Abstract
Gene number difference among organisms demonstrates that new gene origination is a fundamental biological process in evolution. Exon shuffling has been universally observed in the formation of new genes. Yet to be learned are the ways new exons originate and evolve, and how often new exons appear. To address these questions, we identified 2695 newly evolved exons in the mouse and rat by comparing the expressed sequences of 12,419 orthologous genes between human and mouse, using 743,856 pig ESTs as the outgroup. The new exon origination rate is about 2.71 × 10-3 per gene per million years. These new exons have markedly accelerated rates both of nonsynonymous substitutions and of insertions/deletions (indels). A much higher proportion of new exons have Ka/Ks ratios >1 (where Ka is the nonsynonymous substitution rate and Ks is the synonymous substitution rate) than do the old exons shared by human and mouse, implying a role of positive selection in the rapid evolution. The majority of these new exons have sequences unique in the genome, suggesting that most new exons might originate through “exonization” of intronic sequences. Most of the new exons appear to be alternative exons that are expressed at low levels.
Evolutionary novelties in genomes have recently attracted increasing attention (Lynch and Conery 2000; Prince and Pickett 2002; Long et al. 2003). Studies on young genes have afforded great insight into the mechanism of origin of new genes and their subsequent evolution. Genomic processes of new gene origination involve several fundamental mechanisms, including gene duplication, exon shuffling, retroposition, lateral gene transfer, and transposable element assimilation (Long et al. 2003). These processes sometimes create new variants of genes, but can also yield new genes with novel functions (e.g., Zhang et al. 2002, 2004). Rapid evolution is a common phenomenon in newly evolved genes, often driven by positive Darwinian selection (Long and Langley 1993; Nurminsky et al. 1998; Johnson et al. 2001; Wang et al. 2002; Zhang et al. 2002). Because exon shuffling is widely recognized as important in the generation of new genes (Gilbert 1978; Gilbert et al. 1997; Patthy 1999; Kaessmann et al. 2002), how new exons, the basic units of gene and exon-shuffling, originate and evolve becomes an important question at the genome level.
So far, three processes have been proposed to be involved in the creation of new exons, i.e., exaptation of transposable elements (Brosius and Gould 1992; Makalowski et al. 1994; Nekrutenko and Li 2001; Sorek et al. 2002), exon duplication (Kondrashov and Koonin 2001; Letunic et al. 2002), and exonization of intronic sequences (Gilbert 1978; Kondrashov and Koonin 2003). Makalowski et al. (1994) were the first to describe the integration of an Alu element into the coding portion of the human decay-accelerating factor (DAF) gene, and recently about 4% of human genes were found containing transposable elements in their coding regions (Nekrutenko and Li 2001). Duplication of existing exons has also been reported. About 10% of all genes contain tandemly duplicated exons that might confer further evolutionary potential (Letunic et al. 2002). The most easily conceived mechanism for creating new exons is exonization of intronic sequences due to easy emergence of new splicing sites through mutations. Unfortunately, up to now, only a few potential examples of such a process have been identified (e.g., Kondrashov and Koonin 2003).
The majority of these pioneering reports on the origin of new exons were formulated in the context of alternative splicing (Modrek and Lee 2003; Ast 2004). Many important questions directly related to the general picture of new exon origins are still largely unanswered. For example, how often do new exons emerge? What are the subsequent evolution patterns and driving forces? Do new exons preferentially appear in particular genes? What kinds of sequences contribute the most to the creation of new exons? To address these questions, in this study, we applied genomic and transcriptomic data from four mammalian species, human (Homo sapiens) (International Human Genome Sequencing Consortium 2001), mouse (Mus musculus) (Mouse Genome Sequencing Consortium 2002), rat (Rattus norvegicus) (Rat Genome Sequencing Project Consortium 2004), and pig (BGI, unpubl. EST data), to identify newly evolved exons in mouse and rat (rodents) by using double outgroups (first human and then pig).
Results and Discussion
Identification of newly evolved exons
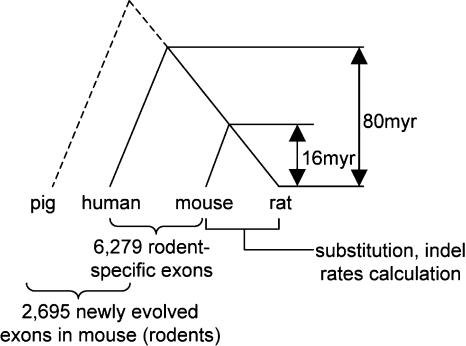
By mapping mouse expressed sequences onto the 12,419 genes that are listed in the HomoloGene database (ftp://ftp.ncbi.nih.gov/pub/HomoloGene/) and have well-defined orthologs with human (see Methods), 79,098 exons could be defined with clear phases in mouse after excluding exons that are short, frame-shifting, or contain UTRs or stop codons. Of these, 71,039 exons were also found in rat, and thus, were used for further analysis. Of those retained 71,039 exons, 6279 are found in mouse but not in human, and are designated as rodent-specific exons. They either have no human homologous sequences (1582 exons), or correspond to human intronic sequences (4697). To exclude those old exons that are lost or intronized in the human lineage, we used 743,856 pig ESTs generated by our Beijing Genomics Institute (BGI, unpubl.) as an outgroup set to fish out those newly evolved exons in rodents, since the ungulate is the outgroup of rodents and primates in the phylogenetic trees for mammals (Murphy et al. 2001; Springer et al. 2003). These pig ESTs were generated from all of the main organs and tissues of pigs. After removing repeats by RepeatMasker, 693,407 pig ESTs were eventually assembled into 50,119 unigenes and 54,125 singletons. With the addition of this outgroup, 3584 of the 6279 rodent-specific exons were found to be shared with the pig. Thus, the remaining 2695 exons found exclusively in the rodent lineage are classified here as newly evolved (Fig. 1). Almost all of these new exons possess typical splicing signals at the boundaries, a few of which are shown in Table 1. We classify these new exons into two categories, based on presence or absence of BLAT hits in the human sequences. The I (for “intron”) category contains exons mapped to human introns and the N (for “none”) category contains exons having no homologous sequences in the human. In a sense, it is an arbitrary classification, because some N exons might originally be from intronic sequences, but the counterparts in human have evolved to be indiscernible or the corresponding sequences have been deleted. It is noteworthy that the mean length of I exons is twice that of N exons (Table 2). This difference is probably because the exons derived from intronic sequences were easier to evolve into functional fragments than inserted sequences like some N exons.
Figure 1.
Phylogenetic relationships of the mammalian species, indicating comparisons used to identify newly evolved exons that are found in rodents, but in neither human nor pig.
Table 1.
Examples of splicing sites of new exons
| Species | RefSeq ID | Exon length | Align at intron/exon boundary | |
|---|---|---|---|---|
| I exon | ||||
| NM_145692_2 | mouse | NM_145692 | 132bp | ctccagAATTTC......ATATAAgtaagt |
| rat | 132bp | ctccagAGTTTC......ACATAAgtaagt | ||
| XM_129746_10 | mouse | XM_129746 | 125bp | tcccagGTTGAG......ACACCTgtaagt |
| rat | XM_237151 | 125bp | tctcagGTAGAG......GCATCTgtaagt | |
| NM_022814_16 | mouse | NM_022814 | 120bp | tttcatATGTCG......GTGCAGgtacag |
| rat | XM_232929 | 120bp | tttcagATGTTG......GTGCAGgtactg | |
| N exon | ||||
| NM_009581_2 | mouse | NM_009581 | 90bp | tttcagACTCAG......GTCATGgtaaga |
| rat | NM_182815 | 87bp | gttcagACCCAA......AAAATGgtaagg | |
| NM_030676_7 | mouse | NM_030676 | 63bp | cctcagCGTCTG......AAAAACgtgagt |
| rat | NM_021742 | 63bp | cttcagCACCTG......AAAAACgtgagt | |
| NM_130904_5
|
mouse | NM_130904 | 87bp | ctccagAGGATG......TGGCAGgtacag |
| rat | XM_344064 | 87bp | ctccagAGGATG......TGGCAGgtacag |
Table 2.
Summary of old exons and new exons (I and N) and their evolution patterns
| Exon no. | Mean len for Ka, Ks | Mean EST no. in mouse | Mean EST no. in human | Indel/kb | Ka | Ks | Ka/Ks | % of Ka/Ks > 1 | |
|---|---|---|---|---|---|---|---|---|---|
| Total exons in mouse/rat | 71,039 | 147.3 | 10.0 | 12.7 | 0.37 | 0.027 | 0.171 | 0.155 | 2.53 |
| Old exons in mouse/rat/human | 64,760 | 142.9 | 10.0 | 12.6 | 0.31 | 0.025 | 0.172 | 0.143 | 2.17 |
| I exons | 1709 | 255.2 | 4.7 | 0 | 0.86* | 0.049* | 0.169 | 0.290* | 6.50* |
| N exons | 986 | 124.5 | 4.2 | 0 | 1.35* | 0.066* | 0.180 | 0.367* | 14.00* |
| I old sister exons | 9951 | 146.0 | 7.2 | 8.3 | 0.35 | 0.028 | 0.170 | 0.166 | 2.67 |
| N old sister exons | 3608 | 147.0 | 6.5 | 7.2 | 0.59 | 0.045 | 0.180 | 0.248 | 5.57 |
See text for I and N.
In Ka indicates significant both in 95% CI and distribution tests. * In other items indicates significant in ϰ2 test. Len refers to length in base pairs.
If we assume that the most recent common ancestor of primates and rodents lived 80 million years ago (Mya) (Murphy et al. 2001; Mouse Genome Sequencing Consortium 2002; Springer et al. 2003), we are able to estimate the rate of new exon origination, based on the new exon number (2695) (Table 2). From our data, the average rate of new exon creation is 2.71 × 10-3 per gene per million years (2695/12,419/80 = 2.71 × 10-3), or 2.71 × 10-3 × 30,000 = 81.3 per genome per million years in the mouse lineage. This represents a conservative estimation of the exon origination rate because we used a very stringent method in identifying new exons. We only used mouse genes with well-defined orthologs in human listed in the HomoloGene database (ftp://ftp.ncbi.nih.gov/pub/HomoloGene/), which represent only one-third of the gene number in the species and are usually conserved throughout human, mouse, and rat. If we were to take numerous other less-conserved, and even unidentified newly created genes into consideration, we would undoubtably see a much higher number of new exons, which would outrival the decrease one would expect to result from adding more outgroup sequences. In fact, the number (1582) of rodent-specific exons without human homologous sequences that we found in this study is less than that (2302) reported in the recent rat genome project (Rat Genome Sequencing Consortium 2004). In addition, the proportion of rodent-specific exons recognized in this study (6279/71,039 = 8.8%) is also lower than a previous estimate (>15%) (Nurtdinov et al. 2003).
Possible functions of new exons
In order to look at what functions the new exons brought to the proteins, we used Interproscan (InterPro Consortium 2001) to annotate the new exons that we identified. We could only obtain annotation information for 587 new exons (Supplemental Table 1), including 125 N exons and 462 I exons. Interestingly, the functional domains appearing at highest frequencies are involved in extracellular binding processes (e.g., immunoglobulin-like and EGF-like domains) and protein-protein interactions (e.g., proline-rich regions and ankyrin domain) (Supplemental Table 1). Although it is difficult to statistically infer a general pattern because the exons are usually short, this result implies that the appearance of new exons might have been subject to selection for adaptation to variable environments.
To further study whether new exons are more likely to appear in certain functional classes of genes, we examined the gene ontology (GO) classifications of genes acquiring new exons. GO information is available for 4887 of the 12,419 orthologous gene pairs; 981, or 20% of these 4887 genes contain newly evolved exons, while 3906 genes contain only old exons. In none of the GO classes do significantly >20% of the genes have new exons, although we see a few more genes with new exons in the cell adhesion and defence activity categories (Supplemental Fig. 1). This result suggests that exon acquisition is likely unrelated to a functional class of genes as recognized in GO categories. Of course, our gene data set is biased in the sense that we considered only conserved gene pairs; hence, we cannot exclude the possibility that there may be GO functional class bias in less-conserved genes. Future work with a closer outgroup such as a nonmurine rodent should help in clarifying this problem.
Most new exons are from unique intronic sequences
To investigate the sources of the 2695 new exons, we did a BLAST search using each exon against the mouse genome and checked each exon with RepeatMasker. The results are shown in Table 3. Despite the fact that about 40% of a rodent genome consists of transposable element- (TE) derived sequences (Mouse Genome Sequencing Consortium 2002; Rat Genome Sequencing Consortium 2004), only a few new exons are identifiably from TEs. One odd observation is that no new exon traces to a LINE element, despite the fact that LINEs comprise >20% of mouse and rat genomes, with many active elements. Instead, most newly evolved exons are unique sequences in the mouse genome. When the BLAST cut-off of 1E-02 is used, about half of the new exons have only one hit in the genome; when the cut-off was set to 1E-05, 91.3% of the new exons have only one hit. Parsimoniously, these unique exons are most probably derived from unique intronic sequences, which is supported by the observation that most of the new exons belong to the I (intron) category (Table 2). These results suggest that exonization of intronic sequences is a much more important role than exaptation of TEs in the process creating new exons. This conclusion is easily conceivable, because new exons could more easily be created by obtaining new splicing sites in introns through point mutations than by other processes like insertions of external sequences, although some short unrecognizable TE sequences that contain many potential splicing sites might also play a role near exonized sequences and may contribute to exonic sequences.
Table 3.
Number of hits of new exons in BLAST searches and RepeatMasker checks in the mouse genome
| Cut-off | Hits to LTR/MaLR | Hits to SINE (ID, B2, B4, MIR) | 7-21 hits in mouse genome | 3-6 hits in mouse genome | 2 hits in mouse genome | 1 hit in mouse genome | Total |
|---|---|---|---|---|---|---|---|
| 1E-05 | 3 | 16 | 19 | 50 | 147 | 2462 | 2695 |
| 1E-02 | 3 | 16 | 92 | 487 | 772 | 1325 | 2695 |
Rapid nonsynonymous substitution
To look at the general evolutionary patterns of new exons, we first concatenated all of the I exons and (separately) all of the N exons. To control for gene-specific effects, we used the old sister exons, shared by human and mouse, in the same genes as the new exons for comparison. We found that the newly evolved exons have a markedly accelerated rate of nonsynonymous substitution between mouse and rat. The nonsynonymous substitution rates (Ka) in both I and N categories are almost double those of their old sister exons, while the synonymous rates (Ks) are more or less the same in both sets of exons (Table 2). The Ka values are significantly different. The 95% confidence interval (CI) of Ka for I exons (0.04543-0.04691) does not overlap that of their old sister exons (0.03858, 0.03950). The Ka value of the N exons (95% CI: 0.0494, 0.06173) is also significantly higher than that of their sister exons (0.03988, 0.04114). On the other hand, the Ks 95% CIs always overlapped between categories (data not shown).
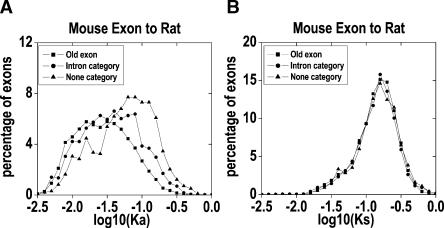
To exclude the possibility that the overall fast nonsynonymous rate is due to a few unusually quickly evolving exons, as observed in the Drosophila orphan genes (Domazet-Loso and Tautz 2003), we compared the distributions of Ka and Ks between the I and N exons and their sister exons, respectively (Fig. 2). The distribution of Ka in the N exons shifts significantly to the right (bigger) compared with that of their sister exons (P < 2 × 10-16), while the rates of synonymous changes between the new and old exons are not statistically different (P = 0.385). In the I category, the Ka distribution is also significantly different from that of their sister exons (P < 2 × 10-16), while the difference of the Ks distributions is significant at the 5% level, but not at the 1% level (P = 0.024). Based on these analyses, we concluded that the newly evolved exons generally have an accelerated amino acid evolution rate compared with old exons.
Figure 2.
Ka (A) and Ks (B) distributions of the newly evolved I and N exons and their sister exons. The bin size for log(Ka) is 0.15, and bin size for log(Ks) is 0.1.
From previous studies by our group and other groups, rapid evolution observed in newly evolved young genes is often driven by positive Darwinian selection (Long and Langley 1993; Nurminsky et al. 1998; Johnson et al. 2001; Wang et al. 2002; Zhang et al 2002). To test whether this observation also applies to the evolution of new exons, we adopted the Ka/Ks ratio test, a frequently used method for detecting positive selection on protein-coding genes (Hughes and Nei 1988; Li 1997), to examine each category of new and old exons. The Ka/Ks ratio for neutrally evolving pseudogenes is expected to be 1, for genes subject to functional constraint is <1, and for genes subject to strong positive selection is higher than 1. In the N category, as many as 14% of exons have Ka/Ks >1, which is about triple the occurrence of that in the sister exons (5.57%), and about seven times that in all old exons identified in this study (2.17%) (Table 2). In the I category, 6.5% exons have Ka/Ks >1, compared with 2.67% in their sister exon. The difference in proportion of Ka/Ks >1 exons between new exons and old exons is highly significant by the χ2 test (P ≈ 0) (Table 2). These results suggest that positive Darwinian selection might have been a considerable force in the evolution of these newly evolved exons.
Rapid insertion/deletion occurrence rate
Another striking evolutionary feature observed in the new exons is their high insertion/deletion (indel) occurrence rate (number of indels per kilobase). We only considered indels that are in three or multiples of three nucleotides, such that the exons stay in frame. Most of the indels are 3 or 6 nucleotides (Supplemental Fig. 2). Between mouse and rat, the indel occurrence rate in both N and I exons is over twice as high as in their respective sister exons; the indel rate for N exons (1.35/kb) is more than four times that of all of the exons shared between mouse and human (0.31/kb) (Table 2). Podlaha and Zhang (2003) recently reported a primate gene bearing an accelerated indel rate even higher than the neutral genomic background rate; they suggested that the high rate might be driven by positive selection on protein length. To estimate the background indel rate in rodents for comparison with the indel rate of newly evolved exons, we randomly picked 51,311 mouse introns longer than 1 kb. A total of 46,584 of these introns, with a total length of 46,287,362 bp, were also found in the rat. Altogether, 739,879 indels were identified in these introns, comprising 2,077,030 bp in total. Therefore, the indel occurrence rate is 15.99/kb (739,879/46,287,362 bp), or 5 × 10-10 per site per year if we assume that mouse and rat diverged 16 Mya (Springer et al. 2003). This rate is five times that in primates (∼1 × 10-10) (Podlaha and Zhang 2003), consistent with the discrepancy in the nucleotide substitution rates between rodents and primates (Li 1997; Rat Genome Sequencing Project Consortium 2004). Nevertheless, this neutral indel rate is 11.8 times bigger than that of the N exons (1.35/kb), the fastest rate in Table 2. Thus, although the indel rate of new exons is much faster than that of old exons, indel occurrence in the new exons is still subject to strong functional constraint. To determine whether the high indel rate in new exons is related to the selective pressure for creating new splicing sites, we noted the position of indels relative to the boundaries of these new exons. However, there was no significant distribution bias relative to the exon boundaries (data not shown). Therefore, the biological significance of this enhanced indel rate remains unclear. It may partially be attributed to the relaxation of negative selection on these new exons. Further work is necessary in order to obtain a conclusion on this issue.
Association with alternative splicing
Alternative splicing of exons makes an important contribution to the complexity of the proteome. For instance, it is believed that 40%-60% of human genes are alternatively spliced (Mironov et al. 1999; International Human Genome Sequencing Consortium 2001; Modrek et al. 2002). A recent comparison between human and mouse-rat alternative splicing patterns suggested that alternative splicing is associated with a large increase in the frequency of recent gain or loss of exons (Modrek and Lee 2002). To explore whether the newly evolved exons identified in our study are correlated with alternative splicing, we classified mouse exons as constitutive (100% inclusion in mouse ESTs and mRNAs), major (≥50% inclusion), minor (<50% inclusion), low (two to five ESTs), or singleton (one EST). New exons constitute a higher fraction of exons in the minor, low, and singleton categories (7.4%, 3.3%, and 11.6%, respectively) than exons in the constitutive and major categories (1.6% and 1.7%, respectively) (Table 4). This result suggests that new exons tend to appear in less-abundant splice forms, consonant with Modrek and Lee's conclusion (Modrek and Lee 2002), and that rise of a new exon does not necessarily mean that the new protein domain has important function and is under strong purifying selection. For genes in general, expression of their new exons is usually relegated to their less-abundant species of mRNA, leading to a lower detectable signal in expression assays. It is noteworthy that many new exons fall into the singleton category. People will wonder whether the evidence for the singleton exons is sufficient enough to prove it is not an artifact. Nevertheless, Johnson et al. (2003) have recently reported that many rare splicing forms are real, based on DNA array data, and we have also collected strong evidence showing that most lowly expressed transcripts and exons are functional (data not shown). Another concern is whether there still is a chance of missing the very rare transcripts in pig and human, even given abundant data set. To prove that this won't affect our basic conclusion, we removed all of the singleton exons from the new exon list, and redid the above analyses. The general patterns (e.g., rapid evolution) still remain the same, and the statistical difference is still significant (Supplemental Table 2). Therefore, we kept these exons in the list of new exons in Table 4, but categorized it as a separated group for better understanding of the data.
Table 4.
Correlation between alternative splicing and newly evolved exons
| Mouse exon category | No. of new exons | Mean no. of ESTs | No. of total exons | % of new exons in each category | % of total new exons |
|---|---|---|---|---|---|
| Constitutive | 360 | 11 | 22,683 | 1.6% | 13.4% |
| Major | 232 | 20 | 13,490 | 1.7% | 8.6% |
| Minor | 60 | 5 | 816 | 7.4% | 2.2% |
| Low | 742 | 3 | 22,846 | 3.3% | 27.5% |
| Singleton | 1301 | 1 | 11,204 | 11.6% | 48.3% |
| Total | 2695 | 5 | 71,039 | 3.8% | 100% |
Conclusions
In summary, our large-scale study of rodent exons revealed that new exons are frequently created in genomes, and that they evolve rapidly at both nucleotide substitution and indel levels. Exonization of intronic sequences may have played the greatest role in the formation of new exons. Due to lack of comprehensive genomic data for closely related species, studies on genomic novelties, such as young genes, still largely rely on case analysis. But, based on the results in this study and conclusions from young gene analyses, it seems that accelerated evolution is far from rare during the evolution of genomic novelties, including new exons, new genes, and other new functional sequences.
Methods
Identification of transcription units
To identify transcription units in human and mouse, we downloaded 12,419 human-mouse orthologous genes in RefSeq format from the HomoloGene database (ftp://ftp.ncbi.nih.gov/pub/HomoloGene/) and the human and mouse mRNA/EST (expression sequence tag) data from UniGene (ftp://ftp.ncbi.nih.gov/repository/UniGene/) with their corresponding mapping information from UCSC (human http://genome.ucsc.edu/goldenPath/10april2003/database/; mouse http://genome.ucsc.edu/goldenPath/mmFeb2003/database/). The RefSeq cDNAs are mapped to their corresponding genomes with the same methods as UCSC (BLAT with default setting).
We deployed a filtration process with a 95% exact match cut-off for the cDNA mapping, in order to exclude paralogous hits and eliminate vector contaminations or sequencing errors in the expression data. Only ∼2% cDNAs had ambiguous hits. For these cDNAs, only the best hit was kept. Based on their mapped genomic loci, we clustered human/mouse RefSeq cDNA and mRNA/EST sequences to define the transcription units. Genomic sequence corresponding to a transcription unit was termed a “genomic transcription unit.” There were, altogether, 12,419 orthologous genomic transcription units identified between the human and mouse genomes.
Exons in genomic transcription units and their inclusion level
We detected alternative splice forms for mouse by mapping mRNA and EST sequences onto genomic transcription units. We used Sim4 to further refine gene structures, in addition to the UCSC mapping data, via BLAT. In order to improve boundary definition of the mRNA/EST coverage regions, we used a P-value test and filtered random mismatches. The two marginal 5′ and 3′ exons within each gene were excluded from further analysis due to their general incompleteness. The inclusion level of each exon (the percentage of all transcripts from a given genomic transcription unit that include this exon) was estimated. Exons with two to five pieces of mRNAs and ESTs data were classed as “low” exons, with only one EST put in “singleton,” and other exons were classed as “constitutive” (100% inclusion), “major” (≥50% inclusion), or “minor” (<50% inclusion).
Exon phase determination
We considered the codon-phase information in the RefSeq cDNAs to be usable to identify an exon's phase. Since earlier, less reliable single-pass expression data may inaccurately indicate more indels than in the nearly finished genomic sequences, we used the genomic counterparts of the mRNA/ESTs rather than the original sequences that we downloaded. The global alignments between the mouse mRNA/EST genomic counterpart and its RefSeq were performed by using CLUSTALW with a parameter GAPEXT = t in consideration of possible alternative splicing. Phase cannot be defined for four kinds of exons as follows: UTR exons, short exons (<60 bp, due to their greater likelilhood of mismatching in the ORF analysis), stop-codon containing exons, and frame-shifted exons. A frame-shifted exon has a frame-shift mutation as compared with its corresponding RefSeq cDNA. Finally, phases of 79,098 mouse exons were determinable, of which 71,039 were also found in the rat; these exons were used in the further analyses.
Search for orthologous exons in human
We attempted to map these 71,039 mouse/rat exons to the corresponding human genomic transcription units by using FASTA. Comparison between the mapped pairs of human-mouse exons within a human genomic transcription unit demonstrated different categories of mouse exons. We defined three categories of mouse exons, i.e., N (“none”) if it has no human homolog; I (“intron”) if >20% or >30 bp of the exon corresponds to a human intronic sequence; or “old exon” if it matches the human exonic region more closely. To further confirm and fish out those exons newly evolved in rodents, we implemented FASTA analysis with a filtration cut-off 1E-2 for N or I category mouse exons by using 743,856 pig ESTs generated by BGI as the outgroup set (BGI, unpubl.) (but the pig EST sequences correspondent to the 71,039 rodent exons are available at http://newexon.genomics.org.cn). If the pig EST coverage of a given mouse exon was >50%, we supposed the exon to be homologous to pig sequence and not newly evolved in the rodent lineage, and vice versa. The phylogeny underlying this strategy for identifying newly evolved exons can be seen in Figure 1.
Calculation and statistics of Ka and Ks
Calculation of Ka and Ks
After annotating ORFs in mouse exons, we identified the orthologous exons in the rat genome via the mouse-rat genome alignment from UCSC (http://genome.ucsc.edu/goldenPath/mmFeb2003/alignments/vsRn2/axtBest/). We calculated Ka and Ks for either individual exons or concatenated exons in each exon category based on mouse-rat alignments, using the Li93 method (Li 1993).
The 95% confidence intervals
Because mouse and rat are closely related species, we treat substitutions as poisson distributed. The maximum-likelihood method was used to compute the 95% confidence intervals for Ks and Ka between mouse and rat orthologous exons. We treat synonymous and nonsynonymous substitutions as poisson processes to estimate the mean value of Ks with the greatest log likelihood (lnLmax), and to estimate the 95% CI of Ks based on lnLmax - 1.92.
Difference of Ka and Ks distributions
We used the generalized linear model (McCullagh and Nelder 1989) to compare the distributions. Usually, we assume that nucleotide substitutions follow a poisson distribution. However, since overdispersion is a possibility, especially for nonsynonymous substitutions, we used a quasipoisson model with its link function log,
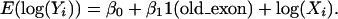 |
For example, to test whether synonymous substitution rates are the same for both old and new exons, Yi and Xi denote the number of synonymous substitutions and sites for each exon, respectively. In the model, log(Xi) is usually called offset; 1(old_exon) is the indication function, taking the value 1 for an old exon or 0 for a new exon. Our goal is to test whether β1 is significantly different from zero.
Gene ontology (GO)
We used Bioverse (McDermott and Samudrala 2003, 2004) to conduct GO annotation to our Ref sequences. Default parameters were used to search databases. Altogether, 7758 RefSeqs (genes) could be given a GO annotation. First, we filtered out those exons with frame shifts or without good ORF-containing homologs in rat, then we excluded those mouse-specific exons found in the pig EST database. Eventually, 3906 genes containing only old exons and 981 genes with newly evolved exons were retained.
Supplementary Material
Acknowledgments
We thank Jian Lu and Hua Tang for their help in the statistics, and Prof. Janice Spofford and Dr. Daniel Neafsey for critically reading the manuscript. W.W. was supported by a CAS-Max Planck Society Fellowship, a CAS key project grant (No. KSCX2-SW-121), a NSFC award (No. 30325016) to distinguished young scientists, and a NSFC key grant (No. 30430400). J.W. and G.K.S.W. were sponsored by the Chinese Academy of Sciences, Commission for Economy Planning, Ministry of Science and Technology (2002AA104250; 2001AA231061; 2004AA231050) and National Natural Science Foundation of China. J.W. was also supported by the Danish National Research Foundation (Danish Platform for Integrative Biology).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3929705. Article published online before print in August 2005.
References
- Ast, G. 2004. How did alternative splicing evolve? Nat. Rev. Genet. 5: 773-782. [DOI] [PubMed] [Google Scholar]
- Brosius, J. and Gould, S.J. 1992. On “genomenclature”: A comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA”. Proc. Natl. Acad. Sci. 89: 10706-10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso, T. and Tautz, D. 2003. An evolutionary analysis of orphan genes in Drosophila. Genome Res. 13: 2213-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, W. 1978. Why genes in pieces? Nature 271: 44. [DOI] [PubMed] [Google Scholar]
- Gilbert, W., de Souza S.J., and Long, M. 1997. Origin of genes. Proc. Natl. Acad. Sci. 94: 7698-7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A.L. and Nei, M. 1988. Patterns of nucleotide substitution at the major histocompatibility complex I loci reveals overdominant selection. Nature 335: 167-170. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- InterPro Consortium 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29: 37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, M.E., Viggiano, L., Bailey, J.A., Abdul-Rauf, M., Goodwin, G., Rocchi, M., and Eichler, E.E. 2001. Positive selection of a gene family during the emergence of humans and African apes. Nature 413: 514-519. [DOI] [PubMed] [Google Scholar]
- Johnson, J.M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P.M., Armour, C.D., Santos, R., Schadt, E.E., Stoughton, R., and Shoemaker, D.D. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302: 2141-2144. [DOI] [PubMed] [Google Scholar]
- Kaessmann, H., Zollner, S., Nekrutenko, A., and Li, W.H. 2002. Signatures of domain shuffling in the human genome. Genome Res. 12: 1642-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, F.A. and Koonin, E.V. 2001. Origin of alternative splicing by tandem exon duplication. Hum. Mol. Genet. 10: 2661-2669. [DOI] [PubMed] [Google Scholar]
- ____ 2003. Evolution of alternative splicing: Deletions, insertions and origin of functional parts of proteins from intron sequences. Trends Genet. 19: 115-119. [DOI] [PubMed] [Google Scholar]
- Letunic, I., Copley, R.R., and Bork, P. 2002. Common exon duplication in animals and its role in alternative splicing. Hum. Mol. Genet. 11: 1561-1567. [DOI] [PubMed] [Google Scholar]
- Li, W.H. 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 36: 96-99. [DOI] [PubMed] [Google Scholar]
- ____ 1997. Molecular evolution. Sinauer Associates, Sunderland, MA.
- Long, M. and Langley, C.H. 1993. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science 260: 91-95. [DOI] [PubMed] [Google Scholar]
- Long, M., Betran, E., Thornton, K., and Wang, W. 2003. The origin of new genes: Glimpes from the young and old. Nat. Rev. Genet. 4: 865-875. [DOI] [PubMed] [Google Scholar]
- Lynch, M. and Conery, J.S. 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151-1155. [DOI] [PubMed] [Google Scholar]
- Makalowski, W., Mitchell, G.A., and Labuda, D. 1994. Alu sequences in the coding regions of mRNA: A source of protein variability. Trends Genet. 10: 188-193. [DOI] [PubMed] [Google Scholar]
- McCullagh, P. and Nelder, J.A. 1989. Generalized linear models. 2nd ed., Chapman & Hall/CRC, London.
- McDermott, J. and Samudrala, R. 2003. BIOVERSE: Functional, structural, and contextual annotation of proteins and proteomes. Nucleic Acids Res. 31: 3736-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2004. Enhanced functional information from protein networks. Trends Biotechnol. 22: 60-62. [DOI] [PubMed] [Google Scholar]
- Mironov, A.A., Fickett, J.W., and Gelfand, M.S. 1999. Frequent alternative splicing of human genes. Genome Res. 9: 1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C. 2002. Alternative splicing in human, mouse, and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 34: 177-180. [DOI] [PubMed] [Google Scholar]
- Modrek, B., Resch, A., Grasso, C., and Lee, C. 2002. Genome-wide detection analysis of alternative splicing using human expressed sequence data. Nucleic Acids Res. 30: 3754-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Murphy, W.J., Eizirik, E., O'Brien, S.J., Madsen, O., Scally, M., Douady, C.J., Teeling, E., Ryder, O.A., Stanhope, M.J., de Jong, W.W., et al. 2001. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294: 2348-2351. [DOI] [PubMed] [Google Scholar]
- Nekrutenko, A. and Li, W.H. 2001. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 17: 619-621. [DOI] [PubMed] [Google Scholar]
- Nurminsky, D.I., Nurminskaya, M.V., De Aguiar, D., and Hartl, D.L. 1998. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature 396: 572-575. [DOI] [PubMed] [Google Scholar]
- Nurtdinov, R.N., Artamonova, I.I., Mironov, A., and Gelfand, M.S. 2003. Low conservation of alternative splicing patterns in the human and mouse genomes. Hum. Mol. Genet. 12: 1313-1320. [DOI] [PubMed] [Google Scholar]
- Patthy, L. 1999. Genome evolution and the evolution of exon shuffling—A review. Gene 238: 103-114. [DOI] [PubMed] [Google Scholar]
- Podlaha, O. and Zhang, J. 2003. Positive selection on protein-length in the evolution of a primate sperm ion channel. Proc. Natl. Acad. Sci. 100: 12241-12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince, V.E. and Pickett, F.B. 2002. Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 3: 827-837. [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493-521. [DOI] [PubMed] [Google Scholar]
- Sorek, R., Ast, G., and Graur, D. 2002. Alu-containing exons are alternatively spliced. Genome Res. 12: 1060-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, M.S., Murphy, W.J., Eizirik, E., and O'Brien, S.J. 2003. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc. Natl. Acad. Sci. 100: 1056-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Brunet, F.G., Nevo, E., and Long, M. 2002. Origin of sphinx, a young chimeric RNA gene in Drosophila melanogaster. Proc. Natl. Acad. Sci. 99: 4448-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Zhang, Y.P., and Rosenberg, H.F. 2002. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 30: 411-415. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Dean, A.M., Brunet, F., and Long, M. 2004. Evolving protein functional diversity in new genes of Drosophila. Proc. Natl. Acad. Sci. 101: 16246-16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.ncbi.nih.gov/pub/HomoloGene/; HomoloGene database for human-mouse orthologous genes in RefSeq format.
- ftp://ftp.ncbi.nih.gov/repository/UniGene/; the human and mouse mRNA/EST data.
- http://genome.ucsc.edu/goldenPath/10april2003/database/; human UniGene database.
- http://genome.ucsc.edu/goldenPath/mmFeb2003/database/; mouse UniGene database.
- http://genome.ucsc.edu/goldenPath/mmFeb2003/alignments/vsRn2/axtBest/; Mouse-rat genome alignment from UCSC.
- http://newexon.genomics.org.cn; the supplemental databases of old, rodent-specific, and newly evolved exons analyzed in this study.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.