Abstract
A unique heterozygous 3-kb microdeletion within STX16, a closely linked gene centromeric of GNAS, was previously identified in multiple unrelated kindreds as a cause of autosomal dominant pseudohypoparathyroidism type Ib (AD-PHP-Ib). We now report a novel heterozygous 4.4-kb microdeletion in a large kindred with AD-PHP-Ib. Affected individuals from this kindred share an epigenetic defect that is indistinguishable from that observed in patients with AD-PHP-Ib who carry the 3-kb microdeletion in the STX16 region (i.e., an isolated loss of methylation at GNAS exon A/B). The novel 4.4-kb microdeletion overlaps with the previously identified deletion by 1,286 bp and, similar to the latter deletion, removes several exons of STX16 (encoding syntaxin-16). Because these microdeletions lead to AD-PHP-Ib only after maternal transmission, we analyzed expression of this gene in lymphoblastoid cells of affected individuals with the 3-kb or the 4.4-kb microdeletion, an individual with a NESP55 deletion, and a healthy control. We found that STX16 mRNA was expressed in all cases from both parental alleles. Thus, STX16 is apparently not imprinted, and a loss-of-function mutation in one allele is therefore unlikely to be responsible for this disorder. Instead, the region of overlap between the two microdeletions likely harbors a cis-acting imprinting control element that is necessary for establishing and/or maintaining methylation at GNAS exon A/B, thus allowing normal Gαs expression in the proximal renal tubules. In the presence of either of the two microdeletions, parathyroid hormone resistance appears to develop over time, as documented in an affected individual who was diagnosed at birth with the 4.4-kb deletion of STX16 and who had normal serum parathyroid hormone levels until the age of 21 mo.
Introduction
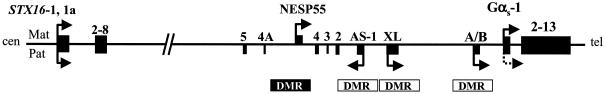
GNAS is a complex imprinted locus that yields—in addition to the α-subunit of the stimulatory G protein (Gαs)—multiple alternative transcripts (fig. 1). These transcripts encode extra-large Gαs (XLαs) (Kehlenbach et al. 1994; Hayward et al. 1998a; Peters et al. 1999) or neurosecretory protein 55 (NESP55) (Ischia et al. 1997; Hayward et al. 1998b; Peters et al. 1999), or they lead to presumably nontranslated RNAs, such as the A/B transcript (also referred to as “1A” or “1′”) (Ishikawa et al. 1990; Swaroop et al. 1991; Liu et al. 2000b) and the antisense (AS) transcript (referred to as “Nespas” in mouse) (Hayward and Bonthron 2000; Liu et al. 2000b; Wroe et al. 2000). NESP55, XLαs, and A/B differ from Gαs only by their first exons, whereas AS transcripts consist of entirely distinct exons. AS, XLαs, and A/B are paternally derived transcripts, whereas NESP55 is a maternally derived transcript. Consistent with their monoallelic expression, the promoters of these imprinted transcripts are located within differentially methylated regions (DMRs) (fig. 1). In contrast, the promoter that gives rise to Gαs is not methylated, and, accordingly, expression of Gαs occurs in most tissues from both parental GNAS alleles (Campbell et al. 1994; Hayward et al. 1998a, 1998b; Zheng et al. 2001). In some tissues (including proximal renal tubules, pituitary, gonads, and thyroid), however, Gαs transcripts are predominantly derived from the maternal allele (Hayward et al. 1998a, 2001; Yu et al. 1998; Li et al. 2000; Mantovani et al. 2002).
Figure 1.
Schematic representation of the human STX16 and GNAS loci (not to scale). Exons are depicted as blackened boxes; arrows indicate direction (sense, antisense) and allelic origin (Mat = maternal [above the line]; Pat = paternal [below the line]) of transcription. Note that Gαs is transcribed from both alleles in most tissues (Campbell et al. 1994; Hayward et al. 1998a, 1998b; Zheng et al. 2001) and that the paternal allele is silenced (dotted arrow) in some tissues, including kidney, adipose tissue, and pituitary (Hayward et al. 1998a, 2001; Yu et al. 1998; Li et al. 2000; Mantovani et al. 2002). Paternal and maternal DMRs are indicated by blackened and unblackened rectangles, respectively.
Pseudohypoparathyroidism (PHP) is a disorder characterized by hypocalcemia, hyperphosphatemia, and parathyroid hormone (PTH) levels that are elevated as a result of end-organ resistance to this hormone. One of the two main subtypes of PHP, PHP type Ia (PHP-Ia [MIM 103580]), is caused by heterozygous maternally inherited loss-of-function mutations in GNAS exons 1–13, which encode Gαs. In addition to PTH resistance, affected individuals typically show clinical abnormalities referred to as “Albright hereditary osteodystrophy” (AHO) and resistance toward several additional hormones, including thyroid-stimulating hormone (TSH) and gonadotropins. When inherited paternally, Gαs loss-of-function mutations are associated with AHO alone, in the absence of hormonal resistance—a variant termed “pseudopseudohypoparathyroidism” (PPHP) (Weinstein et al. 2001; Levine 2002). The other main form of PHP, PHP type Ib (PHP-Ib [MIM 603233]), differs from PHP-Ia and PPHP, in that PHP-Ib patients do not have the AHO phenotype (Weinstein et al. 2001). Furthermore, hormone resistance in PHP-Ib appears to be limited to the renal actions of PTH, although resistance to TSH is also seen occasionally (Bastepe et al. 2001a, 2001b; Liu et al. 2003).
The autosomal dominant variant of PHP-Ib (AD-PHP-Ib) has been linked to a region of chromosome 20q13.3 that comprises the GNAS locus, and, similar to PHP-Ia, this disorder is maternally transmitted (Jüppner et al. 1998). Most individuals affected by AD-PHP-Ib show a methylation defect that is limited to GNAS exon A/B, whereas most apparently sporadic cases of the disease display abnormal methylation at other GNAS DMRs as well (Liu et al. 2000a, 2005; Bastepe et al. 2001b, 2003). An identical 3-kb microdeletion located ∼220 kb upstream of GNAS has been identified in all affected members and obligate carriers of 14 unrelated families with AD-PHP-Ib and in two apparently sporadic cases (Bastepe et al. 2003). All affected individuals share the same epigenetic defect at exon A/B and lack methylation abnormalities at the other DMRs of GNAS. The 3-kb microdeletion lies between two direct repeats and removes intronic as well as exonic sequences of the gene encoding syntaxin-16 (STX16). Consistent with the imprinted inheritance pattern of the disease, transmission of the microdeletion from a female obligate carrier causes PTH resistance, whereas transmission of the same mutation from a male obligate carrier results in normal PTH responsiveness (Bastepe et al. 2003). Recently, additional patients with PHP-Ib and with the isolated GNAS exon A/B methylation defect have been reported to carry the same 3-kb microdeletion (Liu et al. 2005; Mahmud et al. 2005), thereby strengthening the likely causative association between the exon A/B methylation defect and this deletion.
In one family with AD-PHP-Ib (kindred W), the 3-kb microdeletion was not found, despite a loss of methylation at GNAS exon A/B alone in the affected individuals (Bastepe et al. 2001b, 2003), suggesting the presence of another mutation in the STX16 region. We thus searched, in this family, for mutations within STX16 and identified a novel microdeletion in all affected members and unaffected carriers.
Subjects and Methods
Kindred W and Methylation Analysis of GNAS Exon A/B
The pedigree (kindred W) and the clinical and laboratory data of most members of this family have been described elsewhere (Bastepe et al. 2001b) (see also fig. 2A). Genomic DNA was isolated from blood leukocytes of all family members, and a lymphoblastoid cell line was established for patient W-III/3 (Harvard-Partners Center for Genetics and Genomics Biosample Services Facility). Written informed consent was obtained from all investigated individuals (Massachusetts General Hospital institutional review board [#2001p-000648]).
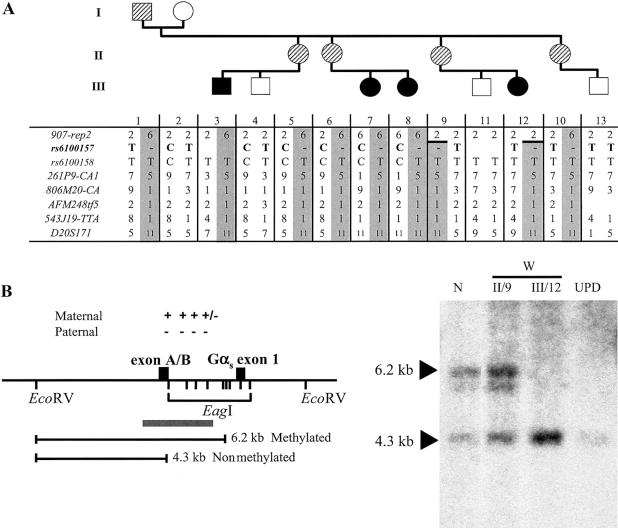
Figure 2.
A, Evidence of allelic loss within the STX16 locus in the obligate carriers W-II/5 and W-II/6. All the affected individuals (blackened symbols) as well as the obligate carriers (striped symbols) share the same disease-associated haplotype (gray columns) at 20q13.3; note that not all investigated markers are shown. A recombination event occurred in individual W-II/9 between markers 907-rep2 and 261P9-CA1 and was transmitted to her affected daughter, W-III/12, thus reducing the critical interval likely containing the genetic lesion that causes AD-PHP-Ib. Evidence of allelic loss was found at SNP rs6100157 (bold typeface) in obligate carriers W-II/5 and W-II/6. B, Loss of methylation at GNAS exon A/B in the newborn W-III/12. The methylation status of GNAS exon A/B was assessed by Southern blot analysis of cord blood DNA from patient W-III/12 and of genomic DNA from her unaffected mother (W-II/9), an individual with paternal uniparental isodisomy of chromosome 20q (“UPD”) (Bastepe et al. 2001a), and a healthy control individual (“N”). These samples were digested with a methylation-sensitive (EagI) and a methylation-insensitive (EcoRV) restriction endonuclease, as described elsewhere (Bastepe et al. 2001b). Locations of restriction sites and their methylation status (“+” = methylated; “−” = nonmethylated), the predicted fragments, and the probe (gray bar) are shown in the left panel. Affected individuals (UPD and W-III/12) show loss of methylation at exon A/B, whereas unaffected individuals (N and W-II/9) have one methylated and one nonmethylated allele. Fragment sizes are indicated by arrowheads. Note that the faint hybridizing band below the 6.2-kb band reflects partial methylation (“+/−”) and, hence, incomplete digestion at one of the EagI sites between exon A/B and Gαs exon 1.
Since 2001, when this family was first reported (Bastepe et al. 2001b), two children have been born: W-III/12 (born to W-II/9) and W-III/13 (born to W-II/10). Genomic DNA was extracted from cord blood of both children. Methylation of GNAS exon A/B was assessed by nucleotide sequence analysis of PCR-amplified bisulfite-treated genomic DNA and by Southern blot analysis with the use of EagI (methylation sensitive) and EcoRV (methylation insensitive) (New England Biolabs), as described elsewhere (Jüppner et al. 1998; Bastepe et al. 2001b, 2003). Total serum calcium, phosphorus, intact PTH (Nichols Institute), and TSH were measured in patient W-III/12 shortly after birth and subsequently at regular intervals.
Analysis of Microsatellite Markers and SNPs
Microsatellite markers D20S102, D20S25, D20S86, D20S164, 907-Rep2, 907-Rep4, 261P9-CA1, 806M20-CA, AFM248tf5, GNAS, 543J19-TTA, D20S171, stAFMa202yb9, and D20S93 were analyzed as described elsewhere (Bastepe et al. 2003). SNPs rs6100157, rs2296524, and rs6100158—which correspond to nucleotides 3959, 4712, and 5326, respectively, of AL139349—were analyzed by nucleotide sequence analysis after PCR amplification, which was performed using conditions and primers reported elsewhere (table 1) (Jüppner et al. 1998; Bastepe et al. 2001b, 2003).
Table 1.
Sequences of the Primers Used in the Present Study
|
Amplification Primers |
Sequencing Primers |
|||
| Use for Primers | Forward | Reverse | Forward | Reverse |
| rs6100157 | TTGGCAGATAACTGCTGTGG | AGGCCCGGGCAGCACTCAC | ACTGCTGTGGACGGATTAA | AATCCAAAATGCTCCAATG |
| rs2296524 | CCTGAAGTCCTCCAGCTGTC | GGGGTGGAGGGAGAGTAAAG | GCTTCTCTCCCAGGCTTGC | AGGCCCGGGCAGCACTCAC |
| rs6100158 | GGGCACCAAGATAGTTCAGC | GGGGTGGAGGGAGAGTAAAG | GTTTCACCCAGCAGACCTT | … |
| Isoform B of STX16 | (G): AGCCCTCTGCATTCACGTAG | (H): TATCGAGACTTCACGCCAAC | … | CCGATGGTAAAGAGTGTTATCGa |
| PCR across the 4.4-kb microdeletion | (A): TGAGGGGAACACATGCATAA | (B): CCGATGGTAAAGAGTGTTATCG | GGAGCAGGAGTGTTGAGGAA | … |
| Multiplex PCR for detection of the 4.4-kb microdeletion | (C): GGAGCAGGAGTGTTGAGGAA; (E): TGTCCACGGATGGTCAGTAA | (D): CAGGGCCAGTGTCATTTCTT; (F): AAAGGTGCCAATCACTCCAC | … | … |
This primer was used to sequence the polymorphic nucleotide 527 of STX16 isoform B, which corresponds to rs2296524.
Southern Blot and PCR Analysis for Detection of the Deletion
For Southern blot analysis, genomic DNA (5 μg) was digested with AflII, BclI, or BglII (New England Biolabs). After separation on a 0.8% agarose gel, DNA fragments were transferred onto a nitrocellulose filter and were hybridized with a 32P-radiolabeled probe (nucleotides 8299–9921 of AL139349), which was generated by PCR; primers are listed in table 1.
PCR amplification of genomic DNA across the predicted deletion was performed using primers 5.9 kb apart from each other (primers A and B) (table 1) and the Expand Long-Template PCR System with Buffer 3 (Roche Molecular Biochemicals). The cycling conditions were an initial 94°C for 3 min; 10 cycles of 94°C for 10 s, 58°C for 30 s, and 68°C for 5 min; 25 cycles of 94°C for 10 s, 58°C for 30 s, and 68°C for 5 min, with an additional 20 s at 68°C after each cycle; and 1 final cycle at 72°C for 10 min. These conditions yielded a 5.9-kb amplicon, which was not consistently amplified in all samples. The same conditions also yielded an ∼1.5-kb amplicon in affected individuals and obligate carriers, which was gel purified and submitted for nucleotide sequence analysis (Massachusetts General Hospital DNA Core Facility).
For multiplex PCR, two pairs of primers (C and D, E and F) were designed, to amplify small fragments across each boundary of the deletion (table 1). Both pairs were used for simultaneous PCR amplification of three different fragments: a 616-bp fragment (primers C and D) and a 252-bp fragment (primers E and F), derived from the centromeric and telomeric ends of the deleted region, respectively, as well as a 4.8-kb fragment (primers C and F), derived from the wild-type allele. As a result of its large size, however, the latter fragment could not be effectively amplified each time.
Analyzing STX16 Transcripts
Total RNA was extracted from lymphoblastoid cells derived from the affected individuals W-III/3, F-III/37, C-II/1, as well as a healthy control individual (Harvard-Partners Center for Genetics and Genomics Biosample Services Facility). Subject F-III/37 carries the previously described 3-kb deletion (Bastepe et al. 2003). Subject C-II/1 has methylation abnormalities at all GNAS DMRs and has been recently shown to carry a novel microdeletion involving exon NESP55 and antisense exons 3 and 4 (Bastepe et al. 2005). Consistent with the conclusion that the STX16 region is intact, subject C-II/1 was furthermore shown to be heterozygous at SNPs rs6100157 and rs2296524, which are located within the centromeric repeat and STX16 exon 4, respectively. STX16 mRNAs were amplified by RT-PCR. One microgram of lymphoblast-derived total RNA was reverse transcribed using Superscript II (Invitrogen) and oligo (dT) primers, in accordance with manufacturer recommendations; the same reactions, without reverse transcriptase, served as negative controls. PCR was then performed using 1 μl of 1:10 dilution of the reverse-transcription mixture, a forward primer located in exon 1 (primer G), and a reverse primer located in exon 8 (primer H) in a total reaction volume of 50 μl (table 1 and fig. 3). The amplification conditions were an initial 94°C for 2 min; 31 cycles of denaturation at 95°C for 30 s, annealing at 61°C for 30 s, and extension at 72°C for 1 min; and 1 final cycle of extension at 72°C for 10 min. Amplicons were separated on a 2% agarose gel, were purified, and were submitted for nucleotide sequence analysis (Massachusetts General Hospital DNA Core Facility), as described elsewhere (Bastepe et al. 2001b). To determine the allelic origin of the STX16 transcripts in the healthy control individual and in subject C-II/1, total RNA was amplified by RT-PCR as described above, and the polymorphic nucleotide at position 527 of STX16 isoform B was analyzed by direct sequence analysis of the amplicon; this corresponds to SNP rs2296524 (table 1).
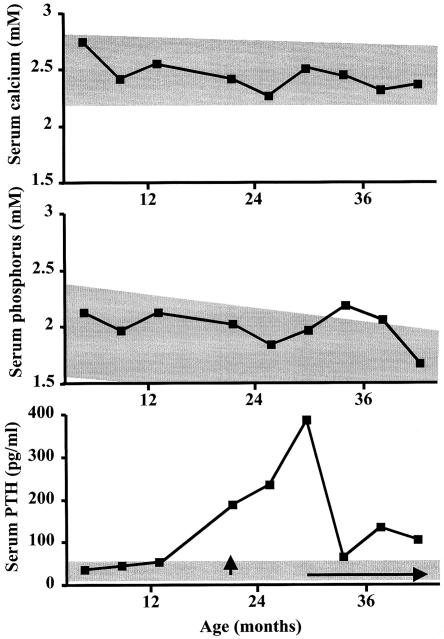
Figure 3.
Delayed onset of PTH resistance in patient W-III/12. Serum calcium, phosphorus, and PTH levels were measured during follow-up visits of the patient—in whom loss of methylation at GNAS exon A/B was identified at birth—and the 4.4-kb deletion was discovered subsequently. Normal ranges are indicated by gray shading. Treatment with an active vitamin D analog (Rocaltrol) was first started at age 21 mo (vertical arrow). However, the parents discontinued the treatment shortly thereafter, and continuous treatment was not resumed until the child was 30 mo old (horizontal arrow).
Results
Loss of Exon A/B Methylation and Development of PTH Resistance
We have reported elsewhere that affected members of kindred W show a loss of exon A/B methylation without additional epigenetic defects of GNAS (Bastepe et al. 2003). Nucleotide sequence analysis of PCR-amplified bisulfite-treated cord blood DNA from the newborn W-III/13 showed a normal methylation pattern at GNAS exon A/B (data not shown). Accordingly, haplotype analysis across the GNAS region revealed that this individual had inherited the grandmaternal haplotype, which is not associated with disease (fig. 2A) (Bastepe et al. 2001b). Thus, two independent approaches indicated that subject W-III/13 would not be affected with AD-PHP-Ib. In contrast, Southern blot analysis of cord blood DNA from his cousin, W-III/12, performed using the restriction endonucleases EcoRV and EagI, showed loss of methylation at GNAS exon A/B (fig. 2B), and haplotype analysis revealed inheritance of the disease-associated grandpaternal haplotype (fig. 2A). She was therefore predicted to develop PTH resistance, which was subsequently documented (see below). Because patient W-III/12 was found to be affected, the maximum LOD score increased from 2.03 (Bastepe et al. 2001b) to 2.43 for genetic linkage to 20q13.3 (D20S171 [θ=0]), increasing the likelihood that the defect leading to AD-PHP-Ib in this family also resides close to the GNAS locus.
Birth weight and height of patient W-III/12 were 4.2 kg (95th percentile [North American reference chart]) and 53 cm (95th percentile), respectively. Her weight, height, and BMI remained above the 95th percentile during infancy and early childhood. Except for being overweight, she had no features suggestive of AHO. PTH resistance was absent at birth but developed by age 21 mo, when serum PTH levels were first noted to be above the normal range (188 pg/ml; normal range 10–65 pg/ml). Both total serum calcium and phosphorous concentrations remained within normal range during this period, although serum phosphorus was close to the upper limit (fig. 3). Because of the genetic findings and the elevation in PTH concentration, treatment with an active vitamin D analog (Rocaltrol) and calcium supplements was first initiated at age 21 mo. However, the parents discontinued these medications shortly thereafter, and it was not until the child was 30 mo old that uninterrupted treatment was started. By that time, the PTH concentration had increased to 386 pg/ml. In addition, mild TSH resistance was suspected at age 30 mo because of elevated TSH (14.5 mUI/l; normal range 0.5–5 mUI/l), normal free T4 (0.9 ng/dl; normal range 0.7–1.7 ng/dl), and undetectable serum anti-thyroglobulin and anti-thioperoxidase antibodies. l-thyroxine treatment was then started, and the TSH level dropped to the normal range (2.89 mUI/l at 41 mo).
Identification of a Novel Deletion within the STX16 Region
All affected individuals of kindred W (W-III/3, W-III/7, W-III/8, and W-III/12) showed a loss of methylation at GNAS exon A/B, without methylation abnormalities involving the NESP55, XL, or AS DMRs (fig. 2B and Bastepe et al. [2001b]). Nevertheless, unlike many other kindreds with AD-PHP-Ib with the same epigenetic defect, the previously described 3-kb deletion (nucleotides 3786–6764 of AL139349) could not be found in this kindred. In fact, all affected individuals and obligate carriers were heterozygous at microsatellite marker 261P9-CA1, and some of them, furthermore, were heterozygous at SNP rs6100158 (figs. 2A and 4A). In addition, PCR analysis across the 3-kb deletion, performed using previously established primers and conditions (Bastepe et al. 2003), revealed no evidence of a smaller deletion within this region.
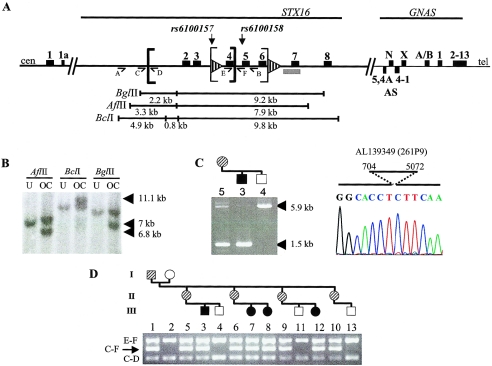
Figure 4.
Identification of a novel 4.4-kb deletion in kindred W. A, Simplified map of the 20q13.3 region and the localization of the two known microdeletions. Sequence of the depicted region is included in AL050327 (clone 907D15) and AL139349 (clone 261P9). Exons are depicted as blackened rectangles, and direct repeats are shown as striped arrowheads. The 3-kb and 4.4-kb deleted regions lie between the thin and thick brackets, respectively. The locations of recognition sites for the restriction endonucleases used in the Southern blot analysis are shown under the gene map. The location of the 1.6-kb probe is marked by the gray bar, and primers A, B, C, D, E, and F, which were used for PCR analyses, are indicated by arrowed lines; cen = centromeric; tel = telomeric; N = NESP55; AS = antisense; X = XLαs; A/B = exon A/B. B, Southern blot analysis of genomic DNA from an unaffected (“U”) subject, W-III/4, and an obligate carrier (“OC”), W-II/5, which was digested with three different restriction endonucleases (AflII, BclI, and BgIII). Hybridization with the 1.6-kb 32P-labeled probe (nucleotides 8299–9921 of AL139349) showed the expected wild-type bands of 7.9 kb, 9.8 kb, and 9.2 kb, respectively. Additional bands (arrowheads) in the obligate carrier (6.8 kb, 11.1 kb, and 7 kb, respectively) are compatible with a 4.4-kb deletion removing one AflII site, two BclI sites, and one BglII site—all of which are centromeric of STX16 exon 2. C, Confirmation and exact localization of the deletion by PCR. Amplification across the predicted location of the deletion, performed using one pair of primers (A [forward] and B [reverse]), yielded a 5.9-kb wild-type fragment in the unaffected subject, the affected individual, and the obligate carrier; both the affected individual and the obligate carrier also showed a shorter fragment (of 1.5 kb). Direct nucleotide sequence analysis of the short amplicon revealed the lack of 4,368 bp extending from nucleotide 704 to nucleotide 5072 of AL139349. D, Amplification of the STX16 region with four different primers, located at nucleotides 404 (primer C [forward]), 1020 (primer D [reverse]), 4938 (primer E [forward]), and 5190 (primer F [reverse]) of AL139349. The sizes of expected wild-type PCR products are 616 bp (primers C and D), 252 bp (primers E and F), and 4.8 kb (primers C and F [not shown]). Obligate carriers and affected individuals show an additional 422-bp amplicon (arrow [primers C and F]). Results are shown for each individual in the family (blackened symbols represent affected individuals, striped symbols represent obligate carriers, and unblackened symbols represent unaffected individuals).
Analysis of additional polymorphisms centromeric of the heterozygous SNP rs6100158 revealed allelic loss at SNP rs6100157 for the obligate carriers W-II/5 and W-II/6, indicating the presence of a deletion comprising this region (figs. 2A and 4A). This deletion was confirmed by Southern blot analysis of genomic DNA from individual W-II/5 and her unaffected child W-III/4, performed using restriction enzymes AflII, BclI, or BglII (fig. 4B). The probe, located telomeric of SNP rs6100158, detected wild-type hybridizing fragments of 7.9 kb, 9.8 kb, and 9.2 kb in both samples. In addition, however, hybridizing AflII, BclI, and BglII fragments of ∼6.8 kb, 11.1 kb, and 7 kb were detected, respectively, in the DNA sample from individual W-II/5 but not in the DNA sample from individual W-III/4. The sizes of these additional bands were compatible with an ∼4.4-kb deletion that removed one centromeric AflII site, two BclI sites, and one BglII site.
To determine the exact location of the deletion, we performed a series of PCRs with the use of primers that were predicted to amplify a region of the mutant allele spanning the deletion breakpoint. One set of primers (primers A and B) amplified a single product of ∼5.9 kb from genomic DNA of the unaffected individual W-III/4, whereas the same primers produced an additional smaller amplicon of ∼1.5 kb from DNA of the affected individual W-III/3 and unaffected carrier W-II/5 (fig. 4A and 4C). The difference between the lengths of the small and the large amplicons was consistent with a 4.4-kb deletion, suggesting that the small amplicon was derived from the mutant allele. Direct nucleotide sequence analysis of this mutant fragment showed that the deletion occurred between nucleotides 704 and 5072 of AL139349, thus eliminating exons 2–4 of STX16 (fig. 4C). The presence of this deletion was also confirmed in the remaining affected individuals or unaffected carriers by multiplex PCR, performed using primers designed to amplify across the deleted region and across the predicted boundaries. All affected individuals and obligate carriers of kindred W showed—in addition to the wild-type products of 616 bp (primers C and D) and 252 bp (primers E and F)—a 422-bp product amplified across the deletion breakpoints (primers C and F) (fig. 4A and 4D). In contrast, genomic DNA from the healthy individuals generated only the two wild-type fragments; note that the 4.8-kb product derived from the wild-type allele (primers C and F) could not be routinely amplified, because of its size (data not shown).
Biallelic Expression of STX16
Although the identified deletion removes coding exons from the gene STX16, a loss-of-function mutation in one STX16 allele could not directly explain the imprinted AD-PHP-Ib phenotype unless STX16 itself is an imprinted gene. We therefore examined allelic expression of STX16 in total RNA extracted from lymphoblastoid cells of a healthy control individual and of three AD-PHP-Ib individuals carrying different mutations. STX16 normally gives rise to at least three differently spliced mRNA transcripts: STX16A, STX16B, and STX16C (AL050327 and AL139349 [fig. 5A]). RT-PCR amplification of total RNA from both the control individual and subject C-II/1 (deletion of the NESP55 DMR [Bastepe et al. 2005]), performed using primers G and H, yielded products of 861 bp and 802 bp, corresponding to isoform B of STX16 and its splice variant (which is missing the alternatively spliced exon 4), respectively (fig. 5B). Subsequent nucleotide sequence analysis of the 861-bp products revealed heterozygosity at position 527 of STX16 isoform B located within exon 4 (SNP rs2296524 [fig. 5C]), indicating that both parental alleles were transcribed and amplified. Furthermore, RT-PCR performed using lymphoblast-derived total RNA of the affected individual W-III/3 (4.4-kb STX16 deletion) showed amplicons from the wild-type splice variants of STX16 isoform B, as well as a shorter amplicon of 449 bp (fig. 5A and 5B). Direct sequence analysis of this short product confirmed the absence of nucleotides derived from STX16 exons 2, 3, and 4. Likewise, transcripts amplified by RT-PCR from individual F-III/37 (3-kb STX16 deletion) included, in addition to the wild-type splice variants of STX16 isoform B, a shortened transcript represented by an amplicon of 462 bp (fig. 5A and 5B). Nucleotide sequence analysis of this shorter product confirmed the deletion of exons 4, 5, and 6. Together, these findings suggested that both parental STX16 alleles are transcribed in lymphoblastoid cells and that the identified deletions do not appear to alter this biallelic expression. We also searched the EST database and identified two ESTs (CN348556 and CN417685) matching the STX16 exon 8 sequence that were derived from the same cDNA library yet differed by one nucleotide (position 937 of STX16 isoform B; position 11538 of AL139349). Since this nucleotide change was also seen in several other ESTs from the database, it indicated its polymorphic nature rather than a sequence error. This finding strongly suggested that STX16 mRNAs were amplified from both parental alleles in that particular library as well.
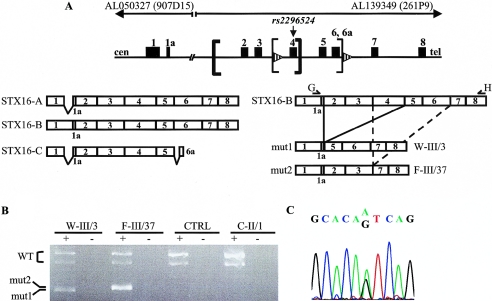
Figure 5.
Biallelic expression of STX16. A, STX16 gives rise to at least three distinct transcripts (left). Isoform A (915 bp) differs from isoform B (965 bp) by a shorter sequence derived from exon 1, whereas isoform C (422 bp) comprises this short exon 1 sequence as well as exons 2–5 and terminates with exon 6a. In addition, exon 4 is alternatively spliced in each isoform, leading to a total of at least six variants. Wild-type STX16 isoform B and the transcripts derived from individuals W-III/3 and F-III/37 (“mut1” and “mut2,” respectively) are depicted (right). The 4.4-kb deletion (thick brackets) removes exons 2–4; the 3-kb deletion (thin brackets) removes exons 4–6. Exons are depicted as blackened boxes, and direct repeats are shown as striped arrowheads. Primers G and H are represented as arrowed lines; cen = centromeric; tel = telomeric. B, RT-PCR analysis of STX16 transcripts in total RNA of immortalized lymphocytes from a healthy control individual (CTRL) and the following individuals with AD-PHP-Ib: W-III/3 (present study), F-III/37 (carrying the 3-kb deletion in STX16 region [Bastepe et al. 2003]), and C-II/1 (carrying a deletion affecting NESP55 but not the STX16 region [Bastepe et al. 2005]). Reactions were performed with (“+”) or without (“−”) reverse transcriptase, as described in the “Subjects and Methods” section. Products derived from the mutant alleles (“mut1” and “mut2”) and from the wild-type alleles (“WT”) are indicated. Note that the wild-type products represent STX16 isoform B with or without sequences derived from exon 4. C, Direct nucleotide sequence of STX16 isoform B amplified from RNA of a healthy control individual shows heterozygosity at the polymorphic nucleotide 527, corresponding to SNP rs2296524.
Discussion
We report a novel STX16 microdeletion of 4.4 kb in all affected individuals and obligate carriers of a large kindred with AD-PHP-Ib. This 4.4-kb microdeletion was present in the unaffected mothers of the affected individuals (n=4) and in the father of these obligate carriers (figs. 2 and 4). This mode of inheritance is concordant with the paternal imprinting of the disease in this and other families with AD-PHP-Ib. The 4.4-kb microdeletion is the first STX16 mutation identified in patients with AD-PHP-Ib that differs from the 3-kb microdeletion present in multiple unrelated kindreds with this disorder (Bastepe et al. 2003; Liu et al. 2005; Mahmud et al. 2005). Our findings thus provide further evidence supporting the conclusion that STX16 harbors an important element involved in the pathogenesis of PHP-Ib. As the two deletions overlap by 1,286 bp, it is likely that this element is contained in this overlap region. Thus far, of the 31 unrelated individuals with PHP-Ib that we have investigated, 18 (58%) carry a deletion within STX16 (Bastepe et al. 2003; Mahmud et al. 2005). Recently, the 3-kb deletion has also been found in 5 of 20 unrelated individuals with PHP-Ib (Liu et al. 2005). Thus, mutations in this region are frequently the cause of PHP-Ib. Affected individuals who carry the 3-kb and 4.4-kb microdeletions exhibit loss of methylation only at GNAS exon A/B; the methylation pattern is not altered at other GNAS DMRs. Conversely, individuals with PHP-Ib who lack such STX16 deletions display methylation abnormalities at all GNAS DMRs (Bastepe et al. 2003; Liu et al. 2005). On the basis of these findings, we conclude that PHP-Ib comprises at least two distinct conditions sharing the same clinical phenotype: one is associated with the loss of exon A/B methylation alone—and, in most of the cases, with a heterozygous microdeletion in the STX16 region—and the other is associated with methylation abnormalities at all GNAS DMRs, including the DMR at exon A/B. In two unrelated kindreds with AD-PHP-Ib with broad methylation defects of GNAS, we have recently identified deletions affecting the NESP55 DMR (Bastepe et al. 2005), and it remains to be determined whether the sporadic cases with this epigenotype also carry an as-yet-undefined microdeletion within or close to this region.
Deletion of STX16 exons 2–4, caused by the mutation presented in the present study, results in a frameshift and a premature termination codon following 20 new amino acids, whereas deletion of STX16 exons 4–6, caused by the previously described 3-kb microdeletion (Bastepe et al. 2003), maintains the ORF. Thus, the syntaxin-16 proteins generated by the 4.4-kb or 3-kb deleted alleles either are shortened by 276 amino acids at the carboxyl-terminus or have a deletion of 133 amino acids in the midportion, respectively. Because of these severe alterations, both products are predicted to be nonfunctional, raising the question of whether syntaxin-16 deficiency may be responsible for the epigenetic abnormality at GNAS exon A/B and the resulting PTH resistance. However, this possibility seems unlikely for two reasons. First, the previously reported lack of differential methylation at the STX16 promoter (Bastepe et al. 2003)—as well as our present demonstration of biallelic expression of STX16 isoform B—strongly argues that this gene is not imprinted. Although not entirely ruled out by our analysis, selective imprinting of the other STX16 transcripts seems unlikely, given that all the known transcripts apparently use the same promoter. This lack of parental origin–specific STX16 expression is inconsistent with the imprinted mode of inheritance in kindreds with AD-PHP-Ib. Second, PTH resistance can develop despite the presence of two intact copies of STX16, as shown in a case of paternal uniparental isodisomy of chromosome 20q (Bastepe et al. 2001a). On the other hand, maternal inheritance of STX16 mutations is associated with loss of methylation at maternal GNAS exon A/B, and it is therefore more likely that these deletions disrupt a cis-acting imprinting control element (ICE) necessary for the methylation imprint at exon A/B.
The novel 4.4-kb microdeletion overlaps with the former 3-kb deletion and therefore reduces the size of the critical region containing the putative ICE for GNAS exon A/B to 1,286 bp. This region encompasses the centromeric direct repeat, and the flanking telomeric sequence of 896 bp, which includes exon 4 of STX16 and intronic sequences of 244 bp and 344 bp on the centromeric and telomeric side, respectively. We could not identify, within the 1,286-bp overlapping region, sequences or elements homologous to those previously identified in other ICEs. For example, microdeletions at the ICE of the IGF2/H19 locus abolish two CTCF binding sites required for the H19 DMR and thus result in the Beckwith-Wiedemann syndrome (Sparago et al. 2004). Sequence analysis of the overlapping region encompassing exon 4 of STX16 did not reveal such known CTCF sites. Another feature of ICEs is that they are mostly located within CpG-rich sequences; in addition, they display differential methylation (Spahn and Barlow 2003). Although the overlapping region includes a 140-bp CpG-rich sequence within exon 4 (nucleotides 4572–4711 of AL139349), we have shown elsewhere that this region is not differentially methylated (Bastepe et al. 2003). It is therefore possible that the mechanisms governing the function of the identified ICE are different from those of most other ICEs in the genome (Spahn and Barlow 2003). Further investigations are necessary to identify the specific features required for the proper activity of the putative ICE.
Two candidate ICEs have been previously identified in the mouse Gnas cluster: one comprising exon 1A (homolog of human A/B) and the other comprising the Gnasxl (homolog of human XLαs) and Nespas promoters (Liu et al. 2000b; Coombes et al. 2003). Mice with a paternal ablation of exon 1A exhibit loss of Gαs silencing in cis, leading to expression of the Gαs transcript from the paternal allele in brown fat and renal proximal tubules (Williamson et al. 2004)—two tissues in which Gαs is normally expressed from the maternal allele alone (Yu et al. 1998). This ablation does not alter the imprinting of the other alternative transcripts, and it therefore appears that a cis-acting element at exon 1A specifically regulates Gαs expression in brown fat, kidney, and perhaps other tissues. Findings in patients with PHP-Ib indicate that this element allows active transcription from the Gαs promoter in the kidney only when it is methylated (Liu et al. 2000a; Bastepe et al. 2001b). However, although the impact of this element is confined to certain tissues, its methylation on the maternal GNAS allele appears to be ubiquitous (Liu et al. 2000b). Our findings in patients with AD-PHP-Ib provide evidence of another cis-acting element within the overlapping region encompassing STX16 exon 4. The latter appears to specifically control the methylation of GNAS exon A/B and, thus, of the cis-acting element regulating the tissue- and parent-specific expression of Gαs. Deletion of this putative STX16 ICE results in a switch from a maternal to a paternal epigenotype at exon A/B, thereby causing derepression of maternal A/B transcription. Because the epigenetic changes associated with the deletion of this putative element have been identified in lymphocytes, its impact seems ubiquitous rather than restricted to kidney and brown fat. In addition to this putative ICE that controls methylation on a restricted area of GNAS (i.e., exon A/B), the identification of deletions of the NESP55 DMR in two unrelated families with AD-PHP-Ib has recently provided evidence of an additional ICE that is involved in methylation of the entire GNAS cluster (Bastepe et al. 2005). Thus, in humans, maternal methylation at GNAS exon A/B (and, hence, Gαs transcription) requires at least two ICEs: one at the STX16 locus, which affects exon A/B, and the other at the NESP55 DMR, which affects all the maternally methylated promoters within the GNAS cluster.
Genetic analysis of cord blood (i.e., exon A/B methylation analysis and multiplex PCR) was particularly useful, allowing us to rule out the diagnosis of AD-PHP-Ib in one newborn and to establish the diagnosis in another. The child with the genetic diagnosis of AD-PHP-Ib was subsequently monitored with respect to development of PTH resistance, and treatment eventually was started to prevent hypocalcemia and to avoid long-term elevation of serum PTH; the latter can lead to increased bone resorption, resulting from the apparent absence of PTH resistance in this tissue. The observed delay in the onset of PTH resistance in this patient is consistent with past studies of individuals affected with PHP-Ib (Marguet et al. 1997; Bastepe et al. 2001b) and was also noted in patients with loss-of-function mutations of Gαs (i.e., PHP-Ia) (Weinstein et al. 2001). PTH signaling through the PTH/PTHrP receptor, therefore, seems intact in the renal proximal tubule during the early postnatal period and infancy. One could hypothesize that Gαs is biallelically expressed in this tissue during this specific period of development. Consistent with this hypothesis, biallelic expression of Gαs has been shown by RT-PCR in whole kidneys and renal cortices from mouse and human fetuses (Hayward et al. 1998a; Zheng et al. 2001). However, these findings—which contradict those showing the exclusive maternal expression of Gαs in renal cortices of adult mice (Yu et al. 1998)—do not rule out the possibility that Gαs expression is monoallelic in renal proximal tubules of fetal or newborn kidney, especially considering that proximal tubules constitute a small portion of the entire kidney. Thus, there may be alternative possibilities to explain the delay in the onset of PTH resistance. One of those possible explanations may involve XLαs, the paternally expressed variant of Gαs. XLαs is expressed in multiple fetal and adult tissues (Kehlenbach et al. 1994; Pasolli et al. 2000), including murine kidney at postnatal day 4 (Plagge et al. 2004). Furthermore, on the basis of data from transfected cells lacking endogenous Gαs and XLαs, XLαs is capable of functionally coupling to the PTH/PTHrP receptor (Bastepe et al. 2002). It is thus possible that PTH actions during early postnatal period are mediated through XLαs.
In conclusion, in the present study, we have (1) identified a novel deletion in the STX16 locus as a cause of AD-PHP-Ib and therefore established a strong association between a unique epigenotype (i.e., loss of A/B methylation alone) and a genotype (i.e., microdeletion involving STX16), (2) refined the boundaries of a putative cis-acting ICE crucial for the establishment/maintenance of methylation at GNAS exon A/B, (3) demonstrated biallelic expression of STX16 mRNAs in lymphoblastoid cells, and (4) established a molecular diagnosis that was based on cord blood DNA in a patient, which helped in the subsequent clinical management of that patient.
Acknowledgments
This work was supported by grants from National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases (RO1 46718-10 [to H.J.] and KO1 DK-062973-01 [to M.B.]) and by a Research Fellowship Grant from the European Society for Paediatric Endocrinology (to A.L.).
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
References
- Bastepe M, Fröhlich LF, Hendy GN, Indridason OS, Josse RG, Koshiyama H, Korkko J, Nakamoto JM, Rosenbloom AL, Slyper AH, Sugimoto T, Tsatsoulis A, Crawford JD, Jüppner H (2003) Autosomal dominant pseudohypoparathyroidism type Ib is associated with a heterozygous microdeletion that likely disrupts a putative imprinting control element of GNAS. J Clin Invest 112:1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastepe M, Fröhlich LF, Linglart A, Abu-Zahra HS, Tojo K, Ward LM, Jüppner H (2005) Deletion of the NESP55 differentially methylated region causes loss of maternal GNAS imprints and pseudohypoparathyroidism type Ib. Nat Genet 37:25–27 [DOI] [PubMed] [Google Scholar]
- Bastepe M, Gunes Y, Perez-Villamil B, Hunzelman J, Weinstein LS, Jüppner H (2002) Receptor-mediated adenylyl cyclase activation through XLαs, the extra-large variant of the stimulatory G protein α-subunit. Mol Endocrinol 16:1912–1919 [DOI] [PubMed] [Google Scholar]
- Bastepe M, Lane AH, Jüppner H (2001a) Paternal uniparental isodisomy of chromosome 20q—and the resulting changes in GNAS1 methylation—as a plausible cause of pseudohypoparathyroidism. Am J Hum Genet 68:1283–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastepe M, Pincus JE, Sugimoto T, Tojo K, Kanatani M, Azuma Y, Kruse K, Rosenbloom AL, Koshiyama H, Jüppner H (2001b) Positional dissociation between the genetic mutation responsible for pseudohypoparathyroidism type Ib and the associated methylation defect at exon A/B: evidence for a long-range regulatory element within the imprinted GNAS1 locus. Hum Mol Genet 10:1231–1241 [DOI] [PubMed] [Google Scholar]
- Campbell R, Gosden CM, Bonthron DT (1994) Parental origin of transcription from the human GNAS1 gene. J Med Genet 31:607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes C, Arnaud P, Gordon E, Dean W, Coar EA, Williamson CM, Feil R, Peters J, Kelsey G (2003) Epigenetic properties and identification of an imprint mark in the Nesp-Gnasxl domain of the mouse Gnas imprinted locus. Mol Cell Biol 23:5475–5488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward B, Barlier A, Korbonits M, Grossman A, Jacquet P, Enjalbert A, Bonthron D (2001) Imprinting of the Gsα gene GNAS1 in the pathogenesis of acromegaly. J Clin Invest 107:R31–R36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward B, Bonthron D (2000) An imprinted antisense transcript at the human GNAS1 locus. Hum Mol Genet 9:835–841 [DOI] [PubMed] [Google Scholar]
- Hayward B, Kamiya M, Strain L, Moran V, Campbell R, Hayashizaki Y, Bonthron DT (1998a) The human GNAS1 gene is imprinted and encodes distinct paternally and biallelically expressed G proteins. Proc Natl Acad Sci USA 95:10038–10043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward BE, Moran V, Strain L, Bonthron DT (1998b) Bidirectional imprinting of a single gene: GNAS1 encodes maternally, paternally, and biallelically derived proteins. Proc Natl Acad Sci USA 95:15475–15480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischia R, Lovisetti-Scamihorn P, Hogue-Angeletti R, Wolkersdorfer M, Winkler H, Fischer-Colbrie R (1997) Molecular cloning and characterization of NESP55, a novel chromogranin-like precursor of a peptide with 5-HT1B receptor antagonist activity. J Biol Chem 272:11657–11662 [DOI] [PubMed] [Google Scholar]
- Ishikawa Y, Bianchi C, Nadal-Ginard B, Homcy CJ (1990) Alternative promoter and 5′ exon generate a novel Gsα mRNA. J Biol Chem 265:8458–8462 [PubMed] [Google Scholar]
- Jüppner H, Schipani E, Bastepe M, Cole DEC, Lawson ML, Mannstadt M, Hendy GN, Plotkin H, Koshiyama H, Koh T, Crawford JD, Olsen BR, Vikkula M (1998) The gene responsible for pseudohypoparathyroidism type Ib is paternally imprinted and maps in four unrelated kindreds to chromosome 20q13.3. Proc Natl Acad Sci USA 95:11798–11803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehlenbach RH, Matthey J, Huttner WB (1994) XLαs is a new type of G protein. Nature 372:804–809 (erratum 375:253) [DOI] [PubMed] [Google Scholar]
- Levine MA (2002) Pseudohypoparathyroidism. In: Bilezikian JP, Raisz LG, Rodan GA (eds) Principles of bone biology. Vol 2. Academic Press, New York, pp 1137–1163 [Google Scholar]
- Li T, Vu TH, Zeng ZL, Nguyen BT, Hayward BE, Bonthron DT, Hu JF, Hoffman AR (2000) Tissue-specific expression of antisense and sense transcripts at the imprinted Gnas locus. Genomics 69:295–304 [DOI] [PubMed] [Google Scholar]
- Liu J, Erlichman B, Weinstein LS (2003) The stimulatory G protein α-subunit Gsα is imprinted in human thyroid glands: implications for thyroid function in pseudohypoparathyroidism types Ia and Ib. J Clin Endocrinol Metab 88:4336–4341 [DOI] [PubMed] [Google Scholar]
- Liu J, Litman D, Rosenberg M, Yu S, Biesecker L, Weinstein L (2000a) A GNAS1 imprinting defect in pseudohypoparathyroidism type Ib. J Clin Invest 106:1167–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Nealon JG, Weinstein LS (2005) Distinct patterns of abnormal GNAS imprinting in familial and sporadic pseudohypoparathyroidism type Ib. Hum Mol Genet 14:95–102 [DOI] [PubMed] [Google Scholar]
- Liu J, Yu S, Litman D, Chen W, Weinstein L (2000b) Identification of a methylation imprint mark within the mouse gnas locus. Mol Cell Biol 20:5808–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud FH, Linglart A, Bastepe M, Jüppner H, Lteif AN (2005) Molecular diagnosis of pseudohypoparathyroidism type Ib in a family with presumed paroxysmal dyskinesia. Pediatrics 115:e242–e244 [DOI] [PubMed] [Google Scholar]
- Mantovani G, Ballare E, Giammona E, Beck-Peccoz P, Spada A (2002) The Gsα gene: predominant maternal origin of transcription in human thyroid gland and gonads. J Clin Endocrinol Metab 87:4736–4740 [DOI] [PubMed] [Google Scholar]
- Marguet C, Mallet E, Basuyau JP, Martin D, Leroy M, Brunelle P (1997) Clinical and biological heterogeneity in pseudohypoparathyroidism syndrome: results of a multicenter study. Horm Res 48:120–130 [DOI] [PubMed] [Google Scholar]
- Pasolli H, Klemke M, Kehlenbach R, Wang Y, Huttner W (2000) Characterization of the extra-large G protein α-subunit XLαs. I. Tissue distribution and subcellular localization. J Biol Chem 275:33622–33632 [DOI] [PubMed] [Google Scholar]
- Peters J, Wroe SF, Wells CA, Miller HJ, Bodle D, Beechey CV, Williamson CM, Kelsey G (1999) A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc Natl Acad Sci USA 96:3830–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagge A, Gordon E, Dean W, Boiani R, Cinti S, Peters J, Kelsey G (2004) The imprinted signaling protein XLαs is required for postnatal adaptation to feeding. Nat Genet 36:818–826 [DOI] [PubMed] [Google Scholar]
- Spahn L, Barlow DP (2003) An ICE pattern crystallizes. Nat Genet 35:11–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparago A, Cerrato F, Vernucci M, Ferrero GB, Silengo MC, Riccio A (2004) Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith-Wiedemann syndrome. Nat Genet 36:958–960 [DOI] [PubMed] [Google Scholar]
- Swaroop A, Agarwal N, Gruen JR, Bick D, Weissman SM (1991) Differential expression of novel Gsα signal transduction protein cDNA species. Nucleic Acids Res 19:4725–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein LS, Yu S, Warner DR, Liu J (2001) Endocrine manifestations of stimulatory G protein α-subunit mutations and the role of genomic imprinting. Endocr Rev 22:675–705 [DOI] [PubMed] [Google Scholar]
- Williamson CM, Ball ST, Nottingham WT, Skinner JA, Plagge A, Turner MD, Powles N, Hough T, Papworth D, Fraser WD, Maconochie M, Peters J (2004) A cis-acting control region is required exclusively for the tissue-specific imprinting of Gnas. Nat Genet 36:894–899 [DOI] [PubMed] [Google Scholar]
- Wroe SF, Kelsey G, Skinner JA, Bodle D, Ball ST, Beechey CV, Peters J, Williamson CM (2000) An imprinted transcript, antisense to Nesp, adds complexity to the cluster of imprinted genes at the mouse Gnas locus. Proc Natl Acad Sci USA 97:3342–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Yu D, Lee E, Eckhaus M, Lee R, Corria Z, Accili D, Westphal H, Weinstein LS (1998) Variable and tissue-specific hormone resistance in heterotrimeric Gs protein α-subunit (Gsα) knockout mice is due to tissue-specific imprinting of the Gsα gene. Proc Natl Acad Sci USA 95:8715–8720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Radeva G, McCann JA, Hendy GN, Goodyer CG (2001) Gαs transcripts are biallelically expressed in the human kidney cortex: implications for pseudohypoparathyroidism type Ib. J Clin Endocrinol Metab 86:4627–4629 [DOI] [PubMed] [Google Scholar]