Abstract
The MHC class I gene, PD1, has neither functional TATAA nor Initiator (Inr) elements in its core promoter and initiates transcription at multiple, dispersed sites over an extended region in vitro. Here, we define a novel core promoter feature that supports regulated transcription through selective transcription start site (TSS) usage. We demonstrate that TSS selection is actively regulated and context dependent. Basal and activated transcriptions initiate from largely nonoverlapping TSS regions. Transcripts derived from multiple TSS encode a single protein, due to the absence of any ATG triplets within ∼430 bp upstream of the major transcription start site. Thus, the PD1 core promoter is embedded within an “ATG desert.” Remarkably, extending this analysis genome-wide, we find that ATG deserts define a novel promoter subclass. They occur nonrandomly, are significantly associated with non-TATAA promoters that use multiple TSS, independent of the presence of CpG islands (CGI). We speculate that ATG deserts may provide a core promoter platform upon which complex upstream regulatory signals can be integrated, targeting multiple TSS whose products encode a single protein.
Regulation of gene expression is mediated by specific interactions of transcription factors with promoter DNA sequences, resulting in the assembly of the transcription machinery and onset of transcription (Chen et al. 1994; Roeder 1996; Berk 1999; Gill 2001; Kadonaga 2004). RNA pol II promoters are conceptually divided into two domains, upstream regulatory and core promoter regions. Although the diversity of transcription factor binding sites and the complexity of their organization in upstream regulatory regions has been long recognized (Struhl 2001), it is increasingly apparent that core promoter regions are also highly diverse and complex (Burke and Kadonaga 1997; Lagrange et al. 1998; Smale et al. 1998; Kutach and Kadonaga 2000; Willy et al. 2000; Smale 2001; Butler and Kadonaga 2002). Core promoters can be grouped according to the presence of specific DNA sequence elements such as TATAA box (Singer et al. 1990; Butler and Kadonaga 2002), Inr (Smale and Baltimore 1989; Zenzie-Gregory et al. 1993; Kaufmann and Smale 1994; Lo and Smale 1996; Smale et al. 1998), TFIIB response element (BRE) (Lagrange et al. 1998; Littlefield et al. 1999), the downstream promoter element (DPE) (Burke and Kadonaga 1997; Burke et al. 1998; Kutach and Kadonaga 2000; Butler and Kadonaga 2001; Kadonaga 2002), or the MED-1 element (Ince and Scotto 1995).
Another sequence feature common to many promoters is the presence of CpG islands (CGI) (Bird 1986; Gardiner-Garden and Frommer 1987; Cross and Bird 1995; Antequera 2003; Wang and Leung 2004). Although the presence of CGI has been used to localize promoters, not all CGI are associated with promoter regions. In general, CGI associated with promoters are distinguished from CGI not associated with promoters by their greater size (≥500 bp) and a higher G+C content (>0.55) and observed/expected CpG ratio (>0.65) (Takai and Jones 2002). In the human genome, it is estimated that there are 41,468 CGI based on NCBI's Build 34 genome annotation (Takai and Jones 2002) and 37,000 in the mouse (Antequera and Bird 1993). Further, 90% of all housekeeping genes and 40% of all tissue-specific genes fall within CGI. For many genes a CGI is the only identifiable core promoter structure, but little is known about how CGI directly contribute to transcription initiation (Butler and Kadonaga 2002).
The sequence elements in the core promoter and its structure can both contribute to the regulation of gene expression. In yeast, it has been shown that these different classes of core promoters subserve different functions. While only about 20% of promoters in the yeast genome have TATAA elements, 50% of stress-responsive genes are TATAA promoters (Basehoar et al. 2004; Zanton and Pugh 2004). In Drosophila, differential usage of two closely linked promoter elements of the ADH gene is developmentally regulated (Hansen and Tjian 1995). In mammalian cells, the usage of promoters associated with the CIITA gene is tissue specific (Wong et al. 2002).
Core promoter regions also differ in their patterns of transcription start sites (TSS). Recent genome-wide analyses have reported that the majority of genes initiate transcription at multiple sites distributed over the core promoter region (Suzuki et al. 2004). The observed TSS range from unique to tightly clustered to highly dispersed among the different promoters examined. Based on an analysis of 276 genes, Suzuki and colleagues suggested that the presence of a TATAA promoter in 42 genes correlated with tightly clustered start sites. The functional significance of multiple TSS in a promoter is unknown. However, the diversity of TSS suggests that initiation at individual promoters is surprisingly complex and may be a target for transcriptional regulation. A major challenge is to understand the degree to which differential TSS utilization contributes to the regulation of gene expression.
We have begun to address this challenge by characterizing the core promoter structure and patterns of expression of an MHC class I gene. The MHC class I gene family encodes cell-surface molecules that provide immune surveillance against intracellular pathogens. The classical class Ia genes HLA-A, B, and C in human and PD1 in miniature swine are ubiquitously expressed, however, their expression is actively regulated in a tissue-specific fashion (Singer and Maguire 1990; Le Bouteiller 1994; Girdlestone 1995; Howcroft and Singer 2003). The highest levels of class I gene expression are found in the cells and tissues of the immune system. The promoter region of the MHC class I gene, PD1, is contained within a CGI extending from -556 to +1452 bp relative to a YTCA+1GYY Inr-like sequence that is conserved among class I genes. Our in vitro transcription studies revealed that initiation occurs at multiple TSS within the core promoter (Howcroft et al. 2003). Indeed, individual TSS usage in vitro reflects the prior exposure history of cells to modulatory cytokines such as γ-interferon (IFNγ) that regulate class I expression.
Here we report that differential transcription start site usage within the core promoter occurs in vivo in basal and activated transcription, demonstrating that transcription start-site selection is actively regulated. The regulation of class I transcription through the use of multiple TSS is made possible by the absence of any ATG codons within ∼460 bp upstream of the translation initiation codon of the class I gene. The presence of this “ATG desert” ensures that only a single protein product is made, regardless of the TSS selected.
Importantly, we identify a subclass of promoters in the human, mouse, and rat genomes that contain ATG deserts, thereby defining a novel core promoter feature. The ATG desert is a DNA segment that has a lower frequency of occurrence of the ATG trinucleotide than the surrounding sequences and spans a region of ∼1 kB both upstream and downstream of the major transcription start site. ATG deserts are an intrinsic feature of core promoters that do not contain canonical TATAA elements, independent of the presence of a CGI. We further document a significant correlation between the presence of ATG deserts and the use of multiple transcription start sites among non-TATAA promoters.
A consequence of the presence of ATG deserts is that they enable the use of multiple TSS whose products all encode a single protein, thereby permitting the core promoter to serve as a platform where complex upstream regulatory signals are integrated through selective transcription start site usage.
Results
MHC class I gene transcription initiates at multiple start sites whose usage can be actively regulated
In vitro transcription from a truncated promoter construct of the MHC class I gene, PD1, initiates at multiple start sites within a region of about 50 bp (Fig. 1A) (Howcroft et al. 2003), suggesting that transcription of native MHC class I genes in vivo also might initiate at multiple start sites.
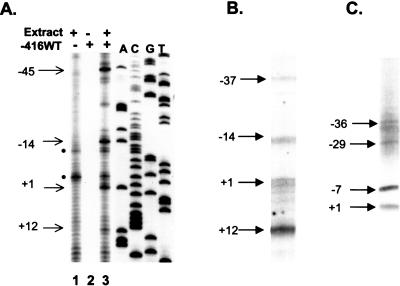
Figure 1.
Determination of TSSs utilized by the PD1 class I gene in vitro and in vivo. (A) In vitro transcription of the PD1 class I promoter/reporter construct, -416WT. (Lane 1) HeLa extract alone control; nonspecific background bands are indicated by black filled circles; (lane 2) -416WT template alone; (lane 3) -416WT template in the presence of HeLa extract. Specific TSS are indicated by arrows. A sequence ladder of the class I promoter is provided. (B) Analysis of in vivo TSS by 5′RACE analysis of isolated mRNA from spleen of PD1 class I transgenic mice. (C) TSS generated by the endogenous H-2Kb class I gene in spleens as determined by 5′RACE, using a H-2Kb gene-specific primer with the same mRNA samples as in B. The major TSS regions are indicated by arrows.
To examine this possibility, the in vivo TSS of a genomic MHC class I transgene (PD1), as well as an endogenous MHC class I gene (H-2Kb) were determined in splenocytes by 5′RACE. As shown in Figure 1B, basal transcription in vivo of the PD1 transgene initiates over a 49-bp region at four predominant start sites at +12, +1, -14, and -37 and at a multiplicity of additional minor sites further upstream that are observed reproducibly. (The +1 assignment was arbitrarily chosen, as a reference point. Translation initiates at +32.) The usage of multiple start sites is also observed at the endogenous H-2Kb promoter that displays a similar pattern of start-site usage, the predominance of a few sites with a multiplicity of minor sites further upstream (Fig. 1C). Thus, the use of multiple transcription initiation sites is a naturally occurring in vivo phenomenon, which is likely to be common among MHC class I genes.
To determine the functional 5′ boundary of transcription initiation, we utilized a translational knock-out approach in which out-of-frame ATG codons (relative to a downstream reporter) were introduced at various upstream sites in the class I promoter, ligated to a CAT reporter gene (see schematic in Fig. 2A). Any transcription that initiates upstream of the out-of-frame ATG in these constructs would be translated into a nonsense product. The extent of the resulting decreased CAT reporter activity provides an assessment of the relative contribution of each TSS region to basal core promoter activity. Promoter constructs with an out-of-frame ATG codon inserted upstream of one of the three principle start sites were analyzed. Surprisingly, insertion of an out-of-frame ATG at position -6 bp dramatically reduced reporter activity, while a control TAG mutation (Cont-6) at the same position had no effect (Fig. 2A). The steady-state levels of CAT reporter RNA generated by the out-of-frame ATG-6 mutation and wild-type constructs were indistinguishable (Fig. 2B), demonstrating that altering the sequence at this position does not affect overall levels of transcription. Therefore, the majority of TSS occur upstream of -6 bp in basal transcription.
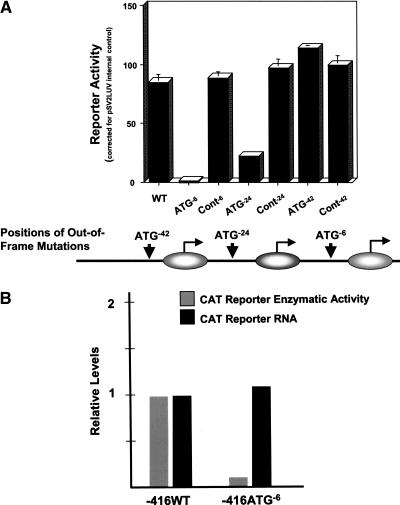
Figure 2.
Determination of the functional PD1 class I core promoter region by mapping of TSSs that contribute to basal promoter activity. (A) HeLa epithelial cells were transiently transfected with wild-type (WT) and class I promoter mutants with out-of-frame ATGs located at positions -6 (ATG-6), -24 (ATG-24), and -42 (ATG-42). The positions of the ATG mutations relative to the determined TSS regions are provided at the bottom of the figure. Constructs with a TAG triplet at positions -6, -24, or -42 served as controls. Data are expressed as relative percentages of acetylation corrected to an internal transfection control, pSV2LUC. Error bars indicate standard error derived from four independent experiments, each performed in triplicate. (B) HeLa cells were transiently transfected with either -416WT or out-of-frame mutant -416ATG-6 reporter constructs. After 48 h, RNA was prepared from half of the sample and used in Northern analysis of CAT reporter RNA (black bars) to quantitate steady-state transcription levels; functional reporter CAT enzyme activity was determined in the other half (gray bars). Results are presented as the relative amount of either CAT reporter activity or RNA, after correcting for transfection efficiency with an internal control pSV2LUC and correcting for RNA loading by tubulin RNA levels, respectively.
To further map the functional 5′ boundary of transcription initiation, we examined the effect of additional out-of-frame ATG's inserted at either -24 bp or -42 bp (Fig. 2A). The construct with an out-of-frame ATG at position -24 generated modest reporter activity (relative to the wild-type control), while the construct with an out-of-frame ATG at -42 was indistinguishable from the wild-type control. Thus, basal transcription in HeLa epithelial cells initiates from multiple, distinct start sites located largely between -42 and -6 bp.
Although many promoters are known to use multiple TSS (Suzuki et al. 2001), relatively little is known about whether their usage is regulated. To determine whether start-site selectivity reflects active regulation of functional transcripts, the ability of the translation knock-out promoter constructs to respond to the IFNγ-induced coactivator, CIITA, was examined. HeLa cells were cotransfected with the knock-out promoter constructs and either a CIITA expression vector or control. Remarkably, CIITA was able to activate both the ATG-6 and ATG-24 promoters, which have nearly undetectable basal levels of promoter activity (Table 1). The fold activation of these translation knock-out constructs by CIITA was markedly higher than those of either the ATG-42 or wild-type promoter (14.8 and 15.5-fold vs. 4.3 and 5.6-fold, respectively). If CIITA activated uniformly at all TSS, then equivalent levels of activation would have occurred. Therefore, CIITA-mediated activation altered relative start site usage, preferentially increasing transcription initiation at sites downstream of -24 bp. These findings indicate that start-site usage can be regulated and varied under different conditions.
Table 1.
CIITA activates core promoter mutants
| Co-transfection with:
|
|||
|---|---|---|---|
| Promoter construct | Control vector (%) | CIITA expression vector (%) | Fold activation |
| −416WT | 17.5 ± 1.4 | 97.5 ± 7.5 | 5.6 |
| −416ATG−6 | 2.2 ± 0.5 | 32.5 ± 3.5 | 14.8 |
| −416ATG−24 | 1.3 ± 0.4 | 20.2 ± 3.4 | 15.5 |
| −416ATG−42 | 19.9 ± 2.3 | 92.4 ± 2.3 | 4.3 |
The ability of CIITA to activate wild type (WT) or mutant (out-of-frame ATG) −416CAT reporter constructs was examined in transiently transfected HeLa cells, which were co-transfected with either a control or CIITA expression vector. Data are expressed as CAT activity, corrected for the pSV2LUC internal transfection control; standard errors derive from triplicate samples.
The MHC class I core promoter resides in an “ATG desert”
The PD1 core promoter region contains no ATG codons within 42 bp of the major TSS, which enabled the use of the translation knock-out strategy. Indeed, there is a remarkable distribution of ATG codons in the extended promoter. There are no ATG codons, in any reading frame, within the first 459 bp upstream of the initiator ATG, in striking contrast to the occurrence of 17 ATG codons in the next 631 bp of DNA (extending 5′ from -459) (Supplemental Fig. 1A). Thus, there is a sharp discontinuity in the ATG codon frequency between the promoter proximal and distal segments. Similarly, in the mouse class I H-2Kb gene, no ATG sequence occurs within ∼500 bp of its promoter, which also uses multiple transcription start sites. The absence of an ATG sequence within the extended promoter is not limited to the PD1 and H-2Kb genes, but is also a common feature of the human MHC class I genes, which reveal a relative paucity of ATG sequences in the 200-400 bp of the core promoter (Supplemental Figure 1B).
Since the first ATG sequence to occur in any MHC class I transcript is the correct translation initiation codon, all transcripts from multiple TSS generate a common product. We propose that the absence of ATG sequences enables the extended core promoter to support multiple TSS, whose usage can be regulated. We have termed the absence of ATG codons in the extended promoter an “ATG desert.”
The ATG desert defines a novel promoter class
The finding of ATG deserts within the MHC class I promoter family raised the question of whether the presence of ATG deserts is a general feature of promoters in the genome. To address this question, we retrieved a total of 17,718 human genes from GenBank (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/) and analyzed sequences 2 kb upstream and 2 kb downstream of transcription initiation for the presence of ATG triplets, by scanning 100-bp windows. For this analysis, the genes were sorted according to the three promoter subgroups that are associated with the PD1 promoter, i.e., (1) the absence of a canonical TATAA box, (2) the presence of a CGI, and (3) the usage of multiple TSS, as shown above. Whether any of these features correlates with the presence of an ATG desert was examined.
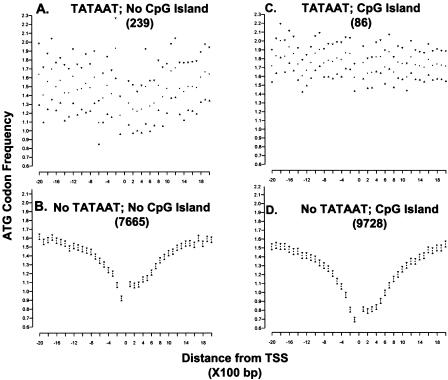
To determine whether TATAA boxes correlated with the ATG deserts, promoters were sorted based on their classification as either TATAA or non-TATAA (TATAA box was defined as the sequence TATAAT occurring within 200 bp of transcription initiation); each group was analyzed for the presence of ATG deserts. Remarkably, we found ATG deserts are a common feature of promoters, in particular, promoters that do not contain a TATAAT element (Fig. 3, cf. C and D with A and B). Among non-TATAA promoters, the ATG deserts are uniform V-shaped curves, symmetrically placed around the major TSS, such that the frequency of ATG codons within 1-2 Kb of transcription initiation plummets relative to the surrounding sequences, reaching a minimum within 100 bp of the TSS. (The same results are obtained if the definition of a TATAA box is expanded to include sequences TATA(A/T)(A/T) located within 40 bp of transcription initiation [Supplemental Fig. 2]).
Figure 3.
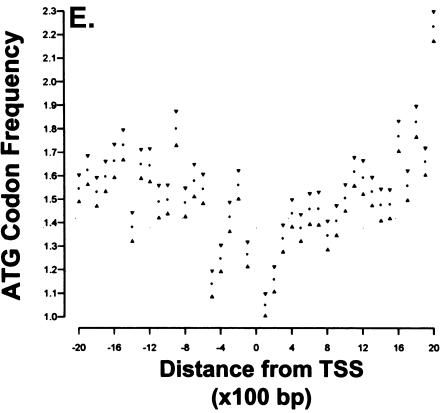
An ATG desert is specifically correlated with the absence of a TATAA box. ATG codon frequencies were analyzed in the region 2 Kb upstream and 2 Kb downstream from transcription initiation using a data set of 17,718 unique genes. In this analysis, TATAA was defined as the sequence TATAAT occurring within 200 bp of transcription initiation. CGI were identified as sequences with a CpG content >0.55, a length of >500 bp, and observed/expected ratio >0,65. The major TSS for any given gene is located at position 0 on the X axis. Each point represents the frequency of observed ATG codons within a 100-bp window; 95% confidence intervals are indicated by the triangles. The number of genes represented in each group is in parenthesis. The groups are designated (A) TATAAT, no CGI; (B) No TATAAT, no CGI; (C) TATAAT, CGI; (D) No TATAAT; CGI. (E) Noncoding genes 75 rRNA, 119 snoRNA, and 1604 tRNA genes.
To control for possible distortions due to the differences in sample size between TATAA and non-TATAA promoter sets, we performed a permutation analysis. One thousand sets of randomly sampled genes, at a pool size equivalent to the TATAA group, were analyzed for the presence of ATG deserts. Even with the smaller pool size, ATG deserts were observed, indicating that the absence of ATG deserts in the TATAA-containing gene set is not due to the small sample size (data not shown).
We conclude that ATG deserts are a novel structural feature of a subclass of promoters that is strongly correlated with non-TATAA promoters in the human genome.
ATG deserts do not correlate with CGI
The presence of the ATG deserts is consistent with a region enriched for CpG and raises the possibility that the ATG deserts are a trivial consequence of the presence of a CGI around non-TATAA promoters. This concern was strengthened by the finding that an analysis of the distribution of codons containing an A or T in two positions showed a similar pattern to the ATG desert (data not shown); deserts were observed among non-TATAA, but not TATAA, promoters. Furthermore, all human, pig, and mouse classical (Ia) MHC class I genes examined are contained within CGI of lengths >1200 bp (Supplemental Table I) (Pontarotti et al. 1988; McQueen et al. 1997).
To determine whether ATG deserts are restricted to those promoters residing within CGI, promoters were sorted into four classes, according to the presence or absence of a CGI and the presence or absence of a TATAAT sequence within 200 bp of transcription initiation (as defined by annotation in RefSeq NT_xxxxxx for the gene). No correlation was observed between the presence of an ATG desert and the location of the promoter within a CGI (Fig. 3, cf. C and D).
The only correlation found with CGI is that a more extreme ATG desert was observed in those non-TATAA promoters that resided within a CGI than those that did not (minimum count of 0.6 vs 1.0), presumably due to their location within a region with an overall high GC content. Furthermore, it is clear that the ATG desert is not simply the result of a nucleotide bias in CGI, since an ATG desert is also observed among non-TATAA promoter genes that lack CGI core promoter structures, but not in TATAA promoters in CGI. (We also considered the possibility that the ATG reflected a chance skewing of the GC content in the set of genes analyzed. However, the GC content in the 4-Kb region surrounding the non-TATAA promoters is 0.45, which is not different from the 0.44 value for the entire set; the GC content of the entire human genome is 0.41.) Among TATAA promoters, the frequency of ATG triplets more closely approximated that of the surrounding sequences, whether associated with a CGI or not. Thus, ATG deserts define a novel promoter region enriched in CpG dinucleotides relative to surrounding sequences, independent of the presence of a CGI.
ATG deserts do not occur around promoters of noncoding genes
The genes analyzed above for the presence of ATG deserts were limited to those encoding proteins. To investigate whether the ATG desert is unique to genes that encode mRNA and are transcribed by pol II, we also analyzed the ATG distribution around the promoters of regulatory RNAs. The regulatory RNA data set contained 1798 regulatory RNAs including 75 rRNAs, 119 snoRNAs, and 1604 tRNAsequences. As shown in Figure 3E, no defined ATG desert is associated with the promoters of regulatory RNAs. Thus, the presence of ATG deserts correlates with non-TATAA promoters that are transcribed by pol II.
Identification of ATG deserts in the genomes of other species
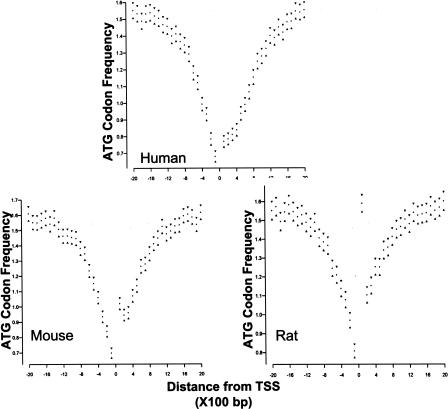
The identification of ATG deserts as a new class of promoters in the human genome was achieved based on an analysis of characterized promoters with known TSS. To determine how large this class is and whether it exists in other species, we developed an algorithm to detect ATG deserts. Because a straightforward ATG scan of a single genome does not have the sensitivity/power to detect the relatively small change in codon frequency that defines an ATG desert associated with a single gene, the algorithm was based on a multispecies alignment of genomic DNA sequences (described in the Methods). Genomic sequences of human, mouse, and rat were aligned, and sequences 1 Kb upstream of the aligned segments were scanned in 100-bp windows for the frequency of ATG triplets. A gene was scored as having an ATG desert if the following criteria were met: the median frequency of ATG triplets in the interval between -1 and -600 bp was ≤1.4 in at least two of the three aligned species and the median ATG frequency in the interval between -601 and -1000 bp was ≥1.6 in at least two of three species. As shown in Table 2A, 7138 of the 8003 genes analyzed were predicted to have promoters that reside in ATG deserts. Further, 6595, or 90%, of the predicted ATG desert genes have non-TATA promoters. The ATG desert prediction was validated by analysis of the ATG frequency around the aligned regions in each of the species (Fig. 4). The algorithm identified clear ATG desert profiles among the identified human, rat, and mouse genes. This algorithm thus provides a tool to identify ATG deserts in the genome through multispecies alignment.
Table 2A.
Identified ATG deserts based on multispecies alignment of mouse, rat, and human RefSeq genes
| ATG desert | Non-ATG desert | |
|---|---|---|
| Non-TATA promoters | 6595 (90%) | 724 |
| TATA promoters | 543 (79%) | 141 |
All mouse, rat, and human RefSeq genes for which an alignment was possible were analyzed for the presence of ATG codons, as follows. Following alignment of genes for the three species, the segment 1-kb upstream of the promoter (as defined by RefSeq) was analyzed in two segments, from − 1 to − 600 and − 601 to − 1000 bp. Within each segment, 100-bp windows were scanned for the presence of an ATG triplet. An ATG desert was scored if the scan met the criteria that (1) the − 1 to − 700-bp window had a median ATG frequency of ≤1.4 in at least two of the three species, and that the − 601 to − 1000-bp window had a median ATG frequency of ≥1.6 and that this pattern was observed in at least two of these species. P-value <2.2 × 10−16.
Figure 4.
A subclass of promoters with ATG deserts is identified in human, mouse, and rat genomes by a computational algorithm. Analysis of 8003 genes that could be aligned among the human, rat, and mouse genomes identified 6595 genes as having potential ATG desert. Validation of the prediction was by plotting ATG frequencies within 2 kb of the major TSS, for each of the species. The major TSS for any given gene is located at position 0 on the X axis. Each point represents the frequency of observed ATG codons within a 100-bp window; 95% confidence intervals are indicated by the triangles. (The high ATG count in window 1 in rat and moderately high ATG count in mouse are the result of genome annotations, which defined the first ATG of an open reading frame as the transcription start site for many of the rat genes and some of the mouse genes.)
Surprisingly, yeast promoters also have associated ATG deserts, although these are shorter than those in mammalian genomes and are associated with non-TATAA promoters (Supplemental Fig. 3). The shorter deserts are consistent with the smaller genome of yeast. There is some association of ATG deserts with TATAA promoters in the yeast genome, perhaps a reflection of the observed regulated usage of tandem TATAA elements associated with a single gene (Iyer and Struhl 1995).
Thus, not only are ATG deserts a common feature in human genes, but are conserved in other species.
Multiple TSS usage correlates with non-TATAA promoters and ATG deserts
The third salient feature of the MHC class I genes is the regulated usage of multiple TSS. The absence of ATGs within the promoter ensures that only a single protein product is generated, regardless of the transcription start site utilized. This finding led us to speculate that ATG deserts may be a general characteristic of non-TATAA promoters using multiple TSS, thus allowing integration of various upstream signals.
A correlation between non-TATAA promoters and multiple TSS usage has been generally accepted but not exhaustively demonstrated, although Suzuki et al. (2001) reported in an analysis of 276 genes that TATAA promoters are highly associated with tightly clustered TSS. To directly determine the correlation between non-TATAA promoters and multiple TSS, 47,368 full-length transcripts (FLJ) from the DBTSS database (http://dbtss.hgc.jp/) were analyzed for their transcription start sites. Of these transcripts, we could correlate start-site usage with TATAA promoter status from RefSeq for 1019 genes. For this analysis, promoters with multiple TSS were defined as those with a standard deviation (SD) of distance among transcript start sites within 5 bases, as described by Suzuki et al. (2001). We observed a bimodal distribution of standard deviation of distance (the first mode was near SD = 2 and the second mode was near SD = 12; data not shown), justifying the definition of single versus multiple TSS classification with a cutoff at SD = 5. As expected, non-TATAA promoters are significantly correlated with multiple TSS usage (Table 3A) with a P-value of 2.67e-08 (χ2 test). Among the non-TATAA promoters with multiple TSS, the average distribution of TSS was 15.4 bp SD, while that of the TATAA promoters with multiple TSS was 9 bp SD. Thus, even where TATAA promoters use multiple TSS, their distribution is significantly tighter than among the non-TATAA promoters (P < 0.001). These findings extend the earlier ones of Suzuki et al. (2001) who reported that among all genes they examined, the distance (mean ± S.D.) between the most 5′ and 3′ TSS was 61.7 ± 19.5 nt. However, when segregated into promoters with either highly variable or tightly clustered TSS (85% and 15%, respectively) those promoters that contained multiple, but tightly clustered TSS, were correlated with the presence of a TATAA box (Suzuki et al. 2001, 2004).
Table 3A.
The use of multiple transcription start sites correlates with non-TATAA promoters
| Transcription start sites
|
|||
|---|---|---|---|
| Promoter type | Single | Multiple | Relative usagea (multiple/single) |
| TATAA | 48 | 77 | 1.6 |
| Non-TATAA | 151 | 743 | 4.92 |
The difference in usage between TATAA and non-TATAA is significant to a value of P < 2.67 × 10−8.
To determine whether there is a direct correlation between the presence of an ATG desert around a promoter and the use of multiple TSS, human full-length transcripts (FLJ) from the DBTSS database (http://dbtss.hgc.jp/) were analyzed for the presence of ATG deserts. Of the set of human genes characterized by Suzuki et al. (2004), 383 could be aligned with mouse and rat genes. The predicted ATG deserts in this group correlated with non-TATAA promoters, as observed for the larger data set (Table 2B). Significantly, there is a correlation between ATG deserts and non-TATAA promoters that use multiple TSS (Table 3A; Supplemental Fig. 4). To extend this analysis, ATG desert predictions were determined by alignment of the human FLJ from the DBTSS database with the rat and mouse genomes and application of the algorithm. In this analysis, a gene was scored as having an ATG desert if the median frequency of ATG triplets in the interval between -1 and -1000 bp was ≤1.5 in at least one of three species. Of the 382 genes that could be analyzed, 360 were identified as being in ATG deserts (Table 3B). Consistent with the hypothesis that multiple TSS promoters are highly associated with ATG deserts, 293 of 360, or 81% of the predicted ATG deserts genes contain multiple TSS promoters. This is in contrast to 13 of 22, or 59%, of the predicted non-ATG desert promoters with multiple TSS. This is statistically significant with a P value of 0.023 and an odds ratio of 3. The presence of ATG deserts predicted to be associated with promoters with multiple TSS in the three species was verified by plotting ATG frequency as a function of TSS usage (Supplemental Fig. 5). Thus, there is a correlation between the presence of an ATG desert around a promoter and its use of multiple TSS, suggesting that among non-TATAA promoters that are located within an ATG desert, multiple TSS could be used as a regulatory mechanism.
Table 2B.
Identified ATG deserts among human genes with mapped transcription start sites
| ATG desert | Non-ATG desert | |
|---|---|---|
| Non-TATA promoters | 305 (94.4%) | 18 |
| TATA promoters | 48 (81%) | 11 |
The same algorithm was applied to the set of human genes characterized by Suzuki et al. (2001) for their TSS for which alignments with rat and mouse genes were possible. P-value = 0.001289.
Table 3B.
The use of multiple transcription start sites in non-TATAA correlates with ATG deserts
| Predicted
|
||
|---|---|---|
| ATG desert | Non-ATG desert | |
| Multiple TSS | 293 (81%) | 13 (59%) |
| Single TSS | 67 | 9 |
Genes were assessed as either in ATG deserts or not and were assessed for the presence of single or multiple TSS among the non-TATAA promoters in the data set. There is an enrichment of multiple TSS among promoters residing in ATG deserts. P-value, 0.0233; odds ratio, 3.028; accuracy, 0.791; sensitivity, 0.118; specificity, 0.958.
Discussion
The present studies identify a new class of promoters characterized by the presence of a novel core promoter feature, the ATG desert. ATG deserts are found in the genomes of human, mouse, and rat, extend symmetrically up to 1 kB upstream and downstream of the major regions of transcription initiation, and are largely associated with promoters that do not contain a canonical TATAA element. Although the function of the ATG desert remains to be clearly established, we speculate that two distinct properties are conferred on ATG desert promoters as follows: (1) the ATG deserts establish a platform for the integration of regulatory signals through alternative transcription start site usage, and (2) the ATG desert region acquires a structural feature that focuses the transcription initiation complex on the core promoter in the absence of the canonical TATAA box.
The class of ATG desert promoters is distinct from other characterized promoters that are defined by distinct DNA sequence motifs that are present in various combinations in a sub-population of core promoters and function to focus transcription initiation to one or a limited number of tightly clustered start sites (Chen et al. 1994; Emami et al. 1995; Verrijzer and Tjian 1996; Reinberg et al. 1998; Butler and Kadonaga 2001). Another promoter element that has been associated with non-TATAA promoters is the MED-1 element (GCTCCC/G), which occurs downstream of transcription initiation in a small set of non-TATAA promoters with multiple TSS (Ince and Scotto 1995). No correlation was observed between ATG deserts and the presence of the MED-1 element or any known core promoter elements. Furthermore, promoters of genes transcribed by RNA polymerase I or III do not display prominent ATG deserts. Only the lack of a TATAA element in a Pol II promoter appears to correlate with the ATG deserts.
Another DNA structure that is highly correlated with core promoter regions is the CGI (Bird 1986; Gardiner-Garden and Frommer 1987; Cross and Bird 1995; McQueen et al. 1997; Antequera 2003; Wang and Leung 2004). However, there is no correlation between the presence of classically defined CGI and ATG deserts; ATG deserts are found even within CGI.
The presence of ATG deserts correlates with non-TATAA promoters that use multiple TSS, leading to a model in which the ATG deserts provide a platform for the integration of complex regulatory networks at multiple TSS within an extended promoter. The organization and regulation of the promoter of the MHC class I PD1 gene provides an example of regulated TSS usage. Like other class I genes, the PD1 gene is ubiquitously expressed, requiring that the promoter be continually operational. In addition, its level of transcription varies among the different cell types and is further activated or repressed by hormonal or cytokine signals (Singer and Maguire 1990; Singer et al. 1997; Howcroft et al. 2003). The ATG desert associated with the PD1 promoter extends ∼450 bp upstream of the major site of transcription initiation and 530 bp downstream through the second intron; a single in-frame ATG occurs at the site of translation initiation within a consensus Kozak box (Kozak 1987). The functional core promoter contains multiple TSS whose usage is actively regulated. Basal transcription initiates at a set of sites distinct from those utilized in response to the IFNγ-mediator, CIITA. Similar differential TSS usage occurs in vivo and in vitro, where activation of transcription by the inflammatory cytokine, IFNγ, markedly shifts transcription to downstream sites (Howcroft et al. 2003). Thus, transcription start-site selection within the MHC class I promoter is actively regulated. The absence of all but the ATG at the site of translation initiation ensures that the same protein product will be generated under either basal or activated conditions.
The regulated use of TSS has been previously reported only for the yeast his3 gene (Iyer and Struhl 1995). The his3 promoter contains two TATA elements, TC and TR that are differentially utilized under basal or activated conditions, respectively. The upstream TC element, which is a noncanonical TATAA box, directs transcription initiation to the +1 bp site, whereas the downstream canonical TATAA box, TR element directs transcription initiation to the +13 bp site. Whether complex promoters—such as his3 and PD1—are considered a single promoter, or multiple promoters, the net effect is to permit fine-tuning of transcription rates by multiple TSS usage, which is strongly correlated with promoters in ATG deserts. In this regard, it is interesting to note that there is a short ATG desert immediately upstream of the his3 promoter (data not shown).
The presence of ATG deserts may also define structural boundaries of the core promoter, thus providing a recognition site for the transcription preinitiation complex (PIC). The TATAA sequence in those promoters that contain it nucleates the PIC by serving as a binding site for TBP; the Inr sequence serves to stabilize the interaction of TFIID with the promoter (Aso et al. 1994). In the absence of either of these elements, it is not clear how the promoter is distinguished from surrounding sequences by the transcription machinery. It has been suggested that the preferential accessibility of yeast promoters, such as his3, to restriction enzymes, reflects an intrinsic property of the promoter DNA sequence that determines its organization in the genome, allowing the preferential recognition and binding of transcription factors (Mai et al. 2000). A variety of unusual DNA structures are known to exist within the genome that could be generated by intrinsic DNA sequences. For example, the presence of Z DNA has been documented in mammalian genomes (Herbert and Rich 1996; Rich and Zhang 2003). Zhao and colleagues have identified a long stretch of Z DNA in the 5′ region of the CSF promoter (Liu et al. 2001). Unusual DNA structures have been associated with sites of chromosomal translocation. ATG deserts may confer upon the DNA a structure that creates a docking site for the transcription initiation complex.
The ATG deserts described in this report differ from the previously reported negative selection against ATG triplets near start codons, which examined only 5′ regions' mRNAs and speculated a role in translation regulation (Saito and Tomita 1999). The ATG deserts extend both 5′ and 3′ to the most abundant TSS, correlate with non-TATAA promoters and multiple TSS usage. We speculate that they may provide an extended transcriptional platform that can accommodate diverse transcriptional complexes with distinct physicochemical requirements and whose constituent subunit structure varies depending upon developmental stage, cell cycle, activation status, and tissue origin. These various transcription complexes may utilize distinct transcription initiation sites within the ATG desert and may be differentially regulated. Future experiments will attempt to address these issues.
Methods
Plasmids and cloning strategies
The MHC class I promoter used in these studies derived from the swine class I gene, PD1 (Singer et al. 1982; Frels et al. 1985). The PD1 promoter reporter construct -416WT, containing 416 bp of core promoter and promoter proximal upstream sequence ligated to the CAT reporter, was previously described (Howcroft et al. 1999). To generate the ATG mutants -416ATG-6, -416ATG-24, and -416ATG-42, -416WT was digested with NarI and HindIII, which cut at positions -50 and +14. Synthetic double-stranded oligonucleotides were inserted into the NarI/HindIII-digested -416WT to generate derivative ATG mutant and control TAG reporter constructs. The sense-strand sequences of the oligonucleotides synthesized (from -50 to +14) are provided in the Supplemental material.
Transfections
Culture of HeLa epithelial cells, transient transfections, and CAT assays were performed as previously described (Howcroft et al. 1993, 2003). Reporter activity was corrected by cotransfecting an internal control plasmid control, either pSV2LUC or CMV-β-gal.
Isolation of RNA and 5′RACE analysis of transcription start sites
The PD1 transgenic mouse strain used for in vivo start-site analysis, B10.PD1, was described previously (Singer et al. 1982); the analysis is detailed in Supplemental materials.
In vitro transcription/coupled primer extension
Analysis of in vitro start sites for -416WT was by primer extension as previously described (Weissman et al. 1998).
Genome-wide analysis for ATG deserts and for TSS
Two TATAA search criteria were used, TATAAT in the 200-bp upstream or TATA(A/T)(A/T) in the 40-bp upstream sequences. ATG counts were computed in the sliding window of 100 bases. The -1 indicates the first 100 bases upstream the transcript start site (TSS) where +1 marks the 100 bases downstream from the TSS. Details of the criteria are provided in Supplemental materials.
ATG desert predictor using multiple-species sequence alignments
Multiple species sequence alignments among human, mouse, and rat were derived from GoldenPath (http://hgdownload.cse.ucsc.edu/goldenPath/hg16/humor/). We extracted 2 kb of promoter sequence from the conserved regions. The ATG count in each of ten 100-bp windows was calculated for mouse and rat sequences as described for the human sequence. We set out to develop an ATG desert gene algorithm using conserved features among the three species. Two related algorithms were developed and are detailed in the Supplemental materials.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Dr. Ying Hu's assistance in the analysis of TSS and regulatory RNAs. We also thank David Levens, Ranjan Sen, Alfred Singer, and Jocelyn Weissman for helpful discussions and critical reading of the manuscript and Eric Lander for helpful discussions and suggestions.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3873705. Article published online before print in August 2005.
References
- Antequera, F. 2003. Structure, function and evolution of CpG island promoters. Cell. Mol. Life Sci. 60: 1647-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera, F. and Bird, A. 1993. Number of CpG islands and genes in human and mouse. Proc. Natl. Acad. Sci. 90: 11995-11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso, T., Conaway, J.W., and Conaway, R.C. 1994. Role of core promoter structure in assembly of the RNA polymerase II preinitiation complex. A common pathway for formation of preinitiation intermediates at many TATA and TATA-less promoters. J. Biol. Chem. 269: 26575-26583. [PubMed] [Google Scholar]
- Basehoar, A.D, Zanton, S.J., and Pugh, B.F. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699-709. [DOI] [PubMed] [Google Scholar]
- Berk, A.J. 1999. Activation of RNA polymerase II transcription. Curr. Opin. Cell. Biol. 11: 330-335. [DOI] [PubMed] [Google Scholar]
- Bird, A.P. 1986. CpG-rich islands and the function of DNA methylation. Nature 321: 209-213. [DOI] [PubMed] [Google Scholar]
- Burke, T.W. and Kadonaga, J.T. 1997. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes & Dev. 11: 3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, T.W., Willy, P.J., Kutach, A.K., Butler, J.E., and Kadonaga, J.T. 1998. The DPE, a conserved downstream core promoter element that is functionally analogous to the TATA box. Cold Spring Harb. Symp. Quant. Biol. 63: 75-82. [DOI] [PubMed] [Google Scholar]
- Butler, J.E. and Kadonaga, J.T. 2001. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes & Dev. 15: 2515-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes & Dev. 16: 2583-2592. [DOI] [PubMed] [Google Scholar]
- Chen, J.L., Attardi, L.D., Verrijzer, C.P. Yokomori, K., and Tjian, R. 1994. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell 79: 93-105. [DOI] [PubMed] [Google Scholar]
- Cross, S.H. and Bird, A.P. 1995. CpG islands and genes. Curr. Opin. Genet. Dev. 5: 309-314. [DOI] [PubMed] [Google Scholar]
- Emami, K.H., Navarre, W.W., and Smale, S.T. 1995. Core promoter specificities of the Sp1 and VP16 transcriptional activation domains. Mol. Cell. Biol. 15: 5906-5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frels, W.I., Bluestone, J.A., Hodes, R.J., Capecchi, M.R., and Singer, D.S. 1985. Expression of a microinjected porcine class I major histocompatibility complex gene in transgenic mice. Science 228: 577-580. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden, M. and Frommer, M. 1987. CpG islands in vertebrate genomes. J. Mol. Biol. 196: 261-282. [DOI] [PubMed] [Google Scholar]
- Gill, G. 2001. Regulation of the initiation of eukaryotic transcription. Essays Biochem. 37: 33-43. [DOI] [PubMed] [Google Scholar]
- Girdlestone, J. 1995. Regulation of HLA class I loci by interferons. Immunobiology 193: 229-237. [DOI] [PubMed] [Google Scholar]
- Hansen, S.K. and Tjian, R. 1995. TAFs and TFIIA mediate differential utilization of the tandem Adh promoters. Cell 82: 565-575. [DOI] [PubMed] [Google Scholar]
- Herbert, A. and Rich, A. 1996, The biology of left-handed Z-DNA. J. Biol. Chem. 271: 11595-11598. [DOI] [PubMed] [Google Scholar]
- Howcroft, T.K. and Singer, D.S. 2003. Expression of nonclassical MHC class Ib genes: Comparison of regulatory elements. Immunol. Res. 27: 1-30. [DOI] [PubMed] [Google Scholar]
- Howcroft, T.K., Richardson, J.C., and Singer, D.S. 1993. MHC class I gene expression is negatively regulated by the proto-oncogene, c-jun. EMBO J. 12: 3163-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howcroft, T.K., Murphy, C., Weissman, J.D., Huber, S.J., Sawadogo, M. and Singer, D.S. 1999. Upstream stimulatory factor regulates major histocompatibility complex class I gene expression: The U2DeltaE4 splice variant abrogates E-box activity. Mol. Cell. Biol. 19: 4788-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howcroft, T.K., Raval, A., Weissman, J.D., Gegonne, A., and Singer, D.S. 2003. Distinct transcriptional pathways regulate basal and activated major histocompatibility complex class I expression. Mol. Cell. Biol. 23: 3377-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince, T.A. and Scotto, K.W. 1995. A conserved downstream element defines a new class of RNA polymerase II promoters. J. Biol. Chem. 270: 30249-30252. [DOI] [PubMed] [Google Scholar]
- Iyer, V. and Struhl, K. 1995. Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol. Cell. Biol. 15: 7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga, J.T. 2002. The DPE, a core promoter element for transcription by RNA polymerase II. Exp. Mol. Med. 34: 259-264. [DOI] [PubMed] [Google Scholar]
- ____. 2004. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116: 247-257. [DOI] [PubMed] [Google Scholar]
- Kaufmann, J. and Smale, S.T. 1994. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes & Dev. 8: 821-829. [DOI] [PubMed] [Google Scholar]
- Kozak, M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15: 8125-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutach, A.K. and Kadonaga, J.T. 2000. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 20: 4754-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange, T., Kapanidis, A.N., Tang, H., Reinberg, D., and Ebright, R.H. 1998. New core promoter element in RNA polymerase II-dependent transcription: Sequence-specific DNA binding by transcription factor IIB. Genes & Dev. 12: 34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bouteiller, P. 1994. HLA class I chromosomal region, genes, and products: Facts and questions. Crit. Rev. Immunol. 14: 89-129. [DOI] [PubMed] [Google Scholar]
- Littlefield, O., Korkhin, Y., and Sigler, P.B. 1999. The structural basis for the oriented assembly of a TBP/TFB/promoter complex. Proc. Natl. Acad. Sci. 96: 13668-13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, R., Liu, H., Chen, X., Kirby, M., Brown, P.O., and Zhao, K. 2001. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106: 309-318. [DOI] [PubMed] [Google Scholar]
- Lo, K. and Smale, S.T. 1996. Generality of a functional initiator consensus sequence. Gene 182: 13-22. [DOI] [PubMed] [Google Scholar]
- Mai, X., Chou, S., and Struhl, K. 2000. Preferential accessibility of the Yeast his3 promoter is determined by the general property of the DNA sequence, not by specific elements. Mol. Cell. Biol. 20: 6668-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen, H.A., Clark, V.H., Bird, A.P., Yerle, M., and Archibald, A.L. 1997. CpG islands of the pig. Genome Res. 7: 924-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontarotti, P., Chimini, G., Nguyen, C., Boretto, J., and Jordan, B.R. 1988. CpG islands and HTF islands in the HLA class I region: Investigation of the methylation status of class I genes leads to precise physical mapping of the HLA-B and -C genes. Nucleic Acids Res. 16: 6767-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg, D., Orphanides, G., Ebright, R., Akoulitchev, S., Carcamo, J., Cho, H., Cortes, P., Drapkin, R., Flores, O., Ha, I., et al. 1998. The RNA polymerase II general transcription factors: Past, present, and future. Cold Spring Harb. Symp. Quant. Biol. 63: 83-103. [DOI] [PubMed] [Google Scholar]
- Rich, A. and Zhang, S. 2003. Timeline: Z-DNA: The long road to biological function. Nat. Rev. Genet. 4: 566-572. [DOI] [PubMed] [Google Scholar]
- Roeder, R.G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21: 327-335. [PubMed] [Google Scholar]
- Saito, R. and Tomita, M. 1999. On negative selection against ATG triplets near start codons in eukaryotic and prokaryotic genomes. J. Mol. Evol. 48: 213-217. [DOI] [PubMed] [Google Scholar]
- Singer, D.S. and Maguire, J.E. 1990. Regulation of the expression of class I MHC genes. Crit. Rev. Immunol. 10: 235-257. [PubMed] [Google Scholar]
- Singer, D.S., Camerini-Otero, R.D., Satz, M.L., Osborne, B., Sachs, D., and Rudikoff, S. 1982. Characterization of a porcine genomic clone encoding a major histocompatibility antigen: Expression in mouse L cells. Proc. Natl. Acad. Sci. 79: 1403-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, V.L., Wobbe, C.R., and Struhl, K. 1990. A wide variety of DNA sequences can functionally replace a yeast TATA element for transcriptional activation. Genes & Dev. 4: 636-645. [DOI] [PubMed] [Google Scholar]
- Singer, D.S., Mozes, E., Kirshner, S., and Kohn, L.D. 1997. Role of MHC class I molecules in autoimmune disease. Crit. Rev. Immunol. 17: 463-468. [PubMed] [Google Scholar]
- Smale, S.T. 2001. Core promoters: Active contributors to combinatorial gene regulation. Genes & Dev. 15: 2503-2508. [DOI] [PubMed] [Google Scholar]
- Smale, S.T. and Baltimore, D. 1989. The “initiator” as a transcription control element. Cell 57: 103-113. [DOI] [PubMed] [Google Scholar]
- Smale, S.T., Jain, A., Kaufmann, J., Emami, K.H., Lo, K., and Garraway, I.P. 1998. The initiator element: A paradigm for core promoter heterogeneity within metazoan protein-coding genes. Cold Spring Harb. Symp. Quant. Biol. 63: 21-31. [DOI] [PubMed] [Google Scholar]
- Struhl, K. 2001. A paradigm for precision. Science 293: 1054. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., Taira, H., Tsunoda, T., Mizushima-Sugano, J., Sese, J., Hata, H., Ota, T., Isogai, T., Tanaka, T., Morishita, S., et al. 2001. Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2: 388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y., Yamashita, R., Sugano, S., and Nakai, K. 2004. DBTSS, dataBase of transcriptional start sites: Progress report 2004. Nucleic Acids Res. 32: D78-D81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai, D. and Jones, P.A. 2002. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. 99: 3740-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrijzer, C.P. and Tjian, R. 1996. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem. Sci. 21: 338-342. [PubMed] [Google Scholar]
- Wang, Y. and Leung, F.C. 2004. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics 20: 1170-1177. [DOI] [PubMed] [Google Scholar]
- Weissman, J.D., Brown, J.A., Howcroft, T.K., Hwang, J., Chawla, A., Roche, P.A., Schiltz, L., Nakatani, Y., and Singer, D.S. 1998. HIV-1 tat binds TAFII250 and represses TAFII250-dependent transcription of major histocompatibility class I genes. Proc. Natl. Acad. Sci. 95: 11601-11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willy, P.J., Kobayashi, R., and Kadonaga, J.T. 2000. A basal transcription factor that activates or represses transcription. Science 290: 982-985. [DOI] [PubMed] [Google Scholar]
- Wong, A.W., Ghosh, N., McKinnon, K.P., Reed, W., Piskurich, J.F., Wright, K.L., and Ting, J.P. 2002. Regulation and specificity of MHC2TA promoter usage in human primary T lymphocytes and cell line. J. Immunol. 169: 3112-3119. [DOI] [PubMed] [Google Scholar]
- Zanton, S.J. and Pugh, B.F. 2004. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc. Natl. Acad. Sci. 101: 16843-16848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenzie-Gregory, B., Khachi, A., Garraway, I.P., and Smale, S.T. 1993. Mechanism of initiator-mediated transcription: Evidence for a functional interaction between the TATA-binding protein and DNA in the absence of a specific recognition sequence. Mol. Cell. Biol. 13: 3841-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/; GenBank.
- http://dbtss.hgc.jp/; DBTSS database.
- http://hgdownload.cse.ucsc.edu/goldenPath/hg16/humor/; GoldenPath.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.