Abstract
Linguistic and archaeological evidence about the origins of the Malagasy, the indigenous peoples of Madagascar, points to mixed African and Indonesian ancestry. By contrast, genetic evidence about the origins of the Malagasy has hitherto remained partial and imprecise. We defined 26 Y-chromosomal lineages by typing 44 Y-chromosomal polymorphisms in 362 males from four different ethnic groups from Madagascar and 10 potential ancestral populations in Island Southeast Asia and the Pacific. We also compared mitochondrial sequence diversity in the Malagasy with a manually curated database of 19,371 hypervariable segment I sequences, incorporating both published and unpublished data. We could attribute every maternal and paternal lineage found in the Malagasy to a likely geographic origin. Here, we demonstrate approximately equal African and Indonesian contributions to both paternal and maternal Malagasy lineages. The most likely origin of the Asia-derived paternal lineages found in the Malagasy is Borneo. This agrees strikingly with the linguistic evidence that the languages spoken around the Barito River in southern Borneo are the closest extant relatives of Malagasy languages. As a result of their equally balanced admixed ancestry, the Malagasy may represent an ideal population in which to identify loci underlying complex traits of both anthropological and medical interest.
The island of Madagascar lies in the Indian Ocean, ∼250 miles from the African coast and ∼4,000 miles from Indonesia. Paleoecological and archaeological evidence suggest that, by 1,500–2,000 years ago, Madagascar had become the last great island landmass to be settled (Dewar and Wright 1993; Burney et al. 2004). The Malagasy language shares 90% of its basic vocabulary with Maanyan, a language spoken in the Barito River region of southern Borneo, which indicates that the predominant ancestry of the Malagasy language most likely derives from Borneo (Dahl 1951; Adelaar 1995). Malagasy also contains linguistic borrowings from the Bantu languages spoken in East Africa (Dahl 1988). Furthermore, substantial components of Malagasy material culture (e.g., cattle pastoralism) could be derived only from African sources.
At the time of the first Madagascan settlement, the entire Indian Ocean was a vast trading network connecting China with the Mediterranean and all societies in between (Vérin and Wright 1999). There is substantial evidence of Islamic influence and limited evidence of Indian influence on the Malagasy, in both language and culture.
In contrast to these cultural and linguistic traces of Malagasy ancestry, the genetic origins of the Malagasy are relatively poorly understood, and conflicting signals of African, Asian, and Pacific origin have appeared from studies of different loci (Migot et al. 1995; Soodyall et al. 1995; Hewitt et al. 1996). These contradictions result, in part, from being able to identify the likely origins of only a subset of lineages present at any single locus.
In the present study, we employed the detailed phylogenetic and geographic resolution of paternally inherited Y-chromosomal lineages and maternally inherited mtDNA lineages to apportion Malagasy lineages to ancestral populations. In this way, the contributions of the different ancestral populations to the modern Malagasy gene pool can be estimated directly, and likely geographic origins can be pinpointed with precision.
We assayed mtDNA and Y-chromosomal diversity in a Malagasy population sample comprising four different ethnic populations: Bezanozano (n=6), Betsileo (n=18), Merina (n=10), and Sihanaka (n=3). Ten potential ancestral populations (n=327) representing major population groups within Island Southeast Asia and Oceania were also analyzed with Y-chromosomal markers. To type all these samples for the required number of Y-chromosomal and mitochondrial (mt) markers, it was necessary to perform whole-genome amplification. Degenerate oligonucleotide-primed PCR (Nrich [Genetix]) (Telenius et al. 1992) performed better than multiple displacement amplification (Molecular Staging) (Dean et al. 2002) in early trials and, consequently, was used throughout.
We selected 44 binary markers in the present Y-haplogroup phylogeny (Y Chromosome Consortium [YCC] 2002; Jobling and Tyler-Smith 2003) that were predicted to be particularly informative in this study. These markers were typed using a combination of single-plex PCRs described elsewhere (Hurles et al. 2002; YCC 2002) and nine novel PCR multiplexes, each analyzing between three and seven SNPs. These multiplexes were designed to facilitate hierarchical typing, which minimizes the amount of genomic DNA required to define lineages at high resolution. These multiplexes use locus-specific primers tagged with universal primers to enable a two-step amplification protocol that equalizes the simultaneous amplification of multiple loci (Belgrader et al. 1996; Paracchini et al. 2002). SNPs lying within these PCR products were subsequently genotyped by single-base extension (SNaPshot [Applied Biosystems]) and capillary electrophoresis. Primer extension reactions were performed in half the recommended reaction volume but were otherwise processed in accordance with the manufacturer’s instructions. The amplification and extension primers used in the present study are detailed in table 1.
Table 1.
Primers for Y Binary and mt Variant Marker Multiplexes
|
Primer |
|||
| Amplificationb |
|||
| Multiplex and Markera | Forward | Reverse | Extensionc |
| Y binary A multiplex: | |||
| M130 | ggagcacgctatcccgttagacTTGTGTTTTGGTGGGATGTTG | cgctgccaactaccgcacatgTACTCTGCCCACAGAGATGGT | tgactgaGCCCTTTCCCCTGGGCAG |
| M145 | ggagcacgctatcccgttagacTATTCAGCAAGAGTAAGCAAGAGG | cgctgccaactaccgcacatgATCCTTTTTGGATCATGGTTCTT | actgactgactTTAGGCTAAGGCTGGCTCT |
| M89 | ggagcacgctatcccgttagacTCCTATGAGGTGCCATGAAA | cgctgccaactaccgcacatgGGATCACCAGCAAAGGTAGC | ctgactgactgactgactgactgactCTCAGGCAAAGTGAGAGAT |
| M9 | ggagcacgctatcccgttagacTCTGCAAAGAAACGGCCTAAG | cgctgccaactaccgcacatgACCGATTAAAAAGAGGCATTTTG | ACGGCCTAAGATGGTTGAAT |
| M45 | ggagcacgctatcccgttagacAGCTGGCAAGACACTTCTGAG | cgctgccaactaccgcacatgTAATATGTTCCTGACACCTTCC | gactgactgactgactgactCCTCAGAAGGAGCTTTTTGC |
| M96 | ggagcacgctatcccgttagacAGTTGCCCTCTCACAGAGCAC | cgctgccaactaccgcacatgAAAGGTCACTGGAAGGATTGC | ctgactgactgactGAAAACAGGTCTCTCATAATA |
| M168 | ggagcacgctatcccgttagacAGGATTCATGATGAAATCTGCTT | cgctgccaactaccgcacatgAAATCTCATAGGTCTCTGACTGTTC | ctgactgactgactgactgactgactgactGTATGTGTTGGAGGTGAGT |
| Y binary B multiplex: | |||
| M60 | ggagcacgctatcccgttagacAGAGCCCTGATGTGGACTCAA | cgctgccaactaccgcacatgACGCCAGTGCATTGAACACTA | gactgactgactgactgactgTAACCACTGTGTGCCTGAT |
| M91 | ggagcacgctatcccgttagacCACCCGTTAAGCAAAAATCC | cgctgccaactaccgcacatgTGCAGTGCCCTTCCAAATAAA | gactgactgactgGTAGTGAACTGATTAAAAAAAA |
| Y binary C multiplex: | |||
| LLy22g | ggagcacgctatcccgttagacTGATGTTGGCCTTTACAGCTC | cgctgccaactaccgcacatgTTTGGCTGAGAGACTGCGGG | gactgactgactgactgaAATTATTGTTTAAGCCACTAAG |
| M5 | ggagcacgctatcccgttagacGGGGTCCTATCAGGGGTTTA | cgctgccaactaccgcacatgTTTGTCTATTACCAAAGGTTTGTG | gactgaCTTGCACTCTTCTCCTTCT |
| M122 | ggagcacgctatcccgttagacTGGTAAACTCTACTTAGTTGCCTTT | cgctgccaactaccgcacatgATCAGCGAATTAGATTTTCTTGC | gactgactgacTTTTTTTTCCCCTGAGAGC |
| M134 | ggagcacgctatcccgttagacTAGAATCATCAAACCCAGAAGG | cgctgccaactaccgcacatgTCTTTGGCTTCTCTTTGAACAG | gactgactgactgactgactgactgTACTTTTGATCCCCACCAAT |
| M175 | ggagcacgctatcccgttagacTATCAGGCACATGCCTTCTCAC | cgctgccaactaccgcacatgATGGTCGAGTGTAGTGCATTGG | CACATGCCTTCTCACTTCTC |
| PN31 | ggagcacgctatcccgttagacTTAAGGCTGCGTGTTCCCTAT | cgctgccaactaccgcacatgTTGCACCTGACCTGTTCTTAC | gactgactgacATAAATAAGGTTTTTTTTTGGTTG |
| Y binary D multiplex: | |||
| M3 | ggagcacgctatcccgttagacTAATCAGTCTCCTCCCAGCA | cgctgccaactaccgcacatgAAATTGTGAATCTGAAATTTAAGG | gactgactgactgactgGGTCACCTCTGGGACTGA |
| MEH2 | ggagcacgctatcccgttagacTCGTTTTCTGATAGAAGATATAAATG | cgctgccaactaccgcacatgATACCATGAAAATTCATAATCCACA | gactgacTTTATGTAATTTAAAGCATAGTG |
| PN25 | ggagcacgctatcccgttagacAGCTATGCCTACAAAATGACAC | cgctgccaactaccgcacatgTAAAGGCTAAAGCAAAAAAGAAAC | gactgacCTGCCTGAAACCTGCCTG |
| M207 | ggagcacgctatcccgttagacAAGGAAAAATCAGAAGTATCCCTG | cgctgccaactaccgcacatgTTGGGATCTAATTTCTTCATTAG | gactgactgactgactgaTGTAAGTCAAGCAAGAAATTTA |
| Y binary E multiplex: | |||
| M35 | ggagcacgctatcccgttagacTATAAGCCTAAAGAGCAGTCAGAG | cgctgccaactaccgcacatgAGGTGAATGAACAACTAATCCAT | gactgactgactgactCGGAGTCTCTGCCTGTGTC |
| M123 | ggagcacgctatcccgttagacTACACAGAGCAAGTGACTCTCAAA | cgctgccaactaccgcacatgAAGTTGCCCAGGAATTTGCAT | GTATCTGAACTAGCATATCA |
| PN2 | ggagcacgctatcccgttagacTCTTGATGCAAATGAGAAAGAACT | cgctgccaactaccgcacatgACTCTAAAAACTGGAGGGAGAAA | gactgactgactgTGCCCCTAGGAGGAGAA |
| M2 | ggagcacgctatcccgttagacTCCCAGGAAGGTCCAGTAACA | cgctgccaactaccgcacatgAAAATGGAAAATACAGCTCCCC | gactgaTTATCCTCCACAGATCTCA |
| Y binary F multiplex: | |||
| M27 | ggagcacgctatcccgttagacTCATGCCCAGCTGAAACAATA | cgctgccaactaccgcacatgTCATGATTTGTCTTCTATTTCG | gGGAATCGAGGTTCAGGACA |
| M61 | ggagcacgctatcccgttagacTATTGGATTGATTTCAGCCTTC | cgctgccaactaccgcacatgTATTTTATTTTCTGTGTTCCTTGC | gactgacAGCTTCTCCTCTGGAGTC |
| M70 | ggagcacgctatcccgttagacTGGCACCATCTGTGAAAACAC | cgctgccaactaccgcacatgTTATCTTTATTCCCTTTGTCTTGCT | gactgactgacTCTGTTGTGGTAGTCTTAG |
| M147 | ggagcacgctatcccgttagacTCACTCTGGAGGCCAAGGTAG | cgctgccaactaccgcacatgTTATTCTGGGGCAATTTTAGGG | gactgactgactgGTCTCTGAAAGAAAAAAACAAA |
| SRY9138 | ggagcacgctatcccgttagacTGTTGATATGATATTATAGAGGC | cgctgccaactaccgcacatgTCCCAGATGCATATATTACAGG | gactgactgactgactgactgGCAAATTTAATGCTCTCGG |
| M214 | ggagcacgctatcccgttagacTTAGGCTGATTTTGCTGCTGA | cgctgccaactaccgcacatgTGAAATGCCACTTCACTCCAG | gactgactgactgactgactgactGAGACACTGTCTGAAAACAAC |
| Y binary G multiplex: | |||
| SRY465 | ggagcacgctatcccgttagacTGCCGAAGAATTGCAGTTTGC | cgctgccaactaccgcacatgTGTTGATGGGCGGTAAGTGGC | gaGTTGTCCAGTTGCACTTC |
| 47Z | ggagcacgctatcccgttagacTTCACCGTCTTAGCCAGGATG | cgctgccaactaccgcacatgTTAGTTACGCCTTGCATAAC | gactgactCTGGACTTGGTGGCTCA |
| M88 | ggagcacgctatcccgttagacATTCTAGGGTCAGGCAACTAGG | cgctgccaactaccgcacatgTTGTTTGTTCTATTCTATGGTCTTCC | gactgactgacTTATTCCTGCTTCTTCTGC |
| M95 | ggagcacgctatcccgttagacGAGTGGAAATCAAGATGCCAAG | cgctgccaactaccgcacatgTGCACCTGTTTTGTGTAAGAG | gactgactgactgacGAAAGACTACCATATTAGTG |
| Y binary H multiplex: | |||
| M50 | ggagcacgctatcccgttagacCGGCAACAGTGAGGACAGT | cgctgccaactaccgcacatgTGGTCCAAGGGCTGCTGGAG | gaAAAGGGCTCTGGTAAGAC |
| M101 | ggagcacgctatcccgttagacTGCCTCTTGCTTACTCTTGCT | cgctgccaactaccgcacatgTTGCAATCGGAAGCCTCAATCT | gactgGGAGATTTACTGAATCAGTG |
| M119 | ggagcacgctatcccgttagacGGGAAATGCCAAGGTAAATG | cgctgccaactaccgcacatgTTATGGGTTATTCCAATTCAG | gactgactgactCCAATTCAGCATACAGGC |
| Y binary I multiplex: | |||
| M33 | ggagcacgctatcccgttagacTTTGAGATAAGCCGCTAAACTTATTG | cgctgccaactaccgcacatgTTAGCCCCCAAGAGAGACAACT | gacTTATCTCATAAGTTACTAGTTA |
| M41 | ggagcacgctatcccgttagacTAGTATAATAGGCTGGGTGCTG | cgctgccaactaccgcacatgACATGAGTTCAAATGATTCTTC | gaGCCAACATGGTGAAACTG |
| M44 | ggagcacgctatcccgttagacTGCAGGAATCCCTGAGCATAA | cgctgccaactaccgcacatgCATGGCTGACAGCTAGGAAA | gactgactgacCTAACCTTCTAGTACACTG |
| M54 | ggagcacgctatcccgttagacAAGACTGAGGCCTCCTCTGGT | cgctgccaactaccgcacatgACCATCTCCTCACCTCTCCAA | gactgactgactgactgaCCCTCAGGCAGCCGCAC |
| M75 | ggagcacgctatcccgttagacTGCTAACAGGAGAAATAAATTACAGAC | cgctgccaactaccgcacatgATATTGAACAGAGGCATTTGTGA | gactgactgactgactgacGACAATTATCAAACCACATCC |
| mt Variant multiplex: | |||
| 10400 | ggagcacgctatcccgttagacTTGATCTAGAAATTGCCCTCCT | cgctgccaactaccgcacatgTCATAATTTAATGAGTCGAAATCAT | gactgactTGTTTAAACTATATACCAATTC |
| 15043 | ggagcacgctatcccgttagacTTCATCCGCTACCTTCACGC | cgctgccaactaccgcacatgTGTTGTTTGATCCCGTTTCGTG | gaCCTCTTCCTACACATCGG |
| 10398 | ggagcacgctatcccgttagacTTGATCTAGAAATTGCCCTCCT | cgctgccaactaccgcacatgTCATAATTTAATGAGTCGAAATCAT | gactgactgactgactgacCTACAAAAAGGATTAGACTGA |
| 15301 | ggagcacgctatcccgttagacTTCATCCGCTACCTTCACGC | cgctgccaactaccgcacatgTGTTGTTTGATCCCGTTTCGTG | gactgactgactgactgactgactCTTTACCTTTCACTTCATCTT |
| 10310 | ggagcacgctatcccgttagacTTGATCTAGAAATTGCCCTCCT | cgctgccaactaccgcacatgTCATAATTTAATGAGTCGAAATCAT | gactgactgactgactgactgactgactgaGCCCTACAAACAACTAACCT |
| 6455 | ggagcacgctatcccgttagacTAGGAACAGGTTGAACAGTCTA | cgctgccaactaccgcacatgTGAAAAATCAGAATAGGTGTTGG | gactgaAATACCAAACGCCCCTCTT |
| 9824 | ggagcacgctatcccgttagacTCCATTTCCGACGGCATCTAC | cgctgccaactaccgcacatgTATTAAGGCGAAGTTTATTACTC | gactgactgactgactgCACAGGCTTCCACGGACT |
All but “Variant” markers are Y-binary markers.
Lowercase letters indicate universal (ZIP) primers; letters in italics indicate spacer primers; bold uppercase letters indicate locus-specific primers.
Lowercase letters indicate variable-length tag primers; uppercase letters indicate locus-specific primers.
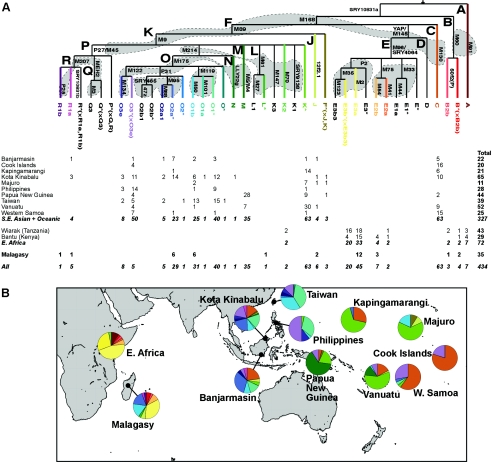
Together, these markers define 41 Y-chromosomal lineages, of which 10 are found in the Malagasy, 16 are found within Island Southeast Asia and Oceania, and 8 are found in East African populations (Luis et al. 2004). The Y-chromosomal lineages in East Africa are nonoverlapping with those found in Island Southeast Asia and Oceania (see fig. 1). As a consequence of this population differentiation, it is simple to apportion lineages found in the Malagasy to either an African or an Asian origin. All but two Malagasy lineages can be found in either East African or Southeast Asian populations. The two unaccounted-for lineages are single chromosomes belonging to haplogroups L* and R1b. Haplogroup L* is found at appreciable frequencies only in populations bordering the northern Indian Ocean, and haplogroup R1b reaches highest frequencies in northwestern Europe (Jobling and Tyler-Smith 2003). We believe these two lineages most likely reflect recent admixture events as a result of Indian Ocean trading links (Duplantier et al. 2002) and European colonization, respectively.
Figure 1.
A, Y-chromosomal haplogroup frequencies in the Malagasy and in potential ancestral populations. The maximum parsimony phylogeny relating the 41 Y-chromosomal haplogroups defined in the present study is shown above the absolute frequencies of those lineages in the different populations. The phylogeny is labeled with single-letter clades, and branches are labeled with the markers that define them. The lineage nomenclature is the updated version of that proposed by the YCC (YCC 2002; Jobling and Tyler-Smith 2003). The gray shading on the phylogeny indicates the nine sets of markers typed together in multiplexes. B, Pie charts illustrating the relative frequencies of the different haplogroups (colored to agree with the coloring of the phylogeny) shown on a map of the Indian Ocean. The Taiwanese population sample represents individuals pooled from four different aboriginal groups. Samples in these Island Southeast Asian and Pacific populations that were identified elsewhere as representing recent paternal European admixture in published lower-resolution Y-chromosomal marker typing of the same populations (Hurles et al. 2002) were not typed in the present study. East African data come from Luis et al. (2004).
To identify which Island Southeast Asian or Oceanic population represents the most likely source population for the Asian lineages found in the Malagasy, we computed pairwise FST distances, using the Arlequin software, to determine the closest populations, in terms of genetic distance to the Malagasy. This analysis indicates that, among the populations we sampled, the two populations from Borneo are the best candidates for the likely source of these lineages (table 2). This genetic proximity between the Malagasy and Borneo populations reflects the presence of appreciable frequencies of lineages O1b and O2a* in both populations, as well as a relative lack of chromosomes belonging to O3 lineages. The closest single Island Southeast Asian or Oceanic population to the Malagasy is that from Banjarmasin.
Table 2.
Pairwise Genetic Distances (FST) to the Malagasy on the Basis of Y-Haplogroup Frequencies
| Population Location | Distance (FST) |
| East Africaa | .083 |
| Banjarmasin | .094 |
| Kota Kinabalu | .102 |
| Taiwan | .170 |
| Majuro | .237 |
| Philippines | .249 |
| Vanuatu | .276 |
| Western Samoa | .283 |
| Papua New Guinea | .313 |
| Kapingamarangi | .316 |
| Cook Islands | .386 |
From the publication by Luis et al. (2004).
To explore the statistical significance of these observations, we devised a permutation test to assess whether the genetic distance between the Malagasy (A) and one population (B) is significantly smaller than that between the Malagasy and another population (C). In this test, the individual haplotypes observed in populations B and C are pooled and are randomly reassigned 10,000 times into two simulated populations (B′ and C′) with the same sample sizes as B and C. The P value of the difference in genetic distance—FST(A:B)-FST(A:C)—is then calculated as the fraction of simulated population pairs in which the difference in genetic distance between each of the populations and the Malagasy is greater than that observed in the real data—(FST[A:B′]-FST[A:C′])>(FST[A:B]-FST[A:C]). By use of this test, it was observed that there is no significant difference between the two Borneo populations (P=.8374) but that the resultant pooled Borneo population is significantly closer to the Malagasy than any other Island Southeast Asian population (P<.001).
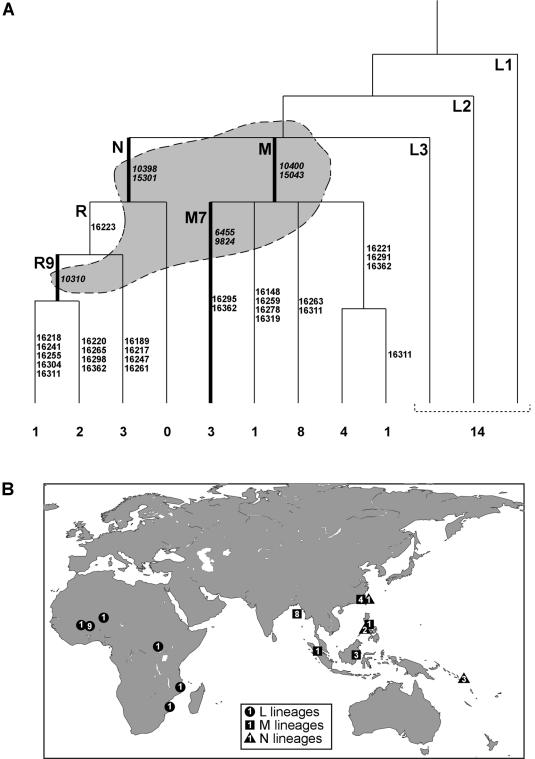
The phylogeny of mtDNA variation present in modern humans can be crudely characterized as comprising L lineages, present almost exclusively in Africa, and M and N lineages, present almost exclusively outside of Africa. Thus, classifying mt genomes into these major clades has significant power for discriminating between African and Asian origins. We devised a novel multiplex using the single base–extension method described above to type seven coding-region base substitutions (transitions at positions 15043, 10400, 10398, 15301, 6455, 9824, and 10310) that define the M and N lineages, as well as the R9 sublineage within haplogroup N and the M7 sublineage within haplogroup M (Kivisild et al. 2002), both of which are known to be present in Island Southeast Asian populations (primers used in this multiplex assay are detailed in table 1). Among 37 Malagasy mt genomes, we found 23 that belong to M and N lineages and 14 that belong to L lineages (fig. 2 and table 3).
Figure 2.
A, Phylogeny of mt sequence types found in the Malagasy and their CoGs. The maximum parsimony phylogeny of the 14 maternal lineages defined in the present study builds on the phylogeny constructed by Kivisild et al. (2002). The tips of the phylogeny are labeled with the number of Malagasy mt genomes found in each lineage. The phylogeny is also labeled with the major clades and the variable sites that define individual branches. The gray shading on the phylogeny indicates the seven coding variants that are typed together in a single multiplex. B, A map of the Old World, showing the positions of the CoGs of the 14 different mt sequence types (blackened circles, triangles, and squares) and the lineage group to which that sequence type belongs. Strictly speaking, mtDNA type “L” encompasses mtDNA types “M” and “N,” which are Eurasian subgroups of “L”; however, for simplicity, “L” here denotes “L” lineages excluding “M” and “N” types. The number within each shape indicates the frequency of that lineage within the data set. For most mt types, it was sufficient to enter only their HVSI sequences. However, to obtain monophyletic hits in the geographic database for two Malagasy HVSI sequence types found in paraphyletic mt lineages, it was also necessary to consider their coding SNP haplotypes, to eliminate spurious matches in geographical regions in which these haplotypes are known to be absent. Full HVSI sequence types are given in table 3.
Table 3.
mt Haplotypes Found in Study Populations[Note]
|
Allele at Position |
Variant Sites in HVSI |
||||||||||||
| Population and Haplotype | Frequency | 15043 | 6455 | 10400 | 9824 | 10398 | 15301 | 10310 | Lineage | 16085–16362 | 16085–16350 | Mutation Distance to Best Match | Weighted Average Deviation of Best Matches from CoG(miles) |
| Malagasy: | |||||||||||||
| 1 | 1 | G | C | G | T | G | A | G | L | 16223, 16265T | 0 | 3,423 | |
| 2 | 1 | G | C | G | T | G | A | G | L | 16209, 16223, 16311 | 0 | 1,551 | |
| 3 | 1 | G | C | G | T | G | A | G | L | 16182C, 11683C, 16189, 16223, 16278, 16290, 16294, 16309 | 0 | ,819 | |
| 4 | 1 | G | C | G | T | G | A | G | L | 16223, 16278, 16362 | 0 | 1,217 | |
| 5 | 9 | G | C | G | T | G | A | G | L | 16093, 16223, 16278, 16362 | 0 | 1,238 | |
| 6 | 1 | G | C | G | T | A | A | G | L | 16185, 16223, 16327 | 0 | 2,349 | |
| 7 | 1 | A | C | A | T | G | A | G | M(xM7) | 16086, 16148, 16223, 16259, 16278, 16319 | 0 | 2,800 | |
| 8 | 8 | A | C | A | T | G | A | G | M(xM7) | 16223, 16263, 16311 | 1 | 2,200 | |
| 9 | 4 | A | C | A | T | G | A | G | M(xM7) | 16221, 16223, 16291, 16362 | 1 | 2,500 | |
| 10 | 1 | A | C | A | T | G | A | G | M(xM7) | 16221, 16223, 16291, 16311, 16362 | 1 | 0 | |
| 11 | 3 | A | T | A | C | G | A | G | M7 | 16223, 16295, 16362 | 0 | 1,200 | |
| 12 | 3 | G | C | G | T | A | G | G | N(xR9) | 16189, 16217, 16247, 16261 | 0 | 2,700 | |
| 13 | 1 | G | C | G | T | A | G | A | R9 | 16218, 16241, 16255, 16304, 16311 | 2 | 40 | |
| 14 | 2 | G | C | G | T | A | G | A | R9 | 16220C, 16265, 16298, 16362 | 1 | 360 | |
| Banjarmasin: | |||||||||||||
| 1 | 1 | 16093, 16319 | |||||||||||
| 2 | 1 | 16311 | |||||||||||
| 3 | 2 | 16093, 16311 | |||||||||||
| 4 | 1 | 16129, 16263 | |||||||||||
| 5 | 1 | 16129, 16185, 16260, 16298 | |||||||||||
| 6 | 1 | 16129, 16234, 16290, 16311 | |||||||||||
| 7 | 1 | 16172, 16173, 16278, 16311 | |||||||||||
| 8 | 1 | 16093, 16220C, 16223, 16298 | |||||||||||
| 9 | 2 | 16108, 16111, 16129, 16162, 16172, 16183C, 16189, 16223, 16304 | |||||||||||
| 10 | 1 | 16111, 16168, 16172, 16183C, 16189, 16311 | |||||||||||
| 11 | 1 | 16129, 16172, 16223, 16294, 16304 | |||||||||||
| 12 | 2 | 16136, 16183C, 16189, 16217, 16223 | |||||||||||
| 13 | 1 | 16182C, 16183C, 16189, 16217, 16223, 16261 | |||||||||||
| 14 | 1 | 16192, 16223, 16234, 16288, 16304, 16309 | |||||||||||
| 15 | 1 | 16223, 16249, 16288, 16295, 16304 | |||||||||||
| 16 | 1 | 16086, 16147, 16183C, 16184A, 16189, 16217, 16223 | |||||||||||
| 17 | 1 | 16086, 16148, 16259, 16278, 16319 | |||||||||||
| 18 | 1 | 16093, 16184A, 16278 | |||||||||||
| Kota Kinabalu: | |||||||||||||
| 1 | 1 | 16111, 16129, 16223, 16266, 16304 | |||||||||||
| 2 | 1 | 16111, 16129, 16235, 16300 | |||||||||||
| 3 | 1 | 16111, 16168, 16172, 16183C, 16189, 16263, 16286, 16311 | |||||||||||
| 4 | 1 | 16126, 16129, 16183C, 16189, 16223, 16278 | |||||||||||
| 5 | 1 | 16126, 16129, 16297 | |||||||||||
| 6 | 1 | 16129, 16172, 16192A, 16223, 16294, 16304 | |||||||||||
| 7 | 1 | 16129, 16172, 16223, 16304 | |||||||||||
| 8 | 1 | 16129, 16209, 16272 | |||||||||||
| 9 | 1 | 16140, 16182C, 16183C, 16189, 16217, 16223, 16274, 16335 | |||||||||||
| 10 | 1 | 16140, 16182C, 16183C, 16189, 16223, 16266A | |||||||||||
| 11 | 1 | 16140, 16183C, 16189, 16223, 16243, 16294 | |||||||||||
| 12 | 1 | 16140, 16183C, 16189, 16223, 16266A | |||||||||||
| 13 | 1 | 16157, 16223, 16256, 16304, 16311, 16335 | |||||||||||
| 14 | 4 | 16185, 16291 | |||||||||||
| 15 | 1 | 16189, 16192, 16294G, 16297 | |||||||||||
| 16 | 1 | 16220C heteroplasmy | |||||||||||
| 17 | 1 | 16220C, 16223, 16258C, 16265, 16298 | |||||||||||
| 18 | 2 | 16223 | |||||||||||
| 19 | 1 | 16223, 16304 | |||||||||||
| 20 | 2 | 16278, 16295 | |||||||||||
| 21 | 2 | 16291 | |||||||||||
| 22 | 2 | 16295 | |||||||||||
| 23 | 1 | 16295, 16346C | |||||||||||
| 24 | 1 | 16311 | |||||||||||
| 25 | 1 | 16093, 16129, 16209, 16272 | |||||||||||
| 26 | 1 | 16093, 16136, 16295, 16337 | |||||||||||
| 27 | 1 | 16093, 16148, 16182C, 16183C, 16189 | |||||||||||
| 28 | 1 | 16093, 16295 | |||||||||||
| 29 | 1 | ||||||||||||
| Philippines: | |||||||||||||
| 1 | 1 | 16129, 16172, 16223, 16304, 16311 | |||||||||||
| 2 | 2 | 16111, 16129, 16140, 16183C, 16189, 16223, 16234, 16243 | |||||||||||
| 3 | 1 | 16126, 16223, 16231, 16284, 16311 | |||||||||||
| 4 | 2 | 16126, 16223, 16231, 16311 | |||||||||||
| 5 | 1 | 16129, 16172, 16223, 16243, 16304, 16311 | |||||||||||
| 6 | 2 | 16129, 16172, 16223, 16304, 16311 | |||||||||||
| 7 | 1 | 16140, 16183C, 16189, 16223, 16243 | |||||||||||
| 8 | 1 | 16145, 16176, 16223, 16224, 16233, 16311 | |||||||||||
| 9 | 1 | 16182C, 16183C, 16189, 16217, 16223, 16261, 16293 | |||||||||||
| 10 | 1 | 16192, 16223, 16278, 16325 | |||||||||||
| 11 | 1 | 16220C, 16223, 16240, 16265, 16298, 16335 | |||||||||||
| 12 | 3 | 16220C, 16223, 16265, 16298, 16335 | |||||||||||
| 13 | 1 | 16269, 16271 | |||||||||||
| 14 | 1 | 16291, 16311 | |||||||||||
| 15 | 1 | 16291 | |||||||||||
| 16 | 1 | 16295 | |||||||||||
| 17 | 2 | ||||||||||||
| 18 | 2 | 16093, 16182C, 16183C, 16189, 16217, 16223, 16261, 16293 | |||||||||||
Note.— Most variant sites are transitions; transversions are indicated by a letter given after the variant site, which indicates the derived state.
To further localize the geographical origins of Asian mtDNA lineages found in the Malagasy, we studied the hypervariable segment I (HVSI) sequence of the mt genome, for which a large volume of comparative data is available, we amplified and sequenced HVSI, using primers TTAACTCCACCATTAGCACC and GAGGATGGTGGTCAAGGGAC (Forster et al. 2002a) (between positions 16093 and 16362) in mtDNA from these 37 Malagasy individuals, and, by combining these data with the coding SNP haplotypes described above, we defined 14 distinct maternal lineages in the Malagasy (fig. 2).
A recently developed method for identifying the likely ancestry of a set of mt sequences is to perform a “center of gravity” (CoG) analysis of individual sequence types observed within a population (Röhl et al. 2001; Forster et al. 2002b). In our CoG analysis, the best matches to an HVSI sequence type were identified within a manually curated database of HVSI sequences associated with a precise geographical location. A CoG was then calculated by weighted interpolation of all best-match locations (see fig. 2). The relative lack of published Island Southeast Asian HVSI data could hamper a CoG analysis. To counteract this sampling bias, we added 82 HVSI sequences from Banjarmasin (n=21), Kota Kinabalu (n=36), and the Philippines (n=25) to the analysis. These sequence types are given in table 3. Exact matches within our database of 19,371 HVSI sequences can be found for all six maternal lineages in the Malagasy that appear to be Africa derived. By contrast, exact matches can be found for only three of eight Asia-derived maternal lineages.
The CoGs observed in the Malagasy fall within either Island Southeast Asia or sub-Saharan Africa. These CoGs accord exactly with the lineage classifications: all sequence types that belong to L haplogroups are found in Africa, and all sequence types that belong to M and N haplogroups are found in Island Southeast Asia. The relatively broad distribution of the Asian CoGs suggests that the present level of geographical resolution afforded by a CoG analysis is not sufficient to enable us to identify a single likely source population in Island Southeast Asia. It does, however, allow us to exclude the possibility that a Pacific Island population was the sole source of these mt lineages.
We calculated Nei’s gene diversity (using the Arlequin software) in HVSI sequences from the Malagasy and compared it with diversity apparent in the three Island Southeast Asian populations described above, as well as in published data on Mozambique (Pereira et al. 2001) and Oceanic populations (Hurles et al. 2003b). The Malagasy appear to have diversity that is significantly lower than that seen in Island Southeast Asia and Mozambique populations (Pereira et al. 2001) but that is higher than that seen in Pacific islands colonized within the past 3,500 years (table 4).
Table 4.
Diversity of mt HVSI Sequences in Different Populations
| HVSI Sequence Source | Nei’s Gene Diversity ± SE |
| Malagasy | .884 ± .031 |
| Malagasy Asian lineagesa | .838 ± .053 |
| Malagasy African lineagesa | .582 ± .150 |
| Mozambiqueb | .960 ± .008 |
| Kota Kinabalu | .989 ± .010 |
| Banjarmasin | .986 ± .019 |
| Philippines | .973 ± .018 |
| Samoac | .805 ± .053 |
| New Zealand Maoric | .239 ± .096 |
“Malagasy Asian” and “Malagasy African” refer, respectively, to the HVSI gene diversity among the Asia-derived and Africa-derived mt lineages in the Malagasy.
Mozambique mtDNA data are from the publication by Pereira et al. (2001).
Samoan and New Zealand Maori gene-diversity values are from the publication by Hurles et al. (2003b).
The amount of genetic diversity observed in a population is heavily influenced by demography and thus gives insights into settlement patterns. We might expect that the presence of HVSI sequences from two diverse ancestral populations would inflate HVSI sequence diversity; however, the lower genetic diversity in the Malagasy compared with both ancestral populations suggests either that early migrations were relatively restricted in numbers, duration, and origin or that subsequent population bottlenecks resulted in a postsettlement reduction of diversity. Recently colonized islands often exhibit reduced genetic diversity as a result of a combination of founder events and elevated genetic drift due to lower population sizes. However, this impact does not appear to be as severe in the Malagasy as it is for Pacific Island populations with a similarly recent settlement (reviewed by Hurles et al. [2003a]). This observation holds true even when only Asia-derived lineages are considered. This suggests that the sequential founder events and bottlenecks that were a feature of Pacific Island settlement were not paralleled in the colonization of Madagascar from the East and provides support for a direct rather than multistep process of migration from Indonesia. Alternatively, successive waves of migration from Asia may have brought different sets of lineages to Madagascar.
If we calculate gene diversity separately for Asia-derived and Africa-derived maternal lineages in the Malagasy, we find that the Asian lineages are significantly more diverse. These observations are largely explained by the predominance of a single African HVSI sequence type (found in 9 of 14 Africa-derived mt lineages). This sequence type is found in all four Malagasy ethnic populations sampled in the present study, so its predominance does not result from genetic drift in a single Malagasy subpopulation. Intuitively, one might expect fewer founders and therefore lower genetic diversity from the more geographically distant ancestral population. However, this does not appear to be the case in this situation. Given that the diversity apparent within the two ancestral populations is comparable, this implies that migrations from Africa may have been more limited than those from Indonesia.
In principle, it would be interesting to test whether these differences in Africa-derived and Asia-derived lineage diversity are replicated in the paternal lineages of the Malagasy. However, it is well documented that estimates of diversity that are based on genotyping known SNPs are biased by the markers selected for genotyping and the geographic distribution of the initial screening set used to identify these markers (Jobling and Tyler-Smith 2003). As a consequence, it is not appropriate to compare apparent SNP diversity between African and Asian Y-chromosomal lineages.
The above analyses demonstrate that we can consider the Malagasy to be an admixed population derived from two ancestral populations, one African and the other Indonesian; we can now estimate the admixture proportions of these populations. The mutual exclusivity of Y-chromosomal and mtDNA lineages between these two ancestral populations means that we can obtain a point estimate of admixture proportions simply by counting lineages. Of Malagasy mtDNA lineages, 38% (14/37) can be traced to Africa, whereas 51% (18/35) of Y-chromosomal lineages have an African origin. This increases to 55% (18/33) when the two putative recently admixed Y chromosomes are removed.
When estimating admixture proportions, we are estimating the cumulative contributions made by different ancestral populations to a hybrid population (Chakraborty 1986). We do not know the true frequencies of the different lineages in these three populations at the time that admixture occurred, and we can only infer these frequencies from sampling the contemporary populations that best approximate these ancient populations. Various factors influence the accuracy and precision of estimates of admixture proportions from contemporary populations, including sampling errors, genetic drift in all populations, the degree of population differentiation between the ancestral populations, and mutations. Various statistical methods that take into account some of these factors are available for estimating admixture proportions (reviewed by Jobling et al. [2004]). Using the software LEADMIX (Wang 2003), we employed three different statistical methods to estimate the proportion of African ancestry in Malagasy paternal lineages; in order of increasing complexity, these estimators are RH62 (Roberts and Hiorns 1962), L91 (Long 1991), and W03 (Wang 2003), the last of which is a recently derived likelihood estimator that estimates rates of genetic drift simultaneously in the ancestral and hybrid populations. These three methods all gave very similar estimates of African admixture proportions, which do not differ greatly from the estimate obtained from lineage counting: 58% for RH62, 56% for L91, and 56% for W03. There are broad confidence limits (19%–82%) for the latter likelihood estimate of paternal African admixture, which encompasses the point estimate of the proportion (38%) of African ancestry from maternal lineages. Consequently, the paternal and maternal estimates of the proportion of African ancestry in the Malagasy are statistically indistinguishable; there is no evidence of ancient sex-biased admixture. Further microgeographic sampling within Madagascar will be required to explore how admixture proportions vary among different Malagasy ethnic populations.
Characterization of genetic ancestry in the Malagasy has hitherto remained partial and imprecise. By contrast, in this study, because we generated comparative data from a wide range of potential ancestral populations, we have been able to identify the likely origins of all paternal and maternal lineages found in four different Malagasy ethnic populations.
We have confirmed the presence of the mt “Polynesian motif” among maternal Malagasy lineages, as was reported elsewhere (Soodyall et al. 1995). However, direct migration from Polynesia can be discounted, since the predominant Y-chromosomal haplogroups found in Polynesians, O3 and C, are not found at all among Malagasy paternal lineages.
Among the 10 potential ancestral populations in Island Southeast Asia and Oceania that we sampled, the Borneo populations had Y-chromosomal haplogroup distributions that were the most similar to those observed among the Malagasy. This observation is in striking agreement with the linguistic evidence that the Malagasy language is most closely related to the Maanyan language from the Barito River Valley in southern Borneo. Now that we have identified the region of origin for this Asian migration to Madagascar, further microgeographic sampling within Indonesian islands may pinpoint more precisely the origins of the Malagasy. Populations that possess both the paternal (e.g., O1b and O2a*) and maternal lineages (e.g., Polynesian motif) that are common in the Malagasy would be of particular interest. It is intriguing that the majority of Asia-derived mtDNA types present in the Malagasy do not have exact matches in an extensive database of HVSI sequences, and identification of these specific mtDNA sequence motifs within potential ancestral populations in Indonesia should be a priority. However, it must be remembered that genetic diversity in contemporary populations is an imperfect proxy for variation within ancient populations. The ongoing processes of population fission and fusion as well as genetic drift may prohibit the identification of a precise contemporary population that exactly represents the ancient population from which migrants departed. In addition, the possibility remains that migration either occurred from several genetically distinct sources within Indonesia or was kin structured (Fix 1999), such that no ancient population ever had the same lineage distribution as that of the migrants to Madagascar.
Admixture between two highly differentiated populations generates long-range allelic associations that decay over time (Chakraborty and Weiss 1988). The amount of linkage disequilibrium (LD) exhibited by an admixed population depends on a number of factors, including proportions of admixture, differentiation between ancestral populations, time since admixture, and demography (Pfaff et al. 2001). It has been proposed that it will be possible to efficiently map genes underlying complex traits by focusing on association studies in admixed populations, and a range of potentially informative populations has been identified (Halder and Shriver 2003). Although the time since admixture in the Malagasy is comparatively long, the high degree of differentiation between the two ancestral populations and the even balance of their contributions suggest that excess LD might still exist. Admixture mapping of genes underlying complex traits is predicated on the observation that the trait itself is differentially manifested among the ancestral populations. Therefore, although most attention has focused on European and African admixture in African Americans (Halder and Shriver 2003), it would be of interest to identify a range of admixed populations that are derived from various different ancestral combinations. The admixture of Indonesian and African lineages present in the Malagasy may be uniquely informative. Further characterization of LD in the Malagasy will be necessary for determining whether the Malagasy can be added to the list of admixed populations suitable for the identification of genes underlying complex traits that are of interest to anthropologists and medical geneticists alike.
Acknowledgments
The authors thank John Clegg, for his generosity in donating samples for this project, and Jinliang Wang, for his assistance with the LEADMIX software. We are grateful to Robert Dewar, for his comments on an earlier manuscript, and to Sue Adams, for support with genotyping. This work was funded by the Royal Society Research grant “Prehistory in Oceanic Populations,” the Wellcome Trust, and the McDonald Institute for Archaeological Research. M.A.J. was supported by a Wellcome Trust Senior Fellowship in Basic Biomedical Science (grant 057559).
References
- Adelaar A (1995) Asian roots of the Malagasy: a linguistic perspective. Bijdragen tot de Taal-Land en Volkenkunde 151:325–356 [Google Scholar]
- Belgrader P, Marino MM, Lubin M, Barany F (1996) A multiplex PCR-ligase detection reaction assay for human identity testing. Genome Sci Technol 1:77–87 [Google Scholar]
- Burney DA, Burney LP, Godfrey LR, Jungers WL, Goodman SM, Wright HT, Jull AJ (2004) A chronology for late prehistoric Madagascar. J Hum Evol 47:25–63 [DOI] [PubMed] [Google Scholar]
- Chakraborty R (1986) Gene admixture in human populations: models and predictions. Yearb Phys Anthropol 29:1–43 [Google Scholar]
- Chakraborty R, Weiss KM (1988) Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc Natl Acad Sci USA 85:9119–9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl OC (1951) Malgache et Maanyan: une comparison linguistique. Egede Intitutett, Oslo [Google Scholar]
- ——— (1988) Bantu substratum in Malagasy. Études Océan Indien 9:91–132 [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, Driscoll M, Song W, Kingsmore SF, Egholm M, Lasken RS (2002) Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA 99:5261–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar RE, Wright HT (1993) The culture history of Madagascar. J World Prehistory 7:417–466 [Google Scholar]
- Duplantier JM, Orth A, Catalan J, Bonhomme F (2002) Evidence for a mitochondrial lineage originating from the Arabian peninsula in the Madagascar house mouse (Mus musculus). Heredity 89:154–158 [DOI] [PubMed] [Google Scholar]
- Fix AG (1999) Migration and colonisation in human microevolution. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- Forster L, Forster P, Lutz-Bonengel S, Willkomm H, Brinkmann B (2002a) Natural radioactivity and human mitochondrial DNA mutations. Proc Natl Acad Sci USA 99:13950–13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P, Cali F, Röhl A, Metspalu E, D’Anna R, Mirisola M, De Leo G, Flugy A, Salerno A, Ayala G, Kouvatsi A, Villems R, Romano V (2002b) Continental and subcontinental distributions of mtDNA control region types. Int J Legal Med 116:99–108 [DOI] [PubMed] [Google Scholar]
- Halder I, Shriver MD (2003) Measuring and using admixture to study the genetics of complex diseases. Hum Genomics 1:52–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt R, Krause A, Goldman A, Campbell G, Jenkins T (1996) β-globin haplotype analysis suggests that a major source of Malagasy ancestry is derived from Bantu-speaking Negroids. Am J Hum Genet 58:1303–1308 [PMC free article] [PubMed] [Google Scholar]
- Hurles ME, Matisoo-Smith E, Gray RD, Penny D (2003a) Untangling Oceanic settlement: the edge of the knowable. Trends Ecol Evol 18:531–540 [Google Scholar]
- Hurles ME, Maund E, Nicholson J, Bosch E, Renfrew C, Sykes BC, Jobling MA (2003b) Native American Y chromosomes in Polynesia: the genetic impact of the Polynesian slave trade. Am J Hum Genet 72:1282–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles ME, Nicholson J, Bosch E, Renfrew C, Sykes BC, Jobling MA (2002) Y chromosomal evidence for the origins of Oceanic-speaking peoples. Genetics 160:289–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling MA, Hurles ME, Tyler-Smith C (2004) Human evolutionary genetics: origins, peoples and disease. Garland Science, New York [Google Scholar]
- Jobling MA, Tyler-Smith C (2003) The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 4:598–612 [DOI] [PubMed] [Google Scholar]
- Kivisild T, Tolk HV, Parik J, Wang Y, Papiha SS, Bandelt HJ, Villems R (2002) The emerging limbs and twigs of the East Asian mtDNA tree. Mol Biol Evol 19:1737–1751 [DOI] [PubMed] [Google Scholar]
- Long JC (1991) The genetic structure of admixed populations. Genetics 127:417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis JR, Rowold DJ, Regueiro M, Caeiro B, Cinnioğlu C, Roseman C, Underhill PA, Cavalli-Sforza LL, Herrera RJ (2004) The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am J Hum Genet 74:532–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migot F, Perichon B, Danze PM, Raharimalala L, Lepers JP, Deloron P, Krishnamoorthy R (1995) HLA class II haplotype studies bring molecular evidence for population affinity between Madagascans and Javanese. Tissue Antigens 46:131–135 [DOI] [PubMed] [Google Scholar]
- Paracchini S, Arredi B, Chalk R, Tyler-Smith C (2002) Hierarchical high-throughput SNP genotyping of the human Y chromosome using MALDI-TOF mass spectrometry. Nucleic Acids Res 30:e27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Macaulay V, Torroni A, Scozzari R, Prata MJ, Amorim A (2001) Prehistoric and historic traces in the mtDNA of Mozambique: insights into the Bantu expansions and the slave trade. Ann Hum Genet 65:439–458 [DOI] [PubMed] [Google Scholar]
- Pfaff CL, Parra EJ, Bonilla C, Hiester K, McKeigue PM, Kamboh MI, Hutchinson RG, Ferrell RE, Boerwinkle E, Shriver MD (2001) Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium. Am J Hum Genet 68:198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DF, Hiorns RW (1962) The dynamics of racial admixture. Am J Hum Genet 14:261–277 [PMC free article] [PubMed] [Google Scholar]
- Röhl A, Brinkmann B, Forster L, Forster P (2001) An annotated mtDNA database. Int J Legal Med 115:29–39 [DOI] [PubMed] [Google Scholar]
- Soodyall H, Jenkins T, Stoneking M (1995) “Polynesian” mtDNA in the Malagasy. Nat Genet 10:377–378 [DOI] [PubMed] [Google Scholar]
- Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A (1992) Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics 13:718–725 [DOI] [PubMed] [Google Scholar]
- Vérin P, Wright H (1999) Madagascar and Indonesia: new evidence from archaeology and linguistics. Indo Pac Prehist Assoc Bull 18:35–42 [Google Scholar]
- Wang J (2003) Maximum-likelihood estimation of admixture proportions from genetic data. Genetics 164:747–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y Chromosome Consortium (YCC) (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]