Abstract
P450 oxidoreductase (POR) is the obligatory flavoprotein intermediate that transfers electrons from reduced nicotinamide adenine dinucleotide phosphate (NADPH) to all microsomal cytochrome P450 enzymes. Although mouse Por gene ablation causes embryonic lethality, POR missense mutations cause disordered steroidogenesis, ambiguous genitalia, and Antley-Bixler syndrome (ABS), which has also been attributed to fibroblast growth factor receptor 2 (FGFR2) mutations. We sequenced the POR gene and FGFR2 exons 8 and 10 in 32 individuals with ABS and/or hormonal findings that suggested POR deficiency. POR and FGFR2 mutations segregated completely. Fifteen patients carried POR mutations on both alleles, 4 carried mutations on only one allele, 10 carried FGFR2 or FGFR3 mutations, and 3 patients carried no mutations. The 34 affected POR alleles included 10 with A287P (all from whites) and 7 with R457H (four Japanese, one African, two whites); 17 of the 34 alleles carried 16 “private” mutations, including 9 missense and 7 frameshift mutations. These 11 missense mutations, plus 10 others found in databases or reported elsewhere, were recreated by site-directed mutagenesis and were assessed by four assays: reduction of cytochrome c, oxidation of NADPH, support of 17α-hydroxylase activity, and support of 17,20 lyase using human P450c17. Assays that were based on cytochrome c, which is not a physiologic substrate for POR, correlated poorly with clinical phenotype, but assays that were based on POR’s support of catalysis by P450c17—the enzyme most closely associated with the hormonal phenotype—provided an excellent genotype/phenotype correlation. Our large survey of patients with ABS shows that individuals with an ABS-like phenotype and normal steroidogenesis have FGFR mutations, whereas those with ambiguous genitalia and disordered steroidogenesis should be recognized as having a distinct new disease: POR deficiency.
Introduction
P450 oxidoreductase (POR) is the single flavoprotein that transfers electrons from reduced nicotinamide adenine dinucleotide phosphate (NADPH) to all microsomal (type 2) cytochrome P450 enzymes. The human genome contains 57 genes for cytochrome P450 enzymes, of which 50 encode microsomal P450s, including 15 genes encoding hepatic drug-metabolizing P450s; 20 genes encoding P450s involved in the biosynthesis of cholesterol, steroid hormones, fatty acids, and eicosanoids; and 15 genes encoding “orphan” P450 enzymes whose catalytic roles are unclear (Guengerich 2004). As all of these microsomal enzymes require POR for catalysis, one would predict that disruption of POR would have devastating consequences. Not surprisingly, Por knockout mice are embryonic lethal (Shen et al. 2002; Otto et al. 2003), apparently due to disruption of extrahepatic P450 enzymes, as liver-specific knockout of Por yields phenotypically and reproductively normal mice that accumulate hepatic lipids and have a remarkably diminished capacity for hepatic drug metabolism (Gu et al. 2003; Henderson et al. 2003). Similarly, globally hypomorphic Por mice also have reduced hepatic drug metabolism (Wu et al. 2005).
Despite the dire consequences in Por knockout mice, we have recently identified five missense mutations and a splicing mutation in the POR genes of four patients who had hormonal evidence for partial, combined deficiencies of two steroidogenic microsomal P450 enzymes: P450c17, which catalyzes steroid 17α-hydroxylation and 17,20 lyase activity, and P450c21, which catalyzes steroid 21-hydroxylase activity (Flück et al. 2004). Combined deficiency of these enzymes was first reported in 1985 (Peterson et al. 1985), and we had suggested that mutation of POR might explain the hormonal phenotype (Miller 1986). However, the involvement of POR with so many P450 enzymes, the lack of a published POR gene sequence, and the severity of the Por knockout mouse phenotype had dissuaded investigators from pursuing POR as a candidate gene (Miller 2004). Three of the four index cases also had the Antley-Bixler skeletal malformation syndrome (ABS [MIM 207401]), which is characterized by craniosynostosis, radioulnar or radiohumeral synostosis, bowed femora, and other bony deformities (Antley and Bixler 1975; DeLozier et al. 1980; Hassell and Butler 1994; Crisponi et al. 1997).
Work in the 1990s showed that four phenotypically related craniosynostosis syndromes—Apert, Crouzon, Jackson-Weiss, and Pfeiffer syndromes—were caused by mutations in the gene for fibroblast growth factor receptor 2 (FGFR2) (Jabs et al. 1994; Muenke et al. 1994; Reardon et al. 1994; Gorry et al. 1995; Park et al. 1995a, 1995b; Schell et al. 1995; Wilkie et al. 1995; Meyers et al. 1996) and that the same FGFR2 missense mutation, S351C, could be found in Crouzon and Pfeiffer syndromes (Pulleyn et al. 1996; Gripp et al. 1998; Okajima et al. 1999), suggesting that each eponymic syndrome is a phenotypic variant of a single genetic disorder. The heterozygous gain-of-function mutation C342Y is sufficient to cause craniosynostosis and skeletal abnormalities in knock-in mice (Eswarakumar et al. 2004), consistent with the dominant inheritance of these craniosynostosis syndromes. FGFR2 mutations were also reported in patients with ABS without genital ambiguity or other evidence of disordered steroidogenesis (Chun et al. 1998; Reardon et al. 2000). However, FGFR2 mutations were not found in a series of patients with ABS who had evidence of disordered steroidogenesis (Reardon et al. 2000), and FGFR1 and FGFR3 mutations were not found in a similar patient (Roth et al. 2000). We (Flück et al. 2004)—and subsequently others (Adachi et al. 2004; Arlt et al. 2004; Fukami et al. 2005)—have found POR mutations in all examined patients who had the ABS phenotype with evidence of abnormal steroidogenesis, indicating that what has been termed “ABS” is, in fact, two distinct disorders, as was first suggested by Reardon and Winter (Reardon et al. 2000). Thus, ABS without disordered steroidogenesis is a variant of the autosomal dominant group of craniosynostosis syndromes caused by mutant FGF receptors, usually FGFR2, and ABS with disordered steroidogenesis is a novel autosomal recessive disorder caused by mutations in POR.
We have now examined the POR genes in 32 additional patients, including 31 patients in whom ABS was initially diagnosed, and in another 6 patients with the Beare-Stevenson craniosynostosis syndrome, which can be associated with genital anomalies. Fifteen of the 19 patients having abnormal genitalia and disordered steroidogenesis were homozygotes or apparent compound heterozygotes for POR mutations that destroyed or dramatically inhibited POR activity; only one mutant allele was identified in the other 4 patients. Of the 12 patients with normal genitalia and normal steroidogenesis diagnosed as having ABS, 9 carried FGFR2 or FGFR3 mutations and none carried POR mutations on both alleles. These data are consistent with the emerging view that patients with ABS fall into two distinct groups that should now be categorized as two distinct diseases (Miller 2004).
Subjects and Methods
Clinical Material
Samples 1–32 were from patients identified by various medical geneticists as having ABS on the basis of dysmorphic features or identified by pediatric endocrinologists as having combined 17α-hydroxylase and 21-hydroxylase deficiencies. All samples were collected in accordance with local institutional review board or medical ethics committee guidelines. Not all clinical data are available for all subjects. Exons 8 and 10 of FGFR2 were sequenced in all patients; only the FGFR sequencing in addition to exons 8 and 10 of FGFR2 is specifically mentioned below. The available clinical histories and hormonal data are summarized below, and the available dysmorphological data are summarized in table 1; craniosynostosis and radioulnar or radiohumeral synostosis were confirmed radiographically.
Table 1.
Physical Findings in Patients[Note]
|
Dysmorphologic Feature |
||||||||||||||
| PatientNo. | Patient ID | Craniosynostosis | Brachycephaly | RH/RUaSynostosis | FemoralBowing | FemoralFractures | MidfaceHypoplasia | Proptosis | Rocker-Bottom Feet | Arachnodactyly | Camptodactyly | Choanal Stenosis/Atresia | Abnormal Steroids | Abnormal Genitalia |
| 1 | IRL1 | Y | N | Y | Y | N | Y | Y | N | N | N | Y | Y | |
| 2 | AUS2 | Y | Y | Y | N | N | Y | Y | Y | Y | Y | Y | Y | Y |
| 3 | PL1 | Y | N | Y | N | N | Y | N | N | N | Y | Y | Y | |
| 4 | CF1 | Y | Y | Y | Y | |||||||||
| 5 | 337 | Y | Y | Y | Y | Y | ||||||||
| 6 | 259 | Y | Y | Y | Y | |||||||||
| 7 | 699 | Y | Y | Y | Y | |||||||||
| 8 | JPSZ | Y | Y | Y | Y | N | Y | Y | N | Y | Y | Y | ||
| 9 | JPSS | Y | Y | Y | Y | Y | Y | |||||||
| 10 | JPSB | Y | Y | Y | Y | Y | Y | Y | Y | Y | ||||
| 11 | MC25 | Y | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | |
| 12b | 07942 | Y | Y | Y | Y | Y | Y | Y | Y | |||||
| 13b | 50081 | Y | Y | Y | Y | |||||||||
| 14b | 11314 | Y | Y | Y | N | Y | Y | N | N | N | N | N | ||
| 15b | 14506 | Y | Y | Y | Y | Y | Y | Y | N | N | Y | |||
| 16b | 14683 | Y | Y | Y | Y | N | Y | Y | Y | Y | Y | |||
| 17b | 15441 | Y | Y | Y | Y | N | Y | Y | Y | Y | ||||
| 18b | 16666 | Y | Y | Y | Y | Y | Y | Y | ||||||
| 19b | 20554 | Y | Y | Y | Y | Y | ||||||||
| 20b | 24019 | Y | Y | Y | Y | Y | Y | |||||||
| 21b | 07862 | Y | Y | Y | Y | N | Y | Y | Y | |||||
| 22b | 24872 | Y | Y | N | N | N | Y | Y | N | Y | ||||
| 23b | 14837 | Y | Y | N | N | Y | Y | Y | ||||||
| 24 | 56661 | Y | Y | Y | Y | Y | Y | Y | ||||||
| 25 | JPAS | Y | Y | Y | Y | Y | Y | Y | ||||||
| 26 | 1110 | Y | Y | |||||||||||
| 27 | 1115 | N | N | Y | Y | Y | ||||||||
| 28 | 1323 | Y | Y | Y | Y | |||||||||
| 29 | 1434 | Y | Y | N | Y | Y | N | N | Y | |||||
| 30 | 2430 | Y | N | Y | Y | Y | ||||||||
| 31 | STJ1 | Y | Y | Y | Y | N | Y | Y | N | N | N | Y | Y | Y |
| 32 | ISRL1 | N | N | N | N | N | N | N | N | N | N | Y | Y | |
Note.— “Y” (yes) means the patient had this finding; “N” (no) means the patient did not have the finding. Blank cells indicate that the case reports did not mention whether the patient had this finding.
RH = radiohumeral; RU = radioulnar.
These patients were described by Reardon et al. (2000). Patient 12 is MF; patient 13 is RS; patient 14 is DW; patient 15 is HD; patient 16 is SP; patient 17 is MN; patient 18 is NC; patient 19 is ID; patient 20 is JH; patient 21 is KM; patient 22 is BW; patient 23 is MR.
Patient 1 (IRL1), who has been described recently (Hurley et al. 2004), was born at 38 wk, weighed 2.98 kg, and had a head circumference of 31 cm, with midface hypoplasia. He had fused lambdoid and sagittal sutures, radiohumeral synostosis, and femoral bowing. He was 46,XY, with a stable 8;12 translocation that was also seen in the father. The phallus was small, and the labioscrotal folds were fused. At age 2 wk, the patient had low serum cortisol (2.3 μg/dl), but it rose normally to 16.5 μg/dl after stimulation with adrenocorticotropic hormone (ACTH [corticotropin]). ACTH was normal, at 28 pg/ml. At age 2 wk, the patient had normal values for androstenedione (1.2–2.5 nmol/L), dehydroepiandrosterone sulfate (DHEAS) (0.1 μmol/L), and testosterone (1.8 nmol/L). Urinary steroids included decreased amounts of C19 steroid sulfates and increased C21 steroids. No mutations were found in exons 7 and 10 of FGFR3, but the patient was heterozygous for the FGFR1 mutation I300T. This mutation has been reported in a single case of trigonocephaly (Kress et al. 2000).
Patient 2 (AUS2), a 46,XY male, was delivered at 37 wk by emergency cesarean section for abruptio placenta. Height, weight, and head circumference were at the 50th percentile for gestational age. There was proptosis, brachycephaly, frontal and temporal bossing, and ridging of the posterior sutures, but the metopic, sagittal, and coronal sutures were open. There was limitation of supination and elbow extension but no radiologically apparent synostosis. There also was camptodactyly, arachnodactyly, rocker-bottom feet, choanal stenosis, several hemivertebrae, bronchomalacia, and an absent auditory meatus and absent ossicular chain on one side. There was a horseshoe kidney, micropenis, hooded prepuce, hypospadias, chordee, bilaterally descended testes, and anteriorly placed anus. At age 1 year, the patient had normal cortisol (594 nmol/L), but ACTH was elevated (137 pg/ml; normal [nl] <50 pg/ml). Progesterone (143 nmol/L; nl <1.6 nmol/L) and 17-hydroxyprogesterone (17OHP) (71.6 nmol/L; nl <3 nmol/L) were elevated and were hyperresponsive to stimulation with ACTH, whereas DHEA (0.31 nmol/L; nl >0.7 nmol/L) and androstenedione (0.3 nmol/L; nl >0.4 nmol/L) were low and were unresponsive to ACTH. Sequencing of exons 7, 8, and 10 of FGFR1, FGFR2, and FGFR3 was normal.
Patient 3 (PL1), born at 38 wk with severe oligohydramnios, had a large forehead, hypertelorism, abnormal ears, conductive hearing loss, craniosynostosis requiring ventriculoperitoneal shunting, radiohumeroulnar synostosis, and camptodactyly. He was 46,XY, with micropenis, hypospadias, and bilateral cryptorchidism. The 17OHP on newborn screening was 120 nmol/L at age 3 d and 193 nmol/L at age 5 d (nl <10 nmol/L) and was 3,780 μg/dl by radioimmunoassay at age 12 d (nl <200 μg/dl). These 17OHP data led to a presumptive diagnosis of congenital adrenal hyperplasia due to 21-hydroxylase deficiency (despite the genital findings), but sequencing of the CYP21 gene was normal. The patient was treated with replacement doses of glucocorticoids and mineralocorticoids but died of “septic shock” at another hospital at age 3 years; no hormonal evaluation was done at that time.
Patient 4 (CF1) was a 38-wk-old, 46,XY, SRY-positive phenotypic female who weighed 2,055 g at birth. ABS was diagnosed at birth on the basis of craniosynostosis, radiohumeral synostosis with shortened humeri, midface hypoplasia, a narrow thoracic cage with pectus excavatum, and an absent corpus callosum (all confirmed by radiographic studies). She had normal female external genitalia; ultrasonography showed no uterus or ovaries, small adrenals, and normal kidneys; and a radiographic contrast study confirmed the presence of a blind vaginal pouch. She had profound respiratory distress and frequent apneic episodes and died at age 3 wk. A limited hormonal evaluation included a Müllerian inhibiting substance of 34.9 pmol/L (normal for a female but indicating Sertoli cell failure in a male); slightly elevated aldosterone (126 ng/dl; nl <100 ng/dl); high 11-deoxycortisol (6271 ng/dl; nl <150 ng/dl); and normal plasma renin content (27 ng/L), ACTH (26 pg/ml), cortisol (205 nmol/L), 17OHP (1.4 nmol/L), DHEA (25 nmol/L), DHEAS (8.9 μmol/L), and androstenedione (5.4 nmol/L). The mother also had two spontaneous abortions, and a maternal uncle died soon after birth with ambiguous genitalia, but no hormonal testing was done, and no samples are available for DNA sequencing.
Patient 5 (337) was a phenotypically female infant with cloverleaf skull, craniosynostosis, bowed femora, radiohumeral synostosis, and arachnodactyly, who also had an imperforate anus, clitoromegaly, hypoplastic labia majora, and fused labia minora; a karyotype was not done. Although these findings suggest hyperandrogenism, no hormonal studies were done. She died at age 4 years from pneumonia, complicated by severe kyphoscoliosis. Sequencing of all FGFR2 exons was normal.
Patient 6 (259), a boy born to parents from Senegal, had craniosynostosis, cloverleaf skull, radiohumeral synostosis, bowed femora, arachnodactyly, and micropenis. No hormonal data are available. He is now 9 years old and has severe kyphoscoliosis. Sequencing of all FGFR2 exons was normal.
Patient 7 (699) was the product of a therapeutic abortion performed at 22 wk gestation because of ultrasonographic detection of craniosynostosis, bilateral radiohumeral synostosis, bowing of long bones, and arachnodactyly. Karyotype was 46,XY, and the external genitalia were normal. Amniotic fluid steroids were not measured, but, because of the suspicion of ABS, gas chromatographic analysis of liver tissue was performed and showed “substantially increased” lanosterol and dihydrolanosterol.
Patient 8 (JPSZ) had craniosynostosis, bowed femora, elbow synostosis, Arnold-Chiari malformation, scrotal hypoplasia, and a “small” penis. No steroids were measured other than normal total serum cholesterol.
Patients 9 (JPSS) and 10 (JPSB) were siblings. Both had craniosynostosis and elbow synostosis. Patient 9 had bowed femurs and fractures; this information is not known for patient 10. Patient 9 was said to have an undescribed genital anomaly, and patient 10 had undescended testes. No other data are available.
Patient 11 (MC25), who has been described elsewhere (Hassell and Butler 1994), was small for gestational age and had brachycephaly, midface hypoplasia, proptosis, radiohumeral synostosis, bowed femora, and choanal atresia or stenosis. She was 46,XX, with clitoromegaly, no vaginal orifice, and a vesicovaginal fistula. No hormonal data are available.
Patients 12–23 were described elsewhere (Reardon et al. 2000) (see table 1). Patients 17, 18, and 22 were 46,XY; the others were 46,XX. Patient 12 was originally described by DeLozier et al. (1980); genital anomalies were not described. At age 18 years, patient 12 had normal excretion of urinary free cortisol (67 μg/24 h), 17-hydroxycorticosteriods (7.5 mg/24 h), and 17-ketosteroids (8.5 mg/24 h). Patient 13 had low excretion of free cortisol (3.7 μg/24 h), 17-hydroxycorticosteroids (0.2 mg/24 h), and 17-ketosteroids (1.9 mg/24 h). Patient 16, illustrated in figure 1 of Reardon et al. (2000), had genital ambiguity (clitoromegaly and hooded prepuce), fused labia, elevated 17OHP (42.8 nmol/L; nl <12 nmol/L), and a cortisol of 220 nmol/L (nl 80–800), which rose to 534 nmol/L in response to ACTH. Patient 20, initially described by Kumar and Masel (1997), reportedly had an “abnormal steroid profile,” which was not described further, and had a urogenital sinus, vestigial uterus, and a posteriorly placed septated vagina. For patient 22, gas chromatographic analysis of urinary steroids at age 18 mo showed elevated excretion of 11β-hydroxyandrosterone plus 17-hydroxypregnanolone (80 μg; nl <50 μg), pregnanetriol (60 μg; nl <50 μg), tetrahydrocortisone (320 μg; nl 50–250 μg), allotetrahydrocortisol (380 μg; nl 20–300 μg), α-cortolone (380 μg; nl <50 μg), α-cortol (120 μg; nl <20 μg), and β-cortolone plus β-cortol (80 μg; nl <50 μg).
Patient 24 (56661) had craniosynostosis with cloverleaf skull, brachycephaly, midface hypoplasia, elbow synostosis, bilateral choanal atresia, proptosis, femoral fractures, and only 11 pairs of ribs. The corpus callosum was “small,” and the cerebral gyri were “abnormal.” A randomly obtained cortisol in infancy was 3.9 μg/dl, but an ACTH test was not done; hence, this value is not informative.
Patients 25–30 initially had a clinical diagnosis of ABS, which has been reconsidered in some cases. None of these patients had genital malformations or known disorders of steroidogenesis. Patient 25 (JPAS), a 46,XY male with unilateral agenesis of one adrenal, kidney, ureter, testis, and spermatic cord, had a classic ABS phenotype (table 1), but sequencing of all exons of FGFR2 was normal. Gas chromatographic analysis of urinary steroids at age 4 years showed normal 24-h excretions of tetrahydrocortisone (690 μg), tetrahydrocortisol (240 μg), allotetrahydrocortisol (180 μg), α-cortolone (260 μg), α-cortol (110 μg), and β-cortolone plus β-cortol (240 μg). Patient 26 (1110) had a family history of unspecified renal disease. Patients 28 (1323) and 29 (1434), reported as cases 1 and 2 by Okajima et al. (1999), are both heterozygous for FGFR2 mutation S351C. Patient 28 had a 2.5-cm phallus, contractures of the elbows and knees without synostosis and Peters anomaly, and was subsequently considered to have Pfeiffer syndrome. Patient 29 had radioulnar—but not radiohumeral—synostosis, knee contractures without synostosis and Peters anomaly, and was subsequently considered to have Crouzon syndrome. Patient 30 (2430) developed acanthosis nigricans 4–5 years after the initial diagnosis of ABS and was then considered to have Crouzonodermoskeletal syndrome; the patient was found to be heterozygous for FGFR3 mutation A391E.
Patient 31 (STJ1), a 46,XY male identified as having ABS at birth, had craniosynostosis, brachycephaly, radioulnar synostosis, femoral bowing, midface hypoplasia, and bilateral choanal stenosis. There was no genital ambiguity, but the patient had microphallus and bilaterally descended testes. At age 9 mo, an afternoon 17OHP was 360 nmol/L (nl <2 nmol/L at this age), but 8 a.m. values for cortisol (371 nmol/L), androstenedione (1.7 nmol/L), DHEAS (0.33 μmol/L), testosterone (<0.17 nmol/L), and aldosterone (2.8 nmol/L) were all normal for the patient's age. The elevated 17OHP in the presence of normally low C19 steroids suggested combined partial deficiencies of P450c17 and P450c21. At age 32 mo, the 17OHP remained greatly elevated (72.6 nmol/L).
Patient 32 (ISRL1), who has been described elsewhere (Augarten et al. 1992), was the 46,XY, 2,620-g, 48-cm product of a 38-wk pregnancy; the patient had no dysmorphic features suggestive of ABS but had a 1-cm microphallus, a bifid scrotum, glandular hypospadias, and bilaterally palpable testes. He had some limitation of elbow extension attributed to arthrogryposis; X-rays of the elbow joints were not done. Hormonal stimulation of the adrenal with ACTH and of the testes with human chorionic gonadotropin showed a pattern consistent with partial deficiencies of the activities of both P450c17 and P450c21 (Augarten et al. 1992). He entered puberty spontaneously but had hypergonadotropic hypogonadism and required ongoing supplementation with testosterone. He is now 22 years old and has required no glucocorticoid, mineralocorticoid, or salt supplementation.
Patients 33–38 had Beare-Stevenson syndrome, which can result in skeletal and genital anomalies (Przylepa et al. 1996). Patients 33, 36, and 37 had rugated labia majora; patient 36 had hypoplastic labia majora. Patients 35, 36, 37, and 38 had anteriorly placed anus. No endocrine data are available for these patients.
PCR and Sequencing
Specific primers in the intronic regions flanking the 15 exons of POR were designed on the basis of National Center for Biotechnology Information (NCBI) sequences GI 4508114 and GI 11181841 (see table 2 ). These primers amplified each exon with at least 100 bases of flanking intronic DNA; the sizes of the PCR products are listed in table 3. Genomic DNA samples (10 ng) were used as templates for PCR amplification in a 5-μl or 10-μl reaction. PCR amplification of exons 1, 2, 3, 5, 7, and 8–9 was performed using AmpliTaq Gold (Applied Biosystems) under high touchdown cycling conditions, starting at 95°C for 5 min, followed by 14 touchdown cycles of 94°C for 20 s, 65°C–58.5°C for 20 s (the annealing temperature for each subsequent cycle was decreased by 0.5°C), and 72°C for 1 min. This was followed by 35 cycles of PCR amplification at 94°C for 20 s, at 58°C for 20 s, and at 72°C for 1 min; the final extension was held at 72°C for 10 min, followed by 4°C to stop the reaction. PCR amplification of exons 3, 6, and 10–11 was performed using Platinum Taq (Invitrogen), and amplification of exons 12–15 was performed using Platinum Taq with N,N,N-trimethylglycine (betain) (Invitrogen) at 95°C for 1 min, followed by 14 touchdown cycles, 35 cycles of PCR amplification, and a 10-min final extension at 72°C, as described above. When updated databases revealed SNPs in the sites recognized by some primers, additional primers were designed (see table 2) and used as described for exons 12–15. FGFR2 exons 8 and 10 were sequenced as described elsewhere (Meyers et al. 1996), and FGFR2 exon 11 was sequenced in the patients with Beare-Stevenson syndrome, as described by Przylepa et al. (1996). The PCR products were subjected to automated direct sequencing with the use of the primers listed in table 2. All cycling for PCR and sequencing was performed on ABI GeneAmp PCR System 9700 thermocyclers. The sequencing was performed with ABI BigDye terminator, version 3.1, and was displayed on ABI 3730×1 DNA Analyzer. Genomic DNA from four normal males and four normal females was used as a reference. Mutations were analyzed with Mutation Surveyor 2.02 (see the Softgenetics Web site). All identified mutations were confirmed by forward and reverse sequencing. Nucleotide mutations are numbered in accordance with the NCBI reference sequence NM_000941.1, and protein sequence mutations are numbered in accordance with the NCBI reference sequence NP_000932.1.
Table 2.
Oligonucleotide Primers for PCR, Sequencing, and Site-Directed Mutagenesis[Note]
| Purpose and Primer Name | Sequence (5′ → 3′) | Start |
| PCR and sequencing: | ||
| POR-1Fa | TGCAGTGACCATTTCCTGCAG | 2568 |
| POR-1Ra | AATGCTACAAGGAGCCTTGCT | 2168 |
| POR-2Fb | GTGGCACGGGACAAAGCTGGAA | 80266 |
| POR-2Rb | GGTTAGGCAAGAATGACTCCCA | 79936 |
| POR-3Fb | TTTGCCACAGTGGCTGTGACAGT | 73264 |
| POR-3Rb | GGGAAGGCAACTTCCGAGGACGT | 72841 |
| POR-4Fb | GTGTTGTTACTTCTCTCTGATCCCA | 72343 |
| POR-4Rb | AAGTGGCCTGCCCTGGCTGCTGAG | 71938 |
| POR-5Fb | TGGAACGGAGGCCTGCAGGTGTT | 71615 |
| POR-5Rb | GAGTCTCAGCTGACACAATGGTG | 71288 |
| POR-6Fb | AGGGTGCACAGTCCTGAGCTTTG | 71133 |
| POR-6Rb | AACGTGGCAGAGTGAGTCCTTGG | 70784 |
| POR-7Fb | TTGTGCATCTGCAGCAGGGGCT | 70466 |
| POR-7Rb | GAAGGGCCCTGTGGGGATATG | 70134 |
| POR-8–9Fb | GCCCTTGATGTAACCGGTGAGATT | 69152 |
| POR-8–9Rb | AACCGTGGAGTCTCGGTGCAAGG | 68563 |
| POR-10–11Fb | GCACCTGTTGCCGCAGAGCTGG | 67866 |
| POR-10–11Rb | GGAACTGGAGCTCAGGGCTCAGA | 67235 |
| POR-12–15Fb | AGGGGGCCTCTGAGGTTTGGGTGCCA | 67092 |
| POR-12–15Rb | ATGCAGGCCCTGGGCTGTGCTGCCAG | 65866 |
| POR seq 12Fb | GTGCCAGGTGGGCTGGAAGA | 67078 |
| POR seq 12Rb | CGTAGTACAGCAGCGTCTCC | 66601 |
| POR seq 13Fb | ATCCAGGAGCGGGCCTGG | 66720 |
| POR seq 13Rb | GCTCTCGGTCTTGCTTTAGC | 66361 |
| POR seq 14Fb | GTGAGACGGGCGGGCACCCA | 66468 |
| POR seq 15Rb | CGGTGGACCTCACCTGGCCT | 65896 |
| POR_x10Fb | AGGGAGGCATCAGAGAGCATAG | 67912 |
| POR_x11Rb | GGCTGGACAGATGCTGAGAA | 67153 |
| POR_x12Fb | GAGGGGGCCTCTGAGGTTTG | 67099 |
| POR_x13Rb | ACAGGTGCTCTCGGTCTTGCTT | 66355 |
| POR_x14Fb | GGGAGACGCTGCTGTACTACG | 66601 |
| POR_x15Rb | GCCCAGAGGAGTCTTTGTCACT | 65941 |
| Site-directed mutagenesis: | ||
| POR A115Vc | GCGAGGCATGTCAGTGGACCCTGAGGAGT | 345 |
| POR T142Ac | TTTCTGCATGGCCGCCTACGGTGAGGGA | 426 |
| POR Q153Rc | CACCGACAATGCCCGGGACTTCTACGACT | 459 |
| POR Y181Dc | TTGGGAACAAGACCGACGAGCACTTCAAT | 542 |
| POR P228Lc | GAGCAGTTCTGGCTGGCCGTGTGTGAAC | 685 |
| POR M263Vc | CGGCCAAGGTGTACGTGGGGGAGATGGGC | 788 |
| POR A287Pc | CCAAGAATCCGTTCCTGCCTGCAGTCACCACC | 857 |
| POR R316Wc | CGGACTCCAAAATCTGGTATGAATCTGGGG | 947 |
| POR G413Sc | GCCTCCTCCTCCAGCGAGGGCAAGGAG | 1240 |
| POR R457Hc | GCGCCTGCAGGCCCACTACTACTCCATCG | 1371 |
| POR Y459Hc | TGCAGGCCCGCTACCACTCCATCGCCTCA | 1376 |
| POR V492Ec | GCATCAACAAGGGCGAGGCCACCAACTGGC | 1475 |
| POR A503Vc | GCCAAGGAGCCTGTCGGGGAGAACGGC | 1510 |
| POR G504Rc | CAAGGAGCCTGCCAGGGAGAACGGCGGC | 1512 |
| POR G539Rc | TGGGCCCCGGCACCAGGGTGGCACCCTTC | 1616 |
| POR L565Pc | GGGAGACGCTGCCGTACTACGGCTGCCGCC | 1697 |
| POR C569Yc | GCTGTACTACGGCTACCGCCGCTCAGATG | 1707 |
| POR V608Fc | CCCACAAGGTCTACTTCCAGCACCTGC | 1823 |
| POR R616Xc | TGCTAAAGCAAGACTGAGAGCACCTGTGG | 1847 |
| POR V631Ic | GTGCCCACATCTACATCTGTGGGGATGCA | 1892 |
| POR F646delc | GGATGTGCAGAACACCTACGACATCGTGGCTGAG | 1935 |
Table 3.
Primers for PCR and Sequencing, and Product Sizes of POR Amplicons
|
Primer Pair(s) for |
|||
| Amplicon(s) | PCR | Sequencing | PCRProduct Size(bp) |
| Exon 1 | POR-1F/POR-1R | POR-1F/POR-1R | 420 |
| Exon 2 | POR-2F/POR-2R | POR-2F/POR-2R | 351 |
| Exon 3 | POR-3F/POR-3R | POR-3F/POR-3R | 445 |
| Exon 4 | POR-4F/POR-4R | POR-4F/POR-4R | 429 |
| Exon 5 | POR-5F/POR-5R | POR-5F/POR-5R | 349 |
| Exon 6 | POR-6F/POR-6R | POR-6F/POR-6R | 371 |
| Exon 7 | POR-7F/POR-7R | POR-7F/POR-7R | 353 |
| Exons 8–9 | POR-8–9F/POR-8–9R | POR-8–9F/POR-8–9R | 612 |
| Exons 10–11 | POR-10–11F/POR-10–11R | POR-10–11F/POR-10–11R | 652 |
| Exons 12–15 | POR-12–15F/POR-12–15R | POR seq 12F/POR seq 12R; POR seq 13F/POR seq 13R; POR seq 14F/POR seq 15R | 1251 |
| Exons 10–11 | POR_x10F/POR_x11R | POR_x10F/POR_x11R | 781 |
| Exons 12–13 | POR_x12F/POR_x13R | POR_x12F/POR_x13R | 764 |
| Exons 14–15 | POR_x14F/POR_x15R | POR_x14F/POR_x15R | 681 |
Sequence Analysis and Molecular Modeling of Human POR
The POR sequences from human, rat, Xenopus, Drosophila, and yeast were aligned with ClustalW (Thompson et al. 1994), and sequences were displayed and edited using GeneDoc. Molecular modeling of human POR was performed by MODELLER (Sali and Blundell 1993; Fiser et al. 2000), which uses satisfaction of spatial restraints for comparative modeling of proteins. The model was based on the X-ray crystal structure of rat POR (Wang et al. 1997), which shares 94% sequence identity with human POR. Energy minimizations were performed with GROMOS96 (see the GROMOS Home Page), and the model was examined and edited using DeepView (Guex and Peitsch 1997) and POVRAY (see the Persistence of Vision Raytracer Web site).
Expression of Wild-Type and Mutant POR in Bacteria
Human POR cDNA lacking codons for 27 N-terminal residues (Dierks et al. 1998) was subcloned into pET22b for bacterial expression. Mutant cDNA expression vectors were generated by PCR-based, site-directed mutagenesis with the use of the primers shown in table 2 . The resulting mutated plasmids were selected by digestion with DpnI and were propagated in Escherichia coli DH5α. All POR mutations were confirmed by direct sequencing.
E. coli BL21DE3(pLysS) transformed with pET22b were grown in terrific broth (Dierks et al. 1998) supplemented with 40 μM FeCl3, 4 μM ZnCl2, 2 μM CoCl2, 2 μM Na2MoO4, 2 μM CaCl2, 2 μM CuCl2, 2 μM H3BO3, 10% (v/v) potassium phosphate solution (0.17 M KH2PO4, 0.72 M K2HPO4 pH 7.4), 0.5 mg/ml riboflavin, 50 μg/ml ampicillin, and 34 μg/ml chloramphenicol at room temperature to an optical density (OD) 600 nm of 0.4 and were induced with 0.4 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) overnight. The bacteria were washed with PBS and were resuspended in 100 mM Tris-acetate (pH 7.6), 0.5 M sucrose, and 1 mM EDTA. The bacteria were treated with lysozyme (0.5 mg/ml) and EDTA (0.1 mM [pH 8.0]) to generate spheroplasts. The spheroplasts were pelleted by centrifugation (5,000 × g for 15 min); were resuspended in 100 mM potassium phosphate (pH 7.6), 6 mM MgOAc, 0.1 mM DTT, 20% (v/v) glycerol, 0.2 mM PMSF, and 0.1 mM DNase I; and were disrupted by sonication (30-s on/off cycles for 15 min at 40% power). The lysate was cleared of cellular debris by centrifugation at 12,000 × g for 5 min, and then the membranes in the supernatant were pelleted at 150,000 × g for 45 min at 4°C. Membranes were resuspended in 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 20% (v/v) glycerol; were stored at −70°C; and were used as a source of POR.
Purification and Quantitation of Wild-Type and Mutant POR
Bacterial membranes containing POR were solubilized by stirring for 2 h at 4°C in 20 mM potassium phosphate (pH 7.6) containing 20% glycerol, 0.1 mM EDTA, 1 mM PMSF, 1 mg/L pepstatin, 10 mg/L leupeptin, 0.2% sodium cholate, and 0.2% Triton X-100 (Calbiochem). Insoluble material was removed by centrifugation at 100,000 × g for 1 h, and the supernatant was loaded on a 2′,5′-ADP-Sepharose column (Sigma) equilibrated with 20 mM potassium phosphate (pH 7.6) containing 20% glycerol, 0.1 mM EDTA, and 0.2% sodium cholate. The column was first washed with 5 vol of this buffer; then with 5 vol of 100 mM potassium phosphate (pH 7.6) containing 20% glycerol, 0.1 mM EDTA, 0.2% sodium cholate, and 250 mM sodium chloride; and finally with 5 vol of 20 mM potassium phosphate (pH 7.6) containing 20% glycerol, 0.1 mM EDTA, 0.2% sodium cholate, and 2 mM adenosine. POR was eluted with 20 mM potassium phosphate (pH 7.6) containing 20% glycerol, 0.1 mM EDTA, 0.2% sodium cholate, and 5 mM 2′-AMP. The cholate and 2′-AMP were removed from POR preparations by dialysis against 20 mM potassium phosphate (pH 7.6) containing 10% glycerol.
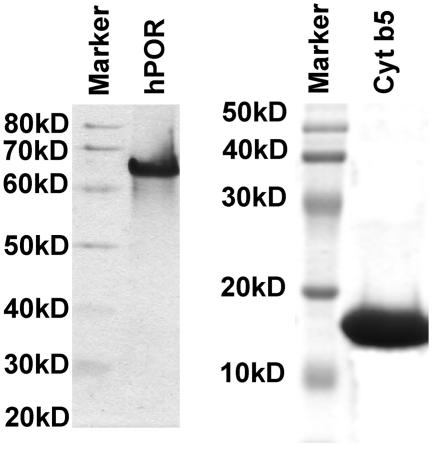
The mutant POR proteins were quantitated by western blotting by comparison with a standard curve of purified wild-type POR protein. Wild-type POR protein was quantitated by Bradford assay, and the purity of the POR preparation was confirmed by SDS-PAGE (fig. 1). The standard curve, using different amounts of wild-type POR protein, and the membrane preparations from bacteria expressing various POR mutants were separated by SDS-PAGE, transferred on polyvinyldifluoride (PVDF) membranes, and probed with a rabbit polyclonal antibody against wild-type human POR (Stressgen) at a dilution of 1:1,000. The blot was developed with a chemiluminescence-based detection reagent (Pierce) by employing goat anti-rabbit antibody, coupled with horseradish peroxidase. Signals were detected on a STORM 860 phosphorimager, operating in blue fluorescence mode, and were quantitated with ImageQuant software. All POR levels were normalized against wild-type POR.
Figure 1.
Purification of POR and cytochrome b5. Left panel, 10% SDS-polyacrylamide gel showing purified preparations of wild-type POR used for quantitation and normalization of POR mutants; right panel, 12% SDS-polyacrylamide gel showing purified cytochrome b5 used in assays of the 17,20 lyase reaction. Both gels were purposely overloaded to permit identification of trace contamination. The gels were stained with Coomassie Brilliant Blue.
Expression of Human P450c17 in Yeast
Yeast strain W(B), which lacks the endogenous yeast Cpr1 gene (Pompon et al. 1995), was propagated and transiently transfected with pYeSF2 expressing human P450c17, as described (Auchus et al. 1998). Yeast microsomes were prepared as described (Auchus et al. 1998), microsomal proteins were quantitated colorimetrically (protein Assay Dye Reagent [Bio-Rad]), and P450 content was measured spectroscopically (Omura and Sato 1964).
POR Assays Based on Cytochrome c
The ability of bacterially expressed POR to reduce cytochrome c or to oxidize NADPH was measured, using a method based on that of Lu et al. (1969), by quantitating the change in A550 when oxidized cytochrome c is reduced (extinction coefficient of 21.1 mM−1) (Van and Slater 1962). The assay system contained an NADPH regeneration system, consisting of 0.1M Tris-HCl (pH 7.8), 2 mM glucose-6-phoshate (Sigma), and 3 U glucose-6-phosphate dehydrogenase (Sigma) in a 1-ml volume. The cytochrome c concentration was 40 μM when NADPH was the variable substrate, and the NADPH concentration was 5 μM when cytochrome c was the variable substrate. Assays were performed with 15–20 μg of bacterial membranes containing 5 pmol of POR, as assessed in comparison to bacterially expressed wild-type POR, as described above. A550 was monitored against time, and velocities were expressed as (nmol reduced cytochrome c) · (nmol POR)−1 · (min−1). Data were analyzed using LEONORA (Cornish-Bowden 1995) (see the Centre National de la Recherche Scientifique Marseilles Group in Enzyme Kinetics and Control Analysis Web site).
POR Assays Based on P450c17
The various POR missense mutants all expressed at similar levels. The ability of bacterially expressed POR to support 17α-hydroxylase and 17,20 lyase activities of P450c17 was then assayed using human P450c17 expressed in yeast. Yeast membrane fractions containing 10 pmol of P450c17 were combined with bacterial membranes containing 20 pmol POR by emulsifying with 20 μg phosphatidylcholine in 100 mM potassium phosphate, 6 mM potassium acetate, 10 mM MgCl2, 1 mM reduced glutathione, 20% glycerol, 3 U glucose-6-phosphate dehydrogenase, 0.1 mM glucose-6-phosphate, and radiolabeled substrate for 20 min at 25°C in a total volume of 200 μl. For the 17α-hydroxylase reaction, the substrate was 14C progesterone; for the 17,20 lyase reaction, the substrate was 3H 17α-hydroxypregnenolone (17OH Preg), with 50 pmol cytochrome b5 (added to facilitate the lyase reaction allosterically) (Auchus et al. 1998). The reactions were started by adding 20 μl 10 mM NADPH; reactions were incubated at 37°C for various lengths of time and were stopped by adding ethyl acetate:iso-octane (3:1) to extract the steroids. Steroids from different reactions were spotted on silica gel 60 F-254 thin-layer chromatography (TLC) plates (Merck) and were developed with ethyl acetate:chloroform (3:1) (Lin et al. 1991). Steroids were quantified by phosphorimaging for P450c17/POR assays, and kinetic behavior was approximated as a Michaelis-Menten system. Curve fitting and calculations of maximum velocity (Vmax) and apparent Michaelis constant (Km) values were performed using LEONORA (Cornish-Bowden 1995).
Expression and Purification of Cytochrome b5
For use in the 17,20 lyase assays, human cytochrome b5 was prepared in E. coli strain BL21(DE3) transformed with plasmid pLW01b5 containing human cytochrome b5 cDNA and was selected over several cycles, as described (Miroux and Walker 1996). A single colony of transformed E. coli was inoculated into Luria-Bertani (LB) medium containing 100 μg/ml carbenicillin, grown to an OD 600 nm of 0.5–0.7, and IPTG was added to a final concentration of 0.4 mM. Cell density was measured every 30 min; when the OD 600 nm started to rise again (90–180 min after induction), 100 μl of cells diluted at a ratio of 1:50 were plated on LB agar plates containing 100 μg/ml carbenicillin and 0.4 mM IPTG and were allowed to grow overnight at 37°C. A single colony was grown the next day, and the process of induction and selection with carbenicillin and IPTG was repeated three more times. After four cycles of selection, the E. coli were transformed with plasmid pLysS/RARE, which encodes lysozyme and six rare tRNAs that assist the expression of mammalian proteins by complementing the codon bias of E. coli. The resultant E. coli A11(DE3) pLysS/RARE cells were used for expression of cytochrome b5. Expression and purification was performed as described (Mulrooney and Waskell 2000), and purity was assessed by SDS-PAGE (fig. 1).
Results
Anatomic Phenotype
A summary of the dysmorphologic features in our patients is presented in table 1. The most consistent features were radiographically demonstrated craniosynostosis, either radioulnar or radiohumeral synostosis, and femoral bowing. Other features generally associated with ABS (brachycephaly, femoral fractures, midface hypoplasia, proptosis choanal atresia, arachnodactyly, camptodactyly, and rocker-bottom feet) were also reported in several patients, and some patients also had conductive hearing loss. A comparison of these clinical findings and the mutation analyses (table 4) shows that, aside from the genital anomalies attributable to disordered steroidogenesis, no morphologic feature distinguished patients with POR mutations from those with FGFR mutations.
Table 4.
Summary of Genetic Findings[Note]
|
Mutation (Parental Ethnicity) at Allele |
||||
| PatientNo. | Patient ID | 1 | 2 | FGFR2(Exons 8 and 10) |
| 1 | IRL1 | G539R (Irish) | Frame 1 (white) | FGFR1 I300T |
| 2 | AUS2 | F646del (Laotian) | A287P (white) | nl |
| 3 | PL1 | A287P (white) | Frame 2 (Moroccan) | nl (all FGFR2) |
| 4 | CF1 | M263V (white) | Not found (white) | nl |
| 5 | 337 | A287P (French) | Not found (French) | nl (all FGFR2) |
| 6 | 259 | R457H (Senegalese) | Not found (Senegalese) | nl (all FGFR2) |
| 7 | 699 | Q153R (Algerian) | Q153R (Algerian) | nl |
| 8 | JPSZ | R457H (Japanese) | R457H (Japanese) | nl |
| 9 | JPSS | R457H (Japanese) | Y459H (Japanese) | nl |
| 10 | JPSB | R457H (Japanese) | Y459H (Japanese) | nl |
| 11 | MC25 | A287P (white) | Frame 3 (Hispanic) | nl |
| 12 | 09742 | A287P (white) | A287P (white) | nl |
| 13 | 50081 | A287P (white) | Frame 4 (white) | nl |
| 14 | 11314 | None found (British) | None found (British) | FGFR2 C342S |
| 15 | 14506 | None found (British) | None found (British) | FGFR2 S351C |
| 16 | 14683 | A287P (British) | None found (British) | nl |
| 17 | 15441 | None found (Pakistani) | None found (Pakistani) | FGFR2 C342R |
| 18 | 16666 | None found (white) | None found (white) | FGFR2 S351C |
| 19 | 20554 | None found | None found | FGFR2 S351C |
| 20 | 24019 | R457H (British) | Frame 6 (British) | nl |
| 21 | 07862 | None found (British) | None found (British) | nl |
| 22 | 24872 | A287P (British) | Frame 7 (British) | nl |
| 23 | 14837 | None found (Hispanic) | None found (Hispanic) | FGFR2 F276V |
| 24 | 56661 | A287P (white) | R616X (white) | nl |
| 25 | JPAS | None found (Japanese) | None found (Japanese) | All FGFR2 nl |
| 26 | 1110 | None found (Arab) | None found | nl |
| 27 | 1115 | None found (white) | None found (white) | nl |
| 28 | 1323 | None found (Black) | None found (Black) | FGFR2 S351C |
| 29 | 1434 | None found (Black) | None found (Black) | FGFR2 S351C |
| 30 | 2430 | None found (Hispanic) | None found | FGFR3 A391E |
| 31 | STJ1 | L565P (white) | R457H (white) | nl |
| 32 | ISRL1 | T142A (Iraqi) | Frame 5 (Yemeni) | nl |
| 33 | MC15 | None found (white) | None found (white) | FGFR2 Y375C |
| 34 | 1095 | None found (Hispanic) | None found | nl |
| 35 | 1603 | None found (Vietnamese) | None found (Vietnamese) | FGFR2 S372C |
| 36 | 1771 | None found (black) | None found (black) | FGFR2 Y375C |
| 37 | 2353 | A115V (white) | None found | FGFR2 Y375C |
| 38 | 1156 | None found | None found | nl |
Note.— Frameshift mutations (base numbers refer to the ORF, with A of the first ATG as base 1): frame 1, 580-581insTACGTGGACAAGC; frame 2, 1551-1552ins TGCCCATGTTCGTGCGC; frame 3, 1619-1620insCCTTCAAGGCCACCACGCCTGTCATCATGATGGGCCCCGGCACCGGGGT; frame 4, 1621-1622insC; frame 5, 1348-1349ins GAGC; frame 6, IVS6-2A→T; and frame 7, IVS7+(2-3)insT.
Hormonal Phenotype
Clear hormonal and genital data are not available for all patients, since most were not seen by a pediatric endocrinologist. Where data are available, a pattern emerges that closely resembles the pattern originally described by Peterson et al. (1985), as well as that described in our previous study (Flück et al. 2004). In all patients in whom it was measured, 17OHP was markedly elevated and was hyperresponsive to stimulation with ACTH, as is typical of defects in steroid 21-hydroxylase (P450c21) (Miller 1994). Basal values for cortisol and other steroids tended to be normal, but such values are not truly informative in the absence of stimulation with ACTH. Despite the elevated values for 17OHP, values for C19 steroids were normal or low, indicating a relatively greater deficiency in the 17,20 lyase activity of P450c17 (Geller et al. 1997). There was no clear evidence of any patient suffering a life-threatening adrenal crisis from glucocorticoid and mineralocorticoid deficiency, but there also was no hormonal evaluation of the several individuals who died in childhood. The possibility of life-threatening adrenal insufficiency remains a concern.
DNA Sequencing
Exons 8 and 10 of FGFR2 are the most frequent sites of mutation in the various craniosynostosis syndromes; these exons were sequenced in all patients. In patients 5, 6, and 26, all exons of FGFR2 were sequenced. Excluding patients 33–38 with Beare-Stevenson syndrome, FGFR2 mutation S351C was found in five patients, other FGFR2 mutations (C342S, C342R, and P276V) were found in three other patients, and one patient carried the FGFR3 mutation A391E (table 4). None of these individuals with FGFR2 or FGFR3 mutations carried a mutation in POR. Patient 1 carried the FGFR1 mutation I300T and was also a compound heterozygote for two POR mutations.
For the POR gene, all 15 exons and at least 50 bases of each intron lying adjacent to the exons were sequenced on both strands in all samples. Additional primers were used at primer sites where the University of California Santa Cruz (UCSC) Genome Browser identified polymorphisms. Allele assignments of identified mutations were made by sequencing parental DNA when such samples were available. SNPs permitted the identification of two alleles in most cases; however, we can not rule out hemizygosity in patients 4–8 and 12 (table 4). The sequencing results are shown in figure 2, and the allele assignments and the available data on ethnic origins are summarized in table 4. The numbering of the nucleotides is based on the NCBI human POR cDNA sequence NM_000941.1; the ORF with A of the first ATG is base 1. The corresponding amino acid numbers in the protein are based on the NCBI POR protein reference sequence NP_000932.1. These sequences correspond to the full-length, 680-aa human POR sequence. Some other investigators (Adachi et al. 2004; Arlt et al. 2004) have used NCBI sequence P16435, which is derived from the SwissProt database and indicates that human POR contains 677 residues (as does rat POR), but the older SwissProt database misses the Met-Ile-Asn sequence that precedes the Met in position 4 (corresponding to rat position 1).
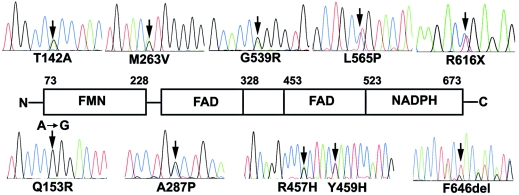
Figure 2.
Sequences of POR mutations. The mutants shown are from the patients in the present study who had disordered steroidogenesis. Each mutant is shown as a heterozygote in association with a wild-type allele, except for Q153R, which was only seen in a homozygously affected patient. The mutated nucleotides, indicated by arrows, are 424A→G, causing T142A; 458A→G, causing Q153R; 787A→G, causing M263V; 859G→C, causing A287P; 1370G→A, causing R457H; 1375T→C, causing Y459H; 1615G→A, causing G539R; 1694T→C, causing L565P; 1845C→T, causing R616X; and 1937-1939delTCT, causing F646del. The location of each mutation is shown on a linear diagram of the POR protein, as described by Wang et al. (1997), which illustrates the crystallographically inferred boundaries of the functional domains of POR.
POR mutations were found in 19 of the 32 patients with ABS (patients 33–38 are excluded because they had Beare-Stevenson syndrome). Mutations were found on only one allele in four samples (patients 4, 5, 6, and 16). The other 15 patients carrying POR mutations included 3 in whom the mutation was homozygous, 4 proven compound heterozygotes in whom allele assignments were made by sequencing parental DNA, and 8 probable compound heterozygotes in whom two heterozygous mutations were found but allele assignments could not be done because parental samples were not available. The 34 affected alleles included 5 unique frameshift mutations, 2 splice-site mutations, 26 missense mutations, and 1 premature stop codon near the carboxyl-terminus of the encoded POR protein. The five frameshift mutations (“Frames 1–5” in table 4) grossly disrupted the flavin mononucleotide (FMN)- and NADPH-binding sites of POR, and four of them (Frames 1, 2, 4, and 5) also caused premature stop codons upstream from residue 616, where the spontaneously occurring nonsense mutation R616X was devoid of activity (described below). The two intronic mutations, termed “Frame 6” and “Frame 7” (table 4), disrupted the canonical splice-acceptor or -donor sites; the splice-site recognition programs FSPLICE (see the Softberry Web site) and NNSplice (see the Berkeley Drosophila Genome Project Web site) indicate that each is very likely to disrupt splicing. Thus, the mutations termed “Frames 1–7” were not tested in vitro. Most missense mutations were found once, but A287P was found on 10 alleles, and R457H was found on 7 alleles. Thus, these two missense mutations accounted for 17 of 34 (50%) of the identified POR missense mutations.
A287P and R457H Mutations
The A287P mutation was found only in samples from subjects described as “white,” “Caucasian,” or “European.” This same mutation was also found on two of eight alleles in our initial study (Flück et al. 2004) and on one of four alleles in a subsequent study (Arlt et al. 2004), all in individuals of European heritage. Thus, A287P is a frequent mutation causing POR deficiency in this group. The nucleotide change 859G→C, which is responsible for all identified cases of A287P, lies in exon 8, changing the sequence from ↓(N)12GCTGC to (N)12CCTGC, destroying the site recognized by restriction endonuclease BbvI. We amplified exons 8 and 9 in a single PCR using primers OR-8-9F and OR-8-9R (table 3), yielding a product of 613 bp. Digestion of this PCR product with BbvI yields four fragments of 250 bp, 167 bp, 101 bp, and 95 bp in the wild type but yields only three fragments of 262 bp, 250 bp, and 101 bp in the mutant.
The R457H mutation was found in four of eight alleles from Japanese patients. This mutation destroyed BsaXI and TstI sites, but these enzymes recognize degenerate 33–37 base sequences and cut both strands twice; hence, this approach did not yield a reliable diagnostic tactic. R457H was also found on 1 of 2 Japanese alleles in our initial study (Flück et al. 2004) and on 2 of 4 alleles (Adachi et al. 2004) and 10 of 16 alleles (Fukami et al. 2005) in two recent studies of Japanese patients. However, other ethnic groups may also carry the R457H mutation. The samples from patient 6, whose parents are from Senegal; patient 21, from the United Kingdom; and patient 31, whose parents are both French Canadian, were heterozygous for the same 1370G→A mutation causing the R457H missense mutation. In addition, Arlt et al. (2004) found R457H in a Polish patient. Thus, R457H is strongly—but not uniquely—associated with patients of Japanese heritage.
Mutations and Polymorphisms
Our previous study described the enzymology of the five missense mutations (A287P, R457H, V492E, C569Y, and V608F) identified in our initial four patients (Flück et al. 2004). The present series adds the mutants or polymorphisms A115V, T142A, Q153R, M263V, Y459H, A503V, G539R, L565P, R616X, V631I, and F646del. In addition, we found the mutants or polymorphisms P228L, R316W, G413S, and G504R by searching a publicly available sequence database (see the BioVentures Web site), and the missense mutation Y181D was reported in another study (Arlt et al. 2004) but was not characterized in a P450 assay. The incidences of these alleles and their ethnic distributions are not yet available in the NCBI, HapMap (see the International HapMap Project Web site), or UCSC Genome Browser databases, although these are being updated regularly. As discussed below, the changes that were found in the patients disrupted steroidogenesis and hence appear to be mutations, whereas the changes that were found in databases had lesser (but variable) effects and hence appear to be polymorphisms. In the discussion of the enzymology below, all of these amino acid changes are referred to as “mutations.”
Enzymology of the Amino Acid Replacement Mutations
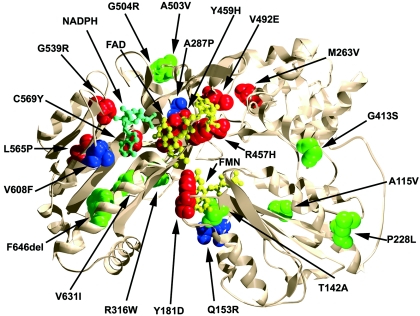
To understand the enzymatic consequences of each amino acid replacement mutation, we built a computer graphic model of the human POR that was based on the crystallographically determined structure of rat POR, which is 94% identical to human POR (Wang et al. 1997). The model of human POR was energetically favorable (total free energy was −33,952 kJ/mol for the human POR model, compared with −27,850 kJ/mol for the rat POR structure) and closely resembled the rat POR structure, as expected (fig. 3). In general, the missense mutations found in patients with ABS mapped to functionally important domains of POR, and the apparent polymorphisms mapped to less crucial regions. Therefore, to assess the enzymatic consequences of each missense mutant, we built bacterial expression vectors for each and characterized their activities in four assays.
Figure 3.
Model of human POR, showing the location of the mutants. The model is based on the X-ray crystal structure of rat POR and was created using the programs DeepView and POVRAY (see the Persistence of Vision Raytracer Web site). The α-carbon backbone is shown as a narrow ribbon, permitting visualization of buried residues. The missense residues identified by sequencing are depicted by charged packed sphere images. Mutations retaining >50% of activity are shown in green, those retaining 25%–50% are shown in blue, and those retaining <25% are shown in red. Ball-and-stick models are used to represent the FMN (yellow), FAD (yellow) and NADPH (cyan) moieties.
From the time of the initial identification and isolation of POR (Lu et al. 1969), the standard POR assay has been the reduction of cytochrome c, which is easily assayed as change in absorption at 550 nm. To perform this assay and also to measure NADPH oxidation, we expressed human POR mutants in bacteria in a form lacking 27 amino-terminal residues (N-27 POR) (Dierks et al. 1998), which is similar to N-24 rat POR, as human POR has a 3-aa amino-terminal extension, compared with rodent POR (NCBI accession numbers NC_000007, NM_000941.1, and NP_000932.1). This form of POR remains associated with membranes, as does wild-type POR, and it is distinct from the soluble N-49 POR used by Arlt et al. (2004). Table 5 shows the Km, Vmax, and Vmax/Km (an estimate of catalytic efficiency) for the capacity of the wild type and the 21 missense mutants of POR to catalyze the reduction of cytochrome c and the oxidation of NADPH. We have previously measured these variables for wild-type POR and for the A287P, R457H, V492E, C569Y, and V608F mutants (Flück et al. 2004); in each case, the values in table 5 are within 20% of the previously measured values. An examination of the Vmax/Km values, shown as percentages of the wild-type values, reveals that there is a very broad range of enzymatic impairment and that there is generally good agreement between the two assays. P228L, R316W, G413S, A503V, and G504R, which were polymorphisms identified from databases, retained 47%–100% of wild-type activity in both assays, but the apparent polymorphism A115V, found in a patient with Beare-Stevenson syndrome with an FGFR2 mutation, had 41%–63% of wild-type activity in the cytochrome c assays (but 70%–80% of wild-type activity in P450c17 assays [table 6]); M263V and V631I, found as heterozygotes during the sequencing of other individuals, had 23%–76% of wild-type activity (table 5).
Table 5.
Kinetics of POR Variants Using Cytochrome c as Substrate[Note]
|
Value of Variable ± SD for |
||||||||
| Cytochrome c |
NADPH |
|||||||
| PORVariant | Km | Vmax | Vmax/Km (% of Wild Type) | Kcat | Km | Vmax | Vmax/Km (% of Wild Type) | Kcat |
| Wild type | 1.4 ± .3 | 91.2 ± 5.1 | 65.1 (100) | 388 | .22 ± .05 | 68.4 ± 2.9 | 310 (100) | 292 |
| A115V | 2.0 ± .4 | 82.5 ± 4.3 | 41.2 (63) | 350 | .29 ± .04 | 37.2 ± 5.9 | 128 (41) | 158 |
| T142A | 1.6 ± .2 | 51.3 ± 6.3 | 32.1 (49) | 218 | .23 ± .03 | 41.7 ± 4.6 | 161 (52) | 178 |
| Q153R | 3.7 ± .4 | 21.4 ± 5.7 | 5.7 (9) | 90 | 1.2 ± .09 | 43.2 ± 3.1 | 36 (11) | 184 |
| Y181D | >50 | … | … | … | >50 | … | … | … |
| P228L | 1.4 ± .3 | 69.2 ± 5.2 | 49.4 (75) | 294 | .24 ± .06 | 53.7 ± 3.8 | 224 (72) | 229 |
| M263V | 1.9 ± .2 | 94.5 ± 3.7 | 49.7 (76) | 402 | .39 ± .07 | 69.1 ± 2.4 | 177 (57) | 294 |
| A287P | 5.9 ± .8 | 37.3 ± 6.4 | 6.3 (9) | 158 | .71 ± .10 | 35.1 ± 4.3 | 49 (16) | 150 |
| R316W | 1.9 ± .4 | 75.9 ± 8.3 | 39.9 (61) | 322 | .29 ± .07 | 69.3 ± 9.7 | 238 (77) | 294 |
| G413S | 1.3 ± .2 | 64.5 ± 7.9 | 49.6 (76) | 274 | .20 ± .05 | 61.7 ± 2.1 | 309 (100) | 262 |
| R457H | 17.2 ± 2.5 | 7.5 ± 2.1 | .43 (.7) | 32 | >50 | … | … | … |
| Y459H | 22.9 ± 6.6 | 6.1 ± .7 | .26 (.4) | 26 | >50 | … | … | … |
| V492E | 29.7 ± 5.1 | 6.8 ± 1.1 | .22 (.3) | 28 | >50 | … | … | … |
| A503V | 1.8 ± .2 | 81.3 ± 6.6 | 45.2 (69) | 344 | .21 ± .06 | 55.7 ± 1.9 | 265 (86) | 238 |
| G504R | 2.1 ± .2 | 72.9 ± 4.1 | 34.7 (53) | 312 | .35 ± .06 | 51.2 ± 3.9 | 146 (47) | 218 |
| G539R | 6.3 ± 1.2 | 37.8 ± 6.7 | 6.0 (9) | 160 | 18.3 ± 2.1 | 9.3 ± 1.2 | .5 (.2) | 40 |
| L565P | 2.8 ± .6 | 26.2 ± 3.2 | 9.3 (14) | 112 | 5.3 ± 1.6 | 23.8 ± 2.3 | 4.5 (1.4) | 102 |
| C569Y | 5.8 ± .2 | 24.3 ± 2.7 | 4.1 (6) | 104 | 1.8 ± .43 | 12.1 ± .7 | 7 (2) | 52 |
| V608F | 5.2 ± 1.3 | 26.3 ± 5.2 | 5.1 (8) | 112 | 3.3 ± .7 | 32.7 ± .6 | 10 (3) | 140 |
| R616X | >50 | … | … | … | >50 | … | … | … |
| V631I | 1.6 ± .4 | 77.3 ± 8.6 | 48.3 (74) | 328 | .47 ± .06 | 32.8 ± 5.3 | 70 (23) | 140 |
| F646del | 3.9 ± .7 | 90.7 ± 5.7 | 23.3 (36) | 384 | .21 ± .13 | 61.2 ± 8.5 | 291 (94) | 260 |
Note.— Km values are in μM (cytochrome c or NADPH); Vmax values are in nmol of cytochrome c reduced/mg POR protein/min. Kcat values were calculated on the basis of the molar content of POR, and, hence, the values are in min−1. POR content of each membrane preparation was measured, and all samples were normalized against wild-type POR.
Table 6.
Kinetics of P450c17 Activities Supported by POR Variants[Note]
|
Value of Variable ± SD for |
||||||||
| 17α-Hydroxylase |
17,20 Lyase |
|||||||
| PORVariant | Km | Vmax | Vmax/Km (% of Wild Type) | Kcat | Km | Vmax | Vmax/Km (% of Wild Type) | Kcat |
| Wild type | 4.3 ± .09 | .149 ± .021 | .035 (100) | .67 | .93 ± .18 | .059 ± .014 | .063 (100) | .27 |
| A115V | 3.5 ± .05 | .097 ± .013 | .028 (80) | .43 | .71 ± .11 | .032 ± .006 | .045 (71) | .14 |
| T142A | 5.1 ± .07 | .106 ± .019 | .021 (60) | .48 | .86 ± .15 | .029 ± .009 | .034 (54) | .13 |
| Q153R | 3.7 ± .09 | .039 ± .017 | .011 (31) | .18 | .97 ± .20 | .017 ± .007 | .017 (27) | .08 |
| Y181D | >50 | … | … | … | >50 | … | … | … |
| P228L | 3.7 ± .06 | .131 ± .013 | .035 (100) | .59 | 1.54 ± .21 | .041 ± .005 | .026 (41) | .18 |
| M263V | 7.2 ± 1.7 | .025 ± .011 | .003 (9) | .11 | 1.17 ± .13 | .009 ± .003 | .008 (13) | .04 |
| A287P | 3.5 ± 1.0 | .049 ± .010 | .014 (40) | .22 | 1.34 ± .09 | .018 ± .002 | .013 (21) | .08 |
| R316W | 3.1 ± .04 | .104 ± .017 | .033 (94) | .47 | .87 ± .18 | .078 ± .003 | .089 (141) | .35 |
| G413S | 5.9 ± 1.1 | .172 ± .015 | .029 (83) | .77 | 1.28 ± .15 | .089 ± .011 | .069 (110) | .40 |
| R457H | 5.1 ± 1.2 | .008 ± .003 | .001 (3) | .04 | … | … | … | … |
| Y459H | 6.5 ± 1.1 | .027 ± .009 | .004 (11) | .12 | … | … | … | … |
| V492E | 7.7 ± 1.5 | .011 ± .004 | .001 (3) | .05 | … | … | … | … |
| A503V | 5.5 ± .03 | .110 ± .018 | .021 (60) | .50 | 1.04 ± .09 | .037 ± .006 | .035 (56) | .17 |
| G504R | 3.9 ± .07 | .124 ± .020 | .032 (91) | .56 | .85 ± .12 | .056 ± .009 | .065 (103) | .25 |
| G539R | 4.5 ± 1.0 | .073 ± .006 | .016 (46) | .33 | 1.95 ± .27 | .009 ± .002 | .005 (8) | .04 |
| L565P | 5.9 ± .65 | .067 ± .008 | .011 (32) | .30 | 1.39 ± .16 | .017 ± .005 | .0120 (19) | .08 |
| C569Y | 9.3 ± 1.9 | .094 ± .009 | .010 (28) | .42 | 1.22 ± .15 | .010 ± .012 | .008 (13) | .05 |
| V608F | 3.5 ± .04 | .099 ± .014 | .028 (80) | .45 | 1.03 ± .17 | .037 ± .012 | .036 (57) | .17 |
| R616X | 4.7 ± .07 | .008 ± .003 | .002 (6) | .04 | … | … | … | … |
| V631I | 4.3 ± .06 | .077 ± .008 | .018 (51) | .34 | 1.32 ± .22 | .033 ± .009 | .025 (40) | .15 |
| F646del | 3.5 ± .07 | .118 ± .011 | .034 (97) | .53 | 1.77 ± .27 | .051 ± .006 | .029 (46) | .23 |
Note.— Km values are in μM progesterone for 17α-hydroxylase and μM 17OH-pregnenolone for 17,20 lyase. Vmax values are in pmol/μg P450c17/min. Kcat values were calculated on the basis of the molar content of P450c17, and, hence, the values are in min−1. POR content of each membrane preparation was measured, and all samples were normalized against wild-type POR.
However, cytochrome c is a soluble protein and is not a physiologic substrate for normal, membrane-bound POR (Lu et al. 1969). Therefore, we supplemented this classic assay with direct assays of the capacity of POR mutants to support catalysis by a cytochrome P450 enzyme. We previously reported assays of POR activity that were based on its ability to support catalysis by P450c17 with the use of doubly transformed yeast (Flück et al. 2004). In this system, a yeast strain lacking the gene for yeast POR was transiently transfected with vectors expressing human P450c17 and POR (Auchus et al. 1998). This system was originally developed to assay P450c17 activities: the transfected human POR was abundant and not limiting, and the amount of P450c17 was measured by difference spectra, yielding reproducible measures of P450c17 enzymology (Auchus et al. 1998; Arlt et al. 2001), but the ratio of P450c17 to POR could not be measured or chosen precisely. However, when POR is limiting, the 17,20 lyase activity of P450c17 is sensitive to changes in the molar ratio of POR to P450c17 (Yanagibashi and Hall 1986; Lin et al. 1993). Therefore, we have developed a modification of this assay using microsomes containing human P450c17 expressed in yeast, as before, combined with a membrane fraction containing human N-27 POR expressed in E. coli (fig. 1). Each component is prepared and assayed separately before mixing, so the P450c17:POR ratio can be set, improving reproducibility and consistency. As the 17,20 lyase activity of P450c17 is facilitated by the allosteric action of cytochrome b5 (Auchus et al. 1998), we also prepared cytochrome b5 in bacteria, to add in support of the lyase reactions (fig. 1). Adding exogenous phosphatidylcholine fostered association of the yeast and bacterial membranes, permitting association of the catalytic components. In this system, POR remains intimately associated with the endoplasmic reticulum and, thus, has a physiologically relevant membrane insertion. As defective androgen synthesis is a hallmark of the patients with POR deficiency and as the 17,20 lyase activity of human P450c17 is exquisitely sensitive to the topology of its interactions with POR (Geller et al. 1997; Auchus and Miller 1999; Geller et al. 1999), we assayed the capacity of the POR mutants to support the 17α-hydroxylase and 17,20 lyase activities of human P450c17.
Table 6 shows the Km, Vmax, and Vmax/Km for the 17α-hydroxylase and 17,20 lyase activities of P450c17 when catalysis is supported by either wild-type POR or each of the missense mutants. We previously measured these values for the A287P, R457H, V492E, C569Y, and V608F mutants (Flück et al. 2004). Those prior data are in excellent agreement with the data in table 6, except for C569Y, which has lower activity in the present study. The polymorphic A115V, P228L, R316W, G413S, A503V, and G504R variants retained 41%–141% of wild-type activity. The V608F mutant, which had 3%–8% of wild-type activity in the cytochrome c assays, had 57%–80% of wild-type activity in the P450c17 assays. Other, lesser, differences can be seen by comparing tables 5 and 6.
Careful comparison of the enzymatic data with the clinical data show that the P450c17 assays provide a much better correlation between genotype and phenotype. Patient 1 was a compound heterozygote for a null (frameshift) mutation and the novel missense mutation G539R. This mutant retained 46% of the 17α-hydroxylase activity but only 8% of the 17,20 lyase activity. Consistent with this, the patient was able to synthesize cortisol adequately (17α-hydroxylase activity) but had poor production of sex steroids (17,20 lyase activity). Patients 2 and 3 were compound heterozygotes, each carrying the previously described A287P mutation on one allele but carrying very different mutations on the other allele. Patient 2 carried F646del, which retained normal hydroxylase activity and about half of normal 17,20 lyase activity, whereas patient 3 carried a null (frameshift) mutation on the second allele. Consistent with this, the hormonal and anatomic phenotype in patient 2 was less severe, with no genital ambiguity and normal cortisol (17α-hydroxylase activity) but with clear evidence of decreased 17,20 lyase activity (elevated 17OHP, low DHEA, and androstenedione). By contrast, patient 3 had moderately severe genital ambiguity, early evidence of disordered steroidogenesis, and a more severe skeletal phenotype. A close correlation between assays of the 17,20 lyase activity of P450c17 and the genital phenotype is to be expected, as 17,20 lyase activity is required for the production of all sex steroids and because the 17,20 lyase activity is much more sensitive than the 17α-hydroxylase activity of P450c17 to minor disturbances in the interactions between P450c17 and POR (Geller et al. 1997, 1999; Auchus and Miller 1999).
Discussion
Assessment of POR Activity
In the present study, we assayed POR activity both by the conventional cytochrome c assays (reduction of cytochrome c and oxidation of NADPH) and by a direct measure of POR-supported catalysis by a microsomal cytochrome P450 enzyme. For this purpose, we used steroidogenic P450c17, as its 17,20 lyase activity is very sensitive to the amount of POR available (Yanagibashi and Hall 1986; Lin et al. 1993) and to the qualitative nature of the interaction between P450c17 and POR (Zhang et al. 1995; Geller et al. 1997, 1999; Auchus et al. 1998; Auchus and Miller 1999; Pandey et al. 2003). Our previous studies of POR mutations (Flück et al. 2004) used a yeast system in which endogenous yeast POR is absent and human P450c17 and human POR are expressed in yeast from transiently transfected plasmids (Auchus et al. 1998). This system works well, but it does not permit direct assessment of the amount of POR present, and each transfection must be tediously assayed for P450c17 content to calculate meaningful enzymatic parameters. In the present study, we modified this system by using yeast-expressed human P450c17, combined in vitro with human N-27 POR, prepared from E. coli. By comparing the enzymatic parameters (Km and Vmax) obtained for the wild type and for five different POR missense mutants (A287P, R457H, V492E, C569Y, and V608F) assessed in the double-transfected yeast system (Flück et al. 2004) with those obtained with the in vitro fused-membrane system (present study), we see that both systems give similar values for Km and that the present system yields slightly lower values for Vmax. Because the present system permits quantitation of all relevant variables, we consider these values to be a more reliable approximation of actual POR activity. Nevertheless, actual activity in vivo may vary as a result of other unknown intracellular factors.
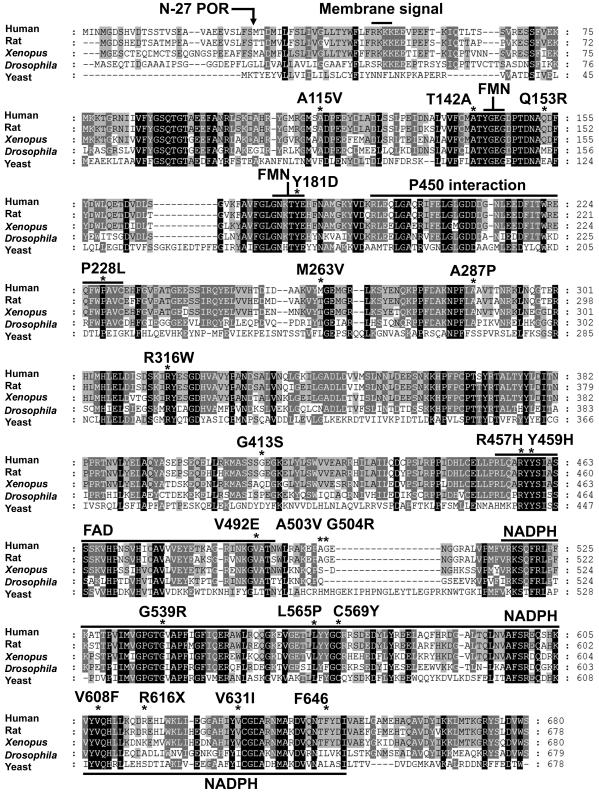
The surface of the electron-donating (FMN) domain of POR is characterized by acidic residues (Shen and Kasper 1995; Estabrook et al. 1996; Wang et al. 1997), whereas the redox-partner-binding site of microsomal P450 enzymes is typically characterized by basic residues (Hasemann et al. 1995; Fisher et al. 1996; Geller et al. 1997; Auchus and Miller 1999; Kondo et al. 1999; Davydov et al. 2000). Thus, electrostatic interactions between oppositely charged residues are obviously important in governing the association of POR with a P450 enzyme. Consistent with this, mutation of arginine residues in the POR-binding site of P450c17 selectively ablates 17,20 lyase activity (Geller et al. 1997, 1999), and chemical modification of acidic residues in the FMN site of POR similarly reduces the activity of P450 (Nadler and Strobel 1988, 1991; Shen and Kasper 1995). In addition, hydrophobic interactions between POR and P450 also play a role (Strobel et al. 1989). The activities assessed in different studies are a function of the specific P450 enzyme used as the readout of the POR activity and are also a function of the form of POR used. Both the full-length POR used in our previous studies (Flück et al. 2004) and the N-27 human POR used here remain associated with membranes and are able to support the reduction of a cytochrome P450. By contrast, when rat POR or human POR is rendered wholly soluble by deleting 54 N-terminal residues, it can still reduce cytochrome c (which is not a physiological substrate for POR), but it cannot reduce P450 (Lu et al. 1969; Roman et al. 2003). Human, rat, and Xenopus PORs have a long hydrophobic domain following the strongly basic sequence RKKK (residues 48–51) that resembles a transmembrane domain (von Heijne 1984), but this sequence is not found in Drosophila or yeast POR (fig. 4). Thus, reliable measurements of POR activity require the use of a membrane-bound form of POR and a sensitive P450-based enzymatic readout.
Figure 4.
Amino acid sequence alignment of POR from five species. The protein sequences used are those of human (NCBI accession number NP_000932.1), rat (NCBI accession number NP_113764.1), Xenopus (NCBI accession number AAH59318.1), yeast (NCBI accession number NP_596046.1) and Drosophila (NCBI accession number NP_477158.1). Alignments were made with ClustalW and are displayed with GeneDoc. Residues that are identical in all species (when the residue exists) are highlighted in black, those identical in four of five species are shown in dark gray, and those identical in three of five species are shown in light gray. Amino acid numbers are shown at the right. Note that the human POR sequence has a three-residue amino-terminal extension. The functional domains and individual residues discussed in the text are shown above the human sequence. The location of the truncation used to create the N-27 POR used in the functional assays is shown.
Structure/Function Correlation of POR Mutants
The mutations examined in this study provide reasonable scanning mutagenesis of POR. Human POR shares highly conserved binding domains for FMN, FAD, and NADPH with the PORs of other species (fig. 4), and the FAD- and NADPH-binding domains are also similar to the FAD- and NADPH-binding domains of ferredoxin reductase (Hanukoglu and Gutfinger 1989; Karplus et al. 1991) and nitric oxide synthase (Bredt et al. 1991). By analogy with the crystallographically determined structure of rat POR (Wang et al. 1997), residues 143–147 and 173–185 of human POR participate in binding the isoalloxazine ring of FMN, with the side chain of Y181 stabilizing the interaction with the flavin. Replacing the rat POR residue Y178 (human Y181) with Asp eliminated 99% of its activity to reduce cytochrome c (Shen et al. 1989), and the corresponding human Y181D mutation caused ABS (Arlt et al. 2004) and reduced activities toward cytochrome c (table 5) and P450c17 (table 6) to undetectable levels. The mutation Q153R, which is near the FMN-binding site, reduced POR activity to ∼30% (table 5). The highly conserved domain that interacts with and donates electrons to the cytochrome P450 comprises residues 194–231, especially residues 178–185 and 210–214 (DDDGN) (Wang et al. 1997) (fig. 4). We found no mutations in this domain; the closest mutations were M263V, which lost ∼90% of activity, and A287P, the mutation commonly found in patients of European ancestry, which lost 60%–80% of activity (table 6). The RY sequence of residues 316–317 is conserved from yeast to human POR, suggesting an important function (fig. 4), yet the nonconservative mutant R316W was found as a polymorphism in a database and retained normal activity, as did the conservative polymorphism G413S (table 6). Thus, the activities of each mutant or polymorphism must be tested experimentally to determine the degree of impairment.
The FAD-binding domain (residues 453–495) is highly conserved, and the sequence RYYSI (457–461) is invariant among human, rat, Xenopus, Drosophila, and yeast POR (fig. 4). X-ray crystallography shows that rat R454 (human R457) forms a hydrogen bond with the pyrophosphate group of FAD and that rat Y456 (human Y459) contacts the FAD isoalloxazine ring and hydrogen bonds with the ribityl 3′-hydroxyl group (Wang et al. 1997; Shen and Kasper 2000). Mutation of each of these residues caused ABS and ablated virtually all measurable activity. Similar results were obtained when these mutations were studied in rat POR (Shen and Kasper 1996, 2000; Shen et al. 1999). Residues 491–497, which lie in helix N, form hydrogen bonds with the FAD pyrophosphate (Wang et al. 1997). The mutation V492E is predicted to disrupt this hydrogen bonding; this mutation causes ABS and is virtually devoid of activity. By contrast, the conservative change A503V and the nonconservative change G504R lie in an unstructured loop, were identified as polymorphisms, and retain >50% activity (table 6).
The NADPH-binding domain comprising the carboxy-terminus of the protein is highly conserved (fig. 4)—especially the sequence GTGVAP (residues 537–542), which contains the consensus GXG sequence typical of NADP+-binding proteins (Hanukoglu and Gutfinger 1989; Scrutton et al. 1990). The mutation G539R lost >90% activity in the 17,20 lyase assay but retained 46% activity in the 17α-hydroxylase assay, indicating that the mutant could still bind some NADPH. Consistent with this, the cytochrome c assays showed that the greatest effect was on NADPH oxidation, for which the Km value was increased ∼100-fold (table 5). Studies in which C569 of human POR was alkylated with iodoacetic acid eliminated activity, implicating a role for cysteine in the binding of NADPH to human POR (Haniu et al. 1989), but mutagenesis of the corresponding C566 to S in rat POR suggested that this residue was not essential, despite a 4.6-fold higher Km value for NADPH (Shen et al. 1991); alkylation of pig POR did not change activity (Haniu et al. 1984). The mutation C569Y in human POR, which we previously found in a patient with disordered steroidogenesis but without ABS (Flück et al. 2004), also had a high Km value for NADPH (table 5) but retained 13% of 17,20 lyase activity and 28% of 17α-hydroxylase activity (table 6). The C569Y mutation was found as a compound heterozygote with V608F (Flück et al. 2004), which retained 57% of 17,20 lyase activity and 80% of 17α-hydroxylase activity, thus explaining that individual’s hormonal profile, which resembled isolated 17,20 lyase deficiency. The crystallographic data indicated that rat W677 (human W679) directly participates in interactions with both FMN and NADPH, suggesting that any carboxy-terminal truncation will have severe consequences. Consistent with this, the premature chain truncation mutant R616X was devoid of activity. The mutations A115V and V631I were detected as heterozygotes only in individuals with normal steroidogenesis, and hence they may be polymorphisms. A115V is a conservative change in a region without apparent function, and it retained full activity with P450c17 (table 6). Although the conservative mutant V631I lies in a highly conserved segment of the NADPH-binding domain, it retained normal activity in the cytochrome c assays (table 5) and 40%–50% of activity in the P450c17 assays. Finally, the in-frame deletion of F646, a residue for which the crystal structure of rat POR does not ascribe a specific function, had no effect on 17α-hydroxylase activity but caused a 54% reduction in 17,20 lyase activity. As POR must interact with all 50 human microsomal P450 enzymes and as the stereochemistry of those interactions with different P450 enzymes will vary with the geometry of the redox-partner-binding site of P450, it is clear that no assay that is based on a single P450 enzyme will reliably forecast all the consequences of a specific POR mutant.
Nosology of ABS and POR Deficiency
ABS was initially described as a skeletal malformation syndrome (Antley and Bixler 1975; DeLozier et al. 1980; Hassell and Butler 1994); descriptions of genital ambiguity were added later, as a finding described in about half of these patients (Crisponi et al. 1997). In contrast to the autosomal dominant craniosynostosis syndromes caused by FGFR mutations, reports of ABS in siblings and in children of consanguineous parents have suggested autosomal recessive inheritance (Reardon et al. 2000). The finding of mutations in receptors for fibroblast growth factors, especially FGFR2, in other craniosynostosis syndromes led to the successful search for FGFR2 mutations in some patients with ABS. These eponymic syndromes resulting from FGFR2 mutations may be regarded as presenting variants of a single disease, as single genotypes often give somewhat varied phenotypes. Reardon et al. (2000) called attention to ambiguous genitalia and/or disordered sterol metabolism as the major feature that appeared to distinguish patients with ABS resulting from FGFR2 mutations from patients with ABS who lacked FGFR2 mutations. We recently reported that three such patients, plus another patient without ABS, carried autosomal recessive mutations in POR, and we provided biochemical and enzymologic support for this observation (Flück et al. 2004). We have now confirmed and expanded that original study by sequencing POR in 32 patients, including 12 of the 16 patients studied by Reardon et al. (2000). Reardon et al. (2000) had suggested that ABS with abnormal genitalia was digenic, resulting from mutations in an FGFR2 gene plus a mutation elsewhere. Although we failed to find mutations in 3 of the 32 patients, we found complete genetic segregation of POR and FGFR mutations in patients with the ABS phenotype. Thus, digenic inheritance appears to be most unlikely, but it cannot be completely excluded, as not all FGFR exons were sequenced.
Patients with ABS with disordered steroidogenesis have been born to mothers who became virilized during gestation and, hence, were thought to have luteomas of pregnancy (Roth et al. 2000; Warmann et al. 2000); it is more likely that these cases reflect feto-placental hyperandrogenism resulting from decreased activity of POR-dependent aromatase (P450aro), which converts androgens to estrogens, as described in our initial study (Flück et al. 2004). Alternatively, it has been suggested that increased androgen synthesis through an alternative “backdoor” pathway (Auchus 2004) may result in the virilization of mothers carrying a POR-deficient fetus (Arlt et al. 2004), but definitive data are lacking. It is noteworthy that, aside from the genital ambiguity, no dysmorphologic feature distinguishes ABS secondary to FGFR2 mutations from ABS secondary to POR mutations; thus, this distinction requires careful hormonal evaluation of adrenal steroidogenesis.
Thus, we propose that the term “ABS” be reserved for those patients with the skeletal dysmorphologic findings initially reported by Antley and Bixler (1975) but who have normal genitalia and normal steroidogenesis, and that patients with the skeletal dysmorphologic phenotype plus evidence of disordered steroidogenesis be referred to as having “POR deficiency.” It is clinically important to view patients with POR mutations as having a disease that is distinctly different from the disease caused by FGFR mutations. Not only are the genes and patterns of inheritance different (dominant FGFR mutations vs. recessive POR mutations), but the patients are also vulnerable to different risk factors and require different management and different teams of physicians. Several of the patients with POR mutations died unexpectedly without clear explanation, and Reardon et al. (2000) noted that several patients required steroid hormone supplementation. Now that the steroidogenic defect is understood, it is apparent that patients with POR mutations may be at risk for adrenal insufficiency and Addisonian crisis, especially at times of severe febrile illness or major surgery. Thus, any patient thought to have ABS should be evaluated by a pediatric endocrinologist for the ability to mount an adequate cortisol response to stress, as well as for the adrenal and gonadal pattern of synthesis of C21 and C19 steroids.
The mechanism(s) by which POR deficiency causes a skeletal disorder resembling ABS remains unclear. Substantial speculation has centered on cholesterol, as some disorders of cholesterol biosynthesis include disorders of skeletal development (Herman 2003). Cholesterol biosynthesis requires two POR-dependent enzymes: squalene epoxidase, a non-P450 enzyme (Ono and Bloch 1975), and lanosterol demethylase (CYP51). Furthermore, cholesterol is required for C-terminal modification of hedgehog proteins, which play multiple roles in morphogenesis (Mann and Beachy 2004). Clinical reports have associated ABS with maternal ingestion of fluconazole, an antifungal agent that acts by preferentially inhibiting yeast CYP51 (Pursley et al. 1996; Aleck and Bartley 1997). Lymphoblastoid cells from a patient having ABS and ambiguous genitalia accumulated lanosterol and dihydrolanosterol, consistent with a lesion in CYP51, but the CYP51 gene was normal (Kelley et al. 2002). We subsequently found that this individual was homozygous for the POR mutation A287P (Flück et al. 2004). Although fluconazole should not inhibit human CYP51, the absence of POR-dependent hepatic enzymes that normally metabolize fluconazole might permit fetal concentrations to rise substantially. However, liver-specific knockout of Por yields morphologically normal mice (Henderson et al. 2003; Wu et al. 2003), whereas the global Por-knockout mouse is embryonic lethal (Shen et al. 2002; Otto et al. 2003). Thus, the absence of a hepatic P450 activity appears to be unrelated to the dysmorphology of POR deficiency in mouse. Similarly, Otto et al. (2003) suggested that the phenotype might be associated with decreased activity of CYP26, which degrades retinoids; decreasing the retinoic acid in the diets of pregnant mice carrying Por-knockout fetuses partially ameliorated the phenotype. However, as CYP26 is also a hepatic enzyme, the answer would appear to lie elsewhere. We speculate that the dysmorphology in POR deficiency is associated with a decrease in one of the numerous extrahepatic enzymes that require electron transfer from POR.
Acknowledgments
This study is dedicated to the memory of Professor Robin M. Winter, who played a central role in associating FGFR mutations and craniosynostosis syndromes. This work was supported by the National Institutes of Health (NIH) Mentored Investigator Grant KO1 DK02939 (to N.H.) and by NIH grants RO1 DK37922, RO1 HD41958, and RO1 GM73020 (to W.L.M.). E.W.J. was supported by NIH grants P60 DE013078 and M01 RR00052. We thank Dr. Jacky Bonaventure and Dr. Martine LeMerrer (Hôpital Necker, Paris), Dr. Marie Gonzalez (Hôpital St. Antoine, Paris), and Dr. Kazuki Okajima (Case Western Reserve University, Cleveland), for contributing DNA samples and clinical information; Dr. Richard Kelly, for growing lymphoblastoid cell lines from several patients; Ms. Izabella Damm, for excellent technical assistance; and all the patients and families who participated in this study.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Berkeley Drosophila Genome Project, http://www.fruitfly.org/seq_tools/splice.html (for the program NNSplice)
- BioVentures, http://www.bioventures.com/products/dmg/por/ (for POR gene polymorphism data)
- Centre National de la Recherche Scientifique Marseilles Group in Enzyme Kinetics and Control Analysis, http://bip.cnrs-mrs.fr/bip10/homepage.htm (for the program LEONORA)
- ClustalW, http://www.ebi.ac.uk/clustalw/index.html
- DeepView, http://www.expasy.org/spdbv/
- GeneDoc, http://www.psc.edu/biomed/genedoc
- GROMOS Home Page, http://igc.ethz.ch/gromos/ (for the program GROMOS96)
- International HapMap Project, http://www.hapmap.org/cgi-perl/gbrowse/gbrowse/hapmap
- MODELLER, http://salilab.org/modeller/modeller.html
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/ (for human POR gene sequence [accession number NC_000007], human POR cDNA sequence [accession number NM_000941.1], human POR protein sequence [accession number NP_000932.1], rat POR protein sequence [accession number NP_113764.1], Xenopus POR protein sequence [accession number AAH59318.1], yeast POR protein sequence [accession number NP_596046.1], Drosophila POR protein sequence [accession number NP_477158.1], mRNA, and SNPs)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ABS) [PubMed]
- Persistence of Vision Raytracer, http://www.povray.org/ (for the program POVRAY)
- Softberry, http://www.softberry.com/berry.phtml?topic=fsplice&group=programs&subgroup=gfind (for the program FSPLICE)
- Softgenetics, http://www.softgenetics.com (for the program Mutation Surveyor)
- University of California Santa Cruz (UCSC) Genome Browser, http://genome.ucsc.edu/ (for SNP information)
References
- Adachi M, Tachibana K, Asakura Y, Yamamoto T, Hanaki K, Oka A (2004) Compound heterozygous mutations of cytochrome P450 oxidoreductase gene (POR) in two patients with Antley-Bixler syndrome. Am J Med Genet A 128A:333–339 [DOI] [PubMed] [Google Scholar]
- Aleck KA, Bartley DL (1997) Multiple malformation syndrome following fluconazole use in pregnancy: report of an additional patient. Am J Med Genet 72:253–256 [PubMed] [Google Scholar]
- Antley R, Bixler D (1975) Trapezoidocephaly, midfacial hypoplasia and cartilage abnormalities with multiple synostoses and skeletal fractures. Birth Defects Orig Artic Ser 11:397–401 [PubMed] [Google Scholar]
- Arlt W, Auchus RJ, Miller WL (2001) Thiazolidinediones but not metformin directly inhibit the steroidogenic enzymes P450c17 and 3β-hydroxysteroid dehydrogenase. J Biol Chem 276:16767–16771 [DOI] [PubMed] [Google Scholar]
- Arlt W, Walker EA, Draper N, Ivison HE, Ride JP, Hammer F, Chalder SM, Borucka-Mankiewicz M, Hauffa BP, Malunowicz EM, Stewart PM, Shackleton CH (2004) Congenital adrenal hyperplasia caused by mutant P450 oxidoreductase and human androgen synthesis: analytical study. Lancet 363:2128–2135 [DOI] [PubMed] [Google Scholar]
- Auchus RJ (2004) The backdoor pathway to dihydrotestosterone. Trends Endocrinol Metab 15:432–438 [DOI] [PubMed] [Google Scholar]
- Auchus RJ, Lee TC, Miller WL (1998) Cytochrome b5 augments the 17,20-lyase activity of human P450c17 without direct electron transfer. J Biol Chem 273:3158–3165 [DOI] [PubMed] [Google Scholar]
- Auchus RJ, Miller WL (1999) Molecular modeling of human P450c17 (17α-hydroxylase/17,20-lyase): insights into reaction mechanisms and effects of mutations. Mol Endocrinol 13:1169–1182 [DOI] [PubMed] [Google Scholar]
- Augarten A, Pariente C, Gazit E, Chayen R, Goldfarb H, Sack J (1992) Ambiguous genitalia due to partial activity of cytochromes P450c17 and P450c21. J Steroid Biochem Mol Biol 41:37–41 [DOI] [PubMed] [Google Scholar]
- Bredt DS, Hwang PM, Glatt CE, Lowenstein C, Reed RR, Snyder SH (1991) Cloned and expressed nitric oxide synthase structurally resembles cytochrome P-450 reductase. Nature 351:714–718 [DOI] [PubMed] [Google Scholar]
- Chun K, Siegel-Bartelt J, Chitayat D, Phillips J, Ray PN (1998) FGFR2 mutation associated with clinical manifestations consistent with Antley-Bixler syndrome. Am J Med Genet 77:219–224 [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A (1995) Analysis of enzyme kinetic data. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- Crisponi G, Porcu C, Piu ME (1997) Antley-Bixler syndrome: case report and review of the literature. Clin Dysmorphol 6:61–68 [PubMed] [Google Scholar]
- Davydov DR, Kariakin AA, Petushkova NA, Peterson JA (2000) Association of cytochromes P450 with their reductases: opposite sign of the electrostatic interactions in P450BM-3 as compared with the microsomal 2B4 system. Biochemistry 39:6489–6497 [DOI] [PubMed] [Google Scholar]
- DeLozier CD, Antley RM, Williams R, Green N, Heller RM, Bixler D, Engel E (1980) The syndrome of multisynostotic osteodysgenesis with long-bone fractures. Am J Med Genet 7:391–403 [DOI] [PubMed] [Google Scholar]
- Dierks EA, Davis SC, Ortiz de Montellano PR (1998) Glu-320 and Asp-323 are determinants of the CYP4A1 hydroxylation regiospecificity and resistance to inactivation by 1-aminobenzotriazole. Biochemistry 37:1839–1847 [DOI] [PubMed] [Google Scholar]
- Estabrook RW, Shet MS, Fisher CW, Jenkins CM, Waterman MR (1996) The interaction of NADPH-P450 reductase with P450: an electrochemical study of the role of the flavin mononucleotide-binding domain. Arch Biochem Biophys 333:308–315 [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Horowitz MC, Locklin R, Morriss-Kay GM, Lonai P (2004) A gain-of-function mutation of Fgfr2c demonstrates the roles of this receptor variant in osteogenesis. Proc Natl Acad Sci USA 101:12555–12560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A, Do RK, Sali A (2000) Modeling of loops in protein structures. Protein Sci 9:1753–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CW, Shet MS, Estabrook RW (1996) Construction of plasmids and expression in Escherichia coli of enzymatically active fusion proteins containing the heme-domain of a P450 linked to NADPH-P450 reductase. Methods Enzymol 272:15–25 [DOI] [PubMed] [Google Scholar]
- Flück CE, Tajima T, Pandey AV, Arlt W, Okuhara K, Verge CF, Jabs EW, Mendonca BB, Fujieda K, Miller WL (2004) Mutant P450 oxidoreductase causes disordered steroidogenesis with and without Antley-Bixler syndrome. Nat Genet 36:228–230 [DOI] [PubMed] [Google Scholar]
- Fukami M, Horikawa R, Nagai T, Tanaka T, Naiki Y, Sato N, Okuyama T, Nakai H, Soneda S, Tachibana K, Matsuo N, Sato S, Homma K, Nishimura G, Hasegawa T, Ogata T (2005) Cytochrome P450 oxidoreductase gene mutations and Antley-Bixler syndrome with abnormal genitalia and/or impaired steroidogenesis: molecular and clinical studies in 10 patients. J Clin Endocrinol Metab 90:414–426 [DOI] [PubMed] [Google Scholar]
- Geller DH, Auchus RJ, Mendonca BB, Miller WL (1997) The genetic and functional basis of isolated 17,20-lyase deficiency. Nat Genet 17:201–205 [DOI] [PubMed] [Google Scholar]
- Geller DH, Auchus RJ, Miller WL (1999) P450c17 mutations R347H and R358Q selectively disrupt 17,20-lyase activity by disrupting interactions with P450 oxidoreductase and cytochrome b5. Mol Endocrinol 13:167–175 [DOI] [PubMed] [Google Scholar]
- Gorry MC, Preston RA, White GJ, Zhang Y, Singhal VK, Losken HW, Parker MG, Nwokoro NA, Post JC, Ehrlich GD (1995) Crouzon syndrome: mutations in two spliceoforms of FGFR2 and a common point mutation shared with Jackson-Weiss syndrome. Hum Mol Genet 4:1387–1390 [DOI] [PubMed] [Google Scholar]
- Gripp KW, Stolle CA, McDonald-McGinn DM, Markowitz RI, Bartlett SP, Katowitz JA, Muenke M, Zackai EH (1998) Phenotype of the fibroblast growth factor receptor 2 Ser351Cys mutation: Pfeiffer syndrome type III. Am J Med Genet 78:356–360 [DOI] [PubMed] [Google Scholar]
- Gu J, Weng Y, Zhang QY, Cui H, Behr M, Wu L, Yang W, Zhang L, Ding X (2003) Liver-specific deletion of the NADPH-cytochrome P450 reductase gene: impact on plasma cholesterol homeostasis and the function and regulation of microsomal cytochrome P450 and heme oxygenase. J Biol Chem 278:25895–25901 [DOI] [PubMed] [Google Scholar]
- Guengerich FP (2004) Cytochrome P450: what have we learned and what are the future issues? Drug Metab Rev 36:159–197 [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 [DOI] [PubMed] [Google Scholar]
- Haniu M, Iyanagi T, Legesse K, Shively JE (1984) Structural analysis of NADPH-cytochrome P-450 reductase from porcine hepatic microsomes: sequences of proteolytic fragments, cysteine-containing peptides, and a NADPH-protected cysteine peptide. J Biol Chem 259:13703–13711 [PubMed] [Google Scholar]
- Haniu M, McManus ME, Birkett DJ, Lee TD, Shively JE (1989) Structural and functional analysis of NADPH-cytochrome P-450 reductase from human liver: complete sequence of human enzyme and NADPH-binding sites. Biochemistry 28:8639–8645 [DOI] [PubMed] [Google Scholar]
- Hanukoglu I, Gutfinger T (1989) cDNA sequence of adrenodoxin reductase: identification of NADP-binding sites in oxidoreductases. Eur J Biochem 180:479–484 [DOI] [PubMed] [Google Scholar]
- Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J (1995) Structure and function of cytochromes P450: a comparative analysis of three crystal structures. Structure 3:41–62 [DOI] [PubMed] [Google Scholar]
- Hassell S, Butler MG (1994) Antley-Bixler syndrome: report of a patient and review of literature. Clin Genet 46:372–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson CJ, Otto DM, Carrie D, Magnuson MA, McLaren AW, Rosewell I, Wolf CR (2003) Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. J Biol Chem 278:13480–13486 [DOI] [PubMed] [Google Scholar]
- Herman GE (2003) Disorders of cholesterol biosynthesis: prototypic metabolic malformation syndromes. Hum Mol Genet 12:R75–R88 [DOI] [PubMed] [Google Scholar]
- Hurley ME, White MJ, Green AJ, Kelleher J (2004) Antley-Bixler syndrome with radioulnar synostosis. Pediatr Radiol 34:148–151 [DOI] [PubMed] [Google Scholar]
- Jabs EW, Li X, Scott AF, Meyers G, Chen W, Eccles M, Mao JI, Charnas LR, Jackson CE, Jaye M (1994) Jackson-Weiss and Crouzon syndromes are allelic with mutations in fibroblast growth factor receptor 2. Nat Genet 8:275–279 [DOI] [PubMed] [Google Scholar]
- Karplus PA, Daniels MJ, Herriott JR (1991) Atomic structure of ferredoxin-NADP+ reductase: prototype for a structurally novel flavoenzyme family. Science 251:60–66 [PubMed] [Google Scholar]
- Kelley RI, Kratz LE, Glaser RL, Netzloff ML, Wolf LM, Jabs EW (2002) Abnormal sterol metabolism in a patient with Antley-Bixler syndrome and ambiguous genitalia. Am J Med Genet 110:95–102 [DOI] [PubMed] [Google Scholar]
- Kondo S, Sakaki T, Ohkawa H, Inouye K (1999) Electrostatic interaction between cytochrome P450 and NADPH-P450 reductase: comparison of mixed and fused systems consisting of rat cytochrome P450 1A1 and yeast NADPH-P450 reductase. Biochem Biophys Res Commun 257:273–278 [DOI] [PubMed] [Google Scholar]
- Kress W, Petersen B, Collmann H, Grimm T (2000) An unusual FGFR1 mutation (fibroblast growth factor receptor 1 mutation) in a girl with non-syndromic trigonocephaly. Cytogenet Cell Genet 91:138–140 [DOI] [PubMed] [Google Scholar]
- Kumar D, Masel JP (1997) A new multiple malformation syndrome of Mullerian dysgenesis and conductive hearing loss with facial hypoplasia, bilateral forearm deformity, brachydactyly, spinal stenosis and scoliosis. Clin Genet 52:30–36 [DOI] [PubMed] [Google Scholar]
- Lin D, Black SM, Nagahama Y, Miller WL (1993) Steroid 17α-hydroxylase and 17,20-lyase activities of P450c17: contributions of serine106 and P450 reductase. Endocrinology 132:2498–2506 [DOI] [PubMed] [Google Scholar]
- Lin D, Harikrishna JA, Moore CCD, Jones KL, Miller WL (1991) Missense mutation serine106 proline causes 17 α-hydroxylase deficiency. J Biol Chem 266:15992–15998 [PubMed] [Google Scholar]
- Lu AY, Junk KW, Coon MJ (1969) Resolution of the cytochrome P-450-containing ω-hydroxylation system of liver microsomes into three components. J Biol Chem 244:3714–3721 [PubMed] [Google Scholar]
- Mann RK, Beachy PA (2004) Novel lipid modifications of secreted protein signals. Annu Rev Biochem 73:891–923 [DOI] [PubMed] [Google Scholar]
- Meyers GA, Day D, Goldberg R, Daentl DL, Przylepa KA, Abrams LJ, Graham JM Jr, Feingold M, Moeschler JB, Rawnsley E, Scott AF, Jabs EW (1996) FGFR2 exon IIIa and IIIc mutations in Crouzon, Jackson-Weiss, and Pfeiffer syndromes: evidence for missense changes, insertions, and a deletion due to alternative RNA splicing. Am J Hum Genet 58:491–498 [PMC free article] [PubMed] [Google Scholar]
- Miller WL (1986) Congenital adrenal hyperplasia. N Engl J Med 314:1321–1322 [PubMed] [Google Scholar]
- ——— (1994) Clinical review 54: genetics, diagnosis, and management of 21-hydroxylase deficiency. J Clin Endocrinol Metab 78:241–246 [DOI] [PubMed] [Google Scholar]
- ——— (2004) P450 oxidoreductase deficiency: a new disorder of steroidogenesis with multiple clinical manifestations. Trends Endocrinol Metab 15:311–315 [DOI] [PubMed] [Google Scholar]
- Miroux B, Walker JE (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol 260:289–298 [DOI] [PubMed] [Google Scholar]
- Muenke M, Schell U, Hehr A, Robin NH, Losken HW, Schinzel A, Pulleyn LJ, Rutland P, Reardon W, Malcolm S, Winter RM (1994) A common mutation in the fibroblast growth factor receptor 1 gene in Pfeiffer syndrome. Nat Genet 8:269–274 [DOI] [PubMed] [Google Scholar]
- Mulrooney SB, Waskell L (2000) High-level expression in Escherichia coli and purification of the membrane-bound form of cytochrome b(5). Protein Expr Purif 19:173–178 [DOI] [PubMed] [Google Scholar]
- Nadler SG, Strobel HW (1988) Role of electrostatic interactions in the reaction of NADPH-cytochrome P-450 reductase with cytochromes P-450. Arch Biochem Biophys 261:418–429 [DOI] [PubMed] [Google Scholar]
- ——— (1991) Identification and characterization of an NADPH-cytochrome P450 reductase derived peptide involved in binding to cytochrome P450. Arch Biochem Biophys 290:277–284 [DOI] [PubMed] [Google Scholar]
- Okajima K, Robinson LK, Hart MA, Abuelo DN, Cowan LS, Hasegawa T, Maumenee IH, Jabs EW (1999) Ocular anterior chamber dysgenesis in craniosynostosis syndromes with a fibroblast growth factor receptor 2 mutation. Am J Med Genet 85:160–170 [DOI] [PubMed] [Google Scholar]
- Omura T, Sato R (1964) The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239:2370–2378 [PubMed] [Google Scholar]
- Ono T, Bloch K (1975) Solubilization and partial characterization of rat liver squalene epoxidase. J Biol Chem 250:1571–1579 [PubMed] [Google Scholar]
- Otto DM, Henderson CJ, Carrie D, Davey M, Gundersen TE, Blomhoff R, Adams RH, Tickle C, Wolf CR (2003) Identification of novel roles of the cytochrome P450 system in early embryogenesis: effects on vasculogenesis and retinoic acid homeostasis. Mol Cell Biol 23:6103–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey AV, Mellon SH, Miller WL (2003) Protein phosphatase 2A and phosphoprotein SET regulate androgen production by P450c17. J Biol Chem 278:2837–2844 [DOI] [PubMed] [Google Scholar]
- Park WJ, Meyers GA, Li X, Theda C, Day D, Orlow SJ, Jones MC, Jabs EW (1995a) Novel FGFR2 mutations in Crouzon and Jackson-Weiss syndromes show allelic heterogeneity and phenotypic variability. Hum Mol Genet 4:1229–1233 [DOI] [PubMed] [Google Scholar]
- Park WJ, Theda C, Maestri NE, Meyers GA, Fryburg JS, Dufresne C, Cohen MM Jr, Jabs EW (1995b) Analysis of phenotypic features and FGFR2 mutations in Apert syndrome. Am J Hum Genet 57:321–328 [PMC free article] [PubMed] [Google Scholar]
- Peterson RE, Imperato-McGinley J, Gautier T, Shackleton CHL (1985) Male pseudohermaphroditism due to multiple defects in steroid-biosynthetic microsomal mixed-function oxidases: a new variant of congenital adrenal hyperplasia. N Engl J Med 313:1182–1191 [DOI] [PubMed] [Google Scholar]
- Pompon D, Perret A, Bellamine A, Laine R, Gautier JC, Urban P (1995) Genetically engineered yeast cells and their applications. Toxicol Lett 82–83:815–822 [DOI] [PubMed] [Google Scholar]
- Przylepa KA, Paznekas W, Zhang M, Golabi M, Bias W, Bamshad MJ, Carey JC, Hall BD, Stevenson R, Orlow S, Cohen MM Jr, Jabs EW (1996) Fibroblast growth factor receptor 2 mutations in Beare-Stevenson cutis gyrata syndrome. Nat Genet 13:492–494 [DOI] [PubMed] [Google Scholar]
- Pulleyn LJ, Reardon W, Wilkes D, Rutland P, Jones BM, Hayward R, Hall CM, Brueton L, Chun N, Lammer E, Malcolm S, Winter RM (1996) Spectrum of craniosynostosis phenotypes associated with novel mutations at the fibroblast growth factor receptor 2 locus. Eur J Hum Genet 4:283–291 [DOI] [PubMed] [Google Scholar]
- Pursley TJ, Blomquist IK, Abraham J, Andersen HF, Bartley JA (1996) Fluconazole-induced congenital anomalies in three infants. Clin Infect Dis 22:336–340 [DOI] [PubMed] [Google Scholar]
- Reardon W, Smith A, Honour JW, Hindmarsh P, Das D, Rumsby G, Nelson I, Malcolm S, Ades L, Sillence D, Kumar D, DeLozier-Blanchet C, McKee S, Kelly T, McKeehan WL, Baraitser M, Winter RM (2000) Evidence for digenic inheritance in some cases of Antley-Bixler syndrome? J Med Genet 37:26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon W, Winter RM, Rutland P, Pulleyn LJ, Jones BM, Malcolm S (1994) Mutations in the fibroblast growth factor receptor 2 gene cause Crouzon syndrome. Nat Genet 8:98–103 [DOI] [PubMed] [Google Scholar]
- Roman LJ, McLain J, Masters BSS (2003) Chimeric enzymes of cytochrome P450 oxidoreductase and neuronal nitric-oxide synthase reductase domain reveal structural and functional differences. J Biol Chem 278:25700–25707 [DOI] [PubMed] [Google Scholar]
- Roth C, Hinney B, Peter M, Steinberger D, Lakomek M (2000) Features of Antley-Bixler syndrome in an infant born to a mother with pregnancy luteoma. Eur J Pediatr 159:189–192 [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815 [DOI] [PubMed] [Google Scholar]
- Schell U, Hehr A, Feldman GJ, Robin NH, Zackai EH, de Die-Smulders C, Viskochil DH, Stewart JM, Wolff G, Ohashi H, Price RA, Cohen MM, Muenke M (1995) Mutations in FGFR1 and FGFR2 cause familial and sporadic Pfeiffer syndrome. Hum Mol Genet 4:323–328 [DOI] [PubMed] [Google Scholar]
- Scrutton NS, Berry A, Perham RN (1990) Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature 343:38–43 [DOI] [PubMed] [Google Scholar]
- Shen AL, Christensen MJ, Kasper CB (1991) NADPH-cytochrome P-450 oxidoreductase: the role of cysteine 566 in catalysis and cofactor binding. J Biol Chem 266:19976–19980 [PubMed] [Google Scholar]
- Shen AL, Kasper CB (1995) Role of acidic residues in the interaction of NADPH-cytochrome P450 oxidoreductase with cytochrome P450 and cytochrome c. J Biol Chem 270:27475–27480 [DOI] [PubMed] [Google Scholar]
- ——— (1996) Role of Ser457 of NADPH-cytochrome P450 oxidoreductase in catalysis and control of FAD oxidation-reduction potential. Biochemistry 35:9451–9459 [DOI] [PubMed] [Google Scholar]
- ——— (2000) Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding and catalysis. J Biol Chem 275:41087–41091 [DOI] [PubMed] [Google Scholar]
- Shen AL, O’Leary KA, Kasper CB (2002) Association of multiple developmental defects and embryonic lethality with loss of microsomal NADPH-cytochrome P450 oxidoreductase. J Biol Chem 277:6536–6541 [DOI] [PubMed] [Google Scholar]
- Shen AL, Porter TD, Wilson TE, Kasper CB (1989) Structural analysis of the FMN binding domain of NADPH-cytochrome P-450 oxidoreductase by site-directed mutagenesis. J Biol Chem 264:7584–7589 [PubMed] [Google Scholar]
- Shen AL, Sem DS, Kasper CB (1999) Mechanistic studies on the reductive half-reaction of NADPH-cytochrome P450 oxidoreductase. J Biol Chem 274:5391–5398 [DOI] [PubMed] [Google Scholar]
- Strobel HW, Nadler SG, Nelson DR (1989) Cytochrome P-450: cytochrome P-450 reductase interactions. Drug Metab Rev 20:519–533 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van GB, Slater EC (1962) The extinction coefficient of cytochrome c. Biochim Biophys Acta 58:593–595 [DOI] [PubMed] [Google Scholar]
- von Heijne G (1984) Analysis of the distribution of charged residues in the N-terminal region of signal sequences: implications for protein export in prokaryotic and eukaryotic cells. EMBO J 3:2315–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Roberts DL, Paschke R, Shea TM, Masters BSS, Kim JJ (1997) Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci USA 94:8411–8416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmann S, Roth C, Glüer S, Fuchs J (2000) Congenital adrenal hyperplasia associated with maternal pregnancy luteoma and the Antley-Bixler syndrome. J Pediatr Surg 35:528–530 [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Slaney SF, Oldridge M, Poole MD, Ashworth GJ, Hockley AD, Hayward RD, David DJ, Pulleyn LJ, Rutland P, Malcolm S, Winter RM, Reardon W (1995) Apert syndrome results from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nat Genet 9:165–172 [DOI] [PubMed] [Google Scholar]
- Wu L, Gu J, Cui H, Zhang QY, Behr M, Fang C, Weng Y, Kluetzman K, Swiatek PJ, Yang W, Kaminsky L, Ding X (2005) Transgenic mice with a hypomorphic NADPH-cytochrome P450 reductase gene: effects on development, reproduction, and microsomal cytochrome P450. J Pharmacol Exp Ther 312:35–43 [DOI] [PubMed] [Google Scholar]
- Wu L, Gu J, Weng Y, Kluetzman K, Swiatek P, Behr M, Zhang QY, Zhuo X, Xie Q, Ding X (2003) Conditional knockout of the mouse NADPH-cytochrome P450 reductase gene. Genesis 36:177–181 [DOI] [PubMed] [Google Scholar]
- Yanagibashi K, Hall PF (1986) Role of electron transport in the regulation of the lyase activity of C21 side-chain cleavage P-450 from porcine adrenal and testicular microsomes. J Biol Chem 261:8429–8433 [PubMed] [Google Scholar]
- Zhang LH, Rodriguez H, Ohno S, Miller WL (1995) Serine phosphorylation of human P450c17 increases 17,20-lyase activity: implications for adrenarche and the polycystic ovary syndrome. Proc Natl Acad Sci USA 92:10619–10623 [DOI] [PMC free article] [PubMed] [Google Scholar]