Abstract
The development of high-throughput screening methods such as array-based comparative genome hybridization (array CGH) allows screening of the human genome for copy-number changes. Current array CGH strategies have limits of resolution that make detection of small (less than a few tens of kilobases) gains or losses of genomic DNA difficult to identify. We report here a significant improvement in the resolution of array CGH, with the development of an array platform that utilizes single-stranded DNA array elements to accurately measure copy-number changes of individual exons in the human genome. Using this technology, we screened 31 patient samples across an array containing a total of 162 exons for five disease genes and detected copy-number changes, ranging from whole-gene deletions and duplications to single-exon deletions and duplications, in 100% of the cases. Our data demonstrate that it is possible to screen the human genome for copy-number changes with array CGH at a resolution that is 2 orders of magnitude higher than that previously reported.
Introduction
The extent to which genomic copy-number polymorphisms (CNPs) contribute to human genetic diversity is not known. Recent studies have demonstrated the presence of CNPs, a proportion of which encompass genes, in the genomes of normal individuals (Iafrate et al. 2004; Sebat et al. 2004). This suggests that these variants may be important in our understanding of phenotypic variation or may predispose to or directly cause disease. On the basis of current knowledge, ∼5%–6% of gene mutations that are causative of inherited disorders are copy-number changes defined as gross deletions or duplications (Armour et al. 2002), although this frequency may represent an underestimate. These disease-causing copy-number changes can range in size from 100 bp to several megabases and can encompass as little as a single exon and as much as entire genes or several genes.
The ability to detect CNPs and pathogenic chromosomal imbalances has been dramatically improved through the use of array-based comparative genome hybridization (array CGH) (Solinas-Toldo et al. 1997; Pinkel et al. 1998). Array CGH offers high sensitivity and dynamic range to quantitatively measure from single copy-number losses or gains to high copy-number amplifications, as well as sufficient resolution and scalability for complete genomewide scans (Snijders et al. 2001; Fiegler et al. 2003; Vissers et al. 2003; Iafrate et al. 2004; Inazawa et al. 2004; Ishkanian et al. 2004). Currently, the most robust array CGH approaches utilize large genomic clone inserts as array elements but have a maximum resolution of ∼40–50 kb (Albertson and Pinkel 2003; Snijders et al. 2003; Mantripragada et al. 2004a). The use of oligonucleotides (Lucito et al. 2000, 2003; Bignell et al. 2004; Carvalho et al. 2004; Sebat et al. 2004), cDNA clones (Pollack et al. 1999, 2002; Heiskanen et al. 2000), or sequence-defined PCR products (Mantripragada et al. 2003, 2004b) for array CGH allow this resolution to be theoretically increased solely on the basis of the size of the genomic region covered by the array element. However, because of limitations of the technologies, the effective resolution for these methods is no greater than 15–30 kb, since they rely on averaging measurements taken from multiple array elements, pooling PCR products to be spotted as single array elements, or reducing the complexity of the human genome to detect and quantitate copy-number changes. Therefore, it has not yet been demonstrated that array elements covering small regions of the genome can deliver substantially higher resolution than that of large genomic clone array CGH to identify small genomic deletions and duplications (100 bp to 15 kb).
The ability to detect copy-number changes encompassing such small regions of the genome has thus far been achievable only by using other types of molecular assays. Apart from the more traditional approaches of Southern analysis, FISH, and quantitative PCR (Armour et al. 2002), more-recent approaches have included the development of multiplex amplifiable probe hybridization (MAPH) (Armour et al. 2000; Sellner and Taylor 2004) and the multiplex ligation-dependent probe amplification (MLPA) (Schouten et al. 2002; Sellner and Taylor 2004). For MAPH and MLPA, the resolution is at the level of individual exons, and they are becoming widely used as research and diagnostic tools, with very high reliability for identifying and quantitating copy-number changes in human disease genes (White et al. 2002, 2003, 2004; Akrami et al. 2003; Sellner and Taylor 2004). However, because both methods rely on multiplexing, the number of measurements obtainable from a single assay is limited. Therefore, the development of approaches that provide robust measurement precision of copy-number changes, scalability, and very high resolution have thus far been unavailable in the field of human molecular genetics.
We describe here an array CGH–based approach that addresses all of these issues for the analysis of copy-number changes in the human genome. We have developed an array platform that allows single strands of DNA derived from double-stranded PCR products to be retained on the surface of a slide through the use of 5′-aminolink chemistry. This platform improves the signal:noise ratio, such that it is possible to detect individual exons in the human genome and to quantify their copy number accurately. We refer to this method as “exon array CGH.” To demonstrate the utility of the approach, we constructed an array containing 162 exons that collectively span five human genes (COL4A5 [MIM 303630], DMD [MIM 300377], NF2 [MIM 607379], PLP1 [MIM 300401], and PMP22 [MIM 601097]) involved in inherited genetic disorders (see table 1). We have analyzed a series of 31 DNA samples from patients affected with these disorders and have characterized copy-number changes that have been validated with other molecular methods for all 31 samples. Our method is 2 orders of magnitude more sensitive than other forms of array CGH and provides resolution and accuracy that are similar to other current methods of screening genes for copy-number changes at the level of the exon. Since exon array CGH is completely scalable, this new molecular tool will provide the means for researchers to screen the human genome at high resolution to identify and annotate novel CNPs and causative mutations in normal and disease states, respectively, as well as to facilitate array-based applications to study other aspects of genome biology.
Table 1.
Human Disease Genes Represented on the Exon Array[Note]
| Gene | ChromosomalLocation | GenomicSize(kb) | No. ofExonsa | Disease | Frequencyin Population | Pathogenic Copy-Number Changesb | Technical Consideration |
| DMD | Xp21.1 | 2,400 | 79 (+1) | Duchenne muscular dystrophy (DMD [MIM 310200])/Becker muscular dystrophy (MIM 300376) | 1/3,500c | 60–70d | Complex gene for diagnostic screening |
| NF2 | 22q12 | 95 | 17 | Neurofibromatosis type 2 (NF2 [MIM 101000]) | 1/40,000e | 20–30e | High repeat content results in quantitation issues for array CGHf |
| COL4A5 | Xq22.3 | 257.6 | 51 (+2) | Alports syndrome (AS [MIM 301050]) | 1/5,000g | 10–15h | Specificity issues, since it is a member of a six-gene triple helical collagen family |
| PMP22 | 17p12 | 35.6 | 5 | Charcot-Marie Tooth type 1 disease (CMT1A [MIM 118220]); hereditary neuropathy with liability to pressure palsy (HNPP [MIM 162500]) | 10/100,000–40/100,000i | 70 dupl (for CMT1A);84 del (for HNPP)j | |
| PLP1 | Xq22.2 | 15.8 | 7 | Pelizaeus-Merzbacher disease (PMD [MIM 312080]) | Extremely rarek | 60–70l |
Note.— Genomic characteristics and clinical/technical relevance for inclusion on the array of the five human disease genes are shown.
The number of exons shown in parentheses represents alternative exons found in some transcripts that were included as array elements.
As a percentage of all mutations.
DMD is the most common inherited neuromuscular disorder (Worton and Thompson 1988).
Bruder et al. 2001.
Mantripragada et al. 2003.
Atkin et al. 1988.
Rautenstrauss et al. 2000.
Nelis et al. 1996.
Heim et al. 1997.
Material and Methods
PCR Amplification and Microarray Fabrication
Primers pairs for exons of the COL4A5, DMD, NF2, PLP1, and PMP22 genes (table 2); for amplicons covering the 3′ UTRs of genes on chromosome 22 (normalization controls); and for other amplicons used here were obtained from primer sets published elsewhere (Strautnieks et al. 1992; Plant et al. 1999; Leiden University Medical Center Web site) or were designed using the relevant genomic sequence and the Primer3 software and Web site (Rozen and Skaletsky 2000; Whitehead Institute). All amplicons used in the present study were repeat free. When possible, primer pairs were designed or obtained for each exon and, ideally, were located in the upstream and downstream intronic sequences flanking the exon. In a few instances, however, primers were designed within the exonic sequence. When two adjacent exons were separated only by a very small intron, a single pair of primers was designed to contain both exons. Primer pairs and all amplicons sequences were compared with the entire human genome sequence by use of e-PCR (Schuler 1997) and BLASTN to identify any potential cross-reacting DNA sequences. In cases in which the PCR product for a given exon did not accurately or reproducibly report copy-number values, additional PCR assays were designed and tested on the array.
Table 2.
Primer Pairs Used to Amplify Exons for Array CGH
|
Primer(5′→3′) |
||||
| Amplicon Namea | 1(Forward) | 2(Reverse) | Sizeb(bp) | Genome Location |
| COL4A5-01 | CTTCCACTCTTAAAAAGCTTC | GGAGGGGAGGAAGGACTTAC | 258 | X:106455055–106455313 |
| COL4A5-01ver2 | CAAGCCTCACTGTCCCTCTC | AAGGACTTACCGCAGCCTCT | 215 | X:106455088–106455303 |
| COL4A5-02 | TGGTGTAGCTTTCCATCTTTAGC | TGCCAATCTAAGACCTCACC | 391 | X:106554520–106554911 |
| COL4A5-03 | TCTCAACCATGCCTGTGCTT | GATGTGACACCTAGTCCCAC | 228 | X:106574085–106574313 |
| COL4A5-03ver2 | AGGGAGAGAGAGGGTTTCCA | CCCGTCATTCCATTGTTTCA | 228 | X:106574148–106574376 |
| COL4A5-04 | TCACAGATGTTTACAGTAGT | GGTCTTTTCCAATTGTCTCA | 237 | X:106578864–106579101 |
| COL4A5-04ver2 | TGTCAGCGATTTCCTTCTCA | GAATTCCATCATCACCCTGAA | 244 | X:106578740–106578984 |
| COL4A5-05/06 | TGGGTTGTCATTTAGTTTAAGGAT | ATGGATGGATCTCACCTTGG | 249 | X:106583674–106583923 |
| COL4A5-07 | GGAAAGTGAAGGCTAATGAA | GCATTGGGCTCTCTCACTAC | 278 | X:106586353–106586631 |
| COL4A5-08 | GAAGTTGGTAATATAGCTTTCTCC | AAGACAGAAGTTTGAGTCATTATCC | 323 | X:106586692–106587015 |
| COL4A5-09 | TTTATTACTGAGATGGCCTAATC | GAGGGATTGTTGTAATCTTCTGG | 278 | X:106588528–106588806 |
| COL4A5-10 | CTGGGCGACACAAGTGAGA | TTAAAGAAGAGAGCTTACCATC | 181 | X:106590896–106591077 |
| COL4A5-10ver2 | CCTACCTGGTCCCACTGGTAT | TCTGAAATGGCCAGAATTGA | 352 | X:106590998–106591350 |
| COL4A5-11/12 | ACCAGGACTTCCAGGACCTAA | AAGAGAAAGCTGAGGGCTGA | 200 | X:106593052–106593252 |
| COL4A5-13 | TTCAGCCCTCAGCTTTCTC | TCCCTACTTACTCACCTGATCTCC | 254 | X:106593231–106593485 |
| COL4A5-14/15 | GATTTCCTTTCCCCTACTACTG | GAATATTAGCAGTTACATCAC | 258 | X:106595592–106595850 |
| COL4A5-16 | GTCCAAGCCAGGAATAAAGC | GCTTCTCTGCCACCTTATGTTATAC | 219 | X:106595972–106596191 |
| COL4A5-17 | TGTGCTGATGTCACCCTATCC | TTTCTGCAACATGGACTGTG | 302 | X:106597935–106598237 |
| COL4A5-18 | AGACACCATCACAGGTTAGGC | AAGTAGGGAGTATGCGATTAAGG | 304 | X:106599391–106599695 |
| COL4A5-18ma | CTCTTATATTCTTGAGGAAATTG | GCGATTAAGGCACAAAAATG | 184 | X:106599498–106599682 |
| COL4A5-19 | TTTTCTTTGGTAATAAAGG | AAGGCCATAAATGCAATCTC | 177 | X:106601683–106601860 |
| COL4A5-20 | TCTTGCTAATGACTTTCCGGTAT | CAACAAATGAGGCCAACCTT | 567 | X:106605843–106606410 |
| COL4A5-21 | TCTGGCTTGTCAGGCTTTC | GTTTGGAGATCAAACCTTTCAC | 179 | X:106606567–106606746 |
| COL4A5-22 | GTTCTTGGCACACAGTAAGC | GAGGCATAATTTACCTGGAGGAC | 290 | X:106610412–106610702 |
| COL4A5-23 | TGTGTTAGGATCTCTTGGTTTCC | CCCTTCAACCCATCCAATAAC | 286 | X:106612076–106612362 |
| COL4A5-24 | CTTTTTTTCCTTACTCATTTC | AACCAAAAATATCAAACCAAC | 240 | X:106612440–106612680 |
| COL4A5-24ver2 | TTCCAGGAATGAAGGGTGAC | AGCATCAGTCCCATCCTTTG | 211 | X:106612533–106612744 |
| COL4A5-24ver3 | CTCATTTCAGGGCATTCCAG | CAGCATCAGTCCCATCCTTT | 292 | X:106612453–106612745 |
| COL4A5-25 | ATATGTTTCTGTATTAAAC | TAAGCACCAAGTTTAAAAC | 239 | X:106613755–106613994 |
| COL4A5-26 | CATCTTGAATCATGGAAGTCCT | ACCATCTCTGCCTGGGTTAC | 311 | X:106616245–106616556 |
| COL4A5-27 | TGGCCCTGATGGCTTCTTTC | TGCTACCCATTCCAACATACC | 185 | X:106616911–106617096 |
| COL4A5-27ver2 | TCCATAGGTGACCCTGGACT | CATGCTAAACTTCTAACCTACTCTGC | 159 | X:106616964–106617123 |
| COL4A5-28 | AATGTGGGTTTGGGAATTTG | GGCTTGGGATGATATCTGAGT | 273 | X:106617971–106618244 |
| COL4A5-29 | TTTGTCATGTGTATGCTC | AGCAAGTTGAGATGCAGTGACAG | 204 | X:106621807–106622011 |
| COL4A5-29ver2 | CCAGGACTTCCAGGTTTCAA | AACAGAATGGCAGCATAGGG | 230 | X:106621867–106622097 |
| COL4A5-30 | TTTTCTTGCTGAATGAATGC | CACTTTATTGATGAGCTAAC | 301 | X:106629880–106630181 |
| COL4A5-31 | ATTGATATTGTATTAACTAG | GAAAACTTTAAACAAATTAC | 208 | X:106635325–106635533 |
| COL4A5-32(ma) | ATAGTTTTCTGGTTGACATC | ATTCTGTACTGACATAAAGC | 250 | X:106636801–106637051 |
| COL4A5-33 | TTGTGTTTTCACACACATTG | CATAAATAAATTCATTCACTC | 200 | X:106637735–106637935 |
| COL4A5-33ver2 | TAGGTGATGATGGCTTGCAG | GCCTTTGTTGGTGAATTCTG | 258 | X:106637759–106638017 |
| COL4A5-34 | TTCGTGTGAGTCCAGTGCTAA | TACCTGGTGGTCCTGGTAATC | 260 | X:106639164–106639424 |
| COL4A5-35 | CCATTCCCATGAAACCAGAC | TGATTCTATCAACACCAGCCTC | 258 | X:106640721–106640979 |
| COL4A5-36 | ATATATCACATATTTTCAACAG | CCTAAAACTATATGCCAAAG | 187 | X:106641274–106641461 |
| COL4A5-37 | TTCAGGGTGAGCCTGGTCT | AGCTGCTCTGTGATACTGGTTG | 203 | X:106670412–106670615 |
| COL4A5-38 | CCAACAAACACCAGCATCTTT | GCTGAACATGATTTGACTTTCC | 303 | X:106680454–106680757 |
| COL4A5-39 | AAACTGGGTGTAACCTGCTG | GGGAAAGTGTGTGGTAGCTTAG | 255 | X:106681547–106681802 |
| COL4A5-40 | TTTGGCCTTGTTTCAGTTTG | ACACTAGGTTTGAGTTTTCTGGGTTC | 250 | X:106682152–106682402 |
| COL4A5-41 | ATCTTCTAATTATACTTTACTTTC | CATTCTCCTACCACTCACGG | 240 | X:106683377–106683617 |
| COL4A5-41A | AATGATGACATTGGGCCTTG | CTTTCCAATAGACAGAAACACTGG | 234 | X:106685272–106685506 |
| COL4A5-41B | AAGAAATGCTGGCAAAGAAC | TTGAGGATAACAGCACTCTCC | 273 | X:106689688–106689961 |
| COL4A5-41ver2 | TGGGTAGATTTGGGATTTGG | GGTTGCCTTTAGGTCCTTCC | 298 | X:106683263–106683561 |
| COL4A5-42 | AATGTCGTCATTTGCTGTGGATTA | CATCAGATATCTACTTCCATTTCC | 190 | X:106692557–106692747 |
| COL4A5-43/44 | GTAACATTAATGATTTTATTTATT | TATAACTATCTTCAGGAATAAGTC | 333 | X:106695738–106696071 |
| COL4A5-45 | GGTCACTTTGGCTTCCATTTC | GGTAAGCCTTGAGGTCCTTG | 195 | X:106696778–106696973 |
| COL4A5-46 | GCATCCCAGAAGCAAATGGA | AGACTTTCCCTCCACCAACAG | 347 | X:106700909–106701256 |
| COL4A5-47 | GTATACTGATTATTTCGTGG | GGAAATTAGATATTGATTATC | 264 | X:106702542–106702806 |
| COL4A5-48 | CTTTACTGTTTTCTCTCCAAATCT | AAAAGTCACAGCTAAATCAATGCC | 236 | X:106707804–106708040 |
| COL4A5-49 | ATTATGTTCCTTCTCCTTTTCCTT | GACAAATGCAAGGAAGAGTG | 173 | X:106709863–106710036 |
| COL4A5-50 | GCGGCACATTTTTCCTTGTC | CTGAATTAAAGCTATAAGCAC | 227 | X:106710325–106710552 |
| COL4A5-51 | GATCTGATTGTCTTATTTCTTATT | TAAAACACAAAAGGAATTCTTCAA | 140 | X:106711353–106711493 |
| COL4A5-51ver2 | TGACCACTAATGGCTCTTAAACC | CGGCAGCAGTAGTAAAGTTGG | 367 | X:106711200–106711566 |
| DMD-01A | TCTGGCTCATGTGTTTGCTCCGAGGTATAG | CTTCCATGCCAGCTGTTTTTCCTGTCACTC | 332 | X:32718801-32719133 |
| DMD-01B | TAGACAGTGGATACATAACAAATGCATG | TTCTCCGAAGGTAATTGCCTCCCAGATCTGAGTCC | 529 | X:32591003–32591532 |
| DMD-01Zma | GAAGGCGGGTCACTTGCTTGTGCGCAG | CAATCTACCTAATTAGTGAGCTTG | 419 | X:32590744-32591163 |
| DMD-02 | CACTAACACATCATAATGG | GATACACAGGTACATAGTC | 233 | X:32399606–32399839 |
| DMD-02ma | TAATTTGGATGCCCCAAACCAGC | GTAACAAACCATTCTTACCTTAGA | 253 | X:32399664-32399916 |
| DMD-03(ma) | TCATCCGTCATCTTCGGCAGATTAA | CAGGCGGTAGAGTATGCCAAATGAAAATCA | 408 | X:32229094–32229502 |
| DMD-04 | AGTAGATTGTCGGTCTCTCTG | CCAAAGCCCTCACTCAAAC | 203 | X:32224255-32224458 |
| DMD-04ma | TTGTCGGTCTCTCTGCTGGT | CAAAGCCCTCACTCAAACATGAAGC | 196 | X:32224256–32224451 |
| DMD-04ver2 | GGAGCAGCCTATCAGGTCAG | ATCCACAAGAGTTCATGCCC | 307 | X:32224082-32224389 |
| DMD-05 | CAACTAGGCATTTGGTCTC | TTGTTTCACACGTCAAGGG | 225 | X:32202758–32202983 |
| DMD-05ma | GCAACTAGGCATTTGGTCTCTTACCT | CCATTCATCAGGATTCTTACCTGCC | 167 | X:32202817-32202984 |
| DMD-06(ma) | GTTTGCATGGTTCTTGCTCA | CTTTCACGCTCCGCTAATGT | 466 | X:32195777–32196243 |
| DMD-06ver3 | CCACATGTAGGTCAAAAATGTAATGAA | GTCTCAGTAATCTTCTTACCTATGACTATGG | 202 | X:32195991–32196193 |
| DMD-07 | GCATGGAAGTAAATCTCATGGAAC | GTGTAGAAATGACAAGTCTCAGATG | 278 | X:32188956–32189234 |
| DMD-07ma | AAGGACTATGGGCATTGGTTGTCA | TGTGTAGAAATGACAAGTCTCAGA | 314 | X:32188955–32189269 |
| DMD-08(ma) | GTCCTTTACACACTTTACCTGTTGAG | GGCCTCATTCTCATGTTCTAATTAG | 360 | X:32078636–32078996 |
| DMD-09(Zma) | CTTCTCTGCAGATCACG | CGAGGAGATAAAAGGCAC | 245 | X:32077307–32077552 |
| DMD-09ver2 | GTAGTCCTTTCGGGTTACTTATGG | AACAAACCAGCTCTTCACGAGGAGA | 321 | X:32077290–32077611 |
| DMD-10 | ATTGTGCAGCATTTGGAAGC | GTTGGAATCCCAAGCACATC | 332 | X:32024372–32024704 |
| DMD-11 | CAAATAAAACTCAAAACCACACC | CTTCCAAAACTTGTTAGTCTTC | 301 | X:32023626–32023927 |
| DMD-12 | GATAGTGGGCTTTACTTACATCCTTC | GAAAGCACGCAACATAAGATACACCT | 332 | X:31993817–31994149 |
| DMD-12ma | GATAGTGGGCTTTACTTACATCCTTC | TAATTCTCTCCCATCAACCATGTCATCT | 405 | X:31993744–31994149 |
| DMD-13(ma) | AATGGGAGTACCTGAGATGTAGCAGAAAT | CTGACCTTAAGTTGTTCTTCCAAAGCAG | 238 | X:31975294–31975532 |
| DMD-14/15 | TTTGCTGATGCTGTGCTTGATTGTC | CAATAGCATAGAAGAGACTAA | 436 | X:31952988–31953424 |
| DMD-16 | TCTATGCAAATGAGCAAATACACGC | GGTATCACTAACCTGTGCTGTACTC | 290 | X:31945232–31945522 |
| DMD-17 | TTTCGATGTTGAGATTACTTTCCC | AAGCTTAAGATGCTCTCACCTTTTCC | 414 | X:31924683:31925096 |
| DMD-17ma | GCTATTTTGATCTGAAGGTCAATCTACC | AAGCTTGAGATGCTCTCACCTTTTCC | 326 | X:31924683:31925008 |
| DMD-18 | GGAGTCTCAGATTGAGAAAAGAATG | CAAGCAGCACAAAATGAGTACAG | 279 | X:31897502–31897781 |
| DMD-18ver2 | GGCATCCCTAGTCAGTCACAG | TTCACAGCTGGATTACTCGC | 314 | X:31897337–31897651 |
| DMD-19(ma) | GATGGCAAAAGTGTTGAGAAAAAGTC | TTCTACCACATCCCATTTTCTTCCA | 459 | X:31881080–31881539 |
| DMD-20 | TGGCTTTCAGATCATTTCTTTC | AAATACCTATTGATTATGCTCC | 357 | X:31870772–31871129 |
| DMD-20ver2 | ATGCTCCAAATGGAAGGAGA | GTGGATCGAATTCTGCCAGT | 222 | X:31870787–31871009 |
| DMD-21 | GCAAAATGTAATGTATGCAAAG | ATGTTAGTACCTTCTGGATTTC | 319 | X:31864382–31864701 |
| DMD-22 | TTGACACTTTGCCACCAATG | GATAAGCGTGCTTTATTGTTTTGAC | 186 | X:31851663–31851849 |
| DMD-23 | GTTTATAACTGATAGAAGATCATC | TTTACAGTTTACAGTGTATCGTTAG | 426 | X:31847964–31848390 |
| DMD-24 | TTGGGCCTGTGTTTAGACATA | AAATCCACCCCAGCTGTAAAA | 291 | X:31844040–31844331 |
| DMD-25 | TGTGGCAGTAATTTTTTTCAG | AGGAAATCTTAGTTAAGTACG | 260 | X:31842925–31843185 |
| DMD-26 | TGTGGAAGGTCTATGCCAGA | CTTCATCTCTTCAACTGCTTTCTGT | 179 | X:31834204–31834383 |
| DMD-27 | AGAGCTAAAGAAGAGGCCCA | CACATATGACCATGTATTGACAT | 238 | X:31827943–31828181 |
| DMD-28 | GAAGTTTTAATAATGAAATGGCAAAA | TGACCTCTTTTAATACTGCATAT | 275 | X:31820665–31820940 |
| DMD-29 | CCAATGTATTTAGAAAAAAAAGGAG | GCAAATTAGATTAAAGAGATTTTTCAC | 243 | X:31817745–31817987 |
| DMD-30 | TACAGAAAAGCTATCAAGAG | AAAAACAAAAGAATGGAAGC | 261 | X:31791254–31791515 |
| DMD-30ma | AGGCTGTAAGGAGGCAAAAGTTGC | GATGTACTTGCCTGGGCTTCCTGAGGC | 175 | X:31791283–31791458 |
| DMD-31 | ACAGGTGTGAACCACCACTC | ACTGGAGTATAATGCCCAACG | 321 | X:31769557–31769878 |
| DMD-32 | GACCAGTTATTGTTTGAAAGGCAAA | TTGCCACCAGAAATACATACCACACAATG | 253 | X:31769023–31769276 |
| DMD-33 | TAGATATTGACCACCGCTGC | TTGCTAAGACTCTAATCATAC | 402 | X:31765756–31766158 |
| DMD-34 | CAGAAATATAAAAGTTCCAAATAAGTG | CATGTTAATACTTCCTTACAAAATC | 338 | X:31759973–31760311 |
| DMD-35 | CCGTTTCATAAGCATTAAATC | AGCTTCTAGCCTTTTCTC | 271 | X:31744540–31744811 |
| DMD-36 | TGTCTAACCAATAATGCCATG | CTGGTGTACAATTTGGACA | 215 | X:31744080–31744295 |
| DMD-37 | CTTTCTCACTCTTCTCGCTCAC | TTCGCAAGAGACCATTTAGCAC | 335 | X:31742278–31742613 |
| DMD-38 | GGTTTATGTTTCTAATAAAAAGTAA | ATTTATTTCCACTCCTAGTT | 231 | X:31727899–31728130 |
| DMD-39 | AAAGAAAGGCTATGAGCACAGT | TTTCTGATGACTAAGTCTGAAGCAG | 404 | X:31725377–31725781 |
| DMD-40 | CTGCAGCCAGAAGTGCACTA | GGAAATGCATCAAATCAAAGA | 340 | X:31722552–31722892 |
| DMD-40ver2 | AATAACTGCAGCCAGAAGTGC | ATCTCTGGGCTCAGGTAGGC | 206 | X:31722691–31722897 |
| DMD-41 | GTTAGCTAACTGCCCTGGGCCCTGTATTG | TAGAGTAGTAGTTGCAAACACATACGTGG | 275 | X:31721593–31721868 |
| DMD-42 | CACACTGTCCGTGAAGAAACGATGATGG | CTTCAGAGACTCCTCTTGCTTAAAGAGAT | 195 | X:31689624–31689819 |
| DMD-42ma | CACACTGTCCGTGAAGAAACGATGATG | TTAGCACAGAGGTCAGGAGCATTGAG | 155 | X:31689664–31689819 |
| DMD-43(ma) | GAACATGTCAAAGTCACTGGACTTCATGG | ATATATGTGTTACCTACCCTTGTCGGTCC | 357 | X:31667054–31667411 |
| DMD-44 | CTTGATCCATATGCTTTTACCTGCA | TCCATCACCCTTCAGAACCTGATCT | 268 | X:31596364–31596632 |
| DMD-44ma | GTTGTGTGTACATGCTAGGTGTGTA | TCCATCACCCTTCAGAACCTGATCT | 426 | X:31596364–31596790 |
| DMD-45 | AAACATGGAACATCCTTGTGGGGAC | CATTCCTATTAGATCTGTCGCCCTAC | 547 | X:31347855–31348402 |
| DMD-45ma | CTTTCTTTGCCAGTACAACTGCATGTG | CATTCCTATTAGATCTGTCGCCCTAC | 307 | X:31347855–31348162 |
| DMD-46 | CCAGTTTGCATTAACAAATAGTTTGAG | AGGGTTAAGAAGAAATAAAGTTGTGAG | 373 | X:31311470–31311843 |
| DMD-46ma | GCTAGAAGAACAAAAGAATATCTTGTC | CTTGACTTGCTCAAGCTTTTCTTTTAG | 148 | X:31311622–31311770 |
| DMD-47 | CGTTGTTGCATTTGTCTGTTTCAGTTAC | GTCTAACCTTTATCCACTGGAGATTTG | 181 | X:31309131–31309312 |
| DMD-48 | TTTGGCTTATGCCTTGAGAA | CGTCAAATGGTCCTTCTTGG | 235 | X:31254735–31254970 |
| DMD-49(ma) | GTGCCCTTATGTACCAGGCAGAAATTG | GCAATGACTCGTTAATAGCCTTAAGATC | 439 | X:31216049–31216488 |
| DMD-50(ma) | CACCAAATGGATTAAGATGTTCATGAAT | TCTCTCTCACCCAGTCATCACTTCATAG | 271 | X:31199434–31199705 |
| DMD-51(ma) | GAAATTGGCTCTTTAGCTTGTGTTTC | GGAGAGTAAAGTGATTGGTGGAAAATC | 388 | X:31153426–31153814 |
| DMD-52 | ATGCATCTTGCTTTGTGTGTC | GGCTGGTCTCACAATTGTAC | 254 | X:31109130–31109384 |
| DMD-52ma | AATGCAGGATTTGGAACAGAGGCGTCC | CATTATGGACTGAAAATCTCAGCAC | 265 | X:31109021–31109286 |
| DMD-53(ma) | TTGAAAGAATTCAGAATCAGTGGGATG | CTTGGTTTCTGTGATTTTCTTTTGGATTG | 212 | X:31058917–31059129 |
| DMD-54 | GTTTGTCCTGAAAGGTGGGTTAC | TTATCGTCTTGAACCCTCCCAAG | 471 | X:31037406–31037877 |
| DMD-55 | AATTTAGTTCCTCCATCTTTCTCT | AAATACATCAGGCTGTATAAAAGC | 409 | X:31007090–31007499 |
| DMD-56 | TTTGTTTGGTAATTCTGC | CTGAAATTGGATGATTTAC | 336 | X:30886706–30887042 |
| DMD-57 | TCTGACATGGTACGCTGCTG | TGACCCTTGGGTGAGAAGAG | 317 | X:30876201–30876518 |
| DMD-58 | TTTTGAGAAGAATGCCACAAGCC | AAAATATGAGAGCTATCCAGACC | 279 | X:30858422–30858701 |
| DMD-59 | CCTGAGGAGAGAGCCCAGA | TCGAGGTGATCTTGGAGA | 251 | X:30857658–30857909 |
| DMD-59ver2 | TTGTGGGAAGATAACACTGCAC | ACTCGGCTTCTACGAAAGCA | 325 | X:30857560–30857885 |
| DMD-60 | TAAATATTCTCATCTTCCAATTTGC | TTACTGTAACAAAGGACAACAATG | 231 | X:30823983–30824214 |
| DMD-60ma | AGGAGAAATTGCGCCTCTGAAAGAGAACG | CTGCAGAAGCTTCCATCTGGTGTTCAGG | 139 | X:30824023–30824162 |
| DMD-61 | ATCCTTTGTGTTTGGCCTTG | AGAAAGTGCTGAGATGCTGG | 279 | X:30728103–30728382 |
| DMD-62 | TAATGTTGTCTTTCCTGTTTGCG | ATACAGGTTAGTCACAATAAATGC | 185 | X:30703086–30703271 |
| DMD-62ma | GTCTTTCCTGTTTGCGATGAATTTGACC | CTCACTTGTGAATATACAGGTTAGTCAC | 191 | X:30703073–30703264 |
| DMD-63 | TACTCATTGTAAATGCTAAAGTC | TAGCAAGTAACTTTCACACTGC | 193 | X:30640412–30640605 |
| DMD-64 | TTCTGATGGAATAACAAATGCT | CATTCTAGGCAAACTCTAGGC | 286 | X:30602421–30602707 |
| DMD-65 | TATGAGAGAGTCCTAGCTAGG | TAAGCCTCCTGTGACAGAGC | 347 | X:30588992–30589339 |
| DMD-66 | GTCAGTAATTGTTTTCTGCTTTG | ATAAGAACAGTCTGTCATTTCCC | 210 | X:30586037–30586247 |
| DMD-66ver2 | GAGGATCCGTGTCCTGTCTT | TTTGACAAGGAATGGCACAA | 241 | X:30585957–30586198 |
| DMD-66ver4 | AAACTGGCATCATTTCCCTG | TTCCGACTTACAGAAAGAGGTT | 364 | X:30585811–30586175 |
| DMD-67 | AATTGCTACTGGAATTGAGTTGG | AAGAATAAATATGTTACCTAGAAGG | 286 | X:30583466–30583752 |
| DMD-68 | TAATCGAACTGATATACACCTCC | ACTAACAGCAACTGGCACAGG | 351 | X:30562185–30562536 |
| DMD-69 | GAACGTGGTAGAAGGTTTATTAAA | CTAACTCTCACGTCAGGCTG | 231 | X:30559879–30560110 |
| DMD-70 | TGGTCATTAGTTTTGAAATCATC | CATCAAACAAGAGTGTGTTCTG | 237 | X:30558172–30558409 |
| DMD-71 | TGATGGCAGTCACACAACTG | GAGCGAATGTGTTGGTGGTAG | 268 | X:30557424–30557692 |
| DMD-72 | AAAGCATTCTAGGCCATGTGT | ATGCACGTTAGAGGGCAAGT | 307 | X:30552937–30553244 |
| DMD-73 | ACGTCACATAAGTTTTAATGAGC | ATGCTAATTCCTATATCCTGTGC | 202 | X:30551795–30551997 |
| DMD-74 | ACCAAAACCTTTGATTTTATTTTCC | TTTCTATGTGTGCAAGTGTATGC | 248 | X:30548923–30549171 |
| DMD-75 | TCTTTTTTACTTTTTTGATGC | AGTGCTCTCTGAGGTTTAG | 344 | X:30526744–30527088 |
| DMD-76 | GTAATTCTGTTTTCTTTTGGATGAC | CTACCTTTCTTCAGACAACAAAAT | 200 | X:30525786–30525986 |
| DMD-77 | TAATCATGGCCCTTTAATATCTG | GATACTGCGTGTTGGCTTCC | 270 | X:30513537–30513807 |
| DMD-78 | TTCTGATATCTCTGCCTCTTCC | CATGAGCTGCAAGTGGAGAGG | 231 | X:30506024–30506255 |
| DMD-79 | AGAGTGATGCTATCTATCTGCAC | TGCATAGACGTGTAAAACCTGCC | 349 | X:30501186–30501535 |
| NF2-01 | GGGCTAAAGGGCTCAGAGTG | AACCTCTCGAGCTTCCACCT | 235 | 22:28324479–28324714 |
| NF2-02 | AAGGAACTGTCCAAGGGATGT | CCCACCAGTTTCATCGAGTT | 398 | 22:28357051–28357449 |
| NF2-03 | TTGCAAAGGCTTCTTTGAGG | TGGTCAACTCTGAGGCCAAC | 280 | 22:28359554–28359834 |
| NF2-04 | AGCCTACACACCTCACTTCCA | GAGTGATCCCATGACCCAAA | 204 | 22:28362685–28362889 |
| NF2-04ver2 | CTCTCCACCTGTCTGCATCA | TCCCATGACCCAAATTAACG | 310 | 22:28362573–28362883 |
| NF2-05 | AATCTCAATCGCCTGCTCTC | TCCTTCAAGTCCTTTGGTTAGC | 184 | 22:28375163–28375347 |
| NF2-06 | ACACGCCCTCTCTCTGTGTG | GCCCATAAAGGAATGTAAACC | 276 | 22:28375983–28376259 |
| NF2-06ver2 | TCCTCCAATAGAATGGTGCC | GCCCATAAAGGAATGTAAACCA | 302 | 22:28375957–28376259 |
| NF2-07 | TAGAGGAATGGCAGGGTCAG | TCGCCTTGGAATGAAGAAGT | 267 | 22:28378633–28378900 |
| NF2-07ver2 | GGATGGGAAATTCTGCTTGA | CACCCGGATTGCAAAGTAGT | 282 | 22:28378528–28378810 |
| NF2-08 | CTTCTACCTGCCCCAATTCAG | CAGGGAAAGATCTGCTGGAC | 361 | 22:28381567–28381928 |
| NF2-08ver2 | TGTGTGCCAGATTCTTTGGA | AACAACCACACCCTCAAAGC | 355 | 22:28381664–28382019 |
| NF2-09 | GATACTGGGAAGCCAGGACA | GAAAGTATGCGCCAAGTGAG | 339 | 22:28385397–28385736 |
| NF2-10 | CTACCTGCAAGAGCTCAAAC | GTAGGCATCGGCAAATGAAG | 366 | 22:28388776–28389142 |
| NF2-11 | CATCTTTGGGCCCTTGTG | CCCAAGTAGCCTCCTGGAAC | 237 | 22:28392294–28392531 |
| NF2-12 | GGGAATGTGGCTTGTCATTTC | CTTCCAGCACCTTCTGCTC | 339 | 22:28393647–28393986 |
| NF2-13 | TTTCCTGCTACCTGCCCTCT | CTCTCTGCACCTCTCATCCA | 297 | 22:28395324–28395621 |
| NF2-14 | AGTTGTGCCCATTGCCTCTG | CCCCAATCACTCAGTCTAGTTC | 294 | 22:28398630–28398924 |
| NF2-15 | TGTCTCACTGTCTGCCCAAG | AGGAAACCAGATGCCAACC | 290 | 22:28401931–28402221 |
| NF2-16 | CCCAGGAACAGTCTCACCAT | GCAGCACCATCACCACATAG | 314 | 22:28403335–28403649 |
| NF2-17A | GGGACTGACAGCCAACTTCT | CCCTATGGATGGCTCTCTTG | 291 | 22:28415142–28415433 |
| NF2-17B | GTCTCAAGCCCAAAGAGCAG | GGCCCTACAGGAAGAAGTGA | 571 | 22:28418465–28419036 |
| PLP-01 | AAAGCAGGCCTGTCCCTTTA | TGTGAATTCCTGTGTCCTCTTG | 260 | X:101803577–101803837 |
| PLP-02 | TGGATGTGCCTGACTGTTTC | TCTGAATGGGCTTCGAATGT | 399 | X:101812330–101812729 |
| PLP-03 | ATTCCCTGGTCTCGTTTGTC | CACCTTGTCGGGATGTCCTAG | 300 | X:101813215–101813515 |
| PLP-04 | CTCCAGGATCTCCCAGTTTG | GCACCCGTACCCTAACTCAC | 267 | X:101814505–101814772 |
| PLP-05 | AGAGATGGAAGAAGGGCTCT | ATTGAAGGCCATGGGTGTAA | 236 | X:101815132–101815368 |
| PLP-06 | AAAGATATCAACACATTCAG | TTGCCTTTCAGAATAGCTGT | 239 | X:101816019–101816258 |
| PLP-07 | CCTCATTCCCAAAGGGATTT | CAGCATTGTAGGCTGTGTGG | 212 | X:101817233–101817445 |
| PMP22-01 | TGGCTTCAGTTACAGGGAGC | TAGGCACACATCACCCAGAG | 224 | 17:15368992–15369216 |
| PMP22-01pma | GTCTTGGCATCACAGGCTTCAGGCA | ACCAGGCTCCCCGAGATGTTCCCTG | 268 | 17:15369166–15369434 |
| PMP22-02 | TCCTCGCAGGCAGAAACT | CAGTCCTGAACCAGCAGGAG | 206 | 17:15364447–15364653 |
| PMP22-02ver4 | ACTCCGCTGAGCAGAACTTG | GCAGATTGCCAGAAACTTCC | 249 | 17:15364389–15364638 |
| PMP22-03 | CTGGCAGAACTGTAGCACCT | CATGGCTCCCTGTCACATC | 205 | 17:15362836-15363041 |
| PMP22-04 | TTCTGCTGCCTGTGAGGAC | TTCTGAGGCCACATCCTTCT | 349 | 17:15343264–15343613 |
| PMP22-05 | CAGGTCTGTGCGTGATGAGT | CCACCTCCACTGCTTTCTGT | 304 | 17:15334662–15334966 |
| PMP22-05uma | CTGCTTTTGTACCTAGCTAGGCTGC | ATGCATCTTAGTCCACACAGTTGG | 306 | 17:15333685–15333991 |
Amplicon names reflect the gene and exons amplified in each assay. Amplicon names ending in the “ma” or “ver2, -3, -4, etc.” designations represent alternative amplicons for the same exon but that differ in sequence. Amplicon names followed by characters in parentheses refer to amplicons that were spotted in two different locations on the array.
Amplicon sizes are shown and genomic locations are given with respect to build NCBI 34 and ENSEMBL version 21.34d.1.
To generate arrays containing single-stranded array elements, all PCR products used in the present study were prepared as follows. To the 5′ end of the forward primer of each pair was added an 8-bp universal sequence (5′-TGACCATG-3′). These primer pairs (final concentration 0.5 μM) were used to amplify exon-containing PCR products in a 20-μl final volume first-round PCR containing 50 mM KCl, 5 mM Tris HCl (pH 8.5), 2.5 mM MgCl2, 10 mM dNTPs (Pharmacia), 0.625 U Taq polymerase (Perkin Elmer–Cetus), and 50 ng of human genomic DNA. Thermocycling was optimized for each primer pair by use of an annealing temperature of 50°C–60°C (cycling conditions available on request). PCR products from these reactions were diluted 1:1000 and were used in second-round reactions containing the 5′-(C6) amino-modified universal primer 5′-GCTGAACAGCTATGACCATG-3′ (Eurogentec) and the reverse gene-specific primer of each set. Reaction conditions were the same as those in the first round, except the reaction volume was 60 μl. For the comparison of single- versus double-stranded DNA array elements, the universal linker was also added to the 5′ end of the reverse gene-specific primer. This allowed a 5′-aminolink to be incorporated on either the forward or the reverse strand in second-round reactions.
To prepare second-round PCR products for arraying, spotting buffer was added at final concentrations of 0.25 M sodium phosphate buffer pH 8.5 and 0.00025% sarkosyl (BDH Laboratory Supplies). The PCR products were then filtered through multiscreen-GV 96-well filter plates (Millipore), were aliquoted into 384-well plates (Genetix), and were arrayed onto Codelink slides (Amersham) in quadruplicate in a 16-block format by use of a Microgrid II arrayer (Biorobotics/Genomic Solutions). Slides were processed to generate single-stranded array elements, as described on the Sanger Institute microarray Web pages, and were stored at room temperature until hybridized.
Patient and Control DNA Samples
Thirty-one patient samples were obtained from a number of laboratories; all samples had been screened for mutations in the relevant disease gene at accredited diagnostic laboratories. Samples from patients with mutations in the COL4A5, DMD, and PMP22 genes were obtained from the Southeast Thames Regional Genetics Centre (at King's, Guy's, and St. Thomas’s Hospitals, London). These samples were all made anonymous for inclusion in this study. DNA samples from patients with NF2 mutations were obtained from the Department of Genetics and Pathology at Uppsala University. DNA samples from patients with mutations in the PLP1 gene were obtained from the Institute of Child Health (London); additional patients with mutations in the PMP22 gene were obtained from the Center for Human Genetics (Leuven). Control DNA was obtained from archives of normal male and female DNA housed at the Sanger Institute (Cambridge). DNA was extracted using a variety of standard methods (salt/chloroform, phenol, commercial kits, etc.) from either peripheral blood or cultured cell lines. Aliquots of patient and control DNA samples were quantitated using a fluorometer (TD-360 [Turner]), were sonicated to an average size of ∼10 kb by use of a water-bath sonicator (VirSonic 300 [Virtis]), and were visualized by agarose-gel electrophoresis. Female and male pool DNA samples that were used as reference DNA controls were derived by combining equal amounts of peripheral blood DNA from five females and five males, respectively. Male (XY) and female (XX) lymphoblastoid cell lines were also used as controls in the validation experiments. None of the control samples had mutations in the genes analyzed in this study.
Fluorescent DNA Labeling, Microarray Hybridization, and Data Analysis
Fluorescent-labeled DNA samples were prepared using a modified Bioprime labeling kit (Invitrogen) in 100-μl reaction volumes containing 600 ng genomic DNA, dNTPs (0.2 mM dATP, 0.2 mM dTTP, 0.2 mM dGTP, and 0.05 mM dCTP), and 0.04 mM Cy5/Cy3 dCTP (GE Healthcare). Reference control samples were labeled with Cy3, and test samples were labeled with Cy5. Labeling reactions were purified using Micro-spin G50 columns (Pharmacia-Amersham) in accordance with the manufacturer's instructions. Reference and test samples were combined and precipitated with 3 M sodium acetate (pH 5.2) in 2.5 volumes of ethanol with 90 μg human Cot DNA (Invitrogen). The DNA pellet was resuspended in hybridization buffer containing 50% deionized formamide, 10% dextran sulphate, 10 mM Tris-HCl (pH 7.4), 2× SSC, 0.1% Tween-20, and 300 μg yeast tRNA (Invitrogen). Similarly, the prehybridization mixture was prepared by precipitating 400 μg herring sperm DNA (Sigma) and 67.5 μg human Cot DNA with 3 M sodium acetate (pH 5.2) and 2.5 volumes of ethanol; this DNA pellet was resuspended in the same hybridization buffer but with no yeast tRNA included.
Microarrays were prehybridized, hybridized, and washed using methods described elsewhere (Fiegler et al. 2003). Microarrays were scanned using a ScanArray 4000 confocal laser-based scanner (Perkin Elmer). Mean spot intensities from images were quantified using QuantArray or ScanArray Express (Perkin Elmer) with background subtraction. Mean ratios and SDs for all exon PCR products in quadruplicate were calculated, and the mean exon ratios were normalized to the chromosome 22 control element mean ratios for each of the 16 blocks independently to within 2% of the theoretical value (1.00±0.02). Ratios were obtained for each exon independently, except in the cases in which more than one exon was contained in a single PCR product. Furthermore, there was no averaging of copy-number data for exons found in different PCR products. Although some exons were represented by more than one PCR product, the data from only the array element that most accurately reported copy number (i.e., that behaved closest to the theoretical values) were retained in the final data set for each sample in all but two instances—for the PMP22 gene, the data from two PCR products representing exon 1 were pooled to derive a mean exon 1 ratio for both products. Similarly, this procedure was also performed for PMP22 exon 5. To visualize the data, the final data set of mean ratios was plotted on a histogram for each exon or exon pair of COL4A5, DMD, NF2, PLP1, and PMP22. Microarray experiments with control samples and patient samples were performed—multiple times, in some cases—to determine the reproducibility of the method. However, the derived mean-ratio data were determined for each hybridization experiment rather than from pooling of data from multiple experiments. For single- versus double-stranded DNA comparisons, mean intensities of quadruplicate spots were calculated with background subtraction in a single channel.
Results
Single-Stranded Technology Allows Detection of Exon-Sized Array Elements
One of the main considerations for array CGH is related to the complexity of the genome, as well as how this complexity is reflected in the signal measurements from which the copy-number changes are derived. Given that a typical exon in the human genome (median and mean sizes of 133 bp and 262 bp, respectively) represents a segment of genomic DNA found at a level of 1–2 in 4×107 sequence equivalents in the diploid human genome, it was necessary to address how to detect such a sequence at this level without reducing genome complexity or compromising the accuracy of the measurements. To this end, we developed an array system that allows single strands of DNA derived from double-stranded PCR products to be retained on the surface of the microarray slide. By incorporating a 5′-aminolink modification onto the end of one strand of a double-stranded DNA molecule during PCR, it is possible to covalently attach this strand to the surface of the slide, and the unmodified strand can be removed (fig. 1). Since single-stranded array elements cannot reanneal to form duplex molecules, we anticipated an increase in the signal:noise ratio in array CGH experiments, since there would be more single-stranded DNA on the surface of the slide capable of hybridizing with labeled sample. We demonstrated increased signal:noise measurements for single-stranded array elements (fig. 1). Test arrays were printed with array elements on which 5′-aminolink modifications were incorporated on either one or both strands. For a series of genomic array elements of 200–400 bp in size that we tested in hybridizations with fluorescently labeled human genomic DNA, single-stranded elements provided an increased signal:noise ratio, with a range of 1.16–2.51-fold increase, depending on the array element. On average, this translated into a 1.79-fold increase in the signal:noise ratio. We had previously demonstrated a similar increase in the signal:noise ratio for cDNA expression analysis (C.L. and D.V., unpublished data). Thus, this array platform facilitated increased signal:noise ratios and thereby provided evidence that we could improve the signal sufficiently to detect human genomic DNA sequences at the resolution of single exons.
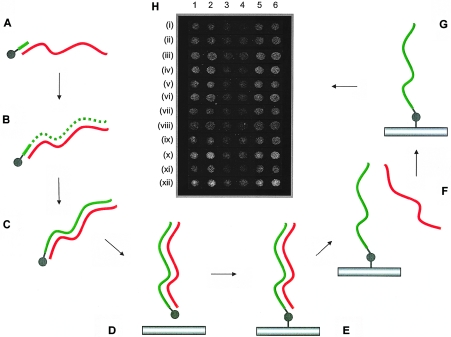
Figure 1.
Development of single-stranded array platform for exon array CGH. Schematic diagram shows the approach adopted to make single-stranded PCR products with the increased sensitivity on the array platform described in this article. A, Oligonucleotide containing a 5′-C6-aminolink modification (gray ball) used as a primer (short green bar) on a genomic DNA or PCR template strand (red wavy line) in a PCR reaction. B and C, Synthesized double-stranded PCR product containing the 5′-aminolink modification on one strand only. Newly synthesized strand containing the 5′-aminolink modification is shown as a green dotted/solid line. D, PCR product spotted onto the microarray slide. E, Covalent attachment to the surface formed through the 5′-aminolink modification by subjecting the slide to high humidity. F, Removal of reverse strand that is not covalently attached to the surface of the slide by use of physical and chemical denaturation. G, The resultant single-stranded array element. H, Microarray image (gray scale) showing the results of fluorescently labeled genomic DNA hybridized to test arrays containing single-stranded and double-stranded PCR products for a series of genomic array elements (“i–xii”) spotted in duplicate. Single-stranded elements for either one strand (5′-aminolink on forward strand [lanes 1 and 2]) or the other strand (5′-aminolink on the reverse strand [lanes 5 and 6]) or for double-stranded elements (5′-aminolinks on both strands [lanes 3 and 4]) were prepared on the test array. Single-stranded elements gave a higher signal for all genomic products (see the “Results” section).
Assessing the Performance of DNA Elements on Exon Arrays
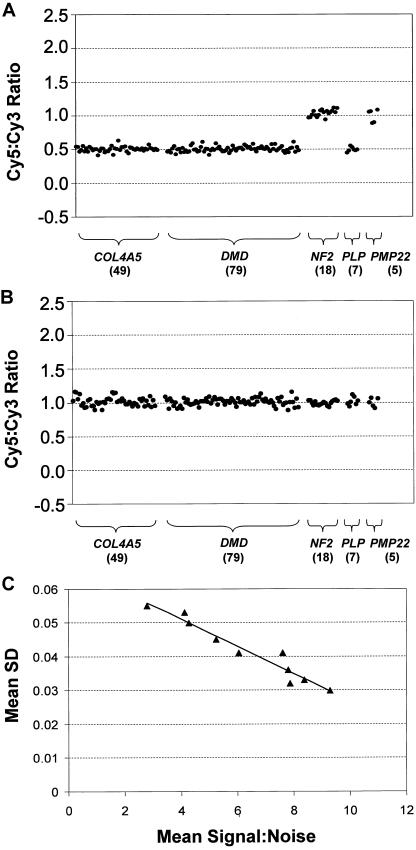
We generated an exon array that included a set of array elements designed for every exon of five genes. The five genes included on this array were chosen to assess technical aspects of the technology and to demonstrate its clinical relevance (table 1) (Atkin et al. 1988; Worton and Thompson 1988; Den Dunnen et al. 1989; Gillard et al. 1989; Nelis et al. 1996; Heim et al. 1997; Lemmink et al. 1997; Sistermans et al. 1998; Mimault et al. 1999; Plant et al. 1999; Rautenstrauss et al. 2000; Bruder et al. 2001). These were the X-linked genes COL4A5, DMD, and PLP1 and the autosomal genes NF2 and PMP22. Mutations and/or copy-number changes in these five genes result in human inherited disorders. For the 162 exons of these genes, 158 array elements were initially designed and spotted on our arrays. We included a second array element for each of 18 exons that we chose at random to help assess sequence context on reporting accuracy, which brought the total number of array elements for these genes to 176. From the 3′ UTRs of genes on chromosome 22, 360 repeat-free amplicons were also spotted on these arrays and were used as normalization controls. These controls were, on average, of a size similar to that of the exon elements. We performed a series of validation experiments of male (XY) versus female (XX), and female (XX) versus female (XX) competitive hybridizations across a series of five batches of array printed at different times, with elements printed in quadruplicate, to assess the performance of the exon array in measuring copy-number changes. From the initial minimal set of 158 array elements, 135 (85.4%) reported copy-number measurements that were within 0.15 copy-number units of the theoretical values. The second array element for each of the 18 exons that had two different elements on the array showed a similar success rate (16 [88.9%] of 18). To obtain accurate copy-number measurements for all remaining exons in the five genes, we designed and tested a second element for each of 23 exons, 2 of which required a third iteration of design. These redesigned elements replaced the original elements for those exons in all subsequent analyses. In total, we analyzed 201 array elements, 174 (86.6%) of which reported accurate copy-number measurements and had a size range of 139–571 bp. On the basis of these experiments, we demonstrated that we could obtain accurate copy-number measurements for ∼85% of the exons in our study by designing a single array element and for 100% of all exons by designing a second or, in a small minority of cases, a third array element. Typical examples of the quality of the data that we obtained from these validation experiments for array elements covering all 162 exons of the five genes are shown in figure 2. The three X-linked genes (DMD, PLP1, and COL4A5) showed easily discernible single copy-number differences in XX versus XY validation experiments, when compared with the two autosomal genes (PMP22 and NF2) (fig. 2A). XX versus XX validation experiments also showed the expected copy-number equivalence for all genes (fig. 2B).
Figure 2.
Validation experiments for exon arrays. A, Fluorescently labeled DNA derived from genomic DNA from a single male (Cy5) hybridized on the exon array with that derived from five females (used as a pool) (Cy3). The plot shows the Cy5 channel:Cy3 channel ratios obtained for exons (represented as black dots) for COL4A5, DMD, NF2, PLP1, and PMP22. Each exon is plotted as a function of its position 5′→3′ for each gene. The number of exon array elements assayed for each gene is shown in parentheses below the gene name. Exons for the three X-linked genes and the two autosomal genes showed ratios centered around the theoretical values of 0.5 and 1, respectively. B, Fluorescently labeled DNA derived from genomic DNA from a single female (Cy5) hybridized on the exon array with that derived from the female pool (Cy3). The plot shows the ratios obtained for each exon, as described above (A), centered around the theoretical value of 1. C, The effect of signal:noise ratio on measurement error determined, using exon arrays, in 10 male versus female-pool validation experiments. The plot shows the relationship between mean signal:noise and mean SD. Both means were derived as the mean of all mean quadruplicate element measurements per experiment. The black triangles represent each of the 10 male/female validation experiments. The correlation coefficient (r) between mean SD and mean signal:noise was −0.97.
Across the validation experiments, signal:noise ratios varied from hybridization to hybridization, which we attributed to variations in cyanine dye quality, DNA quality, labeling efficiencies, array batch, and other technical considerations. However, whereas signal:noise ratio did not appear to dramatically affect the range of mean copy-number values for each exon spotted in quadruplicate, the accuracy of the measurements was noticeably improved as the signal:noise ratio increased (fig. 2C). SDs in the quadruplicate measurements made for each exon showed a strong negative correlation with signal:noise ratio (r=-0.97). This would suggest that the single-stranded array platform we have developed not only helps to improve signal:noise ratio, but, as a consequence, also reduces measurement error. More importantly, our data also demonstrate that even exons that exhibit an extremely low signal:noise ratio on our array (i.e., in the range of 1–1.5) could be quantitated accurately.
We studied further the performance of our single-stranded array elements to identify other features that may contribute to reporting accuracy in array CGH experiments. Using regression analysis on data obtained from all 201 exon array elements, we found no strong correlations between the accuracy of copy-number measurements and inherent features of the array elements, such as G+C content, length, melting temperature, secondary structure, similarity to other genomic sequences, and distance from upstream or downstream repeat sequences in genomic DNA. However, given that we could obtain accurate copy-number measurements for some exons only by redesigning array elements, sequence context is likely to contribute in some way to reporting accuracy. We also assessed whether the location of an element on the array affected reporting accuracy, by spotting independent preparations of 12 of the array elements in alternative locations on the array. All 12 of these array elements reported accurate copy-number measurements irrespective of their array coordinate. This evidence, together with the rest of our empirical data, demonstrates that, although it is unclear which factors may influence the performance of DNA elements on our exon array, this platform is robust within a variety of sequence contexts and technical constraints.
Detection of Copy-Number Changes in Patient Material
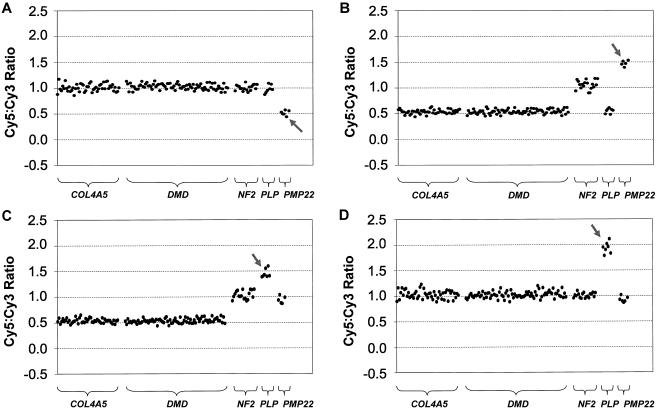
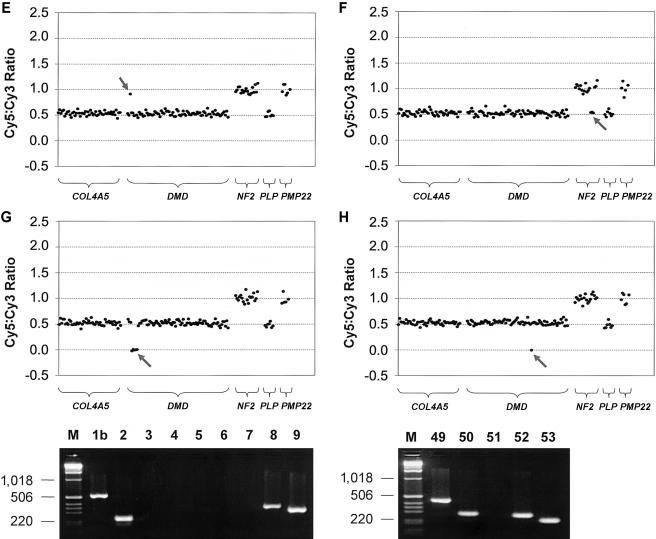
We further tested the performance of the exon array and its potential application as a research and diagnostic tool by detecting copy-number changes in DNA samples from patients with inherited disorders. Thirty-one patient DNA samples (from 24 males and 7 females) were collected that all had known pathogenic copy-number changes in one of the five disease genes represented on the array. We were able to determine copy-number changes in all 31 DNA samples and with equal efficacy in affected males, affected females, and nonsymptomatic carrier females (for X-linked disorders). All of these results were in agreement with the molecular data determined elsewhere for these patients (Ellis and Malcolm 1994; Harding et al. 1995; Woodward et al. 1998, 2000; Bruder et al. 2001; Buckley et al. 2002; Mantripragada et al. 2003; S.A., unpublished data; J.R.V., unpublished data) or as part of the present study. Our results are summarized in table 3 and figure 3. In total, we detected copy-number changes (either a gain or loss) in patient samples for 121 of the 158 array elements containing exons that were present on the array. The demonstrated copy-number changes included 10 whole-gene deletions (fig. 3A), 8 whole-gene duplications (fig. 3B), 2 whole-gene triplications (fig. 3C and 3D), 3 partial-gene duplications (fig. 3E), and 8 partial-gene deletions (fig. 3F, 3G, and 3H).
Table 3.
Copy-Number Changes Detected in 31 Patient Samples[Note]
| Sample | Patient Reference | Sex | Gene Affected | Mutation |
| 1 | JPa,b,c | Male | NF2 | Whole-gene deletion |
| 2 | p7a,b | Male | NF2 | Whole-gene deletion |
| 3 | p130b | Male | NF2 | Deletion (exons 13–15) |
| 4 | EA01(241397) | Male | PMP22 | Whole-gene deletion |
| 5 | EA02(260055) | Female | PMP22 | Whole-gene deletion |
| 6 | EA03(218861) | Male | PMP22 | Whole-gene deletion |
| 7 | EA04(268266) | Male | PMP22 | Whole-gene deletion |
| 8 | EA05(251650) | Male | PMP22 | Whole-gene deletion |
| 9 | EA06(263902) | Female | PMP22 | Whole-gene duplication |
| 10 | EA07(221358) | Male | PMP22 | Whole-gene duplication |
| 11 | EA08(138116) | Male | PMP22 | Whole-gene duplication |
| 12 | EA09(231345) | Male | PMP22 | Whole-gene duplication |
| 13 | EA10(271065) | Male | PMP22 | Whole-gene duplication |
| 14 | EA11 | Male | COL4A5 | Whole-gene deletion |
| 15 | EA12 | Male | DMD | Deletion (exon 51) |
| 16 | EA13 | Male | DMD | Deletion (exon 44) |
| 17 | EA14 | Male | DMD | Deletion (exons 3–7) |
| 18 | EA15 | Female | PMP22 | Whole-gene deletion |
| 19 | EA16 | Male | PMP22 | Whole-gene deletion |
| 20 | EA17 | Male | DMD | Deletion (exons 45–52) |
| 21 | EA18 | Female | DMD | Deletion (exons 45–48) |
| 22 | EA19 | Male | DMD | Duplication (exon 2) |
| 23 | EA20 | Female | DMD | Deletion (exons 17–48) |
| 24 | EA21 | Male | DMD | Deletion (exons 45–50) |
| 25 | EA22 | Male | DMD | Duplication (exons 2–7) |
| 26 | EA23 | Male | DMD | Duplication (exon 3) |
| 27 | DH/PMD2-1d,e,f | Male | PLP1 | Whole-gene triplication |
| 28 | PMD2-2d,e,f | Female | PLP1 | Whole-gene triplication |
| 29 | NO/PMD9-1d,f | Male | PLP1 | Whole-gene duplication |
| 30 | PMD4-2f | Female | PLP1 | Whole-gene duplication |
| 31 | PMD4-1f | Male | PLP1 | Whole-gene duplication |
Note.— Copy-number changes in a collection of 31 patient samples found using the exon array described in the present study. Where relevant, patient references have been included from other published studies.
Mantripragada et al. 2003.
Bruder et al. 2001.
Buckley et al. 2002.
Harding et al. 1995.
Woodward et al. 2000.
Figure 3.
Exon copy-number changes in medically relevant disease genes. Representative results of exon array CGH for patient DNA samples hybridized against a female pool that demonstrates the variety of copy-number changes detected in the present study. All patient samples were fluorescently labeled using Cy5, and all control female-pool samples were fluorescently labeled using Cy3. The features of the plots are as described for figure 2. The gray arrows highlight the copy-number changes. A, Female patient—EA02(260055)—with HNPP showing a deletion of the entire PMP22 gene. B, Male patient—EA07(221358)—with CMT1 showing a duplication of the entire PMP22 gene. C, Male patient (DH/PMD2–1) with PMD showing a triplication of the entire PLP1 gene. D, Female PMD carrier (PMD2–2, mother of patient in panel C) showing a triplication of the entire PLP1 gene. E, Male patient (EA19) with DMD showing a duplication of exon 2 in the DMD gene. F, Male patient (p130) with NF2 showing a deletion of exons 13–15 of the NF2 gene. G, Male patient (EA14) with DMD showing a deletion of exons 3–7 of the DMD gene. Panel also shows the confirmatory PCR results; lanes are numbered according to the exons assayed and size markers (“M”), in base pairs, shown at the left of the gel image. H, Male patient (EA12) with DMD showing the deletion of exon 51 of the DMD gene. Confirmatory PCR is also shown with lanes numbered according to the exons assayed and size markers (“M”), in base pairs, shown at the left.
In the DNA sample of one patient (patient EA11), in which the entire COL4A5 gene was deleted, we detected a copy-number measurement in COL4A5 exon 1 (0.26) that did not suggest a complete absence of this exon (theoretical value of 0). The PCR product for this exon showed an 86% sequence similarity across 212 bp to a sequence on the short arm of chromosome X. This was the highest degree of sequence similarity found for any of the elements on the exon array. Therefore, although exon 1 was completely deleted in patient EA11 (as confirmed by PCR), the cross-hybridizing X-linked sequence was being reported by the exon 1 array element.
Of the 11 partial-gene duplications/deletions, 4 involved only a single exon, 1 of which was a single-copy number gain of DMD exon 2 (fig. 3E). The array element for DMD exon 2 displayed the lowest signal:noise ratio across all array CGH experiments performed for the present study. This not only helps demonstrate the sensitivity of the array system and our ability to quantitate weak signals, but it also shows the utility of the array for accurate prediction of copy-number changes involving only DMD exon 2; a single-exon duplication of exon 2 is the most frequently occurring DMD mutation, according to one study that used the MAPH assay (White et al. 2002), but is also one of the most difficult to determine using hybridization-based approaches, because it is highly AT rich (PCR product is 73.4% AT rich) (White et al. 2002).
Discussion
We report here an array CGH platform that can measure copy-number changes accurately at the resolution of single exons. By developing an array system that results in single-stranded DNA elements being bound to the surface of a slide, we have demonstrated an improved signal:noise ratio that facilitates the accuracy in copy-number measurements for array elements in the size range of 139–571 bp. We have further shown that constitutionally inherited copy-number changes, as either deletions or duplications, were detected in 100% of patient samples analyzed on an exon array containing array elements for 162 exons from five disease genes. The results described here represent the first time, to our knowledge, that array CGH has been shown to perform robustly at the resolution of single exons. This represents an increase in array CGH resolution that is 2 orders of magnitude higher than has been reported elsewhere.
Our study provides compelling empirical evidence that it will be possible to obtain accurate copy-number measurements for virtually all exons in the genome, with ∼85% of exons requiring that a single-array element be designed and tested and the remainder requiring that additional array elements be validated in an iterative process. Indeed, in the present study, we were able to design array elements for 161 (99.4%) of 162 exons that faithfully reported copy number in validation experiments and in the analysis of patient samples. For the one remaining exon, similarity with a cross-hybridizing genomic sequence was an issue, which suggests that this and possibly other sequence-based factors may affect the performance of array elements for other exons in the genome. What these factors are remains unclear. In some instances, it may be necessary to use “surrogate” exons—that is, array elements designed for sequences that lie adjacent to exons and that may perform more robustly in array CGH. Averaging measurements from multiple array elements may also provide the means to improve quantitative measurements for other exons.
The applications of this technology are numerous. Since our arrays are both scalable and sensitive at the resolution of single exons, they fill a technological gap between the high-throughput technologies (e.g., array CGH by use of BAC arrays) and the high-resolution but less scalable assays (e.g., MAPH and MLPA). Therefore, the development of this technology opens the way for identifying copy-number changes at high resolution in both normal and disease states, as a research or diagnostic tool, and in both large-scale and small-scale studies.
The construction of a genomewide exon array would represent an important achievement for the analysis of DNA-copy number in the human genome. To maintain single-exon resolution for genomewide screening, such an array would require at least one array element for each of the ∼245,000 exons that have been annotated in the human genome. On the basis of the data presented here, that would require the design and testing of 280,000–300,000 exon-specific primer pairs and their conversion to array elements to obtain a quantitative working assay for every exon. Furthermore, on the basis of current spotting technologies, multiple microarray slides would be required to spot the entire set of exons, resulting in increased costs per genome assayed. Although this may appear to represent an enormous undertaking, it must be balanced against the tremendous value that such an array set could contribute to human genetics—it would allow the identification and annotation of CNPs and mutations, in normal individuals and in disease, that have previously been undiscovered because of limitations in the resolution of other array CGH technologies.
In more focused approaches, exon arrays would also facilitate positional cloning strategies of novel disease genes in well-characterized patient collections. Given that we have shown here a direct application of this technology to identify copy-number changes in patient material for several single-gene disorders, its use as a diagnostic tool for known disease genes is evident. Furthermore, it should be possible to screen many genes in an exon array CGH format to quickly elucidate copy-number changes for some categories of disease phenotypes that are difficult to distinguish clinically. Exon array CGH could also be applied to cancer genetics. Given that there are several hundreds of genes that have been shown to be mutated in cancer (Futreal et al. 2004), this set serves as an excellent starting point for the construction of cancer-specific exon arrays.
It will also be possible to use our array system at this level of resolution to answer other questions about genome biology, with the one obvious application being chromatin immunoprecipitation (ChIP)-on-chip (Ren et al. 2002; Weinmann et al. 2002). Given the quantifiable signal:noise ratios we have shown here, obtaining detailed maps of protein-binding sites, histone modifications, and other features, such as origins of replications and matrix attachment regions, should also be possible with genomic tiling-path arrays or intergenic arrays at high resolution. This type of approach for identifying transcription-factor binding sites onto human chromosomal regions (Horak et al. 2002) and whole chromosomal tiling paths (Martone et al. 2003; Cawley et al. 2004; Euskirchen et al. 2004) has already been described, although the proportions of sites identified that are false-positive results or false-negative results are not yet known. In particular, our array system is well suited to identify low-affinity DNA-protein interactions that give relatively poor enrichments in ChIP and small quantifiable copy-number increases on arrays. We are currently extending our work in this area by generating high-resolution genomic tiling arrays for regions of interest in the human genome.
Acknowledgments
The authors thank Nigel Carter, Heike Fiegler, and Philippa Carr, for assistance with initial hybridizations; Hazel Arbury and Ruth Bennett, for generating male and female control DNA; David Beare, for designing the primers for the chromosome 22 control elements; and David Bentley, for his critical reading of the manuscript. This work was supported by the Wellcome Trust.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Leiden University Medical Center, http://www.dmd.nl
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for COL4A5, DMD, NF2, PLP1, PMP22, DMD, Becker muscular dystrophy, NF2, AS, CMT1A, HNPP, and PMD)
- Sanger Institute, http://www.sanger.ac.uk/Projects/Microarrays/arraylab/methods.shtml
- Whitehead Institute, http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi (for Primer3)
References
- Akrami SM, Rowland JS, Taylor GR, Armour JA (2003) Diagnosis of gene dosage alterations at the PMP22 gene using MAPH. J Med Genet 40:e123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DG, Pinkel D (2003) Genomic microarrays in human genetic disease and cancer. Hum Mol Genet Spec 2 12:R145–R152 [DOI] [PubMed] [Google Scholar]
- Armour JA, Barton DE, Cockburn DJ, Taylor GR (2002) The detection of large deletions or duplications in genomic DNA. Hum Mutat 20:325–337 [DOI] [PubMed] [Google Scholar]
- Armour JA, Sismani C, Patsalis PC, Cross G (2000) Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res 28:605–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin CL, Hasstedt SJ, Menlove L, Cannon L, Kirschner N, Schwartz C, Nguyen K, Skolnick M (1988) Mapping of Alport syndrome to the long arm of the X chromosome. Am J Hum Genet 42:249–255 [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Huang J, Greshock J, Watt S, Butler A, West S, Grigorova M, Jones KW, Wei W, Stratton MR, Futreal PA, Weber B, Shapero MH, Wooster R (2004) High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res 14:287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruder CE, Hirvela C, Tapia-Paez I, Fransson I, Segraves R, Hamilton G, Zhang XX, et al (2001) High resolution deletion analysis of constitutional DNA from neurofibromatosis type 2 (NF2) patients using microarray-CGH. Hum Mol Genet 10:271–282 [DOI] [PubMed] [Google Scholar]
- Buckley PG, Mantripragada KK, Benetkiewicz M, Tapia-Paez I, Diaz de Ståhl T, Rosenquist M, Ali H, et al (2002) A full-coverage, high-resolution human chromosome 22 genomic microarray for clinical and research applications. Hum Mol Genet 11:3221–3229 [DOI] [PubMed] [Google Scholar]
- Carvalho B, Ouwerkerk E, Meijer GA, Ylstra B (2004) High resolution microarray comparative genomic hybridisation analysis using spotted oligonucleotides. J Clin Pathol 57:644–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, Wheeler R, Wong B, Drenkow J, Yamanaka M, Patel S, Brubaker S, Tammana H, Helt G, Struhl K, Gingeras TR (2004) Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116:499–509 [DOI] [PubMed] [Google Scholar]
- Den Dunnen JT, Grootscholten PM, Bakker E, Blonden LAJ, Ginjaar HB, Wapenaar MC, van Paassen HMB, van Broeckhoven C, Pearson PL, van Ommen GJB (1989) Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am J Hum Genet 45:835–847 [PMC free article] [PubMed] [Google Scholar]
- Ellis D, Malcolm S (1994) Proteolipid protein gene dosage effect in Pelizaeus-Merzbacher disease. Nat Genet 6:333–334 [DOI] [PubMed] [Google Scholar]
- Euskirchen G, Royce TE, Bertone P, Martone R, Rinn JL, Nelson FK, Sayward F, Luscombe NM, Miller P, Gerstein M, Weissman S, Snyder M (2004) CREB binds to multiple loci on human chromosome 22. Mol Cell Biol 24:3804–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, Scott CE, Smith J, Vetrie D, Gorman P, Tomlinson IP, Carter NP (2003) DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer 36:361–374 [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR (2004) A census of human cancer genes. Nat Rev Cancer 4:177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillard EF, Chamberlain JS, Murphy EG, Duff CL, Smith B, Burghes AHM, Thompson MW, Sutherland J, Oss I, Bodrug SE, Klamut HJ, Ray PN, Worton RG (1989) Molecular and phenotypic analysis of patients with deletions within the deletion-rich region of the Duchenne muscular dystrophy (DMD) gene. Am J Hum Genet 45:507–520 [PMC free article] [PubMed] [Google Scholar]
- Harding B, Ellis D, Malcolm S (1995) A case of Pelizaeus-Merzbacher disease showing increased dosage of the proteolipid protein gene. Neuropathol Appl Neurobiol 21:111–115 [DOI] [PubMed] [Google Scholar]
- Heim P, Claussen M, Hoffmann B, Conzelmann E, Gartner J, Harzer K, Hunneman DH, Kohler W, Kurlemann G, Kohlschutter A (1997) Leukodystrophy incidence in Germany. Am J Med Genet 71:475–478 [PubMed] [Google Scholar]
- Heiskanen MA, Bittner ML, Chen Y, Khan J, Adler KE, Trent JM, Meltzer PS (2000) Detection of gene amplification by genomic hybridization to cDNA microarrays. Cancer Res 60:799–802 [PubMed] [Google Scholar]
- Horak CE, Mahajan MC, Luscombe NM, Gerstein M, Weissman SM, Snyder M (2002) GATA-1 binding sites mapped in the β-globin locus by using mammalian chIp-chip analysis. Proc Natl Acad Sci USA 99:2924–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C (2004) Detection of large-scale variation in the human genome. Nat Genet 36:949–951 [DOI] [PubMed] [Google Scholar]
- Inazawa J, Inoue J, Imoto I (2004) Comparative genomic hybridization (CGH)-arrays pave the way for identification of novel cancer-related genes. Cancer Sci 95:559–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP, Snijders A, Albertson DG, Pinkel D, Marra MA, Ling V, MacAulay C, Lam WL (2004) A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet 36:299–303 [DOI] [PubMed] [Google Scholar]
- Lemmink HH, Schroder CH, Monnens LA, Smeets HJ (1997) The clinical spectrum of type IV collagen mutations. Hum Mutat 9:477–499 [DOI] [PubMed] [Google Scholar]
- Lucito R, Healy J, Alexander J, Reiner A, Esposito D, Chi M, Rodgers L, Brady A, Sebat J, Troge J, West JA, Rostan S, Nguyen KC, Powers S, Ye KQ, Olshen A, Venkatraman E, Norton L, Wigler M (2003) Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res 13:2291–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucito R, West J, Reiner A, Alexander J, Esposito D, Mishra B, Powers S, Norton L, Wigler M (2000) Detecting gene copy number fluctuations in tumor cells by microarray analysis of genomic representations. Genome Res 10:1726–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantripragada KK, Buckley PG, Diaz de Ståhl T, Dumanski JP (2004a) Genomic microarrays in the spotlight. Trends Genet 20:87–94 [DOI] [PubMed] [Google Scholar]
- Mantripragada KK, Buckley PG, Jarbo C, Menzel U, Dumanski JP (2003) Development of NF2 gene specific, strictly sequence defined diagnostic microarray for deletion detection. J Mol Med 81:443–451 [DOI] [PubMed] [Google Scholar]
- Mantripragada KK, Tapia-Paez I, Blennow E, Nilsson P, Wedell A, Dumanski JP (2004b) DNA copy-number analysis of the 22q11 deletion-syndrome region using array-CGH with genomic and PCR-based targets. Int J Mol Med 13:273–279 [PubMed] [Google Scholar]
- Martone R, Euskirchen G, Bertone P, Hartman S, Royce TE, Luscombe NM, Rinn JL, Nelson FK, Miller P, Gerstein M, Weissman S, Snyder M (2003) Distribution of NF-κB-binding sites across human chromosome 22. Proc Natl Acad Sci USA 100:12247–12252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimault C, Giraud G, Courtois V, Cailloux F, Boire JY, Dastugue B, Boespflug-Tanguy O, and the Clinical European Network on Brain Dysmyelinating Disease (1999) Proteolipoprotein gene analysis in 82 patients with sporadic Pelizaeus-Merzbacher disease: duplications, the major cause of the disease, originate more frequently in male germ cells, but point mutations do not. Am J Hum Genet 65:360–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelis E, Van Broeckhoven C, De Jonghe P, Lofgren A, Vandenberghe A, Latour P, Le Guern E, Brice A, Mostacciuolo ML, Schiavon F, Palau F, Bort S, Upadhyaya M, Rocchi M, Archidiacono N, Mandich P, Bellone E, Silander K, Savontaus ML, Navon R, Goldberg-Stern H, Estivill X, Volpini V, Friedl W, Gal A (1996) Estimation of the mutation frequencies in Charcot-Marie-Tooth disease type 1 and hereditary neuropathy with liability to pressure palsies: a European collaborative study. Eur J Hum Genet 4:25–33 [DOI] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, Gray JW, Albertson DG (1998) High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet 20:207–211 [DOI] [PubMed] [Google Scholar]
- Plant KE, Green PM, Vetrie D, Flinter FA (1999) Detection of mutations in COL4A5 in patients with Alport syndrome. Hum Mutat 13:124–132 [DOI] [PubMed] [Google Scholar]
- Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO (1999) Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet 23:41–46 [DOI] [PubMed] [Google Scholar]
- Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO (2002) Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA 99:12963–12968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautenstrauss B, Lupski, J, Timmerman V (2000) Draft best practice guidelines for molecular analysis of hereditary motor and sensory neuropathies. European Molecular Genetics Quality Network (http://www.emqn.org/Assets/uploadpdfs/HMSN.pdf) (accessed March 1, 2005)
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD (2002) E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev 16:245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386 [DOI] [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G (2002) Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 30:e57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler GD (1997) Sequence mapping by electronic PCR. Genome Res 7:541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M (2004) Large-scale copy number polymorphism in the human genome. Science 305:525–528 [DOI] [PubMed] [Google Scholar]
- Sellner LN, Taylor GR (2004) MLPA and MAPH: new techniques for detection of gene deletions. Hum Mutat 23:413–419 [DOI] [PubMed] [Google Scholar]
- Sistermans EA, de Coo RF, De Wijs IJ, Van Oost BA (1998) Duplication of the proteolipid protein gene is the major cause of Pelizaeus-Merzbacher disease. Neurology 50:1749–1754 [DOI] [PubMed] [Google Scholar]
- Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, Law S, Myambo K, Palmer J, Ylstra B, Yue JP, Gray JW, Jain AN, Pinkel D, Albertson DG (2001) Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet 29:263–264 [DOI] [PubMed] [Google Scholar]
- Snijders AM, Pinkel D, Albertson DG (2003) Current status and future prospects of array-based comparative genomic hybridisation. Brief Funct Genomic Proteomic 2:37–45 [DOI] [PubMed] [Google Scholar]
- Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Dohner H, Cremer T, Lichter P (1997) Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer 20:399–407 [PubMed] [Google Scholar]
- Strautnieks S, Rutland P, Winter RM, Baraitser M, Malcolm S (1992) Pelizaeus-Merzbacher disease: detection of mutations Thr181→Pro and Leu223→Pro in the proteolipid protein gene, and prenatal diagnosis. Am J Hum Genet 51:871–878 [PMC free article] [PubMed] [Google Scholar]
- Vissers LELM, de Vries BBA, Osoegawa K, Janssen IM, Feuth T, Choy CO, Straatman H, van der Vliet W, Huys EHLPG, van Rijk A, Smeets D, van Ravenswaaij-Arts CMA, Knoers NV, van der Burgt I, de Jong PJ, Brunner HG, van Kessel AG, Schoenmakers EFPM, Veltman JA (2003) Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am J Hum Genet 73:1261–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann AS, Yan PS, Oberley MJ, Huang TH, Farnham PJ (2002) Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev 16:235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S, Kalf M, Liu Q, Villerius M, Engelsma D, Kriek M, Vollebregt E, Bakker B, van Ommen G-JB, Breuning MH, den Dunnen JT (2002) Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization. Am J Hum Genet 71:365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SJ, Sterrenburg E, van Ommen GJ, den Dunnen JT, Breuning MH (2003) An alternative to FISH: detecting deletion and duplication carriers within 24 hours. J Med Genet 40:e113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SJ, Vink GR, Kriek M, Wuyts W, Schouten J, Bakker B, Breuning MH, den Dunnen JT (2004) Two-color multiplex ligation-dependent probe amplification: detecting genomic rearrangements in hereditary multiple exostoses. Hum Mutat 24:86–92 [DOI] [PubMed] [Google Scholar]
- Woodward K, Kendall E, Vetrie D, Malcolm S (1998) Pelizaeus-Merzbacher disease: identification of Xq22 proteolipid-protein duplications and characterization of breakpoints by interphase FISH. Am J Hum Genet 63:207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward K, Kirtland K, Dlouhy S, Raskind W, Bird T, Malcolm S, Abeliovich D (2000) X inactivation phenotype in carriers of Pelizaeus-Merzbacher disease: skewed in carriers of a duplication and random in carriers of point mutations. Eur J Hum Genet 8:449–454 [DOI] [PubMed] [Google Scholar]
- Worton RG, Thompson MW (1988) Genetics of Duchenne muscular dystrophy. Annu Rev Genet 22:601–629 [DOI] [PubMed] [Google Scholar]