Abstract
The sequencing of entire human mitochondrial DNAs belonging to haplogroup U reveals that this clade arose shortly after the “out of Africa” exit and rapidly radiated into numerous regionally distinct subclades. Intriguingly, the Saami of Scandinavia and the Berbers of North Africa were found to share an extremely young branch, aged merely ∼9,000 years. This unexpected finding not only confirms that the Franco-Cantabrian refuge area of southwestern Europe was the source of late-glacial expansions of hunter-gatherers that repopulated northern Europe after the Last Glacial Maximum but also reveals a direct maternal link between those European hunter-gatherer populations and the Berbers.
Because of maternal transmission and lack of recombination, the sequence differentiation of human mtDNA has been generated by only the sequential accumulation of new mutations along radiating maternal lineages. Over the course of time, this process of molecular divergence has given rise to monophyletic units that are called “haplogroups.” Because this process of molecular differentiation occurred mainly during and after the process of human colonization of and diffusion into the different continents and regions, haplogroups and subhaplogroups tend to be restricted to specific geographic areas and population groups (Wallace 1995; Achilli et al. 2004).
Only the founders of the sister superhaplogroups M and N (which includes haplogroup R) (Quintana-Murci et al. 1999) participated in the “out of Africa” exit (Cann et al. 1987; Stringer and Andrews 1988; Cavalli-Sforza et al. 1994; Underhill et al. 2000; Forster et al. 2001) and were successful in colonizing the rest of the Old World. Superhaplogroup N is globally distributed outside Africa, encompassing virtually all of the western Eurasian mtDNA variation, and embraces haplogroup U, nested in haplogroup R. Haplogroup U has an extremely broad geographic distribution that ranges from Europe and North Africa to India and Central Asia and has a very high overall frequency (15%–30%) (Richards et al. 2000; Kivisild et al. 2003; Quintana-Murci et al. 2004)
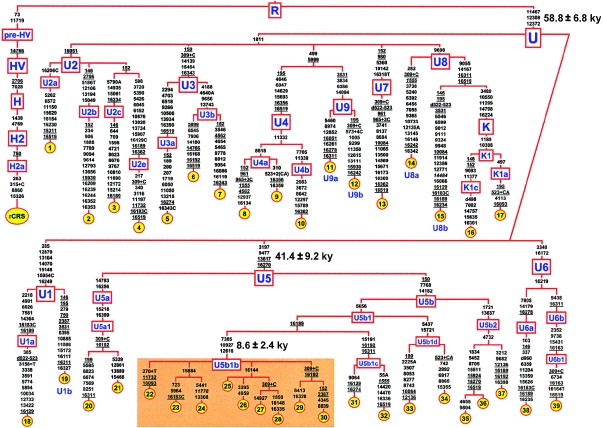
To assess the nature and extent of haplogroup U variation, we initially sequenced 28 entire U mtDNAs (see authors' Web site and GenBank) from a wide range of populations. These were selected through a preliminary sequence analysis of the mtDNA control region to include the widest possible range of haplogroup U internal variation. A tree of the mtDNA sequences (fig. 1) reveals that haplogroup U first splits into two major subsets, distinguished by the mutation at nt 1811, and that there is a very large number of independent basal branches. Among these, representatives of all known subhaplogroups (U1–U9) were included. However, subhaplogroup U5 provided a rather intriguing result. A Yakut from northeastern Siberia (27 in fig. 1) and a Fulbe from Senegal (29 in fig. 1) harbored mtDNAs that differed at only two coding-region nucleotide positions.
Figure 1.
Tree of 39 mtDNA sequences belonging to haplogroup U. The tree, rooted using the reference sequence (rCRS) (Andrews et al. 1999) as an outgroup, illustrates subhaplogroup affiliations. The sequencing procedure and phylogeny construction were performed as described elsewhere (Torroni et al. 2001b; Achilli et al. 2004). For phylogeny construction, the highly variable site 16519 and the length variation in the poly-C stretch at nts 309–315 were not used, and half the weight was assigned to the control-region mutations, relative to that assigned to coding-region mutations. Mutations are shown on the branches; they are transitions, unless a base change is explicitly indicated. Insertions are suffixed with a plus sign (+) and the inserted nucleotide(s), and deletions have a “d” prefix. Recurrent mutations are underlined; pathological mutations are in italics. The ethnic/geographic origins of mtDNAs are as follows: Pakistan (1–3, 12, and 13), Spain (4, 8, 14, 23, 36, and 39), Yemen (5 and 7), Adygei (6, 10, 18, and 20), Italy (9, 15–17, 19, 21, 22, 24, 31–33, 35, and 37), Ethiopia (11 and 38), Berber (30 and 34), Saami (25, 26, and 28), Yakut (27), and Fulbe from Senegal (29).
To investigate this striking similarity, the portion of the tree encompassing these two mtDNAs was enriched by sequencing 11 additional mtDNA sequences (22–26, 28, 30, 32, 34, 36, and 37 in fig. 1) bearing the control-region motif 16270-150, a motif found generally at low frequencies (<2%) in Berber populations and in other African groups (such as the Fulbe) known to have intermingled with Berbers (Rosa et al. 2004). The motif also shows similarly low frequencies in virtually all European populations, except the Saami of northern Scandinavia, in which it reaches ∼48% (Tambets et al 2004). Because virtually all Saami mtDNAs with 16270-150 harbor the transitions at nts 16189 and 16144 seen in the Yakut mtDNA, three Saami mtDNAs were included among the additional samples.
Seven of the new sequences (one Berber from Algeria, two Italian, one Spanish, and three Saami) clustered into U5b1b, the subclade encompassing the Yakut and Fulbe mtDNAs. The Saami and the Yakut mtDNAs formed a minor branch distinguished only by the transition at nt 16144, the Berber and the Fulbe mtDNAs clustered in a second minor branch also characterized only by control-region mutations, and the Italian and Spanish mtDNAs formed other minor branches.
The average sequence divergence (± SE, computed as per Saillard et al. [2000]) of the 39 coding-region sequences from the root of haplogroup U was 11.4 ± 1.3 substitutions (disregarding indels and pathological mutations), a value which corresponds—according to the mutation-rate estimate of Mishmar et al. (2003)—to a coalescence time estimate of 58.8 ± 6.8 thousand years (ky) for the entire haplogroup U. This value agrees well with the corresponding estimate of 61.6 ± 12.5 ky, based on the hypervariable segment I (HVS-I) mutation rate (Forster et al. 1996), for these 39 mtDNAs. An age of ∼60 ky indicates that haplogroup U arose very soon after the “out of Africa” exit. As for U5, its sequence divergence was 8.1 ± 1.8 substitutions, corresponding to 41.4 ± 9.2 ky, a time estimate in full agreement with its proposed proto-European origin (Richards et al. 2000). It is striking that the sequence divergence of U5b1b, the subclade encompassing mtDNAs from the Saami, Yakut, Berbers, and Fulbe, was 1.7 ± 0.5 substitutions, thus corresponding to only 8.6 ± 2.4 ky.
Such a recent common ancestry of maternal lineages found in populations living as far as 9,000 miles apart and whose anthropological affinities are not at all obvious is, to say the least, unexpected. Can we provide a reasonable explanation? The recent molecular dissection of other mtDNA haplogroups reveals some clues. H1 and H3, two frequent subhaplogroups of H, display frequency peaks centered in Iberia and surrounding populations, including the Berbers of Morocco, and coalescence ages of ∼11 ky (Achilli et al. 2004). Furthermore, their frequency patterns and ages resemble those reported for haplogroup V (Torroni et al. 2001a)—which, similar to U5b1b, is extremely common only in the Saami (together, U5b1b and V encompass almost 90% of the Saami mtDNAs) (Torroni et al. 1996; Tambets et al. 2004). Thus, although these previous studies have highlighted the role of the Franco-Cantabrian refuge area as a major source of the hunter-gatherer populations that gradually repopulated much of central and northern Europe when climatic conditions began to improve ∼15 ky ago, the identification of U5b1b now unequivocally links the maternal gene pool of the ancestral Berbers to the same refuge area and indicates that European hunter-gatherers also moved toward the south and, by crossing the Strait of Gibraltar, contributed their U5b1b, H1, H3, and V mtDNAs to modern North Africans.
In conclusion, this study is a paradigmatic example of the power of genetic inference in human-origin and evolutionary studies. It shows that mtDNA data—in this case, at the highest possible level of molecular resolution—can be used not only to evaluate models proposed by other disciplines and based on the direct survey of ancient material but also to identify previously unknown links between populations and geographic areas. Thus, the study of human genetics directly fosters the development of new research avenues in paleontology, archaeology, linguistics, and history.
Acknowledgments
This research was supported by CNR-MIUR Genomica Funzionale-Legge 449/97, Fondo Investimenti Ricerca di Base 2001, the Istituto Pasteur Fondazione Cenci Bolognetti, and Progetti Ricerca Interesse Nazionale 2003.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Author’s Web site, http://ipvgen.unipv.it/docs/projects/torroni_data/torroni_sequences.html (for the complete mtDNA sequences)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the complete mtDNA sequences [accession numbers AY882379–AY882417])
References
- Achilli A, Rengo C, Magri C, Battaglia V, Olivieri A, Scozzari R, Cruciani F, Zeviani M, Briem E, Carelli V, Moral P, Dugoujon JM, Roostalu U, Loogväli EL, Kivisild T, Bandelt HJ, Richards M, Villems R, Santachiara-Benerecetti AS, Semino O, Torroni A (2004) The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am J Hum Genet 75:910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 10.1038/13779 [DOI] [PubMed] [Google Scholar]
- Cann RL, Stoneking M, Wilson AC (1987) Mitochondrial DNA and human evolution. Nature 325:31–36 10.1038/325031a0 [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt H-J (1996) Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet 59:935–945 [PMC free article] [PubMed] [Google Scholar]
- Forster P, Torroni A, Renfrew C, Rohl A (2001) Phylogenetic star contraction applied to Asian and Papuan mtDNA evolution. Mol Biol Evol 18:1864–1881 [DOI] [PubMed] [Google Scholar]
- Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J, Metspalu E, Adojaan M, Tolk HV, Stepanov V, Golge M, Usanga E, Papiha SS, Cinnioglu C, King R, Cavalli-Sforza L, Underhill PA, Villems R (2003) The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet 72:313–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, Sukernik RI, Olckers A, Wallace DC (2003) Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA 100:171–176 10.1073/pnas.0136972100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Chaix R, Wells RS, Behar DM, Sayar H, Scozzari R, Rengo C, Al-Zahery N, Semino O, Santachiara-Benerecetti AS, Coppa A, Ayub Q, Mohyuddin A, Tyler-Smith C, Qasim Mehdi S, Torroni A, McElreavey K (2004) Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. Am J Hum Genet 74:827–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt HJ, Passarino G, McElreavey K, Santachiara-Benerecetti AS (1999) Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23:437–441 10.1038/70550 [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, et al (2000) Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 67:1251–1276 [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Brehm A, Kivisild T, Metspalu E, Villems R (2004) MtDNA profile of West Africa Guineans: towards a better understanding of the Senegambia region. Ann Hum Genet 68:340–352 (erratum 68:658) 10.1046/j.1529-8817.2004.00100.x [DOI] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt H-J, Nørby S (2000) mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 67:718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer CB, Andrews P (1988) Genetic and fossil evidence for the origin of modern humans. Science 239:1263–1268 [DOI] [PubMed] [Google Scholar]
- Tambets K, Rootsi S, Kivisild T, Help H, Serk P, Loogväli EL, Tolk HV, et al (2004) The western and eastern roots of the Saami—the story of genetic “outliers” told by mitochondrial DNA and Y chromosomes. Am J Hum Genet 74: 661–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Bandelt H-J, Macaulay V, Richards M, Cruciani F, Rengo C, Martinez-Cabrera V, et al (2001a) A signal, from human mtDNA, of postglacial recolonization in Europe. Am J Hum Genet 69:844–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus ML, Wallace DC (1996) Classification of European mtDNAs from an analysis of three European populations. Genetics 144:1835–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Rengo C, Guida V, Cruciani F, Sellitto D, Coppa A, Calderon FL, Simionati B, Valle G, Richards M, Macaulay V, Scozzari R (2001b) Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet 69:1348–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 10.1038/81685 [DOI] [PubMed] [Google Scholar]
- Wallace DC (1995) Mitochondrial DNA variation in human evolution, degenerative disease and aging. Am J Hum Genet 57:201–223 [PMC free article] [PubMed] [Google Scholar]