Abstract
22q11.2 microduplications of a 3-Mb region surrounded by low-copy repeats should be, theoretically, as frequent as the deletions of this region; however, few microduplications have been reported. We show that the phenotype of these patients with microduplications is extremely diverse, ranging from normal to behavioral abnormalities to multiple defects, only some of which are reminiscent of the 22q11.2 deletion syndrome. This diversity will make ascertainment difficult and will necessitate a rapid-screening method. We demonstrate the utility of four different screening methods. Although all the screening techniques give unique information, the efficiency of real-time polymerase chain reaction allowed the discovery of two 22q11.2 microduplications in a series of 275 females who tested negative for fragile X syndrome, thus widening the phenotypic diversity. Ascertainment of the fragile X–negative cohort was twice that of the cohort screened for the 22q11.2 deletion. We also report the first patient with a 22q11.2 triplication and show that this patient's mother carries a 22q11.2 microduplication. We strongly recommend that other family members of patients with 22q11.2 microduplications also be tested, since we found several phenotypically normal parents who were carriers of the chromosomal abnormality.
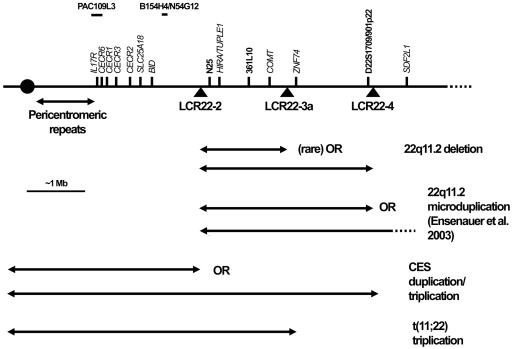
Chromosomal rearrangements of 22q11.2 result in several genomic disorders, including the 22q11.2 deletion syndrome (also known as “DiGeorge syndrome” [MIM 188400] and “velocardiofacial syndrome” [MIM 192430]), and partially overlapping duplications result in cat eye syndrome (CES [MIM 115470]) and der(22) syndrome (reviewed by McDermid and Morrow [2002]) (fig. 1). These rearrangements are thought to be the result of nonallelic homologous recombination between low-copy repeats found in the region (LCR22s) (Edelmann et al. 1999b; Stankiewicz and Lupski 2002). In particular, the 22q11.2 deletion syndrome represents the most commonly ascertained deletion syndrome in humans, with a frequency estimated at 1/4,000–6,000 live births (Shaffer and Lupski 2000; Botto et al. 2003). Features of the 22q11.2 deletion syndrome include cleft palate, velopharyngeal insufficiency, hypernasal speech, characteristic facial features, heart defects, learning disabilities, and behavioral disorders (Shprintzen et al. 1978; Swillen et al. 1999). More-severely affected patients may have a reduced or absent thymus, as well as hypocalcemia (DiGeorge 1965). Since this microdeletion results from unequal crossing over—usually between the LCR22s separated by 3 Mb and usually on different chromosome 22 homologues (Saitta et al. 2004)—one would expect the reciprocal 3-Mb microduplication of the region to be present with a frequency equal to that of the 22q11.2 deletion. However, there are few reports of microduplications of this region of 22q11.2 (Edelmann et al. 1999b; Papenhausen et al. 2002; Ensenauer et al. 2003; Hassed et al. 2004). A small number of patients with larger duplications that include the critical regions for both CES and 22q11.2 deletion syndrome have also been reported (Reiss et al. 1985; Knoll et al. 1995; Lindsay et al. 1995), but these patients have features typical of CES. These duplications also overlap with critical regions in patients with der(22) syndrome, who have partial trisomy for the 22pter-q11 regions, including half of the region typically deleted in 22q11.2 deletion syndrome (Funke et al. 1999; Kurahashi et al. 2000) (fig. 1). Similarly, Smith-Magenis syndrome (SMS) is a microdeletion syndrome associated with 17p11.2 (Smith et al. 1986) and is characterized by mental retardation, neurobehavioral abnormalities, sleep disorders, speech and motor delays, midface hypoplasia, short stature, and brachydactyly. The reciprocal microduplication of 17p11.2 has only recently been identified. It has a phenotype that is milder than that of the deletion (Potocki et al. 2000), including behavioral problems, mild-to-borderline mental retardation, short stature, dental anomalies, normal facies, and a lack of major-organ malformations.
Figure 1.
Duplications and deletions on chromosome 22q11.2. Arrows indicate the size of the various deletions and duplications. Triangles below the line representing the chromosome show the location of the LCR22s. Probes located above the line were used in the various FISH and dosage techniques.
Microduplications of 22q11.2 may be largely undetected, as a result of a less-distinct, unpredictable, and/or milder phenotype, which leads not only to problems with ascertainment and choosing the patient cohort to search but also to technical difficulties involved in identifying microduplications. A recent report described the clinical, cytogenetic, and molecular findings of 10 independent patients with 22q11.2 microduplications. Seven of the microduplications include only the 22q11.2 deletion syndrome region (Ensenauer et al. 2003). These individuals were ascertained from 653 patients tested for the 22q11.2 deletion; the latter syndrome was diagnosed in 40 of 653 patients. The patients with 22q11.2 microduplications had a variable phenotype, including heart defects, velopharyngeal insufficiency with and without cleft palate, hearing loss, growth delay, cognitive deficits, motor delay, learning disabilities, behavioral problems, and mild dysmorphic features. Whereas the phenotype of some individuals with a 22q11.2 microduplication may overlap with that of individuals with the 22q11.2 deletion syndrome, this overlap may (as a result of ascertainment bias) represent only one part of this syndrome’s phenotypic spectrum. If these reciprocal chromosomal rearrangements are of equal frequency, then the difference in frequency between patients with 22q11.2 deletion syndrome (6.1%) and those with 22q11.2 microduplications (1.5%) in the test group used by Ensenauer et al. (2003) indicates that the majority of patients with microduplications do not fall within this test group.
In this report, we present seven unrelated patients/families (one reported elsewhere [Edelmann et al. 1999b]) with increased copy number in the 22q11.2 deletion syndrome region. Table 1 compares the clinical features of these patients with the features of patients published elsewhere. Figure 2 illustrates features of two patients. The microduplications in patients 1–5 were originally identified during tests for the 22q11.2 deletion syndrome at five different centers. Patients 6 and 7 were ascertained by screening 275 female patients referred for diagnostic fragile X syndrome testing. The seven patients show a remarkable variation in phenotype, with some showing little-to-no resemblance to the phenotype of 22q11.2 deletion syndrome.
Table 1.
Clinical Characterization of Eight Patients with Increased Dosage of 22q11.2[Note]
|
Finding for Patient |
|||||||||
| Feature | 1 | 2 | 3 | 4 | 4M | 5 | 6 | 7 | Results(No. Affected/No. Tested)of Ensenaueret al. (2003) |
| 22q11.2 copy number | 3 | 3 | 3 | 4 | 3 | 3 | 3 | 3 | |
| Age at last evaluation | 8 years | 4 years | 1 mo | 8 years | 4 mo | 3 years | 34 years | ||
| Sex | M | F | M | F | F | F | F | F | |
| Heart defect | − | − | +a | − | +b | − | − | 2c,d/6 | |
| Velopharyngeal insufficiency | − | + | − | − | − | 5/5 | |||
| Palatal defect | A | − | − | − | − | 4/7 | |||
| Hearing impairment | +e | − | − | +e | + | +f | − | 4/6 | |
| Failure to thrive | − | + | − | − | + | + | − | ||
| Sleep apnea | + | − | − | ||||||
| Absent thymus/asplenia | − | − | − | − | − | − | 1/6 | ||
| Urogenital abnormality | − | − | − | − | − | − | 2/5 | ||
| Hypotonia | − | + | − | − | + | + | − | 1/5 | |
| Gastrointestinal abnormality | − | − | − | − | + | − | |||
| Cognitive deficits | +g,h | +g,h | +g,i | +h | +i | 5/5 | |||
| Behavioral problems | +j | +k | − | +l | 3/3 | ||||
| Seizures | + | + | − | + | 1/6 | ||||
| Dysmorphic features: | 6/6 | ||||||||
| Broad nasal bridge | + | + | + | + | |||||
| Hand/foot abnormality | +m | +n | +o | +o | − | − | − | 4p,q/5 | |
| Hypertelorism | + | + | + | ||||||
| Epicanthal folds | + | + | 2/5 | ||||||
| Micrognathia | + | − | + | + | 3/5 | ||||
| Microcephaly | − | + | + | 1/6 | |||||
| Additional features | +r | +s | +t | +u | +v | +w | 6/6 | ||
| Other | +x | +y | +z | +aa | +bb | ||||
Note.— “+” = feature present; “−” = feature absent; blank cells indicate unknown or patient too young to determine; A = arched.
Hypoplastic left heart.
Tetrology of Fallot and right-sided aortic arch.
Tetrology of Fallot.
Hypoplastic left heart and interrupted aortic arch.
Impairment secondary to recurrent otitis media.
Mild conductive deafness.
Speech delay.
Developmental delay.
Learning difficulties.
Impulsivity and aggression.
Short concentration span and social immaturity.
Childhood aggression and childhood ADD.
Clinodactyly of the fifth fingers.
Hypoplastic fifth fingernail and toenails smaller than normal.
Broad hands, with square tipped fingers and prominent fetal finger pads.
Abnormal palmar creases.
Long fingers and/or toes.
High forehead, round face, flat supraorbital ridge, short nose, thin upper lip, and long, smooth philtrum.
Narrow face and downslanting palpebral fissures.
Prominent eyes, medial deficiency of the eyebrows, and prominent chin and lower lip.
Eversion of the lateral eye lids and bulbous nasal tip.
Facial asymmetry.
Mild synophrys, tented peak in the middle of each eyebrow, facial asymmetry, and small, downslanting mouth.
Maternal half brother (aged 16 years) with developmental delay, paternal cousin (aged 9 years) with behavioral problems, and another paternal cousin with cleft lip and palate.
Gross motor delay and poor fine-motor skills.
Nystagmus and myopia; prominent metopic suture, with an anterior fontanel of a width of three fingers; sagittal sutures widely separated; and open posterior fontanel.
Left ear pit.
Chorioretinal coloboma, visual impairment, and preauricular skin tags.
Figure 2.
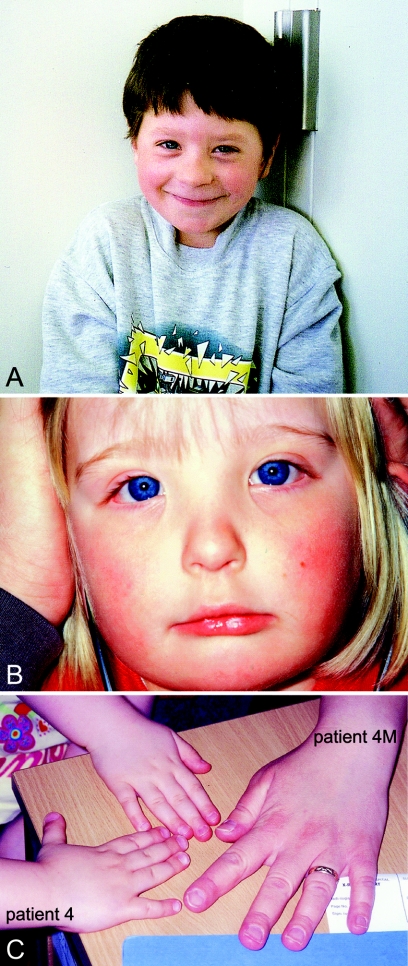
Photographs of patients with increased dosage of 22q11.2. A, Patient 1. B, Patient 4. C, Hands of patient 4 and her mother (patient 4M). The unlabeled hand is an unaffected sister of patient 4.
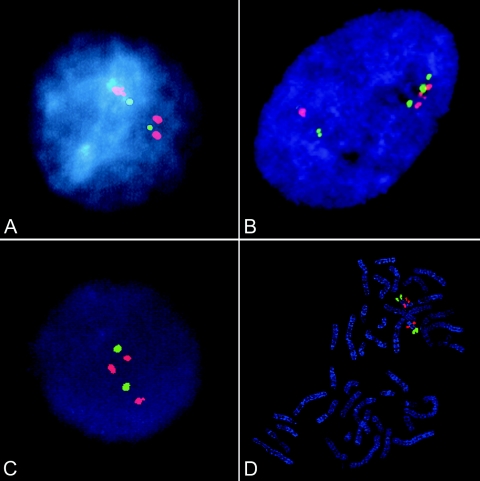
Most patients in this study were analyzed by several different methods to confirm an increased copy number within 22q11.2. Patients 1–5 were originally identified by FISH. Analysis of interphase nuclei of patients 1 and 3 showed a microduplication of the Vysis TUPLE1 probe (fig. 3A [patient 1]). To narrow the extent of microduplication, cells from patient 1 were further tested using a number of BAC clones (obtained from the Wellcome Trust Sanger Institute and M. Rocchi) within and surrounding the 22q11.2 deletion syndrome region. Probes proximal (109L3, N54G12, and BK154H4) and distal (BA24N11, BCR, and BA297B9) to the 22q11.2 deletion syndrome region were not duplicated. Only probes D22S75/N25 and TUPLE1, located within the 22q11.2 deletion syndrome region, were duplicated. Duplications of the 22q11.2 deletion syndrome region were shown in patient 2 by use of Oncor probe N25 (Edelmann et al. 1999b). Interphase FISH analysis was performed on patient 4 with the use of two BAC probes located within the 22q11.2 deletion syndrome region. Probe 361L10 (green) was clearly triplicated and showed four hybridization spots (fig. 3B), whereas 901P22 (red) gave ambiguous results (three copies in most cells, but four copies in some cells [see fig. 3B]). Analysis could not be repeated, as a result of a lack of sample. The consistent clustering of three of the four copies of 361L10 indicates the presence of three copies on one chromosome 22 and one copy on the other homologue. FISH on interphase nuclei of patient 5 showed three TUPLE1 (red) signals, compared with two ARSA (green) signals (fig. 3C). On review of signals for metaphase chromosomes, a TUPLE1 signal on one chromosome 22 was subjectively larger (fig. 3D), supporting a tandem duplication of this region, rather than a cryptic translocation. FISH studies of the parents of patient 5 revealed the same microduplication in the infant’s father, who was healthy but had a history of significant learning difficulties, unilateral hearing loss, and poor vision in one eye. No FISH analysis was done for patients 6 and 7.
Figure 3.
FISH analyses. A, Patient 1 test probe TUPLE1 (red) shows three copies for an interphase cell; control probe ARSA (green) shows two copies. B, Patient 4 test probes 361L10 and 901P22 (green and red, respectively) show four copies for this cell. C, Patient 5 test probe TUPLE1 (red) shows three copies; control probe ARSA (green) shows two copies. D, Metaphase spread from patient 5 shows the duplication of the TUPLE1 probe.
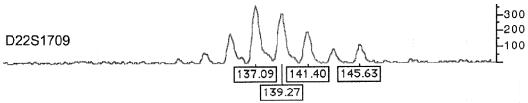
The triplication in patient 4 was analyzed using the microsatellite marker D22S1709, which is located in the distal portion of the 22q11.2 deletion syndrome region (fig. 1). Four different alleles are present at this locus, confirming four copies of the region (fig. 4). This probe is located within BAC 901P22, which gave ambiguous FISH results of three or four copies. The microsatellite analysis confirms the involvement of four different chromosome 22 homologues in the production of this triplication. Microsatellite analyses were also done on patients 3, 5, 6, and 7. Several loci within the 22q11.2 deletion syndrome region were informative for the presence of three distinct alleles in all four patients (results not shown).
Figure 4.
Microsatellite analysis of probe D22S1709, showing four different alleles in patient 4, with sizes (in bp) marked below the peaks. Unmarked peaks are due to stutter bands, an artifact of PCR. The method has been described elsewhere (Edelmann et al. 1999a).
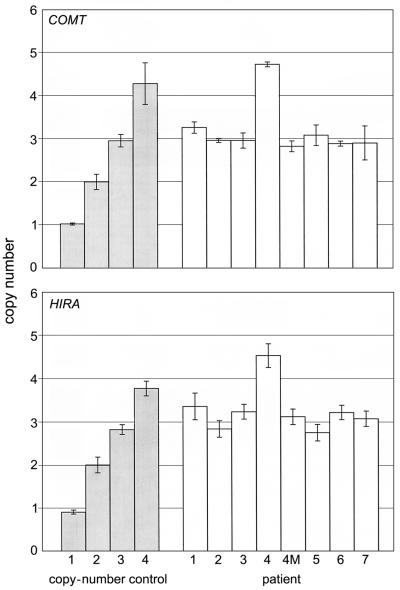
Since it is cumbersome, expensive, and labor-intensive to screen for 22q11.2 interstitial microduplications by use of interphase FISH, we sought a more efficient screening method. Real-time PCR analysis of two genes in the proximal 1.5 portion (fig. 1) of the 22q11.2 deletion syndrome region (HIRA and COMT) was performed on all patients (fig. 5) with the use of a multiplexed array of two quantitative reactions (Thiel et al. 2003). A TaqMan GJA5 gene Pre-Developed Assay Reagent (PDAR [Applied Biosystems]) was multiplexed as a two-copy control in both reactions. Reactions from patients with 22q11.2 deletion syndrome, from wild-type DNA, and from “no template” controls were included in each analysis. All real-time PCR amplifications were performed on an ABI Prism 7900 HT Sequence Detection System (Applied Biosystems) with the use of a 384-well plate. HIRA and COMT were assayed in separate reactions, and each assay was performed in triplicate. Sequence Detection Software (SDS, version 2.0 [Applied Biosystems]) was used to process raw fluorescence data and to produce a threshold cycle number (Ct) for each sample. The Ct value was the cycle number at which the fluorescence emission of the reporter dye passed a fixed threshold on the amplification plot. The default setting for the threshold was 10 SDs above the mean baseline emission. All Ct settings were manually checked and adjusted within the logarithmic curve between background and plateau levels. The ABI 7900 HT data were exported to a Microsoft Excel macro in which all Ct values were normalized to the multiplexed PDAR and were compared with the wild-type and positive-control values.
Figure 5.
Real-time PCR analysis of probes COMT and HIRA (also known as “TUPLE1”). Copy number was calculated by multiplying the ΔΔCt ratio by 2. A two-copy control was run for each set of experiments, but only the most variable two-copy control is shown in each graph. Error bars represent 1 SD above and below the mean. Each reaction mixture consisted of 200 nmol/L of each primer, 50 nmol/L of TaqMan probe, 40 ng of genomic DNA, 0.3 μl of PDAR (Applied Biosystems), and 7.5 μl of 2× TaqMan mix (Applied Biosystems), in a total volume of 15 μl. Thermal cycling conditions included a presoak for 2 min at 50°C and for 10 min at 95°C. Samples were amplified for 40 cycles at 95°C for 15 s and at 60°C for 1 min.
Patients 1, 2, 3, and 5 have three copies of HIRA and COMT, indicating a microduplication of the 22q11.2 deletion syndrome region of 22q11.2. Patient 4 was found to have four copies of HIRA and COMT, confirming a triplication of the 22q11.2 deletion syndrome region. Interestingly, the mother of patient 4 (patient 4M) was found to have only three copies of 22q11.2, indicating an expansion of the 22q11.2 deletion syndrome region from three to four copies between mother and child. This was confirmed cytogenetically. In addition to FISH analysis, patients 3 and 5 were independently ascertained by real-time PCR among 671 22q11.2 deletion syndrome referrals (0.3%). In this same cohort, 26 cases of 22q11.2 deletion were detected (3.9%).
The efficiency of the real-time PCR analysis allowed us to quickly screen for microduplications in a set of 275 unrelated females who were negative for fragile X syndrome; the individuals in this set were collected from multiple referrals, with little or no clinical data available. However, since common features of patients with the 22q11.2 microduplication are behavioral and cognitive problems, the fragile X–negative cohort would be a reasonable group to screen. Only females were screened, to minimize the contribution of X-linked conditions to the analysis. Our screen resulted in the discovery of two patients (0.7%) with duplications in the 22q11.2 deletion syndrome region (fig. 5 [patients 6 and 7]). Permission from the Health Research Ethics Board of the University of Alberta Health Sciences Faculties allowed us to access the clinical data for these two patients (table 1). As controls, 520 unrelated individuals referred for hemochromatosis testing were also screened for microduplication of 22q11.2. None of the control individuals had a microduplication of 22q11.2 (P<.00003), indicating that this chromosomal abnormality is not a polymorphism. Interestingly, one patient (0.4%) in the fragile X–negative female group had a deletion of 22q11.2.
Although real-time PCR allowed the rapid analysis of a large number of individuals, it did not allow us to define the size of the microduplication, since only two probes in the 22q11.2 deletion syndrome region were examined. Sizing of microduplications requires the examination of multiple sites along 22q11.2, which is expensive to set up for real-time PCR and is time-consuming to test by FISH and microsatellites. We therefore analyzed the patients by use of our CES/22q11.2 deletion syndrome multiplex amplifiable probe hybridization (MAPH) probe set that covered regions both distal and proximal to the 22q11.2 deletion syndrome region (table 2). Seven probes were located within the CES region, one probe was within the 3-Mb 22q11.2 deletion syndrome region (ZNF74 [position 19073420 bp in the UCSC Genome Browser May 2004 Assembly Map]), and one probe was ∼600 kb distal to the 22q11.2 deletion syndrome region (SDF2L1 [position 20321160 bp in the UCSC Genome Browser May 2004 Assembly Map]) (fig. 1). Control probes, supplied by J. Armour (University of Nottingham), were located on other chromosomes (5, 15, 16, and 17). In addition to patient DNA samples, three control samples were used: the two-copy control was a normal lymphoblast cell line GM03657 (Coriell Institute for Medical Research); the three-copy control was a lymphoblastoid cell line from a patient with a duplication including the CES region and the 22q11.2 deletion syndrome region (Reiss et al. 1985; McDermid et al. 1986; H.E.M., unpublished results); and the four-copy control was patient CM01, who has CES and is known to have four copies of both the CES region and the 22q11.2 deletion syndrome region (McTaggart et al. 1998). Genescan software was used to calculate the peak area for each probe. Peak ratios were calculated by dividing the peak area of a test probe by the sum of the peak areas of the two closest control probes. Each experiment contained four to eight replicates of the two-copy control, the three-copy control, and a patient. Each peak-ratio replicate for all samples was normalized to the mean peak ratio of the two-copy control. The mean and SD of the normalized peak ratios for each probe were calculated for the two-copy control, the three-copy control, and the patient. One experiment also contained a four-copy control.
Table 2.
MAPH Probes, Primer Sequences, and Sizes
|
Primer |
|||
| Probea | Forward | Reverse | Sizeb(bp) |
| IL-17R | 5′-CATCACCGTGGAGACCCTGGA-3′ | 5′-CCGCAGGTATGTGGTGCATGTG-3′ | 146 |
| CECR6 | 5′-CCAGGATCTTCCTCAATACTGC-3′ | 5′-TTTTGCAAGCAGCATCAGTGC-3′ | 158 |
| CECR1 | 5′-TCCATCTGAGCCCTTTCCTA-3′ | 5′-CTCCTTCATCTCAGCGATT-3′ | 253 |
| CECR3 | 5′-CCTGAGAGAGAGACAGAAGCAGC-3′ | 5′-CATGAATCACTCTCTGGTGGTTTTGC-3′ | 122 |
| CECR2 | 5′-AGTGGGCCCGGAGCTCAAAA-3′ | 5′-GCTGAATTCGGGTACTCTAGG-3′ | 390 |
| SLC25A18 | 5′-GAGGTCGTCACTTGCCGGGAA-3′ | 5′-GTGGGTCATTCTGCAGGGGCT-3′ | 188 |
| BID | 5′-CTGGCTAAGCTCCTCACGTA-3′ | 5′-TGCTGCTGGCCAAGAAGGTG-3′ | 115 |
| ZNF74 | 5′-GGCGGTGCCCTCTCAACAGCA-3′ | 5′-GCGTGTGTCCAGCAAGGGTC-3′ | 247 |
| SDF2L1 | 5′-GGAAGACGGCGAGGGCGACGA-3′ | 5′-CTGGGCATGCCGTGGACCTCA-3′ | 177 |
Control probes 2f2 (chromosome 5), pws5 (chromosome 15), pws6 (chromosome 15), e10 (chromosome 17), g8 (chromosome 16), and d11 (pBluescript control) were provided by J. Armour (University of Nottingham).
Indicates the size in bp, when amplified by PCR and run on an agarose gel.
A graph of the normalized mean peak ratio (±1 SD) for each probe, compared with a two-copy and three-copy control, was plotted for patients 1–4 (fig. 6), as well as for patient 6 and the mother of patient 4 (data for the latter two not shown). DNA was not available for MAPH analysis of patients 5 and 7. A four-copy control was included for patient 4. In all cases, a ratio of ∼1:1 with the two-copy control was calculated for all the probes within the CES region and distal to the 22q11.2 deletion syndrome region, indicating no duplication of these probes. Thus, in each patient tested, the region of increased dosage was confined to the 22q11.2 deletion syndrome region, and no cases extended proximally, as in the study by Ensenauer et al. (2003). The size of the duplication could not be determined for patients 5 and 7. In all cases, the previous results showing microduplications and triplications were confirmed. On the basis of the ratios calculated for the ZNF74 probe (∼1.5:1 with the two-copy control), patients 1, 2, 3, and 6 have a microduplication of the 22q11.2 deletion syndrome region. In each experiment, the three-copy control showed a ratio of ∼1.5:1 with the two-copy control. In patient 4 and the four-copy control, a ratio of ∼2:1 with the two-copy control was calculated for ZNF74, confirming that patient 4 has a triplication of the 22q11.2 deletion syndrome region. The mother of patient 4 (patient 4M) was confirmed to have three copies of ZNF74 (data not shown).
Figure 6.
MAPH analysis of the 22q11.2 region for patients 1, 2, 3, and 4. Probes are shown below the four graphs and are listed in order along the chromosome (centromere at left), with the CES region and the 22q11.2 deletion syndrome region indicated. Normalized mean peak ratios (error bars represent 1 SD above and below the mean) are graphed for all probes for each patient (unblackened triangles connected by a line) and control (two-copy control sample represented by blackened diamonds; three-copy control sample represented by squares). The four-copy control sample (circles) has four copies of the CES region and the 22q11.2 deletion syndrome region; however, distal to the 22q11.2 deletion syndrome region, the copy number is 2. Probes and probe mixes, membrane preparation, and hybridization were performed in accordance with the study by Armour et al. (2000) and the Multiplex Amplifiable Probe Hybridization (MAPH) Web site, with the following changes. Genomic DNA was blotted onto a Hybond N membrane (Amersham Biosciences). After hybridization, the membranes were washed six times for 10 min each at 65°C in 1× SSC/1% SDS solution and six times for 10 min each at 65°C in 0.1× SSC/0.1% SDS solution. Membranes were each placed in a 0.2-ml PCR tube with 50 μl of 1× AB gene buffer IV and were heated to 95°C to release the probes; 3.75 μl of this solution was used as a template for a fluorescent PCR containing 1× AB gene buffer IV, 2 mM MgCl2, 0.4 mM dNTPs, 2 μM PZA-FAM, 1 μM PZB, and 2.5 U Taq polymerase (Invitrogen). Finally, 1 μl of the PCR was mixed with 1 μl of formamide loading dye, and 1.2 μl of the mixture was run on a polyacrylamide gel for 3.5 h by use of the ABI Prism 377 DNA sequencer.
In summary, we report seven unrelated patients/families with duplication or triplication of the 3-Mb 22q11.2 deletion syndrome region. Ensenauer et al. (2003) described seven unrelated patients with just the 22q11.2 deletion syndrome region duplicated, as well as an additional three independent patients with duplications that included the 22q11.2 deletion syndrome region and extended distally. All were ascertained from 653 patients referred for 22q11.2 deletion testing. Although distinct dysmorphic features for the microduplication were suggested, the other features of the seven patients have a strong bias toward 22q11.2 deletion syndrome features, including conotruncal heart defects (2/6), velopharyngeal insufficiency (5/5), cleft palate (4/7), cognitive defects (5/5), and absent thymus with T-cell deficiency (1/6). This is not surprising, considering the method of ascertainment. However, although our patients 1–5 were also identified through 22q11.2 deletion testing, they show a less distinct phenotype, with fewer instances of velopharyngeal insufficiency (1/4) and no cases of cleft palate or abnormal thymus. Developmental/speech delay, behavioral problems, and mild dysmorphic features are the major phenotypes observed in our patients 1–5 (table 1). Aggressive behavior and attention deficit disorder (ADD) were seen in patients from both studies.
The true spectrum of 22q11.2 microduplication syndrome will not be apparent until patients are ascertained independently of the 22q11.2 deletion syndrome symptoms. We have initiated this by screening 275 females who were referred for diagnostic fragile X syndrome testing but with no FMR1 expansion detected. We identified two patients with 22q11.2 microduplications in this group and no microduplications in 520 controls (P<.00003). Patient 6 showed an eye defect, as well as dysmorphic features and developmental delay, but her phenotype did not particularly resemble that of 22q11.2 deletion syndrome. In fact, the presence of a coloboma and preauricular tags in this patient suggested an association with the CES duplication, yet the CES critical region was not duplicated. Patient 7 showed only behavioral symptoms, with no similarity to the 22q11.2 deletion syndrome. The analysis of these patients suggests that a large number of individuals with 22q11.2 microduplications do not show similarity to 22q11.2 deletion syndrome and, therefore, are currently undiagnosed, which would explain the imbalance between reported deletions and duplications of this region. The lack of similarity of our patients to each other underscores the remarkable variability in the phenotype of this syndrome, which will make ascertainment of the majority of cases extremely difficult.
A significant number of 22q11.2 microduplications may also be associated with a normal phenotype. The mother and grandmother of patient 2 both carry the microduplication and are normal (Edelmann et al. 1999b). The mother of patient 4 shows a normal phenotype, except for her hands, but has a 22q11.2 microduplication. The father of patient 5 has the microduplication and showed learning difficulties and hearing loss. This suggests that parents of children with 22q11.2 duplications should also be tested for the same microduplication, since it could be responsible for other mild abnormalities within the pedigree and is a potential risk to future children of the parents and of other relatives.
The normal or near-normal phenotype associated with a significant proportion of patients with 22q11.2 microduplications could also explain why there appears to be no correlation between patients with CES with increased dosage of the 22q11.2 deletion syndrome region and those without increased dosage, in addition to triplication of the CES region (McTaggart et al. 1998). Since the sample size of patients with CES was small (10 individuals) and the phenotype of CES is highly variable, small additions to the phenotype from microduplication or triplication of the 22q11.2 deletion syndrome region may not be detectable unless a very large number of patients are analyzed. Interstitial duplications including both the CES and the 22q11.2 deletion syndrome regions have also shown predominantly CES-like features (Reiss et al. 1985; Knoll et al. 1995; Lindsay et al. 1995).
Patient 4 has a triplication of the 22q11.2 deletion syndrome region, which has been confirmed by four independent methods (figs. 3, 4, 5, and 6). To our knowledge, this is the first reported triplication of the 22q11.2 deletion syndrome region. Microsatellite analysis of patient 4 showed the presence of four different alleles of locus D22S1709, located in the distal portion of the 22q11.2 deletion syndrome region. This implies that four different chromosome 22 homologues were involved in the creation of the triplication. FISH analysis showed that the two extra copies are on the same chromosome. This could be explained by an interchromosomal exchange in a maternal heterozygous grandparent—which would have created the microduplication in the mother—followed by a second interchromosomal exchange involving the same chromosome in the heterozygous mother, leading to the tandem triplication in the child. It is unknown whether the presence of the first microduplication predisposed to the second rearrangement. Interestingly, the phenotype of this patient is very mild. The patient’s mother has a microduplication of the 22q11.2 deletion syndrome region and a normal phenotype, except for dysmorphic hands, suggesting that modifier loci in this family are ameliorating the effect of the chromosomal defect.
To ascertain a significant proportion of 22q11.2 microduplications, a very large number of individuals would need to be screened. However, microduplications have generally been more difficult to detect than deletions. Microduplications of 22q11.2 have previously been diagnosed primarily using interphase FISH (Edelmann et al. 1999b; Ensenauer et al. 2003). Although this method is effective, it is time-consuming and is not suited for quickly screening large numbers of patients. However, this technique does give unique information about the position and arrangement of the duplicated segments. In our study, FISH analysis of patient 4 revealed that the two additional copies of the 22q11.2 deletion syndrome region are both located on the same homologue, indicating a tandem triplication (fig. 3). Duplications of 22q11.2 have also previously been seen by use of chromosome banding; however, a resolution greater than what is routinely used was necessary (Ensenauer et al. 2003).
Microsatellite analysis has also been used to detect 22q11.2 microduplications and to define breakpoints (Edelmann et al. 1999a; Ensenauer et al. 2003). However, because dosage analysis with microsatellites is, at best, only semiquantitative, unambiguous results are obtained only when the number of different allele sizes is equal to the copy number of the region. Preferential amplification of the smaller alleles leads to inaccuracies, as do stuttering effects, which produce smaller peaks preceding the major-allele peak (Armour et al. 2002). However, in the case of patient 4, microsatellite analysis gave unique information about possible mechanisms of the formation of the triplication.
Real-time PCR (Thiel et al. 2003) and MAPH (Armour et al. 2000) are two methods used to study changes in dosage that generally give unambiguous results and only require a small amount of DNA from the patient. Real-time PCR is a quick, straightforward, and reproducible test that uses minute amounts of DNA. As many as 185 patients can be analyzed in a single run for the dosage (deletion or duplication) of one probe. The disadvantage of this technique is the cost of both the initial equipment and the individual probes. Alternatively, MAPH was specifically designed to analyze a large number of probes at once and, therefore, is suited to more-complex detection projects, such as breakpoint comparisons. In this study, we scanned for microduplications over seven genes in the CES region, as well as single genes within and distal to the 22q11.2 deletion syndrome region.
All four methods were used in our study, although not all methods were used on each patient. All patients were examined with more than one method, and, in each case, the results concurred. Each technique has advantages and disadvantages. However, for screening large numbers of individuals, real-time PCR is the method of choice; microarray-based comparative genomic hybridization (array CGH) has also been used to detect abnormalities of 22q11.2 (Mantripragada et al. 2004). Large screens of different patient cohorts would aid in defining the full phenotypic spectrum of the 22q11.2 microduplication syndrome. The finding of 22q11.2 microduplications in 2 of 275 females who tested negative for fragile X syndrome implies that these microduplications are not uncommon and that they may be associated more with neurological and behavioral features in many patients. The remaining challenge is to identify these patients, and this syndrome is likely to remain underdiagnosed until this is done. Large-scale real-time PCR screens of neurologically impaired individuals (those with developmental delay, mild dysmorphology, minor behavioral problems, and learning disabilities with mild dysmorphology), such as diagnostic fragile X referrals, followed by family studies to detect relatives with 22q11.2 microduplications, would be a reasonable place to start. Indeed, our success rate for detecting 22q11.2 microduplications among fragile X referrals (0.7%) approaches half of the success rate for detecting fragile X itself in this cohort (2%) and is twice the number of 22q11.2 microduplications found in the 22q11.2 deletion syndrome referrals. By widening the scope of the patients tested, we will gain a better understanding of the phenotype associated with 22q11.2 microduplications.
Acknowledgments
We thank John Armour for providing control MAPH probes and invaluable advice. We also thank John Gaspar, for performing the microsatellite analysis of patient 4; Catherine Kashork (Baylor College of Medicine, Houston, TX), for performing the FISH analysis of patient 4; Lucy Osborne, for helpful discussions; and Jack Scott, for help with the figures. H.E.M. was supported by a grant from the Canadian Institutes of Health Research (MOP11639). This work was also supported by the March of Dimes (1-FY00-768 [to B.E.M.]) and the National Institutes of Health (1 P01 HD39420-01 [to B.E.M. and L.G.S.] and 5 P01 HD34980-05 [to B.E.M.]). This study was approved by the Health Research Ethics Board of the University of Alberta Health Sciences Faculties.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Multiplex Amplifiable Probe Hybridization (MAPH) Web site, http://www.nott.ac.uk/~pdzjala/maph/maph.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DiGeorge syndrome, velocardiofacial syndrome, and CES)
- Wellcome Trust Sanger Institute, http://www.sanger.ac.uk/ (for clones)
References
- Armour JA, Barton DE, Cockburn DJ, Taylor GR (2002) The detection of large deletions or duplications in genomic DNA. Hum Mutat 20:325–337 10.1002/humu.10133 [DOI] [PubMed] [Google Scholar]
- Armour JA, Sismani C, Patsalis PC, Cross G (2000) Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res 28:605–609 10.1093/nar/28.2.605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto LD, May K, Fernhoff PM, Correa A, Coleman K, Rasmussen SA, Merritt RK, O’Leary LA, Wong LY, Elixson EM, Mahle WT, Campbell RM (2003) A population-based study of the 22q11.2 deletion: phenotype, incidence, and contribution to major birth defects in the population. Pediatrics 112:101–107 10.1542/peds.112.1.101 [DOI] [PubMed] [Google Scholar]
- DiGeorge A (1965) A new concept of the cellular basis of immunity. J Pediatr 67:907 [Google Scholar]
- Edelmann L, Pandita RK, Morrow BE (1999a) Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am J Hum Genet 64:1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, Magenis E, Shprintzen RJ, Morrow BE (1999b) A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet 8:1157–1167 10.1093/hmg/8.7.1157 [DOI] [PubMed] [Google Scholar]
- Ensenauer RE, Adeyinka A, Flynn HC, Michels VV, Lindor NM, Dawson DB, Thorland EC, Lorentz CP, Goldstein JL, McDonald MT, Smith WE, Simon-Fayard E, Alexander AA, Kulharya AS, Ketterling RP, Clark RD, Jalal SM (2003) Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am J Hum Genet 73:1027–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke B, Edelmann L, McCain N, Pandita RK, Ferreira J, Merscher S, Zohouri M, Cannizzaro L, Shanske A, Morrow BE (1999) Der(22) syndrome and velo-cardio-facial syndrome/DiGeorge syndrome share a 1.5-Mb region of overlap on chromosome 22q11. Am J Hum Genet 64:747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassed SJ, Hopcus-Niccum D, Zhang L, Li S, Mulvihill JJ (2004) A new genomic duplication syndrome complementary to the velocardiofacial (22q11 deletion) syndrome. Clin Genet 65:400–404 10.1111/j.0009-9163.2004.0212.x [DOI] [PubMed] [Google Scholar]
- Knoll JH, Asamoah A, Pletcher BA, Wagstaff J (1995) Interstitial duplication of proximal 22q: phenotypic overlap with cat eye syndrome. Am J Med Genet 55:221–224 [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Shaikh TH, Hu P, Roe BA, Emanuel BS, Budarf ML (2000) Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22). Hum Mol Genet 9:1665–1670 10.1093/hmg/9.11.1665 [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Shaffer LG, Carrozzo R, Greenberg F, Baldini A (1995) De novo tandem duplication of chromosome segment 22q11-q12: clinical, cytogenetic, and molecular characterization. Am J Med Genet 56:296–299 [DOI] [PubMed] [Google Scholar]
- Mantripragada KK, Tapia-Paez I, Blennow E, Nilsson P, Wedell A, Dumanski JP (2004) DNA copy-number analysis of the 22q11 deletion-syndrome region using array-CGH with genomic and PCR-based targets. Int J Mol Med 13:273–279 [PubMed] [Google Scholar]
- McDermid HE, Duncan AM, Brasch KR, Holden JJ, Magenis E, Sheehy R, Burn J, Kardon N, Noel B, Schinzel A, Teshima I, White BN (1986) Characterization of the supernumerary chromosome in cat eye syndrome. Science 232:646–648 [DOI] [PubMed] [Google Scholar]
- McDermid HE, Morrow BE (2002) Genomic disorders on 22q11. Am J Hum Genet 70:1077–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart KE, Budarf ML, Driscoll DA, Emanuel BS, Ferreira P, McDermid HE (1998) Cat eye syndrome chromosome breakpoint clustering: identification of two intervals also associated with 22q11 deletion syndrome breakpoints. Cytogenet Cell Genet 81:222–228 10.1159/000015035 [DOI] [PubMed] [Google Scholar]
- Papenhausen P, Singh-Kahlon P, Griffin S, Stone P, Rogers KK, Rosini JE, Gadi IK, Tepperberg JH, Mowery PM (2002) Two cases of interstitial duplications detected only by interphase FISH. Am J Hum Genet Suppl 71:302 [Google Scholar]
- Potocki L, Chen KS, Park SS, Osterholm DE, Withers MA, Kimonis V, Summers AM, Meschino WS, Anyane-Yeboa K, Kashork CD, Shaffer LG, Lupski JR (2000) Molecular mechanism for duplication 17p11.2—the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet 24:84–87 10.1038/71743 [DOI] [PubMed] [Google Scholar]
- Reiss JA, Weleber RG, Brown MG, Bangs CD, Lovrien EW, Magenis RE (1985) Tandem duplication of proximal 22q: a cause of cat-eye syndrome. Am J Med Genet 20:165–171 [DOI] [PubMed] [Google Scholar]
- Saitta SC, Harris SE, Gaeth AP, Driscoll DA, McDonald-McGinn DM, Maisenbacher MK, Yersak JM, Chakraborty PK, Hacker AM, Zackai EH, Ashley T, Emanuel BS (2004) Aberrant interchromosomal exchanges are the predominant cause of the 22q11.2 deletion. Hum Mol Genet 13:417–428 10.1093/hmg/ddh041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, Lupski JR (2000) Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet 34:297–329 10.1146/annurev.genet.34.1.297 [DOI] [PubMed] [Google Scholar]
- Shprintzen RJ, Goldberg RB, Lewin ML, Sidoti EJ, Berkman MD, Argamaso RV, Young D (1978) A new syndrome involving cleft palate, cardiac anomalies, typical facies, and learning disabilities: velo-cardio-facial syndrome. Cleft Palate J 15:56–62 [PubMed] [Google Scholar]
- Smith AC, McGavran L, Robinson J, Waldstein G, Macfarlane J, Zonona J, Reiss J, Lahr M, Allen L, Magenis E (1986) Interstitial deletion of (17)(p11.2p11.2) in nine patients. Am J Med Genet 24:393–414 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002) Molecular-evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev 12:312–319 10.1016/S0959-437X(02)00304-0 [DOI] [PubMed] [Google Scholar]
- Swillen A, Devriendt K, Legius E, Prinzie P, Vogels A, Ghesquiere P, Fryns JP (1999) The behavioural phenotype in velo-cardio-facial syndrome (VCFS): from infancy to adolescence. Genet Couns 10:79–88 [PubMed] [Google Scholar]
- Thiel CT, Kraus C, Rauch A, Ekici AB, Rautenstrauss B, Reis A (2003) A new quantitative PCR multiplex assay for rapid analysis of chromosome 17p11.2–12 duplications and deletions leading to HMSN/HNPP. Eur J Hum Genet 11:170–178 10.1038/sj.ejhg.5200920 [DOI] [PubMed] [Google Scholar]