Abstract
Heterochromatin protein 1 (HP1) is a major component of heterochromatin. It was reported to bind to a large number of genes and to many, but not all, transposable elements (TEs). The genomic signals responsible for targeting of HP1 have remained elusive. Here, we use whole-genome and computational approaches to identify genomic features that are predictive of HP1 binding in Drosophila melanogaster. We show that genes in repeat-dense regions are more likely to be bound by HP1, particularly in pericentric chromosomal regions. We also demonstrate that TEs are only bound by HP1 if they are flanked by other repeats, suggesting a cooperative mechanism of binding. Genome-wide DamID mapping of HP1 in larvae and adult flies reveals that repeat-flanked genes typically bind HP1 throughout development, whereas repeat-free genes display developmentally dynamic HP1 association. Furthermore, computational analysis shows that HP1 preferentially binds to transcribed regions of long genes. Finally, we detect low but significant amounts of HP1 along the entire X chromosome in male, but not female, flies, suggesting a link between HP1 and the dosage compensation complex. These results provide insights into the mechanisms of HP1 targeting in the natural genomic context.
Heterochromatin was originally defined cytologically as chromosomal regions that remain condensed throughout the cell cycle (Heitz 1928). By this morphological definition, heterochromatic DNA contains large amounts of repetitive sequences and relatively few genes. Reporter genes inserted within or near heterochromatic regions are typically silenced, which has led to the notion that heterochromatin forms a specialized chromatin structure that represses transcription (for review, see Weiler and Wakimoto 1995; Richards and Elgin 2002). Paradoxically, pericentric regions in Drosophila harbor a number of essential genes (e.g., light [lt] and rolled [rl]), which require a heterochromatic environment for their proper expression (Wakimoto and Hearn 1990; Lu et al. 2000). It is unclear why different genes may respond differently to a heterochromatic environment.
Heterochromatin Protein 1 (HP1) is one of the best-studied components of heterochromatin. HP1 is a small protein containing a chromodomain and a chromoshadow domain. The chromodomain binds to methylated lysine at position 9 in the tail of histone H3 (H3K9me) (Bannister et al. 2001; Lachner et al. 2001; Jacobs and Khorasanizadeh 2002; Nielsen et al. 2002). One of the histone methyltransferases responsible for this methylation is Su(var)3-9 (Czermin et al. 2001; Lachner et al. 2001; Nakayama et al. 2001; Schotta et al. 2002). In Drosophila, Su(var)3-9 can also interact directly with HP1 (Schotta et al. 2002). This set of molecular events may constitute a positive feedback loop that ensures the stability of heterochromatin and could facilitate spreading of heterochromatin in cis from a nucleation site (Jenuwein 2001; Richards and Elgin 2002). Although this model may explain the maintenance of heterochromatin, it does not identify the signals that direct the initiation of its formation. In other words, the mechanisms by which heterochromatin proteins are targeted to specific loci in the genome are poorly understood.
Targeting of HP1 may in part be mediated by sequence-specific DNA binding factors or other chromatin-associated proteins. Several transcriptional regulators have been reported to interact with HP1 and direct it to promoters or other elements (Matsuda et al. 2001; Nielsen et al. 2001; Verdel et al. 2004). Repetitive sequences also appear to play a prominent role in heterochromatin formation. This was originally suggested by the observation that heterochromatic chromosomal regions typically contain a large variety of repetitive sequences. Furthermore, multiple insertions or local duplication of a transgene in a euchromatic environment, causing the formation of a tandem or inverted repeat array, can lead to variegated expression of this transgene, which is a hallmark of heterochromatic silencing (Dorer and Henikoff 1994; Sabl and Henikoff 1996). Staining of polytene chromosomes revealed that HP1 binds to arrays of P-element insertions (Fanti et al. 1998). These and other observations have led to the hypothesis that heterochromatin is formed at repeats irrespective of the DNA sequence (for review, see Henikoff 1998).
Interestingly, the establishment of heterochromatin by at least some repeats appears to involve the RNA interference (RNAi) machinery. In Schizosaccharomyces pombe, it has been shown that the homologs of HP1 and Su(var)3-9 (Swi6p and Clr4p, respectively) are recruited to centromeric repeats and that this recruitment depends on the RNAi pathway (Volpe et al. 2002). Deletion of components of the RNAi pathway in Drosophila also alleviated silencing of a heterochromatic reporter and caused changes in the chromosomal distribution of HP1 (Pal-Bhadra et al. 2004). It is thought that small interfering RNA (siRNA) molecules may direct heterochromatin proteins to the repetitive DNA (Verdel et al. 2004). It is unclear, however, whether other sequence or chromatin features play a role in determining the locus specificity of heterochromatin formation.
Previously we reported genome-wide binding maps for HP1 and Su(var)3-9 in Drosophila Kc167 cells (Greil et al. 2003). Out of more than 6000 probed genes, 152 showed significant binding of HP1. Aside from a modest enrichment of A/T-rich motifs in the target genes, no clear sequence motif was identified that might mediate the targeting of HP1. Thus, the signal(s) responsible for the specific targeting of HP1 to this large set of genes remained unidentified. In addition, association of HP1 was observed at many, but not all (about 50%), of the probed transposable element (TE) sequences. This suggested that an unknown characteristic of TEs determines whether they are bound by HP1 or not. In summary, the genomic signals by which HP1 finds its natural set of target genes and TEs in Drosophila have remained largely elusive.
Here, we used computational analyses and genome-wide mapping experiments in Drosophila to uncover genomic features that are linked to the targeting of HP1. First, we show that genes in repeat-dense regions are more likely to be bound by HP1, particularly in pericentric chromosomal regions. Genome-wide mapping experiments in larvae and adult flies reveal that HP1 binding in repeat-rich regions is stable throughout fly development, whereas HP1 binding in repeat-free regions is developmentally dynamic. Second, we report surprising evidence that HP1 preferentially associates with long genes. Third, we show that HP1 binds along most of the X chromosome in male but not female flies, suggesting a link with the male-specific dosage compensation system. Thus, we have identified three genomic signals that are likely to contribute to the recruitment of HP1 to its natural target genes.
Results
Flanking repeats predict HP1 binding to pericentric but not to non-pericentric genes
Repeats might nucleate the formation of heterochromatin, which could then spread in cis to cover neighboring genes. We first took a bioinformatics approach to investigate whether this model could account for the previously observed HP1 binding to single-copy genes. We used the reported genomic map of HP1 binding (Greil et al. 2003), which was obtained using the DamID technique (van Steensel and Henikoff 2000; van Steensel et al. 2001). This data set, together with the nearly complete sequence of the Drosophila genome, provided a strong basis for statistical analysis of the role of repeats in HP1 targeting to genes in their natural context.
To ensure an unbiased analysis, we broadly defined repeats as all non-unique sequences in the fly genome, irrespective of their copy number, distribution, or orientation. We identified these repeats by aligning the genome of D. melanogaster to itself, using BLAST (Altschul et al. 1990). For each probed gene, we then determined the fraction of repeat sequence within the probed sequence and the flanking sequence (see Methods for more details). We will refer to this measure as Flanking Repeat Index (FRI). The FRI value can theoretically range from 0 (exclusively unique sequences) to 1 (exclusively non-unique sequences). Initially, we chose 20 kb of flanking regions on either side of the probe, which is about twice the distance over which repeats can affect gene expression in fission yeast (Schramke and Allshire 2003).
We found that HP1 target genes are located much more frequently in repetitive regions than non-target genes are (Fig. 1A): 51% of all HP1 target genes have FRI20kb values >0.1 (i.e., more than 10% of the sequence flanking the probes is non-unique sequence), whereas for non-targets this fraction is only 14%. This suggests that about half of the targets of HP1 might be explained by a relatively high density of repetitive sequences in their proximity.
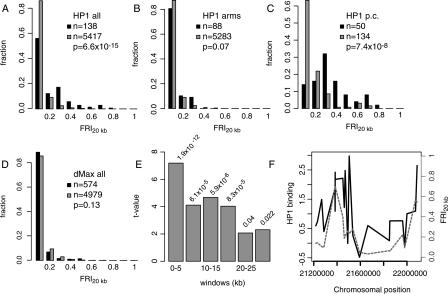
Figure 1.
Binding of HP1 and Su(var)3-9 to genes in pericentric regions is correlated with flanking repeats. (A-D) Histograms of the distributions in FRI20kb for single-copy target genes (black bars) and non-target genes (gray bars) of HP1 (A-C) and dMax (D). (A) All probed genes on the arms and in the pericentric regions; (B) genes on the chromosome arms only, i.e., non-pericentric genes; (C) pericentric genes only. P values in A-D were calculated using the Wilcoxon rank-sum test. (E) The t values for linear regression of HP1 binding vs. FRI for windows at various distances from the probe; corresponding P values are printed above each bar. (F) A detailed view of the pericentric region of chromosome 2L, with HP1 binding (solid line, log2 ratio) and the FRI20kb (dashed line) plotted for all probes in this region.
We tested whether this association between HP1 binding and density of flanking repeats is similar in pericentric and non-pericentric chromosomal regions. For this purpose, we defined pericentric regions as the most centromere-proximal 1 Mb of the sequenced part of each chromosome arm (Greil et al. 2003). Surprisingly, in non-pericentric regions this association between HP1 binding and flanking repeats is virtually absent (Fig. 1B): Here, only 19% of the target genes have an FRI20kb value >0.1. This is not significantly different from the non-pericentric genes that do not bind HP1 (13%). Conversely, the vast majority of probed non-pericentric genes with FRI20kb >0.1 (680/697) do not bind HP1. However, we did identify a locus on the left arm of chromosome 2 that is highly suggestive of repeat-associated binding. In cytogenic region 25D (within the predicted gene CG31916) are four arrays of tandem repeats, to which HP1 is bound (see Supplemental Fig. S1). Thus, although exceptions occur, in non-pericentric regions, flanking repeats do not appear to play a major role in HP1 binding to genes.
This strongly contrasts with the situation in pericentric regions, where 86% of the HP1 target genes have an FRI20kb value >0.1, as opposed to 37% of the non-target genes (Fig. 1C). This trend is illustrated by the pericentric region of chromosome 2L (Fig. 1F), where two regions of high repeat density are interrupted by a repeat-poor region; this pattern of repeat density closely correlates with HP1 binding. As a control, we also carried out this analysis for genome-wide binding data of the transcription factor dMax (Orian et al. 2003). We find that dMax has no preference for genes flanked by repeats, indicating that this phenomenon is specific for HP1 (Fig. 1D). In summary, these results suggest that the majority of HP1 binding to genes in pericentric regions can be explained by the presence of repeats in the flanking regions, whereas in non-pericentric regions most of the HP1 binding occurs irrespective of the presence of repeats.
To estimate the distance over which repeats might contribute to HP1 binding in pericentric regions, we calculated FRI values for windows of 5 kb at various distances from each probed locus. Subsequent linear regression analysis revealed that repeats within 5 kb show the strongest correlation with HP1 binding (Fig. 1E). In windows between 5 and 20 kb, correlations are still highly significant, but beyond 20 kb we find that repeat density has no predictive power for HP1 binding. Thus, in pericentric regions, repeats may positively affect HP1 binding to genes over a range of ∼20 kb, but we could not detect effects over longer distances.
Detailed mapping of HP1 binding in the pericentric gene rolled
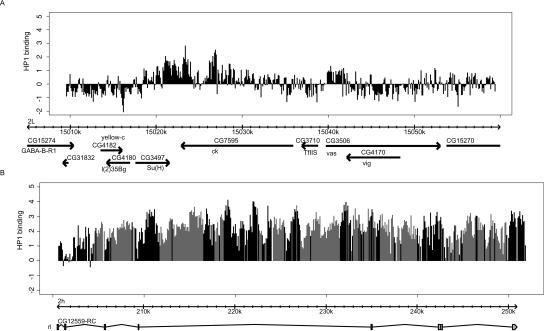
To study the binding of HP1 to repeat-dense regions in more detail we designed a genomic tiling array covering the gene rolled. This gene is located in pericentric heterochromatin and was previously shown to depend on a heterochromatic environment for its correct expression (Eberl et al. 1993). Consistent with our hypothesis that repeats are involved in HP1 recruitment to pericentric genes, rolled contains large numbers of repeats (mostly TEs). Within the entire ∼50-kb gene we chose 60-mer oligonucleotides separated by intervals of 40 bp. For comparison, we selected a 50-kb region surrounding the ck gene, which is located on the arm of chromosome 2L. Previous mapping in this region had shown that HP1 binds to the ck gene but not to most of the surrounding regions (Greil et al. 2003; Sun et al. 2003). Indeed, this observation was confirmed by our new high-resolution oligonucleotide array (Fig. 2A), although HP1 binding appears to extend from the ck gene into the neighboring Su(H) gene. This difference may be explained by slightly different cell culture conditions or by a higher sensitivity obtained with the new array.
Figure 2.
HP1 binding detected by genomic tiling arrays. Data from a tiling array with 60 nt probes every 100 bp (40-bp spacing). (A) HP1 binding in a region surrounding the ck gene. (B) HP1 binding to the rl locus in the pericentric region of chromosome 2. Black bars represent probes that are unique in the genome; gray bars represent probes that gave multiple BLAST results in the fly genome. The height of the bars represents the log2 ratio of HP1-binding (Dam-HP1 over Dam-only). The data has been normalized to the median of the log2 ratios of the probes in the 50-kb ck region.
Strikingly, binding of HP1 to the rolled gene was very strong (Fig. 2B) and encompassed essentially the entire gene. HP1 binding was as prominent in unique sequences as in repeat sequences embedded within this gene. We conclude that there is ubiquitous binding of HP1 to the transcribed part of the rolled gene.
TEs located in repetitive regions are more likely to be bound by HP1
Next, we investigated the association of HP1 with TEs. Our previous DamID study had revealed that HP1 binds to about half of 85 TE probes that were present on our microarray (Greil et al. 2003). To identify the signals that determine HP1 binding to TEs, we performed a detailed computational analysis of these data. We found no association between HP1 binding and general features such as TE class, length, or copy number (data not shown).
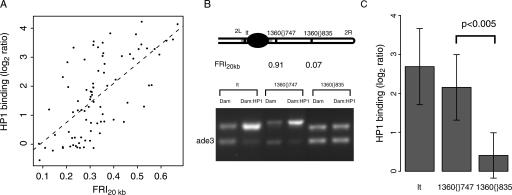
Next, we investigated the link between HP1 binding levels and repeats surrounding the TEs. The reported HP1 binding data for TEs were obtained by hybridizations with probes that cannot discriminate between individual TE copies in the genome (Greil et al. 2003) and thus represent the average binding across all copies of each TE. Accordingly, we calculated the average FRI20kb across all copies of a given TE. Strikingly, we found a strong correlation between the average HP1 binding and the average FRI20kb of TEs (Fig. 3A). Thus, flanking repeats are a strong predictor of HP1 association with TEs.
Figure 3.
HP1 binding to transposable elements is dependent on flanking repeats. (A) HP1 binding to TEs as a function of the FRI20kb. HP1 binding and FRI20kb for cDNAs on the array that contain sequences homologous to any of the consensus TE sequences (Kaminker et al. 2002) are plotted here. The dotted line shows the linear trendline through the data (Spearman rank correlation ρ = 0.62, P < 2.2 × 10-16). (B) Detection of HP1 binding by DamID and duplex PCR performed on two copies of the same transposable element (1360), both located on chromosome 2R, and the pericentric gene lt, a known target of HP1 (Greil et al. 2003). See Methods for a detailed description of the assay. The top bands correspond to the tested sequence, the bottom bands correspond to ade3, which does not bind HP1 (van Steensel and Henikoff 2000) and serves as an internal standard. (C) Quantitation of the HP1 status. Band intensities were quantified and normalized to the internal standard ade3. Average log2 ratios between Dam:HP1 and Dam reference were calculated for the two transposon copies (n = 5 and n = 6, respectively) and lt (n = 5). Difference between 1360{}747 and 1360{}835 is significant (P < 0.005, Student's t-test).
From this it can be hypothesized that for a given TE not all copies have the same degree of HP1 binding; rather, the presence or absence of flanking repeats may determine HP1 binding to a given TE insertion. We tested this hypothesis by measuring HP1 binding at individual copies of the 1360/hoppel transposon (Kaminker et al. 2002). We compared two non-pericentric copies of 1360 on chromosome 2R: 1360{}747, which is in cytological region 42B1-42B2 (roughly 1.5 Mb from the proposed heterochromatin-euchromatin boundary) (Myster et al. 2004) and flanked by many repeats, and 1360{}835 which is located in region 51F11 and has a low FRI20kb (Fig. 3B). To detect HP1 binding at these individual TE copies, we used DamID combined with a semi-quantitative PCR assay (Greil et al. 2003). To ensure that only the TE at the desired location was probed, we selected one primer within the transposon and the second primer in the unique sequence flanking the transposon. The results (Fig. 3, B and C) show that the level of HP1 binding is clearly much higher for the 1360 insertion in repeat-dense DNA than for the copy that is in a repeat-poor region. The level of HP1 binding for 1360{}747 is almost as high as in the classical HP1 target gene lt (Hearn et al. 1991; Greil et al. 2003), whereas HP1 binding at 1360{}835 was essentially as low as in the non-target gene ade3. These results strongly support our computational prediction that not all copies of a given TE are bound by HP1 but that instead the binding of HP1 to TEs is largely dependent on flanking repeats.
HP1 binds preferentially to long genes
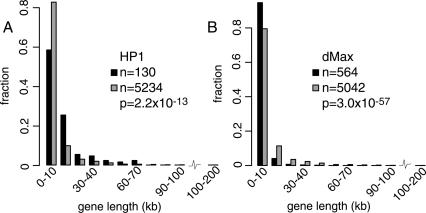
Because repeats can only explain binding of HP1 to ∼50% of its target genes, we reasoned that one or more other signals must exist that are responsible for HP1 recruitment. We therefore searched for other genomic features that correlate with HP1 binding. While inspecting HP1 target genes, we noticed that they often are very long. Indeed, statistical analysis revealed that targets of HP1 showed a clear bias towards longer genes (Fig. 4A). This bias was found both in pericentric and non-pericentric regions (P = 7.8 × 10-6 and P = 3.6 × 10-6, respectively). Multivariate regression revealed that gene length adjusted for repeat density still shows significant association with HP1 binding (data not shown), indicating that the recruitment of HP1 cannot simply be explained by TEs or other repeats in the introns of long genes. We considered the possibility that long genes more often overlap with other genes in the antisense orientation (e.g., in the introns of long genes), which could lead to the formation of double-stranded RNA and subsequent RNAi-mediated HP1 recruitment. However, we found that HP1 has no detectable preference for overlapping genes (data not shown). The unexpected bias of HP1 binding for longer genes, irrespective of chromosomal location, suggests a distinct targeting mechanism, in addition to the repeat-associated recruitment.
Figure 4.
Gene-length distributions for targets and non-targets of HP1. Histograms of the distributions in gene length for target genes (black) and non-target genes (gray) for HP1 (A) and control protein dMax (B). Note that sample size is different from sample size in Figure 1 because it was not possible to obtain gene length information for every clone on the microarray.
This result also sheds additional light on the link between repeats and HP1 binding to genes. Although Figure 2B clearly shows that binding of HP1 occurs within the rolled gene, it was formally possible that at many genes in repeat-dense regions HP1 in fact only binds to repeats located just outside the transcription units, and that DamID signals detected by the gene probes are due to the limited resolution of the DamID method, caused by cis-spreading of targeted dam methylation over 2-3 kb (van Steensel and Henikoff 2000). However, we reasoned that in this scenario, binding of HP1 would be more efficiently detected with cDNA probes representing short genes (e.g, <2 kb) compared with long genes (e.g., >10 kb), because the majority of probed sequence in long genes would lie outside the spreading range of targeted methylation. Indeed, target genes of dMax (a transcription factor that preferentially binds to promoter regions) (Grandori et al. 2000) show a clear bias for short genes (Fig. 4B). The fact that the detected HP1 binding is biased towards long genes argues that HP1 must bind to transcribed regions rather than to flanking repeats only.
Developmental plasticity of HP1 binding
Next, we asked whether the HP1 binding pattern is the same for all cell types or dynamic during development. We used DamID to generate genome-wide maps of HP1 in female third instar larvae and in male and female adult flies. To verify specific targeting of the Dam-fused HP1 we first performed immunofluorescence microscopy of methyl-adenine (m6A) in polytene chromosomes of larvae expressing Dam-HP1 (Supplemental Fig. S2). These experiments revealed specific methylation of regions where HP1 is known to bind, such as pericentric regions and chromosome 4 (Fanti et al. 2003). Thus, Dam-HP1 binds and methylates native HP1 targets in vivo. For larvae and both adult sexes we then performed DamID on four separate individuals each. cDNA microarrays were used for detection of the targeted methylation patterns (for details, see Methods). Comparison of the replicates indicates that DamID in whole animals is highly reproducible, with average pairwise correlations between individual experiments ranging from 0.8 to 0.9.
The general properties of the larval and adult HP1 binding data are similar to those in Kc cells (Greil et al. 2003). First, HP1 is frequently associated with TEs. In larvae, 50 out of 85 TEs are bound by HP1; in adults, 63 of the TEs are bound by HP1. Second, we observe a strong enrichment of target genes on chromosome 4 (larvae: 38/48, adult: 46/48). Third, HP1 is frequently associated with pericentric genes (Supplemental Fig. S3). Fourth, similar to Kc cells, there is an association between genes bound by HP1 in whole animals and the FRI20kb (data not shown). Thus, HP1 shows globally similar binding patterns in Kc cells, larvae, and adults.
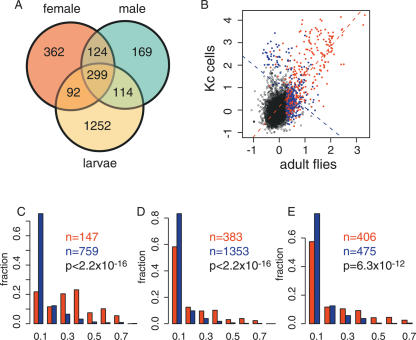
There are, however, also marked differences. In addition to the significant overlap between the HP1 target genes, there are also many genes for which HP1 binding could only be detected in one developmental stage or sex (Fig. 5A). For example, ∼44% of HP1 targets in male adults were not detected as targets in female adults. Similarly, differences were observed between larvae and adults. This indicates that the HP1 binding pattern is subject to considerable developmental regulation.
Figure 5.
Repetitive regions are associated with developmentally stable HP1 binding. (A) Venn diagram showing the number of overlapping and non-overlapping HP1 target genes in adult males, adult females, and female larvae. (B) Scatterplot of the binding log2 ratios of HP1 in whole adult flies versus HP1 binding in Kc cells. Colored circles indicate targets of HP1 either in adult flies or in Kc cells or in both. Blue circles indicate cDNAs that have an FRI20kb < 0.05, red circles are cDNAs that have an FRI20kb > 0.05. The broken lines are the regression lines through the blue and red data points. (C) The distributions of the FRI20kb for the genes that are bound by HP1 in both Kc cells and male adult flies (red) and for the genes that are bound by HP1 in either Kc cells or male adult flies, but not in both (blue). (D) The distributions of the FRI20kb for the genes that are bound by HP1 in both female larvae and in male adult flies (red) and for the genes that are bound by HP1 in either female larvae or male adult flies (blue). (E) The distributions of the FRI20kb for the genes that are bound by HP1 in both male and female adult flies (red) and for the genes that are bound by HP1 in either male or female adult flies, but not in both (blue).
Stable HP1 binding is associated with repetitive DNA
Because repeats serve as a potential HP1 recruitment signal, we investigated their association with HP1 binding in the various developmental stages. In Figure 5B, HP1 binding in male adult flies is plotted against HP1 binding in Kc cells. Two populations of HP1 target genes can be identified: invariant targets, which are bound by HP1 in both Kc cells and in adults, and dynamic targets, which are bound by HP1 in either Kc cells or adult flies, but not in both. Strikingly, the invariant target genes are generally located in repeat-dense regions, whereas the majority of dynamic targets are not flanked by repeats. This difference is illustrated by regression lines through the repeat-rich and repeat-free targets, which have an almost perpendicular slope (Fig. 5B). The distribution of the FRI20kb for the dynamic and invariant target genes is shown in Figure 5C. The vast majority (72%) of the dynamically bound genes have an FRI20kb less than 0.05, whereas for the invariant target genes this is only 17%. Thus, invariant binding by HP1 is associated with repeats, whereas dynamic binding of HP1 occurs independent of repeats.
Kc cells are genotypically different from the fly line that we used for whole-animal DamID. To test whether the differences in the repeat make-up that might exist between these two geno-types could account for the observations, we performed the same comparative analysis for the HP1 binding data from larvae and adults (Fig. 5D) and data from male and female adults (Fig. 5E), which all have the same genetic background. In both comparisons, we observe an enrichment for genes with a higher FRI20kb among the invariant targets compared with the dynamic targets. In conclusion, these results show that developmentally stable HP1 target genes are generally located in repeat-dense regions, whereas developmentally dynamic HP1 targets occur mostly in repeat-free regions.
HP1 is preferentially recruited to the male X chromosome
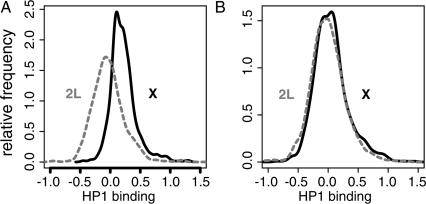
When we compared the binding levels of HP1 in male adults on the various chromosomes we made a striking observation. Along the entire X chromosome in male adults, HP1 binding is moderately elevated compared with HP1 binding in the autosomes, with the exception of chromosome 4 (Fig. 6A and data not shown). This increased binding is statistically highly significant (P < 2.2 × 10-16, Wilcoxon rank-sum test) and appears to be due to a general shift in the “baseline” of HP1 binding rather than to strong binding to a subset of genes on the X chromosome. This phenomenon was observed in four independent experiments. PCR with Y-chromosome specific primers confirmed that all flies in this set were male (data not shown). Interestingly, in adult females no such X-specific enrichment could be detected (Fig. 6B). Thus, these results reveal a surprising male-specific recruitment of HP1 to most of the X chromosome.
Figure 6.
HP1 binding is enriched at the male X chromosomes. Distribution of the HP1 binding levels of all probed genes on the X chromosome (solid line) and on chromosome 2L (dashed line) in adult males (A) and adult females (B).
HP1 binding and gene expression
Finally, we investigated the global link between HP1 binding and gene expression during development. Comparison of our HP1 binding profiles with a recently published developmental expression database (Stolc et al. 2004) reveals that in all three developmental stages, genes bound by HP1 show a lower level of expression than genes not bound by HP1 (Supplemental Fig. S4). Although the differences in expression are less than twofold on average, they are statistically highly significant. Note that these analyses are based on HP1 binding and gene expression profiles that are derived from whole animals. Thus, variations between tissues may occlude stronger links between HP1 binding and gene expression. However, a similarly modest link was previously observed in Kc cells (Greil et al. 2003), which form a homogeneous cell population. Therefore, we suggest that HP1 is primarily involved in fine tuning of gene expression.
Discussion
We have performed a series of whole-genome mapping experiments and computational analyses to identify the genomic signals that are involved in the recruitment of HP1 to its natural targets. We found evidence for three genomic features that are associated with HP1 binding. First, in pericentric regions, HP1 preferentially binds to genes in repeat-dense regions. This binding is generally stable throughout development and between sexes. Second, HP1 binding shows a bias for long genes, both in pericentric regions and on the chromosome arms. Most genes on the chromosome arms show a strong degree of developmental regulation of HP1 binding. Third, we find a surprising association of HP1 with the male but not the female X chromosome. These results suggest that there are several different mechanisms of HP1 recruitment.
The correlation of HP1 binding with repeat density is restricted to pericentric regions and could only be detected for repeats located within ∼20 kb from the genes. On the chromosome arms, however, we did not find a general association between HP1 target genes and repeats. With only a few exceptions, most non-pericentric genes flanked by repeats lack detectable levels of HP1. This is in agreement with the observation that a tandem array of a reporter gene is more strongly silenced when it is integrated close to pericentric heterochromatin (Dorer and Henikoff 1994). Photobleaching experiments in mouse cells also indicate that HP1 is more stably associated with pericentric regions than with non-pericentric regions (Cheutin et al. 2003; Festenstein et al. 2003). It is possible that the centromere itself somehow contributes to an environment that supports HP1 binding. Alternatively, the preference for pericentric regions may be explained by cooperative interactions between heterochromatin complexes. In this model, the high overall density of repeats in pericentric regions would facilitate this cooperative binding. Some non-pericentric repeats may also recruit HP1, provided that the local repeat concentration is high enough to sustain cooperative binding of HP1-complexes. Examples of this include the gene CG31916 (Supplemental Fig. 1), which contains long stretches of repeats, region 42AB (Fig. 2B-C) and the bwD locus, in which ∼1 Mb of satellite repeat sequence is integrated (Platero et al. 1998).
It has been reported that in fission yeast a single TE-derived LTR is able to bind HP1 and control the expression of neighboring genes (Schramke and Allshire 2003). In contrast, our data indicate that in Drosophila a TE by itself (either of the LTR or non-LTR type) cannot serve as a nucleation site for heterochromatin. Instead, a TE must be flanked by other repeats to bind HP1, suggesting that cooperative interactions are required for stable HP1 binding. The 1360 transposable element in Drosophila was proposed to be a nucleation site for heterochromatin formation on the fourth chromosome (Sun et al. 2004). Our results suggest that this can indeed be the case in repeat-rich regions (such as the fourth chromosome) but not in repeat-poor regions.
We provide evidence that developmentally stable recruitment of HP1 is linked to repeats, whereas developmentally dynamic HP1 binding occurs primarily in repeat-free genomic regions. One putative repeat-independent signal for HP1 recruitment is related to gene length: HP1 target genes are significantly longer than non-target genes. Our results also strongly argue that HP1 interacts with transcribed parts of many of its target genes. The reason for the preference of HP1 for long genes remains speculative. In Arabidopsis thaliana, DNA methylation has been proposed to mediate repression of cryptic promoters within transcribed regions (Tran et al. 2005). DNA methylation in Drosophila appears to be limited to the early embryo stage (Lyko et al. 2000), suggesting that a different mechanism may be needed for the repression of cryptic promoters. We suggest that HP1 may be part of this alternative mechanism. Cryptic promoters, if oriented in the antisense direction, could cause the formation of double-stranded RNA and RNAi-mediated recruitment of HP1, creating a negative feedback loop that could help to repress cryptic promoters. By chance, cryptic promoters may occur more frequently in long genes than in short genes, which would explain the preference of HP1 for long genes.
Another striking feature of HP1 that we discovered is its preference for the X chromosome in males. It is possible that this reflects precocious condensation of the X chromosome during spermatogenesis (Lifschytz and Lindsley 1972), which could involve HP1. However, germ cells in meiotic prophase make up only a small fraction of the entire fly body, and it is questionable whether HP1 binding in these cells would be detectable in whole-fly DamID experiments. Alternatively, heterochromatin proteins such as HP1 may have a role in the X chromosome in male somatic cells. This is supported by the observation that mild overexpression of the heterochromatin protein Su(var)3-7 causes male-specific hypercompaction of the X chromosome in salivary gland polytene chromosomes (Delattre et al. 2004). Interestingly, transcription of genes on the Drosophila male X chromosome is increased twofold to compensate for the fact that males have only one copy of this chromosome. This is mediated by the dosage compensation complex (DCC), a protein complex that is specifically associated with most parts of the X chromosome in males only (Akhtar 2003; Gilfillan et al. 2004). The simultaneous binding of both activating (DCC) and repressive protein complexes (HP1) to the male X chromosome would be paradoxical. Possibly, HP1 and the DCC might together be involved in a balancing act to achieve precise twofold elevation of gene expression on the male X chromosome. Further experiments are needed to elucidate the role of heterochromatin proteins on the male X chromosome.
Methods
DamID data and genome annotations
Statistical analyses were performed in the R language (http://www.r-project.org). For all computational analyses we used published DamID data sets (Greil et al. 2003; Orian et al. 2003; van Steensel et al. 2003). The definition of target gene was adopted from these analyses. We used the release 3 sequence of the D. melanogaster genome (Celniker et al. 2002). Annotation data for gene length was extracted from release 3.1 annotation files from the Berkeley Drosophila Genome Project (BDGP) ftp://flybase.net/genomes/Drosophila_melanogaster/dmel_RELEASE3-1/GFF/whole_genome_annotation_dmel_RELEASE3-1.GFF.gz. We defined pericentric regions as the proximal 1 MB of each chromosome arm and the sequences that are annotated as 2h, 3h, Xh, Yh, and U. The remainder of the genome was defined as non-pericentric. The results in Figure 1 were essentially the same if we defined pericentric regions as the proximal 0.8 MB of each chromosome arm (data not shown).
TE classification, length, and copy number within the sequenced parts of the genome were taken from Kaminker et al. (2002), supplemented with data from http://www.ensembl.org. For the results in Figure 2A we did BLAST alignments of the consensus sequences of all TEs (Kaminker et al. 2002) (http://www.fruitfly.org/p_disrupt/datasets/NATURAL_TRANSPOSABLE_ELEMENTS.fa) to the genome (including 2h, 3h, Xh, Yh, and U) to determine the location of the TEs and TE-derived sequences.
Absolute mRNA levels were taken from Stolc et al. (2004). This data set contains probes for every predicted exon in the genome. For every gene we calculated the average log2 expression of its exons, to make it compatible with our data set.
Flanking repeat index
To calculate the FRI, we first identified all non-unique sequences (repeats) in the genome, by performing a BLAST search with the D. melanogaster genome against itself, using MegaBLAST v2.2.5 (NCBI) (Zhang et al. 2000) (minimum alignment length: 28 nt; minimum identity: 78.86%). Next, we also BLASTed the cDNA sequences represented on the microarray against the fly genome. For every BLAST alignment of a given cDNA the fraction of repeats in the alignment and in a window (e.g., 20 kb) 5′ and 3′ of the alignment was determined. A weighted average of these fractions was calculated for the cDNA, using the length of each aligned segment as a weighing factor. Choosing different cut-offs for the minimal repeat length (50 nt and 200 nt) did not significantly alter our results (data not shown).
Whole-animal DamID: Plasmid construction, germline transformation, and DamID
The EcoRI/XbaI fragment encoding the Dam-myc_tag-HP1 fusion protein was excised from pDamM-HP1 (van Steensel and Henikoff 2000) and cloned into pUAST (Brand and Perrimon 1993), resulting into pUDamM-HP1. This vector was microinjected together with transposase plasmid p25.7wcΔ2-3 into w1118 embryos as described (Spradling and Rubin 1982). Survivors were crossed to w1118 flies, and transformants were identified in the progeny by eye pigmentation.
For the DamID experiments, Dam-only expressing flies (line Me4) (Wines et al. 1996) and Dam-HP1 flies were crossed to bwD sp flies. Of the heterozygous progeny, third instar larvae and flies (<24 h after eclosion) were collected and frozen at -80°C. Genomic DNA was isolated by grinding individual flies in 100 μl of 10 mM Tris-HCl, 10 mM EDTA, 100 mM NaCl, 0.5% SDS, followed by incubation for 2 hours at 65°C and subsequent phenol/chloroform/i-amylalcohol extraction. DNA was recovered by ethanol precipitation, dried briefly, and used in the PCR-based DamID protocol as described (Greil et al. 2003). In each microarray hybridization, the experimental sample consisted of amplified DNA from a single Dam-HP1 larva or fly, and the reference sample consisted of pooled amplified DNA from four separately processed sex-matched Dam-only flies or larvae. In each case, four Dam-HP1 animals were analyzed, with two hybridizations in Cy3/Cy5 dye orientation and two in Cy5/Cy3 dye orientation to avoid dye bias. cDNA microarrays have been described (Greil et al. 2003).
DamID of HP1 on tiling arrays
For DamID mapping of HP1 binding to the rl and ck loci, Kc167 cells were cultured in Shields and Sang M3 insect medium, supplemented with 2.5 g/L bacto-peptone, 1 g/L yeast extract, and 5% heat-inactivated fetal calf serum. Cells were transfected with Dam-HP1 or Dam-only by electroporation. After 24 h, genomic DNA was isolated using Qiagen DNeasy tissue kit and processed for DamID as described (Greil et al. 2003). Samples were hybridized to custom 60-mer oligonucleotide arrays (NimbleGen Systems of Iceland, LLC).
Duplex PCR
To monitor the amount of HP1-targeted methylation in specific regions of the genome, we performed duplex PCR on methylated DNA samples, as described (Greil et al. 2003). We used the ade3 gene as an internal standard for the duplex PCR, because this gene is not bound by HP1. As input for our PCR, we use pre-amplified methylated DNA obtained from cells expressing either Dam-HP1 or Dam alone (Greil et al. 2003). We used 1.5-2.2 ng of DNA as starting material for the duplex PCR. A detailed protocol is available on request. We used 1360{}747 forward primer: TTCTCGCATGGTGCCAATTGAT, reverse primer: CGCGAAGTT GTGTGGTTTAAGAGTTG; 1360{}835 forward primer: CCAAAG CACGAATTAAATGAGTATGTTAAG, reverse primer: TCATCTT GCGGCGTGTAAA; lt forward primer TTCCCAAAGGACTTTGT CATTGCCT; reverse primer: TAAGCCACACCCCAATAAA TCGGCT; ade3 forward primer TGGTATGAACTACAAATTAGC TACCACCACGA; and reverse primer TGCTTATGCAATTTC TAAGAATAACCATGCAA.
Supplementary Material
Acknowledgments
We thank C. Moorman for providing DNA samples from transfected Kc cells; R. Lührmann for the m6A-antibody; K. Ahmad, P. Talbert, and S. Henikoff for help with the generation of the fly lines; G. Hart for statistical advice; J. Delrow for providing cDNA arrays; S. Henikoff for suggesting the presence of cryptic promoters in long genes; D. Vermaak, H. Malik, H. Bussemaker, A. Schinkel, and members of the van Steensel laboratory for helpful suggestions. This work was supported by the “Epigenome Network of Excellence” and by an EURYI Award to B.v.S.
Footnotes
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: C. Moorman, R. Lührmann and J. Delrow.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3198905. Article published online before print in August 2005.
References
- Akhtar, A. 2003. Dosage compensation: An intertwined world of RNA and chromatin remodelling. Curr. Opin. Genet. Dev. 13: 161-169. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Bannister, A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., and Kouzarides, T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120-124. [DOI] [PubMed] [Google Scholar]
- Brand, A.H. and Perrimon, N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401-415. [DOI] [PubMed] [Google Scholar]
- Celniker, S.E., Wheeler, D.A., Kronmiller, B., Carlson, J.W., Halpern, A., Patel, S., Adams, M., Champe, M., Dugan, S.P., Frise, E., et al. 2002. Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: research0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin, T., McNairn, A.J., Jenuwein, T., Gilbert, D.M., Singh, P.B., and Misteli, T. 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299: 721-725. [DOI] [PubMed] [Google Scholar]
- Czermin, B., Schotta, G., Hulsmann, B.B., Brehm, A., Becker, P.B., Reuter, G., and Imhof, A. 2001. Physical and functional association of SU(VAR)3-9 and HDAC1 in Drosophila. EMBO Rep. 2: 915-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre, M., Spierer, A., Jaquet, Y., and Spierer, P. 2004. Increased expression of Drosophila Su(var)3-7 triggers Su(var)3-9-dependent heterochromatin formation. J. Cell Sci. 117: 6239-6247. [DOI] [PubMed] [Google Scholar]
- Dorer, D.R. and Henikoff, S. 1994. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell 77: 993-1002. [DOI] [PubMed] [Google Scholar]
- Eberl, D.F., Duyf, B.J., and Hilliker, A.J. 1993. The role of heterochromatin in the expression of a heterochromatic gene, the rolled locus of Drosophila melanogaster. Genetics 134: 277-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti, L., Dorer, D.R., Berloco, M., Henikoff, S., and Pimpinelli, S. 1998. Heterochromatin protein 1 binds transgene arrays. Chromosoma 107: 286-292. [DOI] [PubMed] [Google Scholar]
- Fanti, L., Berloco, M., Piacentini, L., and Pimpinelli, S. 2003. Chromosomal distribution of heterochromatin protein 1 (HP1) in Drosophila: A cytological map of euchromatic HP1 binding sites. Genetica 117: 135-147. [DOI] [PubMed] [Google Scholar]
- Festenstein, R., Pagakis, S.N., Hiragami, K., Lyon, D., Verreault, A., Sekkali, B., and Kioussis, D. 2003. Modulation of heterochromatin protein 1 dynamics in primary Mammalian cells. Science 299: 719-721. [DOI] [PubMed] [Google Scholar]
- Gilfillan, G.D., Dahlsveen, I.K., and Becker, P.B. 2004. Lifting a chromosome: Dosage compensation in Drosophila melanogaster. FEBS Lett. 567: 8-14. [DOI] [PubMed] [Google Scholar]
- Grandori, C., Cowley, S.M., James, L.P., and Eisenman, R.N. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16: 653-699. [DOI] [PubMed] [Google Scholar]
- Greil, F., van der Kraan, I., Delrow, J., Smothers, J.F., de Wit, E., Bussemaker, H.J., van Driel, R., Henikoff, S., and van Steensel, B. 2003. Distinct HP1 and Su(var)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes & Dev. 17: 2825-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearn, M.G., Hedrick, A., Grigliatti, T.A., and Wakimoto, B.T. 1991. The effect of modifiers of position-effect variegation on the variegation of heterochromatic genes of Drosophila melanogaster. Genetics 128: 785-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz, E. 1928. Das Heterochromatin der Moose. Jahrb. Wiss. Bot. 69: 762-818. [Google Scholar]
- Henikoff, S. 1998. Conspiracy of silence among repeated transgenes. BioEssays 20: 532-535. [DOI] [PubMed] [Google Scholar]
- Jacobs, S.A. and Khorasanizadeh, S. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295: 2080-2083. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T. 2001. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 11: 266-273. [DOI] [PubMed] [Google Scholar]
- Kaminker, J.S., Bergman, C.M., Kronmiller, B., Carlson, J., Svirskas, R., Patel, S., Frise, E., Wheeler, D.A., Lewis, S.E., Rubin, G.M., et al. 2002. The transposable elements of the Drosophila melanogaster euchromatin: A genomics perspective. Genome Biol. 3: research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner, M., O'Carroll, D., Rea, S., Mechtler, K., and Jenuwein, T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116-120. [DOI] [PubMed] [Google Scholar]
- Lifschytz, E. and Lindsley, D.L. 1972. The role of X-chromosome inactivation during spermatogenesis (Drosophila-allocycly-chromosome evolution-male sterility-dosage compensation). Proc. Natl. Acad. Sci. 69: 182-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, B.Y., Emtage, P.C., Duyf, B.J., Hilliker, A.J., and Eissenberg, J.C. 2000. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics 155: 699-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko, F., Ramsahoye, B.H., and Jaenisch, R. 2000. DNA methylation in Drosophila melanogaster. Nature 408: 538-540. [DOI] [PubMed] [Google Scholar]
- Matsuda, E., Agata, Y., Sugai, M., Katakai, T., Gonda, H., and Shimizu, A. 2001. Targeting of Kruppel-associated box-containing zinc finger proteins to centromeric heterochromatin. Implication for the gene silencing mechanisms. J. Biol. Chem. 276: 14222-14229. [DOI] [PubMed] [Google Scholar]
- Myster, S.H., Wang, F., Cavallo, R., Christian, W., Bhotika, S., Anderson, C.T., and Peifer, M. 2004. Genetic and bioinformatic analysis of 41C and the 2R heterochromatin of Drosophila melanogaster: A window on the heterochromatin-euchromatin junction. Genetics 166: 807-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, J., Rice, J.C., Strahl, B.D., Allis, C.D., and Grewal, S.I. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110-113. [DOI] [PubMed] [Google Scholar]
- Nielsen, S.J., Schneider, R., Bauer, U.M., Bannister, A.J., Morrison, A., O'Carroll, D., Firestein, R., Cleary, M., Jenuwein, T., Herrera, R.E., et al. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412: 561-565. [DOI] [PubMed] [Google Scholar]
- Nielsen, A.L., Sanchez, C., Ichinose, H., Cervino, M., Lerouge, T., Chambon, P., and Losson, R. 2002. Selective interaction between the chromatin-remodeling factor BRG1 and the heterochromatin-associated protein HP1alpha. EMBO J. 21: 5797-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian, A., van Steensel, B., Delrow, J., Bussemaker, H.J., Li, L., Sawado, T., Williams, E., Loo, L.W., Cowley, S.M., Yost, C., et al. 2003. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes & Dev. 17: 1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra, M., Leibovitch, B.A., Gandhi, S.G., Rao, M., Bhadra, U., Birchler, J.A., and Elgin, S.C. 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669-672. [DOI] [PubMed] [Google Scholar]
- Platero, J.S., Csink, A.K., Quintanilla, A., and Henikoff, S. 1998. Changes in chromosomal localization of heterochromatin-binding proteins during the cell cycle in Drosophila. J. Cell Biol. 140: 1297-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, E.J. and Elgin, S.C. 2002. Epigenetic codes for heterochromatin formation and silencing: Rounding up the usual suspects. Cell 108: 489-500. [DOI] [PubMed] [Google Scholar]
- Sabl, J.F. and Henikoff, S. 1996. Copy number and orientation determine the susceptibility of a gene to silencing by nearby heterochromatin in Drosophila. Genetics 142: 447-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta, G., Ebert, A., Krauss, V., Fischer, A., Hoffmann, J., Rea, S., Jenuwein, T., Dorn, R., and Reuter, G. 2002. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 21: 1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramke, V. and Allshire, R. 2003. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301: 1069-1074. [DOI] [PubMed] [Google Scholar]
- Spradling, A.C. and Rubin, G.M. 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218: 341-347. [DOI] [PubMed] [Google Scholar]
- Stolc, V., Gauhar, Z., Mason, C., Halasz, G., van Batenburg, M.F., Rifkin, S.A., Hua, S., Herreman, T., Tongprasit, W., Barbano, P.E., et al. 2004. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science 306: 655-660. [DOI] [PubMed] [Google Scholar]
- Sun, L.V., Chen, L., Greil, F., Negre, N., Li, T.R., Cavalli, G., Zhao, H., Van Steensel, B., and White, K.P. 2003. Protein-DNA interaction mapping using genomic tiling path microarrays in Drosophila. Proc. Natl. Acad. Sci. 100: 9428-9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, F.L., Haynes, K., Simpson, C.L., Lee, S.D., Collins, L., Wuller, J., Eissenberg, J.C., and Elgin, S.C. 2004. cis-acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four. Mol. Cell. Biol. 24: 8210-8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, R.K., Henikoff, J.G., Zilberman, D., Ditt, R.F., Jacobsen, S.E., and Henikoff, S. 2005. DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr. Biol. 15: 154-159. [DOI] [PubMed] [Google Scholar]
- van Steensel, B. and Henikoff, S. 2000. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 18: 424-428. [DOI] [PubMed] [Google Scholar]
- van Steensel, B., Delrow, J., and Henikoff, S. 2001. Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet. 27: 304-308. [DOI] [PubMed] [Google Scholar]
- van Steensel, B., Delrow, J., and Bussemaker, H.J. 2003. Genomewide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc. Natl. Acad. Sci. 100: 2580-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel, A., Jia, S., Gerber, S., Sugiyama, T., Gygi, S., Grewal, S.I., and Moazed, D. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833-1837. [DOI] [PubMed] [Google Scholar]
- Wakimoto, B.T. and Hearn, M.G. 1990. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics 125: 141-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler, K.S. and Wakimoto, B.T. 1995. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29: 577-605. [DOI] [PubMed] [Google Scholar]
- Wines, D.R., Talbert, P.B., Clark, D.V., and Henikoff, S. 1996. Introduction of a DNA methyltransferase into Drosophila to probe chromatin structure in vivo. Chromosoma 104: 332-340. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Schwartz, S., Wagner, L., and Miller, W. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7: 203-214. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://flybase.net/genomes/Drosophila_melanogaster/dmel_RELEASE3-1/GFF/whole_genome_annotation_dmel_RELEASE3-1.GFF.gz; Annotation files from Flybase.
- http://www.fruitfly.org/p_disrupt/datasets/NATURAL_TRANSPOSABLE_ELEMENTS.fa; Sequences of transposable elements of D. melanogaster.
- http://www.r-project.org; R software package for statistical analysis.
- http://www.ensembl.org; Ensembl genome database.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.