Abstract
About 5% of the human genome consists of segmental duplications or low-copy repeats, which are large, highly homologous (>95%) fragments of sequence. It has been estimated that these segmental duplications emerged during the past ∼35 million years (Myr) of human evolution and that they correlate with chromosomal rearrangements. Williams-Beuren syndrome (WBS) is a segmental aneusomy syndrome that is the result of a frequent de novo deletion at 7q11.23, mediated by large (∼400-kb) region-specific complex segmental duplications composed of different blocks. We have precisely defined the structure of the segmental duplications on human 7q11.23 and characterized the copy number and structure of the orthologous regions in other primates (macaque, orangutan, gorilla, and chimpanzee). Our data indicate a recent origin and rapid evolution of the 7q11.23 segmental duplications, starting before the diversification of hominoids (∼12-16 million years ago [Mya]), with species-specific duplications and intrachromosomal rearrangements that lead to significant differences among those genomes. Alu sequences are located at most edges of the large hominoid-specific segmental duplications, suggesting that they might have facilitated evolutionary rearrangements. We propose a mechanistic model based on Alu-mediated duplicated transposition along with nonallelic homologous recombination for the generation and local expansion of the segmental duplications. The extraordinary rate of evolutionary turnover of this region, rich in segmental duplications, results in important genomic variation among hominoid species, which could be of functional relevance and predispose to disease.
Single base-pair mutations, sequence duplications, small insertions/deletions, and chromosomal rearrangements are the primary forces by which genomes evolve over time. As the differences at the level of genomic DNA sequence are very low among primates (Chen and Li 2001), many relevant phenotypic differences might be due to chromosomal rearrangements and insertion/deletion events (Frazer et al. 2003; Locke et al. 2003). Karyotype analysis of the primate genomes reveals just a few cytogenetic changes between humans and chimpanzees, including nine pericentric inversions and one acrocentric fusion (Yunis and Prakash 1982). The identification of these and additional evolutionary rearrangements provides excellent targets for focused studies of gene expression variation, in order to define the genetic differences responsible for the phenotypic differences between closely related species (Marques-Bonet et al. 2004). Such chromosomal rearrangements could also create genetic barriers leading to stasipatric speciation by facilitating reproductive isolation (Samonte and Eichler 2002).
The identification that ∼5% of the human genome consists of interspersed duplications with a high degree of identity at the nucleotide level (>95%) and covering large genomic distances has raised intense research interest in the dynamic mechanisms of mutation of the human genome and the role of the duplications in evolution (Eichler 2001). Our genome contains many segmental duplications, also called low-copy repeats or duplicons, present in every chromosome with a nonuniform distribution (Lander et al. 2001; Venter et al. 2001). On the basis of the high sequence identity (90%-100%) between paralogous copies, it has been estimated that human segmental duplications emerged during the past ∼35 million years (Myr) of evolution, with special enrichment between 1 and 12 million years ago (Mya) (Samonte and Eichler 2002).
The mechanisms that lead to the generation of segmental duplications are not completely understood. Initial analyses appear to indicate that their distribution in the genome is not random, since they are preferentially located in subtelomeric and pericentromeric regions (Eichler 2001). A significant abundance of segmental duplications in the human genome has been found at the regions of break of synteny with the mouse genome, although it is not clear whether they are the cause or the consequence of the evolutionary rearrangements (Samonte and Eichler 2002; Armengol et al. 2003).
Human chromosome 7 is especially rich in segmental duplications, with an 8.2% overall content and a predominant enrichment of intrachromosomal duplications (7.0% of the sequence) (Hillier et al. 2003; Scherer et al. 2003). Williams-Beuren syndrome (WBS, OMIM#194050) is a segmental aneusomy caused by a common deletion of 1.55 Mb in one of the more challenging genomic regions of this chromosome, 7q11.23 (Pérez Jurado et al. 1996; Robinson et al. 1996). Large and complex segmental duplications flank the commonly deleted interval acting as substrates for nonallelic homologous recombination (NAHR) that mediates the deletion. The region appears to be highly dynamic in humans, since polymorphic variation of block copy number as well as large paracentric inversions between segmental duplications have been found in normal populations (Osborne et al. 2001; Bayés et al. 2003). The entire region has been shown to be single-copy in mouse and other mammals, with evolutionary breaks of synteny located in the regions that harbor the large segmental duplications in humans (DeSilva et al. 1999; Valero et al. 2000).
In the present study, we used computational analyses, FISH, and additional molecular studies to precisely define the structure of the segmental duplications on human 7q11.23 and the copy number and structure of the orthologous regions in other primates: Japanese and rhesus macaques (Macaca fuscata, MFU, and Macaca mulatta, MMU), olive and hamadryas baboons (Papio anubis, PNU, and Papio hamadryas, PHA), orangutan (Pongo pygmaeus, PPY), gorilla (Gorilla gorilla, GGO), and chimpanzee (Pan troglodytes, PTR). Our data support a recent origin of the segmental duplications and a very rapid and divergent evolution of the chromosomal region, including several sequential events of duplication and inversion that led to the complex structure found in the WBS region in humans. We offer a proposal regarding some of the specific molecular mechanisms that have resulted in the generation of these segmental duplications.
Results
Characterization of human 7q11.23 segmental duplications and breakpoint junctions
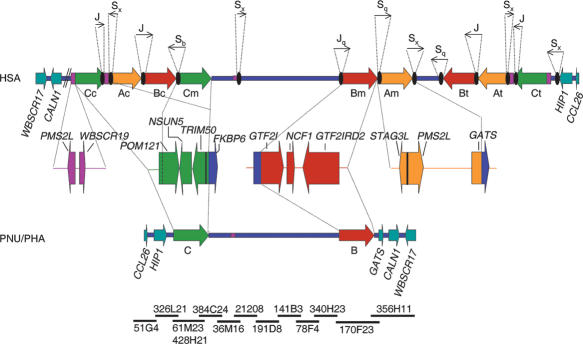
We used our previous assembly of clones of the region (Valero et al. 2000) along with sequence data from the recent human genome assemblies to delineate the edges of the blocks of segmental duplications by sequence alignment with BLAST (http://www.ncbi.nlm.nih.gov/) and/or BLAT (http://www.genome.ucsc.edu/). As previously reported, the 7q11.23 segmental duplications have a modular structure, represented in Figure 1. The specific coordinates of each block with respect to the human chromosome 7 entire sequence along with its gene and pseudogene content based on gene prediction, RT-PCR, and EST data are indicated in Supplemental Table 1. It is worth noting that several transcriptional units of the region appear to be activated from close promoters located in different strands (FKBP6 and TRIM50; PMS2L and STAG3L) or show overlap at the 3′ region (POM121 and NSUN5) (Fig. 1). Alu elements, most of them belonging to the Alu S subfamily, are located at the edges of all the large blocks of segmental duplications in the region (Fig. 1). Additional Alu elements are also found close to many (but not all) intersections of duplicated modules within the blocks (data not shown).
Figure 1.
Schematic representation of the genomic structure of the WBS deletion region with the flanking segmental duplications in humans (HSA), and the homologous region in baboon (PNU/PHA). The large blocks of segmental duplications in the human map (A in yellow, B in red, C in green) are represented by thick arrows to indicate their relative orientation with respect to each other. They are exclusively present in the human map, whereas the baboon's genome contains the ancestral loci as single-copy and no large segmental duplication. The blue line represents the single-copy region, and the genes located immediately outside the region in both directions are represented as light blue arrows indicating the transcriptional direction. Some of the multiple-copy modules present in other chromosome 7 locations are shown in purple. The composition of each duplicated block with the corresponding transcriptional units is shown below the human map. Black ovals represent the Alu repeats located at the edges of the segmental duplications in the human map, with arrows indicating their orientation and approximate size (either partialor full Alu elements) shown on top. Note that the entire region including the ancestral loci of the segmental duplications is inverted in baboon with respect to the flanking genes. To define the baboon genomic structure, a clone contig with sequenced BACs from the RP41 library available in public databases has been assembled (NISC Comparative Sequencing Initiative), shown at the bottom.
Structure of the homologous regions in baboon and other mammals
A clone contig that encompasses the entire region has been assembled based on partially or totally sequenced BAC clones from olive and hamadryas baboons by the NISC Comparative Sequencing Initiative (http://www.nisc.nih.gov) (Fig. 1). Interestingly, there is complete conservation of synteny between mouse and baboon, indicating the absence of any evolutionary chromosomal rearrangement affecting this region since the divergence of rodents and primates, estimated to be ∼80 Mya. In addition, analysis of the available sequence reveals that the region in baboons does not contain any of the large segmental duplications present in humans.
Segmental duplications in hominoids
To determine the genomic structure of this region in other nonhuman primates, we analyzed the absence/presence of segmental duplications and their organization in macaque, orangutan, gorilla, and chimpanzee by several approaches, including microsatellite typing, quantitative PCR of paralogous sequence variants (PSVs), FISH in interphase nuclei and sequencing.
Analysis of block A
The microsatellite marker D7S489 is present in four loci in humans, three within blocks A, and another one within the commonly deleted interval in WBS (Bayés et al. 2003). Genotyping in nonhuman primates indicated the presence of a single locus in macaque (likely corresponding to D7S489B) and two loci in orangutan, gorilla, and chimpanzee. All humans analyzed revealed a total of eight alleles as expected. We also sequenced a PCR fragment (BA/STAG3) of the STAG3 gene (7q22). In humans, the same primers also amplify the three pseudogene copies L1, L2, and L3 located within blocks Ac, Am, and At, respectively (7q11.23). A unique sequence indicative of single copy was obtained in macaque (likely the ancestral gene), while several positions with double peaks, suggesting the presence of at least two copies, were found in orangutan, gorilla, and chimpanzee. The different copies were identified by sequencing of cloned PCR products.
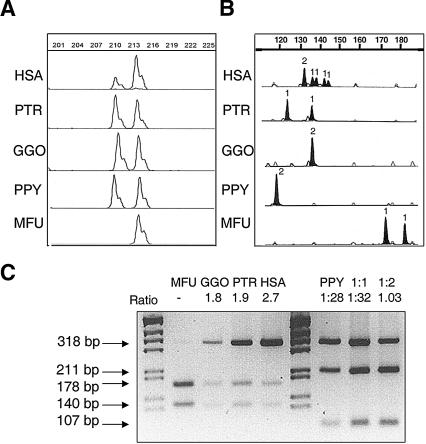
In addition, a 3-base pair (bp) deletion/insertion PSV located in exon 13 of STAG3/L1,L2,L3 allowed us to calculate a dosage ratio of the 7q22 copy versus the 7q11.23 copies. Human samples gave consistent values of 0.47 ± 0.1 (mean ± standard deviation), which were used as reference of a 1:3 ratio. In macaques, we obtained a single peak of the size corresponding to the 7q11.23 copies in humans. In all three hominoids (orangutan, 0.95 ± 0.1; gorilla, 1.04 ± 0.06; chimpanzee, 1.09 ± 0.1), the results were consistent with the presence of an even number of loci, most likely two, one of each size (Fig. 2A).
Figure 2.
Representative assays of copy number quantification in human (HSA), chimpanzee (PTR), gorilla (GGO), orangutan (PPY), and macaque (MFU) DNA, by comparison of paralogous sequence variants (PSVs) and microsatellites located in the segmental duplications. (A) A deletion/insertion PSV in block A distinguishes the ancestral STAG3 gene copy with respect to the pseudogene copies L1, L2, and L3. The STAG3/STAG3L copy ratio calculated for each species was: 0.47 ± 0.1 in HSA, 1.09 ± 0.1 in PTR, 1.04 ± 0.06 in GGO, and 0.95 ± 0.1 in PPY. Numbers on top show the amplimer size (in bp). (B) A microsatellite located between NCF1 and GTF2I in block B (BBSTR1, Bayés et al. 2003). All nonhuman primates displayed one or two alleles indicative of a single locus, whereas humans revealed six alleles corresponding to three different loci. The number of inferred alleles is indicated over each peak. (C)A restriction assay for a PSV of the TRIM50 gene in block C. In all primates but orangutan, there was a differential restriction site for NgoMIV. In orangutan, another assay with MluNI was performed and compared with artificial situations displaying 1:1, 2:1 ratios. Ratios between restriction products are shown at the top of each sample.
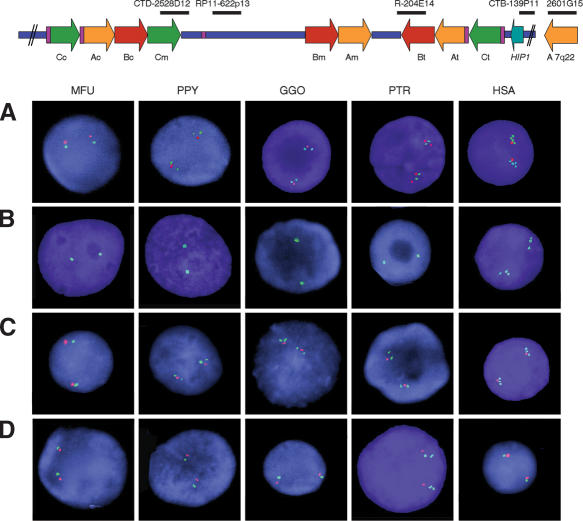
Interphase FISH with BAC CITBI-E1-2601G15 (containing part of the ancestral STAG3 gene and other block A sequences at 7q22) showed eight signals per nucleus in humans (68%/50 nuclei) and two separated locations in metaphases, 7q22 and more intense in 7q11.23, corresponding to the location of the ancestral locus and the three pseudogene copies L1, L2, and L3, respectively. In nonhuman hominoids, four signals per nucleus and two locations in chromosome metaphases were found (72%-80%/100 nuclei), whereas only two signals were visible in macaque (100%/20 nuclei) (Fig. 3A). Therefore, we have detected a duplication event of block A on an ancestral chromosome common to all hominoids, along with two additional recent duplication events exclusively in humans. FISH analysis with BAC RP11-451K15 (containing block Am) and PAC RP1-42M2 (containing PMS2 in 7p22) detected an uncountable number of signals in the nuclei of all species, all located in the homologs to human chromosome 7, indicating that some modules within block A are multiple copy in all primates (data not shown).
Figure 3.
Number of blocks of segmental duplications detected by interphase FISH in the different primates with selected human BAC clones as probes. The location of all probes with respect to a representation of the human map is shown on top. (A) One single signal per chromosome in MFU, two in PPY, GGO, and PTR and four in HSA are detected with CITBI-E1-2601G15 (block A, 7q22, green), whereas CTB-139P11 (HIP1 locus, red) is single-copy in all species except chimpanzee, where it is duplicated. (B) In each species BAC RP11-204E14 (block B, green) displays one signal per chromosome except for humans, where three signals are found. (C) A single signal per chromosome is found in all the nuclei with RP11-622P13 (STX1A locus, red), whereas CTD-2528D12 (block C, green) displays one signal per chromosome in MFU, but two in PPY, GGO, and PTR and three in HSA. The single-copy STX1A locus is located in between the two (hominoids) or three (humans) signals of block C sequences. (D) CTB-139P11 (HIP1 locus, green) shows one signal per chromosome in all interphase nuclei except for PTR, which shows two signals indicating a duplication. Both copies of the HIP1 locus are located telomeric to the STX1A locus (RP11-622P13, red).
Analysis of block B
At BBSTR1, a microsatellite located between NCF1 and GTF2I (Bayés et al. 2003), all nonhuman primates displayed no more than two alleles suggestive of single locus, whereas humans revealed six recognizable alleles corresponding to three different loci (Fig. 2B). The NCF1 variant that distinguishes the nonfunctional pseudogenes (Bc and Bt) from the functional gene (Bm), a GT deletion at exon 2 (Gorlach et al. 1997), was also genotyped. Whereas a 1:2 gene/pseudogenes ratio was observed in humans, no pseudogene-like copies were identified in nonhuman primates, also suggesting that NCF1 is a single-copy gene in all primates but humans.
Furthermore, we sequenced a 2.5-kb PCR product from exon 16 of the GTF2IRD2 gene (BB/GTF2IRD2), that contains 24 predicted PSVs among the three human copies. No double peaks suggestive of more than one copy were found in nonhuman primates.
Finally, we performed FISH analysis with BAC RP11-204E14, containing block Bt (Fig. 3B). Signals indicated the presence of a single locus per chromosome in all nonhuman primates (100%/20 nuclei) and the expected three loci in humans (six signals in 72%/50 nuclei). In conclusion, all these results are consistent with the existence of a single block B in nonhuman primates and the appearance of one entire block B at each segmental duplication after the separation of humans from chimpanzees.
Analysis of block C
Sequencing of two amplicons from the POM121 and TRIM50 genes revealed significant secondary peaks and/or frameshifts in orangutan, gorilla, and chimpanzee as well as in humans, but not in macaque. Relative peak intensities were consistent with two copies in nonhuman hominoids and more than two in humans. We then designed restriction assays based on the putative PSVs detected by sequencing, in order to better define the number of block C copies in each species. NgoMIV is predicted to cut only the functional gene (Cm) but not the pseudogenes (Cc and Ct) in humans, and sequence data suggested that this PSV was also present in chimpanzee and gorilla but not in orangutan. The obtained pseudogenes:gene peak ratios (2.7 ± 0.2) were always consistent with the expected 2:1 ratio in human samples (Fig. 2C). In both gorilla and chimpanzee we observed values compatible with a 1:1 ratio (1.8 ± 0.1 and 1.9 ± 0.2, respectively), indicating the existence of two copies of block C in both species. A different assay was performed to confirm the copy number in orangutan, using another putative PSV detected by MluNI. Since this PSV was not found in other species, clones specific for each of the two orangutan variants were used as restriction controls. To calculate the ratio values, artificial situations displaying 1:1 and 1:2 ratios (actual values: 1.32 and 1.03, respectively) were simulated mixing the adequate quantities of DNA from each clone as a template for the PCR/RFLP analysis. In genomic DNA from orangutan samples, the ratio value was 1.28 ± 0.11, indicating the presence of two copies as well (Fig. 2C). Macaque samples displayed patterns compatible with one single locus at all sites.
Interphase FISH with BAC CTD-2528D12 (containing block Cm and additional 80Kb telomeric to it) along with BAC RP11-622P13 (STX1A) obtained results consistent with one locus in macaque (two signals in 100%/20 nuclei), two in orangutan, gorilla, and chimpanzee (four signals in 76%-84%/100 nuclei), and three in humans were detected as expected (75%/50 nuclei). In orangutan, gorilla, and chimpanzee, the signal corresponding to the STX1A locus was found in between the two copies of block C (Fig. 3C). Specific and colocalized hybridization of all signals was found in metaphase chromosomes in all species. These results indicate the presence of a duplicated block C flanking the orthologous region to the WBS deletion in all three nonhuman hominoids.
Detection of additional segmental duplications external to the region
We analyzed another segmental duplication located 1 Mb telomeric to the WBS region in humans, which contains a part of block C that is absent in Cm including the first three exons of POM121. Within this segmental duplication, the 5′ of POM121 is fused to the 3′ portion of a duplicated ZP3 gene, creating a chimeric transcript (Francke 1999). This fusion gene was detected by PCR only in chimpanzee and humans, suggesting that the duplication took place in a common ancestor to both species after the divergence of gorilla (Supplemental figure).
The HIP1 gene, located next to Ct, is a single copy locus in the human genome, whereas in silico data suggested a duplication in the chimpanzee genome. A 1.1-Kb fragment of the last exon of this gene was PCR-amplified from genomic DNA of the five primate species analyzed. Sequence analysis revealed secondary peaks only in chimpanzee samples. The presence of two sequences corresponding to two different loci in chimpanzees was confirmed by sequencing cloned PCR products. In addition, interphase FISH using BACs CTB-139P11 (HIP1) and RP11-622P13 (STX1A) clearly indicated the duplication of the HIP1 locus in chimpanzee (four signals in 82%/100 nuclei) and its single copy status in all of the other primates analyzed (two signals in 100%/40 nuclei in macaque, orangutan, gorilla, and human; Fig. 3D). Both copies of the HIP1 gene duplication are located at the same side of STX1A, thus the duplication of HIP1 is independent of that of block C. The chimpanzee specific segmental duplication encompasses about 80 Kb of genomic DNA containing 34 kb 5′ and the entire HIP1 gene, along with some of the multicopy modules (with PMS2L and WBSCR19 related sequences) present in block C. Alu repeats are also present at the edges of this segmental duplication. Both HIP1 copies are predicted to encode properly processed transcripts with a 99.52% of identity at the cDNA sequence. One copy encodes the full-size HIP1 protein (995 amino acids), and the second copy contains 12 nucleotidic differences leading to six amino acid changes and a truncation mutation in exon 27 after codon 835.
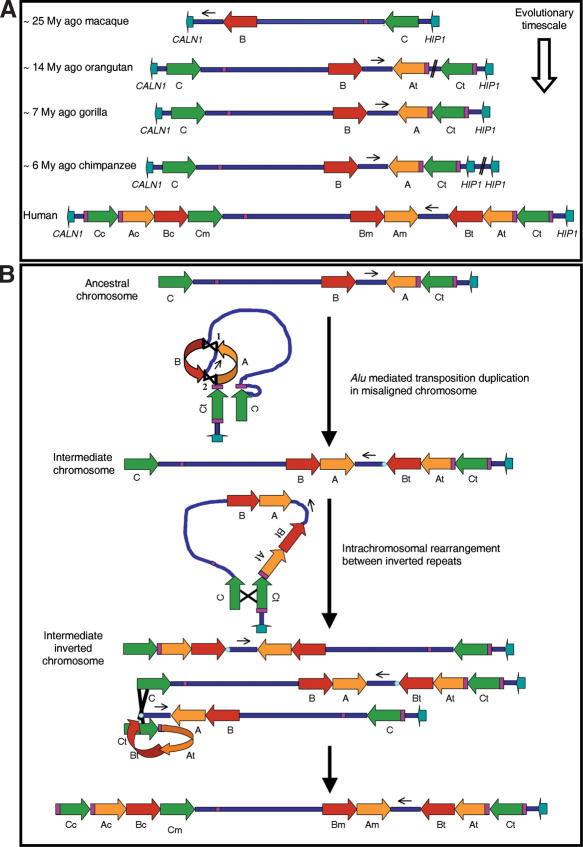
Genomic organization
In order to determine the relative order and organization of the blocks of segmental duplications in primates and predict the evolutionary rearrangements from a putative ancestral chromosome, we tested for the presence or absence of all interblock junctions and block edges by PCR and sequencing. The junctions block B-unique sequence and the block A-unique sequence were detected in all primates analyzed, whereas the C-A interblock junction was only detected in gorilla and chimpanzee, and the A-B and B-A interblock junctions were not detected in any nonhuman primate (Supplemental figure). As in humans, all interblock junctions identified contained Alu sequences.
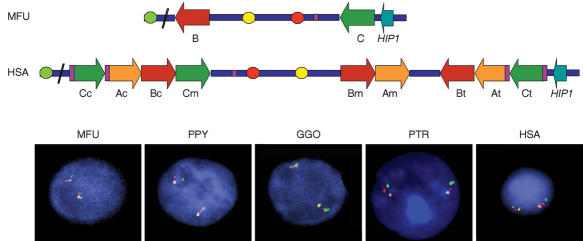
To better define the specific organization of the region in each species, we performed three-color interphase FISH with a combination of human probes: RP11-421B22/CALN1 locus, RP4-665P05/GTF2IRD1 locus, and RP11-622P13/STX1A locus. Gene order in macaque was CALN1-GTF2IRD1-STX1A, identical to mouse and baboon as established by clone contig. Another distribution was found in orangutan, gorilla, chimpanzee, and humans, where the order was CALN1-STX1A-GTF2IRD1 (Fig. 4). Therefore, an inversion of this region occurred in an ancestor to all hominoids after the divergence from the macaque lineage.
Figure 4.
Genomic organization and gene order in the different species by multicolor FISH. Interphase FISH with three probes: BAC RP11-421B22 (CALN1 locus, green), BAC RP11-622P13 (STX1A locus, red), and PAC RP4-665P05 (GTF2IRD1 locus, yellow) showed the relative organization of the two loci within the WBS critical region with respect to an outside gene. A regional inversion is found in all primates but macaque, whose genomic structure is therefore identical to that of baboon and mouse. The location of each probe with respect to the human and predicted macaque maps is shown at the top.
Sequence divergence and test of selection
Analysis of polymorphism was performed for all sequences obtained in our laboratory (BC/POM121; BC/TRIM50; BA/STAG3; BB/GTF2IRD2; HIP1) and from GenBank (Block C Large, Block B Large-1, Block B Large-2, HIP1 Large) (Supplemental Table 2).
Nucleotide diversities calculated using coding and noncoding sequences together were compared with the value obtained using only noncoding sequences. Significant differences were observed only for a single fragment, BB/GTF2IRD2, that showed higher nucleotide diversity when using only noncoding regions (Supplemental Table 2), which may hint at the action of purifying selection upon this fragment.
There was no evidence of transitional saturation in any group of sequences (P values ranging between 0.043 and <0.001). Thus, distances were calculated from both transitions and transversions. The results of the shape parameters that describe the gamma distribution were: for BC/POM121 α = 0.12, for BC/TRIM50 α = 0.16, for BA/STAG3 α = 0.27, for BB/GTF2IRD2 α = 2.63, and for HIP1 α = 0.38.
Overall Ka/Ks calculated for each fragment were lower than 1. In particular, most Ka/Ks values for BB/GTF2IRD2 were significantly lower than 1 which, again, is consistent with the action of purifying selection.
Phylogenetic relationships
Neighbor-joining and Bayesian trees were constructed for each fragment except for BC/TRIM50, which was short. Both trees represented the same phylogenetic relationships in all cases.
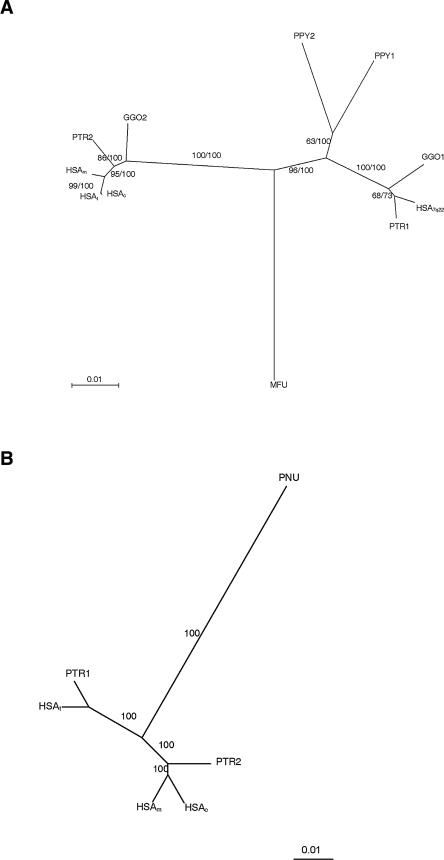
In humans, the first duplication of block A (BA/STAG3) from the ancestral gene in 7q22 occurred between 19.02 and 21.49 Mya. The block Am diverged from Ac and At, which is the ancestral of the 7q11.23 copies, 2.55-2.89 Mya. The calculated divergence time between the two copies of each species was 18.45-20.85 Myr for chimpanzee, 17.89-20.2 Myr for gorilla, and 11.07-12.51 Myr for orangutan (Fig. 5A).
Figure 5.
Phylogenetic trees based on the neighbor-joining method (nucleotide gamma: Tamura Nei) or Bayesian method. Subindexes c, m, t, and 7q22 refer to centromeric, medial, telomeric, and 7q22 chromosomal localization of the human PSVs, respectively. Number 1 refers to the copy more similar to the ancestral human gene, and number 2 to the other copy. (A) Neighbor-joining tree obtained for the STAG3 gene PCR product (BA/STAG3) including HSA, PTR, GGO, PPY, MFU, and PNU. Branch numbers refer to the neighbor-joining bootstrap values/clade credibility values for the Bayesian tree, which was of identical conformation. (B) Bayesian tree obtained for the Block C Large fragment, including HSA, PTR, and PNU. Branch values refer to the clade credibility values.
For block B (BB/GTF2IRD2) the calculated divergence time between two copies (Bt and Bc) with respect to the presumably ancestral (Bm) in humans is 3.6 to 3.76 Myr. The second duplication event giving rise to Bc and Bt occurred 1.44 to 1.5 Mya. Bayesian trees built from the larger fragments of block B (Block B Large-1 and Block B Large-2) showed discordant relationships between them. Block B Large-1, which contains part of the GTF2I gene and the whole sequence of the NCF1 gene, showed Bm as the ancestral copy, whereas Block B Large-2, containing part of the GTF2IRD2 gene, showed Bc as the ancestral copy. This discordance could be due to the nonfunctionality of GTF2IRD2 in Bc, truncated at its 5′ during the evolutionary rearrangement, thus allowing the accumulation of more sequence changes.
For block C (Block C Large), the calculated divergence time in Myr among the different copies was 13-9.6 Myr for human Ct with respect to Cm/Cc, 14.6-10.8 Myr for the two chimpanzee copies, and 6.3-4.6 Myr for human Cm and Cc copies (Fig. 5B). Phylogenetic trees with the smaller fragment (BC/POM121) gave a similar pattern with shorter divergence times, likely due to the shortness of the sequence.
For HIP1, the neighbor-joining and Bayesian trees grouped the two copies of chimpanzee after the divergence of the human lineage. The calculated divergence time between the chimpanzee's copies was between 3.2 and 3.3 Myr.
Discussion
Human chromosome 7 evolution and segmental duplications
Elucidating the steps involved in the generation of segmental duplications may provide new insight into the molecular mechanisms of evolutionary chromosomal rearrangements and their association with speciation, adaptation within species, polymorphic variation, and disease. In hominoids, several cytogenetic rearrangements had been defined on the homologs to human chromosome 7: The gorilla chromosome differs from human and chimpanzee chromosomes by a paracentric inversion with breakpoints in 7q11.23 and 7q22, whereas the orangutan chromosome shows an additional pericentric inversion with breakpoints in 7p22 and 7q11.23 (Yunis and Prakash 1982). More recently, the breakpoints of these paracentric and pericentric evolutionary inversions in hominoids were precisely mapped by FISH to specific segmental duplications in the human genome (Muller et al. 2004). We have studied in further depth the segmental duplications flanking the WBS deleted region previously characterized (Peoples et al. 2000; Valero et al. 2000; Bayés et al. 2003; Hillier et al. 2003; Scherer et al. 2003). Our data indicate that the large blocks of segmental duplications that we previously defined (A, B, and C) apparently evolved as entire blocks during the last evolutionary rearrangements shaping 7q11.23. Two copies of block A are found in all nonhuman hominoids. In orangutan, both are located close to each other near the centromere (chr. 10p), whereas in gorilla and chimpanzee they are separated in distant chromosome 6q bands corresponding to the homologous regions to human 7q22 and 7q11.23. The same number of signals was obtained with block C sequences, but they appear to be close to each other in the same chromosomal location, 10p11 in orangutan and 6q11 in gorilla and chimpanzee. Multiple experimental methods demonstrate that the entire block B is single copy in all other primates but human. In macaque, as well as in the sequence data available from baboon, only the ancestral loci to blocks B and C are identified as single copy, whereas no block A sequences are detected in the homologous region. Previous FISH studies with human BAC RG350L10 in several primates detected multiple loci with signals in the region homologous to human 7q11.23 in all species and additional cross-hybridizing signals in the homologous regions to 7p22 and 7q22 in chimpanzee and 7p22, 7p13, and 7q22 in gorilla (DeSilva et al. 1999). Since this BAC contains the medial blocks B and A, and A is composed of smaller modules with PMS2-related sequences, those modules could be the reason for the multiple signals in all primates. The similar results obtained by FISH with BAC RP11-451K15 and PAC RP1-42M2 further suggest that duplication of PMS2-related sequences might have been one of the first evolutionary events in the generation of the regional segmental duplications.
We also obtained sequence divergence data as an additional method to date the evolutionary appearance of each of the segmental duplications. The results are concordant with the experimental data, although sequence-based divergence times tend to be slightly shorter in most cases, which may be due to a tendency toward homogenization of paralogous segmental duplications through gene conversion events. Evolutionary studies have shown a similar pattern and timing of appearance of the segmental duplications located in other complex and unstable genomic regions involved in human aneusomies, such as 17p11.2-p12 (Stankiewicz et al. 2004), 15q11-q13 (Christian et al. 1999), and 22q11 (Shaikh et al. 2001).
Evolutionary model and intermediate chromosomes
We have shown that the genomic structure of the region in mouse and macaque/baboon is likely representative of the ancestral mammalian chromosome arrangement (Fig. 6A). Therefore, little change, if any, occurred in this region during the >50 Myr of divergence of mouse and the common ancestor of macaques and great apes. The origin of the 7q11 segmental duplications can be dated to ∼25 Mya after the separation of the Cercopithecidae and the Hominidae (Goodman et al. 1998).
Figure 6.
Genomic structure of the orthologous region to human 7q11.23 in the different primates, and hypothetical model for the sequential evolution of the region. (A) Schematic representation of the chromosome region in each primate species. A first inversion of the WBS region must have occurred in an ancestral chromosome to all hominoids. The orangutan and gorilla chromosomes appear identical except for the absence of the block C-block A junction in orangutan, whereas gorilla and chimpanzee chromosomes are identical except for the segmental duplication containing the HIP1 gene in chimpanzee. (B) Predicted human lineage-specific rearrangements from a hypothetical ancestral chromosome identical to that of gorilla. A unique complex intrachromosomal rearrangement from the ancestral chromosome created an intermediate chromosomal structure by two shuffling events between Alu elements, represented as 1 and 2 indicating the order of occurrence. By a similar mechanism of Alu-mediated duplicative transposition, the chimpanzee chromosome could have been generated (data not shown), with a duplication of the HIP1 containing block instead. A putative intrachromosomal paracentric inversion in the intermediate chromosome could have been mediated by the blocks C, which are flanking the region in inverted orientation. Interchromosomal NAHR in an inversion carrier of this intermediate chromosome could have led to duplication of the entire segmental duplication-containing blocks C, A, and B onto the centromeric position. The presence of Alu elements located at the edges of the blocks suggests Alu-mediated genome shuffling in these final steps of the generation of large segmental duplications.
We propose a model for the evolution of the segmental duplications on chromosome 7q11.23 based on our data. Initially, small modules (10-20-Kb) located close but outside the target region, specifically those containing PMS2 and WBSCR19 (RBAK-derived)-related sequences might have duplicated through transcriptional transposition and repair, targeting several regions. Specifically these modules would have been integrated between the HIP1 and POM121 genes and other regions, as they are found in multiple chromosome 7 loci in humans with sequence identities in the 95%-96% range (Osborne et al. 1997). These duplications could have facilitated misalignment and additional rearrangements leading to enlargement of the blocks and novel duplications. Interestingly, some of the genes present in the duplicated modules are actively transcribed during the meiotic division (Nicolaides et al. 1995; Pérez Jurado et al. 1998). Transcriptional activity in both strands is associated with double-strand breaks and has been related to chromosomal recombination in yeast as well as in mammalian cell cultures (Nickoloff 1992; Vedel and Nicolas 1999).
The duplication of the large block A seems to have occurred in a common ancestor to all hominoid lineages after the divergence from macaque, since the divergence time between the two copies of each species is quite similar (12-19 Mya) (Fig. 6A). This block A is located close to both breakpoints of the evolutionary cytogenetic paracentric inversion between q11 and q22 (Muller et al. 2004). Data on sequence divergence for block C would suggest a common duplication in gorilla, chimpanzee, and humans along with an independent duplication in orangutan. However, any model based only in divergence time data may not be completely reliable because of the low accuracy obtained from a short fragment of sequence and variable mutation rates among species. A parsimonious model considering the experimental data might be more consistent with a common origin for the first duplication of block C in all hominoids. Finally, block B has been duplicated only in the human lineage, whereas HIP1 is duplicated only in chimpanzee.
Our model suggests that Alu-mediated duplication transposition could lead to the duplication and flipping of two blocks together (A and B) in a single complex rearrangement involving four strands precipitated by misalignment between the already duplicated blocks C of an ancestral chromosome. Subsequent rearrangements could have been mediated by NAHR between blocks (Fig. 6B). By a similar mechanism of Alu-mediated duplicated transposition of the HIP1-containing block, the chimpanzee chromosome could have been generated.
Therefore, our data support the idea that segmental duplications orchestrate and accelerate the evolution in specific regions of the genome of primates.
Inversions, selection, and evolution
The generation of small paracentric inversions appear to be a common evolutionary event associated with regions rich in segmental duplications. As genomic inversions have a clear effect of suppressing recombination, they are expected to facilitate a faster divergence or negative selection of the genes included in the inverted interval (Navarro and Barton 2003). However, a common inversion has been shown to be under positive selection in recent human populations (Stefansson et al. 2005). We have shown that at least three evolutionary inversions must have occurred in this region, one in hominoids' common ancestor and two during the final shaping of human 7q11.23. In addition, intermediate chromosomes also having regional inversions are hypothesized. These structural variations may contain other targets of selection and may have contributed to speciation as well as to a high degree of genomic variability in human populations. In this regard, inversions in the WBS region have been found in one-third of transmitting progenitors of WBS patients with the deletion (Osborne et al. 2001; Bayés et al. 2003) and are also present in the population with an estimated frequency of 3%-7% (Hobart et al. 2004).
Species-specific gene duplications, novel genes, and pseudogenes
Evolutionary genomic rearrangements leading to segmental duplications generate gene duplications as well as novel fusion/fission genes. In blocks A, there are fusion/fission transcriptional units that do not maintain long open reading frames derived from truncated copies of PMS2 (Osborne et al. 1997) and STAG3 (Pezzi et al. 2000). Blocks C also contain multicopy chimeric transcriptional units along with several genes (Supplemental Table 1). Of those genes, only human POM121 codes for potentially functional proteins in two loci. Another segmental duplication telomeric to the WBS region contains the POM-ZP3 fusion gene, unique to humans and chimpanzees.
Block B is multiple-copy only in humans. Only GTF2IRD2 has two functional copies that code for proteins related to the transcription initiator factor II-I (Tipney et al. 2004). Interestingly, this is under purifying selection, both during primate evolution and after gene duplication. Other genes present in the three blocks B in humans, GTF2I and NCF1, have been shown to be functional only from a single locus (Gorlach et al. 1997; Pérez Jurado et al. 1998). In contrast, the block with HIP1 is duplicated only in the chimpanzee lineage. HIP1 codes for a huntingtin-interacting protein (HIP1), a component of clathrin coats that promotes clathrin assembly. Both chimpanzee copies are putatively functional, since they code for 995- and 835-amino-acid proteins that conserve the internal domains required for clathrin assembly (Legendre-Guillemin et al. 2005).
It is tempting to speculate that some of these species-specific duplicated genes, mostly if they are under purifying selection such as GTF2IRD2, may contribute to the functional differences affecting higher cognitive, behavioral, or other functions related to hominoid evolution.
Alu-driven genome evolution
The human genome is particularly enriched in both number and length of retrotransposons. It grew as a result of a major burst in Alu activity 25-55 Mya and subsequently continued to expand compared to more closely related primates (Liu et al. 2003). Thus, the appearance and propagation of Alu elements is somehow coincidental with the fast evolution of the segmental duplications in the primate genome.
NAHR between Alus has been documented and Alu elements have been found to accumulate at the junction sites of segmental duplications genome-wide (Batzer and Deininger 2002; Bailey et al. 2003). In fact, Alus have been directly implicated in the generation of the segmental duplications in 22q11 (Babcock et al. 2003). The presence of Alu repeats at the sites of integration of the first modules of segmental duplications suggests a mechanism of double-strand breakage followed by repair typically associated with transposition events. In addition, the finding of Alu elements at all the intersections of the large duplicated blocks in 7q11.23 indicates that Alu-mediated genome shuffling may be the main mechanism for the final generation of large segmental duplications.
Although the whole genomic sequence of the chimpanzee genome is released, the multiple assembling errors precludes to precisely elucidate the existing differences in comparison with humans. The annotation and experimental validation of the chimpanzee genome sequence along with the genome sequences of other primates will allow a better definition of all of the species-specific chromosomal evolutionary events to reconstruct the recent and dynamic history of human chromosomes. The extraordinary rate of evolutionary turnover mediated by the segmental duplications points to them as one of the main driving forces for genome evolution in primates.
Methods
Genetic material
Genomic DNA was extracted from peripheral blood or lymphoblastoid cell lines of MFU, MMU, PPY, GGO, PTR, and humans using the Puregene DNA Purification System (Gentra) or standard phenol-chloroform protocols. No primates were sampled for the sake of this project, and the human samples were obtained from volunteers with institutionally approved informed consent. Total RNA was isolated from several human cells and tissues as reported (Pérez Jurado et al. 1998).
Interspecies PCR amplification and sequencing
Primers were designed based on the human sequence in regions of complete identity among the different human segmental duplications using the Primer3 Input Programme (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Nested and/or external primers were designed for those amplicons that failed to amplify with the first set (Supplemental Table 3).
PCR reactions were set up on a final volume of 10-25 μL including 50-100 ng of genomic DNA, standard reagents, and variable cycling conditions (Supplemental Table 3). Products were visualized on 0.8%-2% agarose gels depending on amplimer size and purified either from PCR solution or agarose gel band with the GFX PCR DNA and Gel Band Purification Kit (Amersham Pharmacia Biotech). Microsatellite markers (BBSTR1 and D7S489) as well as the STAG3ex13 deletion/insertion cis-morphism were analyzed using Genescan 3.1 software (PE Applied Biosystems).
Sequencing was done with dRhodamine or Big Dye sequencing kits (Applied Biosystems) and analyzed on a 3100 sequencer (Applied Biosystems). For products larger than 1 Kb, additional internal primers (available upon request) were required in order to sequence the whole fragment. Sequence electropherograms were scrutinized to detect significant double peaks; fragments with a significant number of them (>1/500) suggestive of the existence of PSVs rather than allelic polymorphisms were cloned using the pMOSBlue Blunt Ended cloning kit (Amersham Pharmacia Biotech) for individual allele sequencing.
Expression analysis by RT-PCR
Total RNA (different tissues) was reverse-transcribed with Super-ScriptII Rnase H- and random hexamers following the manufacturer's instructions (Gibco BRL), to obtain cDNA. Analysis of expressed copies was performed by PCR amplification of cDNA with posterior differential restriction assays that could distinguish the copies. Primers located in different exons were chosen to distinguish cDNA from genomic DNA amplicons (Supplemental Table 3).
Copy number quantification analyses
PCR amplification of a PSV in exon 13 of the STAG3 gene/pseudogene sequences (block A) was performed as described above (27 PCR cycles). Gene/pseudogene (7q22/7q11.23) peak ratios were calculated from five human control samples and in artificial situations (1:1, 1:2, 2:1, and 3:1) created by mixing different concentrations of BAC R-248L18 (block At at 7q11.23) and BAC CITBI-E1-2601G15 (block A at 7q22) in the PCR reactions. At least two individuals of each species were analyzed.
A PSV that distinguishes the NCF1 gene at Bm from the pseudogenes located at Bc and Bt (GT deletion) was genotyped and quantified as described (Bayés et al. 2003).
Quantification of block C copy number was performed by analyzing PSVs detected by PCR amplification of the TRIM50 gene followed by restriction analysis with either NgoMIV or MluNI (Roche, NEB). Digital images with nonsaturated bands were captured from a 3% metaphore agarose gel, and the intensities of the bands were quantified using the Volume Tool of the Quantity One software package (Bio-Rad).
Fluorescence in situ hybridization analyses
Selected BAC clones were purchased from the HGMP Resource Center. FISH was performed on metaphase and interphase cells of peripheral blood lymphocytes or Epstein-Barr virus-transformed lymphoblastoid cell lines as described (Bayés et al. 2003). Between 20 and 100 interphase nuclei where all the probes could be identified and had two signals of a control probe were scored. Since physical distances between signals correlate well with genomic distances ranging from 50 Kb to 2 Mb in interphase nuclei (Christian et al. 1999), we could determine the order of probes.
DNA sequence analyses
Human clones corresponding to the different segmental duplications were identified from the existing maps of the region (Bayés et al. 2003; Hillier et al. 2003; Scherer et al. 2003). Chimpanzee and baboon clones were identified from public databases through BLAST (Altschul et al. 1990) on the NCBI site (http://www.ncbi.nlm.nih.gov/) against the high-throughput and the nonredundant sequence databases. Alu sequences were identified using the RepeatMasker program (http://www.repeatmasker.org/). Sequence alignments including repetitive elements were performed with ClustalW (Thompson et al. 1994) or MAVID (http://baboon.math.berkeley.edu/mavid/) (Bray and Pachter 2004) and double-checked visually. Coding and noncoding regions were assigned based on vegaGene or Ensembl databases (http://www.ensembl.org/) information. The DNAsp3.99 programme (Rozas and Rozas 1999) was used to determine the number of polymorphic segregating sites (S), the average number of substitutions per site, the nucleotide diversity (π) (Nei 1987), and the number of synonymous and nonsynonymous substitutions. A sliding window was also obtained at 50-base pair intervals to explore the presence of mutational peaks along the sequences.
To explore the possibility of transitional saturation, we plotted transition and transversion values for each pair of aligned sequences, and we applied the Mantel's test included in Arlequin ver2.000 (Schneider et al. 2000) to obtain a correlation index and P-value. Transition/transversion ratios were calculated for each pair with a gamma-corrected Tamura-Nei model, which assumes substitution rate differences between nucleotides and inequality of nucleotide frequencies (Tamura and Nei 1993) with MEGA c.2-01 (Kumar et al. 2001).
The shape parameters (α-values) that describe the gamma distribution applied to the MEGA program were calculated for each group of sequences with the MrBayes program (Huelsenbeck and Ronquist 2001). Neutrality and selection were estimated with the Modified Nei-Gojobori method included in the MEGA program. Differences between synonymous and nonsynonymous substitutions were examined using Fisher's exact test, and the Sequential Bonferroni Test (Rice 1989) was applied to control over the group-wide type I error rate performing a different test for each DNA fragment analyzed.
We also used the MEGA program to construct a neighbor-joining tree. Bootstrap values were obtained from 1000 replicates. A Bayesian tree was constructed with MrBayes setting the following parameters: nst = 6, site partition = bycodon, rates = gamma, and basefreq = estimated. In the Monte Carlo process, four chains ran simultaneously for 700,000 generations. Trees were sampled every 100 generations. The “stationarity” was determined to have occurred by the 2000th tree, and the first 2000 trees were discarded. The whole procedure was repeated three times starting at random points, and the tree topologies obtained were the same.
The average K-value obtained from the comparison of each cercopithecine against each hominid was used to calculate r (rate of nucleotide substitution), using the formula r = K/2T, (Graur and Li 2000). This rate provided a calibrated value to calculate divergence time between pairs of sequences. We calibrated the rate of substitutions (including noncoding regions and synonymous differences in coding regions) with the estimated time of divergence of 25 Mya for the separation between the families Cercopithecidae and Hominidae (Goodman et al. 1998) based on fossil evidence.
GenBank accession numbers
All the PCR products analyzed were submitted to GenBank, with the following accession numbers:
BB/GTF2IRD2 fragment, MFU (AY882419) and PTR (AY882420). HIP1 fragment, MFU (AY883940), PPY (AY883941), GGO (AY883942), PTR1 (AY883943) and PTR2 (AY883944). BC/TRIM50, MFU (AY883971), PPY1 (AY883972), PPY2 (AY883973), GGO1 (AY883974), GGO2 (AY883975), PTR1 (AY883976), and PTR2 (AY883977). BC/POM121, MFU (AY883962), PPY1 (AY883960), PPY2 (AY883961), GGO1 (AY883958), GGO2 (AY883959), PTR1 (AY883956) and PTR2 (AY883957). BA/STAG3, MFU (AY883970), PPY1 (AY883968), PPY2 (AY883969), GGO1 (AY883966), GGO2 (AY883967), PTR1 (AY883964), and PTR2 (AY883965). POM121 gene-ZP3 gene junction, PTR (AY883963). Block A-unique sequence junction, MFU (AY883946), PPY (AY883945), GGO1 (AY883949), GGO2 (AY883945), PTR1 (AY883947), and PTR2 (AY883948). Block B-unique sequence junction, MFU (AY883951), GGO (AY883952), and PTR (AY883953). Block C-Block A junction, GGO (AY883955) and PTR (AY883954).
Supplementary Material
Acknowledgments
We thank Núria Rivera, Raquel Flores, and Ivon Cuscó for excellent technical assistance and discussion, Victoria Campuzano and Arcadi Navarro for critical reading, Genís Parra for bioinformatic support, and Mariano Rocchi (Università degli Studi di Bari), the Institute of Zoology of London, and the project INPRIMAT for providing primate cell lines and samples. This work was supported by grants from the Spanish Ministries of Science and Technology (SAF2004/6382), Health (Network of Center of Clinical and Molecular Genetics, C03/07), and a joint project from Genoma España-Genome Canada (JLI/038) to L.A.P.J., and the European Commission under contract QLRI-CT-2002-01325 (INPRIMAT project, www.inprimat.org) to X.D.R. Anna Antonell is supported by the Departament d'Universitats, Recerca i Societat de la Informació, Generalitat de Catalunya (2002 FI 00790)
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data described in this paper have been submitted to GenBank under accession nos. AY882419, AY882420, and AY883940-AY883977.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3944605.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Armengol, L., Pujana, M.A., Cheung, J., Scherer, S.W., and Estivill, X. 2003. Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements. Hum. Mol. Genet. 12: 2201-2208. [DOI] [PubMed] [Google Scholar]
- Babcock, M., Pavlicek, A., Spiteri, E., Kashork, C.D., Ioshikhes, I., Shaffer, L.G., Jurka, J., and Morrow, B.E. 2003. Shuffling of genes within low-copy repeats on 22q11 (LCR22) by Alu-mediated recombination events during evolution. Genome Res. 13: 2519-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J.A., Liu, G., and Eichler, E.E. 2003. An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 73: 823-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer, M.A. and Deininger, P.L. 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3: 370-379. [DOI] [PubMed] [Google Scholar]
- Bayés, M., Magano, L.F., Rivera, N., Flores, R., and Pérez Jurado, L.A. 2003. Mutational mechanisms of Williams-Beuren syndrome deletions. Am. J. Hum. Genet. 73: 131-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, N. and Pachter, L. 2004. MAVID: Constrained ancestral alignment of multiple sequences. Genome Res. 14: 693-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F.C. and Li, W.H. 2001. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68: 444-456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian, S.L., Fantes, J.A., Mewborn, S.K., Huang, B., and Ledbetter, D.H. 1999. Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13). Hum. Mol. Genet. 8: 1025-1037. [DOI] [PubMed] [Google Scholar]
- DeSilva, U., Massa, H., Trask, B.J., and Green, E.D. 1999. Comparative mapping of the region of human chromosome 7 deleted in Williams syndrome. Genome Res. 9: 428-436. [PMC free article] [PubMed] [Google Scholar]
- Eichler, E.E. 2001. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 17: 661-669. [DOI] [PubMed] [Google Scholar]
- Francke, U. 1999. Williams-Beuren syndrome: Genes and mechanisms. Hum. Mol. Genet. 8: 1947-1954. [DOI] [PubMed] [Google Scholar]
- Frazer, K.A., Chen, X., Hinds, D.A., Pant, P.V., Patil, N., and Cox, D.R. 2003. Genomic DNA insertions and deletions occur frequently between humans and nonhuman primates. Genome Res. 13: 341-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, M., Porter, C.A., Czelusniak, J., Page, S.L., Schneider, H., Shoshani, J., Gunnell, G., and Groves, C.P. 1998. Toward a phylogenetic classification of Primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 9: 585-598. [DOI] [PubMed] [Google Scholar]
- Gorlach, A., Lee, P.L., Roesler, J., Hopkins, P.J., Christensen, B., Green, E.D., Chanock, S.J., and Curnutte, J.T. 1997. A p47-phox pseudogene carries the most common mutation causing p47-phox-deficient chronic granulomatous disease. J. Clin. Invest. 100: 1907-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur, D. and Li, W-H. 2000. Gene duplication, exon shuffling and concerted evolution. In Fundamentals of molecular evolution, 2nd ed., (eds. D. Graur and W.H. Li). Sinauer Associates, Sunderland, MA.
- Hillier, L.W., Fulton, R.S., Fulton, L.A., Graves, T.A., Pepin, K.H., Wagner-McPherson, C., Layman, D., Maas, J., Jaeger, S., Walker, R., et al. 2003. The DNA sequence of human chromosome 7. Nature 424: 157-164. [DOI] [PubMed] [Google Scholar]
- Hobart, H., Gregg, R., Mervis, C., Robinson, B., Kimberley, K., Rios, C., and Morris, C. 2004. Heterozygotes for the microinversion of the Williams-Beuren syndrome region have an increased risk for affected offspring. In 54th Annual Meeting of the ASHG (ed. ASHG), pp. 177. The American Journal of Human Genetics, Toronto.
- Huelsenbeck, J.P. and Ronquist, F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754-755. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. 2001. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17: 1244-1245. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Legendre-Guillemin, V., Metzler, M., Lemaire, J.F., Philie, J., Gan, L., Hayden, M.R., and McPherson, P.S. 2005. HIP1 (huntingtin interacting protein 1) regulates clathrin assembly through direct binding to the regulatory region of the clathrin light chain. J. Biol. Chem. 280: 6101-6108. [DOI] [PubMed] [Google Scholar]
- Liu, G., Zhao, S., Bailey, J.A., Sahinalp, S.C., Alkan, C., Tuzun, E., Green, E.D., and Eichler, E.E. 2003. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 13: 358-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, D.P., Segraves, R., Carbone, L., Archidiacono, N., Albertson, D.G., Pinkel, D., and Eichler, E.E. 2003. Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome Res. 13: 347-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet, T., Caceres, M., Bertranpetit, J., Preuss, T.M., Thomas, J.W., and Navarro, A. 2004. Chromosomal rearrangements and the genomic distribution of gene-expression divergence in humans and chimpanzees. Trends Genet. 20: 524-529. [DOI] [PubMed] [Google Scholar]
- Muller, S., Finelli, P., Neusser, M., and Wienberg, J. 2004. The evolutionary history of human chromosome 7. Genomics 84: 458-467. [DOI] [PubMed] [Google Scholar]
- Navarro, A. and Barton, N.H. 2003. Chromosomal speciation and molecular divergence—Accelerated evolution in rearranged chromosomes. Science 300: 321-324. [DOI] [PubMed] [Google Scholar]
- Nei, M. 1987. Molecular evolutionary genetics. Columbia University Press, New York.
- Nickoloff, J.A. 1992. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol. Cell. Biol. 12: 5311-5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaides, N.C., Carter, K.C., Shell, B.K., Papadopoulos, N., Vogelstein, B., and Kinzler, K.W. 1995. Genomic organization of the human PMS2 gene family. Genomics 30: 195-206. [DOI] [PubMed] [Google Scholar]
- Osborne, L.R., Herbrick, J.A., Greavette, T., Heng, H.H., Tsui, L.C., and Scherer, S.W. 1997. PMS2-related genes flank the rearrangement breakpoints associated with Williams syndrome and other diseases on human chromosome 7. Genomics 45: 402-406. [DOI] [PubMed] [Google Scholar]
- Osborne, L.R., Li, M., Pober, B., Chitayat, D., Bodurtha, J., Mandel, A., Costa, T., Grebe, T., Cox, S., Tsui, L.C., et al. 2001. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat. Genet. 29: 321-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples, R., Franke, Y., Wang, Y.K., Pérez-Jurado, L., Paperna, T., Cisco, M., and Francke, U. 2000. A physical map, including a BAC/PAC clone contig, of the Williams-Beuren syndrome—Deletion region at 7q11.23. Am. J. Hum. Genet. 66: 47-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez Jurado, L.A., Peoples, R., Kaplan, P., Hamel, B.C., and Francke, U. 1996. Molecular definition of the chromosome 7 deletion in Williams syndrome and parent-of-origin effects on growth. Am. J. Hum. Genet. 59: 781-792. [PMC free article] [PubMed] [Google Scholar]
- Pérez Jurado, L.A., Wang, Y.K., Peoples, R., Coloma, A., Cruces, J., and Francke, U. 1998. A duplicated gene in the breakpoint regions of the 7q11.23 Williams-Beuren syndrome deletion encodes the initiator binding protein TFII-I and BAP-135, a phosphorylation target of BTK. Hum. Mol. Genet. 7: 325-334. [DOI] [PubMed] [Google Scholar]
- Pezzi, N., Prieto, I., Kremer, L., Pérez Jurado, L.A., Valero, C., Del Mazo, J., Martinez, A.C., and Barbero, J.L. 2000. STAG3, a novel gene encoding a protein involved in meiotic chromosome pairing and location of STAG3-related genes flanking the Williams-Beuren syndrome deletion. FASEB J. 14: 581-592. [DOI] [PubMed] [Google Scholar]
- Rice, W. 1989. Analyzing tables of statistical tests. Evolution 43: 223-225. [DOI] [PubMed] [Google Scholar]
- Robinson, W.P., Waslynka, J., Bernasconi, F., Wang, M., Clark, S., Kotzot, D., and Schinzel, A. 1996. Delineation of 7q11.2 deletions associated with Williams-Beuren syndrome and mapping of a repetitive sequence to within and to either side of the common deletion. Genomics 34: 17-23. [DOI] [PubMed] [Google Scholar]
- Rozas, J. and Rozas, R. 1999. DnaSP version 3: An integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174-175. [DOI] [PubMed] [Google Scholar]
- Samonte, R.V. and Eichler, E.E. 2002. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 3: 65-72. [DOI] [PubMed] [Google Scholar]
- Scherer, S.W., Cheung, J., MacDonald, J.R., Osborne, L.R., Nakabayashi, K., Herbrick, J.A., Carson, A.R., Parker-Katiraee, L., Skaug, J., Khaja, R., et al. 2003. Human chromosome 7: DNA sequence and biology. Science 300: 767-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, S., Roessli, D., and Excoffier, L. 2000. Arlequin ver.2.000: A software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland.
- Shaikh, T.H., Kurahashi, H., and Emanuel, B.S. 2001. Evolutionarily conserved low copy repeats (LCRs) in 22q11 mediate deletions, duplications, translocations, and genomic instability: An update and literature review. Genet. Med. 3: 6-13. [DOI] [PubMed] [Google Scholar]
- Stankiewicz, P., Shaw, C.J., Withers, M., Inoue, K., and Lupski, J.R. 2004. Serial segmental duplications during primate evolution result in complex human genome architecture. Genome Res. 14: 2209-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson, H., Helgason, A., Thorleifsson, G., Steinthorsdottir, V., Masson, G., Barnard, J., Baker, A., Jonasdottir, A., Ingason, A., Gudnadottir, V.G., et al. 2005. A common inversion under selection in Europeans. Nat. Genet. 37: 129-137. [DOI] [PubMed] [Google Scholar]
- Tamura, K. and Nei, M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10: 512-526. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipney, H.J., Hinsley, T.A., Brass, A., Metcalfe, K., Donnai, D., and Tassabehji, M. 2004. Isolation and characterisation of GTF2IRD2, a novel fusion gene and member of the TFII-I family of transcription factors, deleted in Williams-Beuren syndrome. Eur. J. Hum. Genet. 12: 551-560. [DOI] [PubMed] [Google Scholar]
- Valero, M.C., de Luis, O., Cruces, J., and Pérez Jurado, L.A. 2000. Fine-scale comparative mapping of the human 7q11.23 region and the orthologous region on mouse chromosome 5G: The low-copy repeats that flank the Williams-Beuren syndrome deletion arose at breakpoint sites of an evolutionary inversion(s). Genomics 69: 1-13. [DOI] [PubMed] [Google Scholar]
- Vedel, M. and Nicolas, A. 1999. CYS3, a hotspot of meiotic recombination in Saccharomyces cerevisiae. Effects of heterozygosity and mismatch repair functions on gene conversion and recombination intermediates. Genetics 151: 1245-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Yunis, J.J. and Prakash, O. 1982. The origin of man: A chromosomal pictorial legacy. Science 215: 1525-1530. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ncbi.nlm.nih.gov/; NCBI home page.
- http://www.nisc.nih.gov/; The NIH Intramural Sequencing Center (NISC).
- http://www.ensembl.org/; Ensembl Genome Browser.
- http://www.genome.ucsc.edu/; UCSC Genome Browser Home.
- http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi; Primer3 Input.
- http://www.repeatmasker.org/; RepeatMasker.
- http://baboon.math.berkeley.edu/mavid/; The MAVID multiple alignment server. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.