human evolution. Proc Natl Acad Sci USA 91:6780–6786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw TL, Li K, Kao L-R, Kaufman TC (1999) The Centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126:2829–2839 [DOI] [PubMed] [Google Scholar]
- Mochida GH, Walsh CA (2001) Molecular genetics of human microcephaly. Curr Opin Neurol 14:151–156 [DOI] [PubMed] [Google Scholar]
- Morwood MJ, Soejono RP, Roberts RG, Sutikna T, Turney CS, Westaway KE, Rink WJ, Zhao JX, van den Bergh GD, Due RA, Hobbs DR, Moore MW, Bird MI, Fifield LK (2004) Archaeology and age of a new hominin from Flores in eastern Indonesia. Nature 431:1087–1091 [DOI] [PubMed] [Google Scholar]
- Moynihan L, Jackson AP, Roberts E, Karbani G, Lewis I, Corry P, Turner G, Mueller RF, Lench NJ, Woods CG (2000) A third novel locus for primary autosomal recessive microcephaly maps to chromosome 9q34. Am J Hum Genet 66:724–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller RF, Bishop DT (1993) Autozygosity mapping, complex consanguinity, and autosomal recessive disorders. J Med Genet 30:798–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neitzel H, Neumann LM, Schindler D, Wirges A, Tonnies H, Trimborn M, Krebsova A, Richter R, Sperling K (2002) Premature chromosome condensation in humans associated with microcephaly and mental retardation: a novel autosomal recessive condition. Am J Hum Genet 70:1015–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellhaus G (1968) Head circumference from birth to eighteen years: practical composite of international abd interracial graphs. Pediatrics 41:106–114 [PubMed] [Google Scholar]
- Opitz JM, Holt MC (1992) Microcephaly: general considerations and aids to nosology. J Craniofac Genet Dev Bio 10:175–204 [PubMed] [Google Scholar]
- Pattison L, Crow YJ, Deeble VJ, Jackson AP, Jafri H, Rashid Y, Roberts E, Woods CG (2000) A fifth locus for primary autosomal recessive microcephaly maps to chromosome 1q31. Am J Hum Genet 67:1578–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon B, Vankerckhove S, Bourrouillou G, Duprez L, Abramowicz MJ (2004) A translocation breakpoint disrupts the ASPM gene in a patient with primary microcephaly. Eur J Hum Genet 12:419–421 [DOI] [PubMed] [Google Scholar]
- Preuss TM (2004) What is it like to be a human? In: Gazzaniga MS (ed) The cognitive neurosciences III. MIT Press, Cambridge, MA [Google Scholar]
- Przeworski M (2002) The signature of positive selection at randomly chosen loci. Genetics 160:1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qazi QH, Reed TE (1973) A problem in diagnosis of primary versus secondary microcephaly. Clin Genet 4:46–52 [DOI] [PubMed] [Google Scholar]
- ——— (1975) A possible major contribution to mental retardation in the general population by the gene for microcephaly. Clin Genet 7:85–90 [DOI] [PubMed] [Google Scholar]
- Rakic P (1988) Specification of cerebral cortical areas. Science 241:170–176 [DOI] [PubMed] [Google Scholar]
- ——— (1995) A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci 18:383–388 [DOI] [PubMed] [Google Scholar]
- Riparbelli MG, Callaini G, Glover DM, Avides MdoC (2001) A requirement for the abnormal spindle protein to organise microtubules of the central spindle for cytokinesis in Drosophila. J Cell Sci 115:913–922 [DOI] [PubMed] [Google Scholar]
- Ripoll P, Pimpinelli S, Valdivia MM, Avila J (1985) A cell division mutant of Drosophila with a functionally abnormal spindle. Cell 41:907–912 [DOI] [PubMed] [Google Scholar]
- Roberts E, Bond J, Springell K, Lizarraga S, Scott S, Higgins J, Hampshire DJ, Morrison E, Leal GF, Silva EO, Costa SM, Karbani G, Rashid Y, Jafri H, Bennett C, Corry P, Walsh CA, Woods CG (2005) A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet 37:353–355 [DOI] [PubMed] [Google Scholar]
- Roberts E, Hampshire DJ, Springell K, Pattison L, Y Crow, Jafri H, Corry P, Kabani G, Mannon J, Rashid Y, Keen J, Bond J, Woods CG (2002) Autosomal recessive primary microcephaly: an analysis of locus heterogeneity and phenotypic variation. J Med Genet 39:718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E, Jackson AP, Carradice AC, Deeble VJ, Mannan J, Rashid Y, Jafri H, McHale DP, Markham AF, Lench NJ, Woods CG (1999) The second locus for autosomal recessive primary microcephaly (MCPH2) maps to chromosome 19q13.1-13.2. Eur J Hum Genet 7:815–820 [DOI] [PubMed] [Google Scholar]
- Roche AF, Mukherjee D, Guo S, Moore WM (1987) Head circumference reference data: birth to 18 years. Pediatrics 79:706–712 [PubMed] [Google Scholar]
- Rosenberg MJ, Agarwala R, Bouffard G, Davis J, Fiermonte G, Hilliard MS, Koch T, Kalikin LM, Makalowska I, Morton DH, Petty EM, Weber JL, Palmieri F, Kelley RI, Schaffer AA, Biesecker LG (2002) Mutant deoxynucleotide carrier is associated with congenital microcephaly. Nat Genet 32:175–179 [DOI] [PubMed] [Google Scholar]
- Ross JJ, Frias JL (1977) Microcephaly. In: Vinken PJ, Bruyn GW (eds) Congenital malformations of the brain and skull, part 1, vol 30. Elsevier Holland Biomedical Press, Amsterdam, pp 507–524 [Google Scholar]
- Saunders RD, Avides MC, Howard T, Gonzalez C, Glover DM (1997) The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J Cell Biol 137:881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells CJ (1977) Microcephaly in a normal school population. Pediatrics 59:262–265 [PubMed] [Google Scholar]
- Shen J, Eyaid W, Mochida GH, Bodell A, Woods CG, Walsh CA (2005) ASPM mutations identified in microcephaly with seizures patients. J Med Genet (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher PK, Brown SB (1975) A longitudinal study of head growth in preterm infants. I. Normal rates of head growth. Dev Med Child Neurol 17:705–710 [DOI] [PubMed] [Google Scholar]
- Stephan H, Baron G, Frahm HD (1988) Comparative size of brain and brain components. Comp Primate Biol 4:1–38 [Google Scholar]
- Stephan H, Frahm HD, Baron G (1981) New and revised data on volume of brain structures in insectivores and primates. Folia Primatol 35:1–29 [DOI] [PubMed] [Google Scholar]
- Sujatha M, Kumari CK, Murty JS (1989) Segregation frequency in microcephaly. Hum Genet 81:388–390 [DOI] [PubMed] [Google Scholar]
- Teebi AS, Al-Awadi SA, White AG (1987) Autosomal recessive nonsyndromal microcephaly with normal intelligence. Am J Med Genet 26:355–359 [DOI] [PubMed] [Google Scholar]
- Terada Y, Uetake Y, Kuriyama R (2003) Interaction of Aurora-A and centrosomin at the microtubule-nucleating site in Drosophila and mammalian cells. J Cell Biol 162:757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmie JL, McNay M, Stephenson JBP (1987) Microcephaly: genetic counselling and antenatal diagnosis after the birth of an affected child. Am J Med Genet 27:583–594 [DOI] [PubMed] [Google Scholar]
- Trimborn M, Bell SM, Felix C, Rashid Y, Jafri H, Griffiths PD, Neumann LM, Krebs A, Reis A, Sperling K, Neitzel H, Jackson AP (2004) Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am J Hum Genet 75:261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncbilek E (2001) Clinical outcomes of consanguineous marriages in Turkey. Turk J Pediatr 43:277–279 [PubMed] [Google Scholar]
- Van den Bosch J (1959) Microcephaly in the Netherlands: a clinical and genetical study. Ann Hum Genet 23:91–116 [DOI] [PubMed] [Google Scholar]
- Wakefield JG, Bonaccorsi S, Gatti M (2001) The Drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. J Cell Biol 153:637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallerman O, Van Eeghen A, Ten Kate LP, Wadelius C (2003) Evidence for a second gene for primary microcephaly at MCPH5 on chromosome 1. Hereditas 139:64–67 [DOI] [PubMed] [Google Scholar]
- Wang YQ, Su B (2004) Molecular evolution of microcephalin, a gene determining human brain size. Hum Mol Genet 13:1131–1137 [DOI] [PubMed] [Google Scholar]
- Warkany J (1981) Microcephaly. In: Warkany J, Lemire RJ, Cohen MM (eds) Mental retardation and congenital malformations of the central nervous system. Year Book Medical, Chicago and London, pp 13–40 [Google Scholar]
- Winter R, Baraitser M (1998) WBDB: The Winter-Baraitser Dysmorphology Database, version 1. London Medical Databases Ltd, London [Google Scholar]
- Woods CG (2004) Human microcephaly. Curr Opin Neurobiol 14:112–117 [DOI] [PubMed] [Google Scholar]
- Xu X, Lee J, Stern DF (2004) Microcephalin is a DNA damage response protein involved in regulation of CHK1 and BRCA1. J Biol Chem 279:34091–34094 [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Jones DL, Fuller MT (2003) Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301:1547–1550 [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski JP (2000) Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15:496–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J (2003) Evolution of the human ASPM gene, a major determinant of brain size. Genetics 165:2063–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]