Abstract
Rice (Oryza sativa L.) chromosome 3 is evolutionarily conserved across the cultivated cereals and shares large blocks of synteny with maize and sorghum, which diverged from rice more than 50 million years ago. To begin to completely understand this chromosome, we sequenced, finished, and annotated 36.1 Mb (∼97%) from O. sativa subsp. japonica cv Nipponbare. Annotation features of the chromosome include 5915 genes, of which 913 are related to transposable elements. A putative function could be assigned to 3064 genes, with another 757 genes annotated as expressed, leaving 2094 that encode hypothetical proteins. Similarity searches against the proteome of Arabidopsis thaliana revealed putative homologs for 67% of the chromosome 3 proteins. Further searches of a nonredundant amino acid database, the Pfam domain database, plant Expressed Sequence Tags, and genomic assemblies from sorghum and maize revealed only 853 nontransposable element related proteins from chromosome 3 that lacked similarity to other known sequences. Interestingly, 426 of these have a paralog within the rice genome. A comparative physical map of the wild progenitor species, Oryza nivara, with japonica chromosome 3 revealed a high degree of sequence identity and synteny between these two species, which diverged ∼10,000 years ago. Although no major rearrangements were detected, the deduced size of the O. nivara chromosome 3 was 21% smaller than that of japonica. Synteny between rice and other cereals using an integrated maize physical map and wheat genetic map was strikingly high, further supporting the use of rice and, in particular, chromosome 3, as a model for comparative studies among the cereals.
As one of the world's most important food crops, rice has emerged as the leading experimental model for functional and evolutionary genomics of cereals. Rice belongs to the genus Oryza, which is composed of 23 species divided into 10 genome types. Cultivated rice is classified as an AA diploid genome (2n = 24) and has two cultivated species, Oryza sativa and Oryza glaberimma, and six wild relatives. The wild AA genome relatives of cultivated rice are rich sources for new alleles for crop improvement, but their usage has been limited because of sterility of the F1 hybrids. O. sativa is thought to have originated from the annual plant type Oryza nivara, which was derived from the perennial Oryza rufipogon. Because of its agronomic importance, compact 430-Mb genome, 50 million years of shared evolutionary history with other larger genome cereals (e.g., maize, wheat, and barley), transformation competency, and abundance of robust molecular genetic resources, the scientific community selected rice (Oryza sativa subsp. japonica cv Nipponbare; hereon referred to as japonica) to be completely sequenced. Currently, whole genome draft sequences of rice subspecies japonica and indica (Goff et al. 2002; Yu et al. 2005) and finished sequences for japonica chromosomes 1, 4, and 10 have been published (Feng et al. 2002; Sasaki et al. 2002; The Rice Chromosome 10 Sequencing Consortium 2003).
As part of the International Rice Genome Sequencing Project (IRGSP), our consortium sequenced japonica chromosome 3 using a clone-by-clone approach. Cytologically, chromosome 3 is the second largest rice chromosome and is one of the most euchromatic chromosomes (Cheng et al. 2001). It is interesting to note that cytologically, the genetically defined “short arm” of chromosome 3 is really the long arm and vice versa for the “long arm” as the arm ratio of chromosome 3 is 1.13 ± 0.12 based on measurements on meiotic pachytene chromosomes (Cheng et al. 2001). Genetically, chromosome 3 is 170 cM in length (Harushima et al. 1998) and has 27 morphological mutants. In addition, more than 133 agronomic genes/traits per QTL and 963 cDNAs (Wu et al. 2002) have been found associated with chromosome 3 (http://www.shigen.nig.ac.jp/rice/oryzabase/top/top.jsp).
Although Oryza separated from maize and sorghum ∼50 million years ago (Mya) and from wheat and barley ∼40 Mya, their common evolutionary history can be traced by the collinear order of genetic markers across their chromosomes. This is particularly true for the short arm of chromosome 3, which shows large stretches of genetic marker collinearity with maize chromosomes 1 and 9, sorghum linkage group L, and barley and wheat chromosomes 4L. In this paper, we report the finished sequence of japonica chromosome 3 and a detailed analysis of its annotated sequence. We show a comprehensive study of the syntenic relationship of rice chromosome 3 with maize and wheat. This study allowed us to construct bacterial artificial chromosome (BAC) clone-based fingerprint contig (FPC) physical maps of the corresponding syntenic maize chromosomes, which will be useful for positional cloning and genome sequencing in maize (Soderlund et al. 2002). In addition, we were able to reconstruct a BAC/BAC-end sequence (BES)-based FPC physical map of chromosome 3 from the progenitor of cultivated japonica rice—Oryza nivara—which allowed us to estimate the level of divergence between these two closely related Oryza species.
Results and Discussion
Structural features of japonica chromosome 3
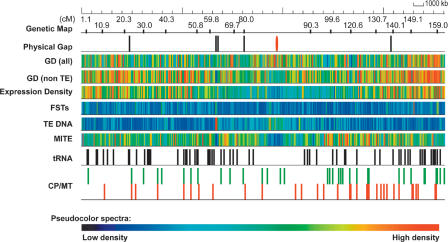
A total of 323 BAC/P1 artificial chromosome (PAC) clones, 44.6 Mb in total, were used to construct a 36.1-Mb pseudomolecule (virtual contig) (Table 1; Fig. 1). The pseudomolecule spans the majority of the chromosome, with five physical gaps present in the two arms, gaps at each telomere, and one gap in the centromere. The centromere of chromosome 3 (CEN3) contains ∼180 kb of the CentO satellite repeats (Cheng et al. 2002). All gaps, including the centromere and telomere gaps, have been sized using fiber or pachytene FISH, and the size missing from the pseudomolecule is estimated to be ∼1.26 Mb. Clones to close one gap in the tiling path have been identified and are currently being sequenced. However, all attempts to identify additional clones from four BAC/PAC libraries failed, suggesting an under-representation of these sequences in existing libraries. In total, the short arm (3S) is 19.4 Mb, whereas the long arm (3L) is 16.7 Mb. The arm lengths are consistent with pachytene chromosomal measurements of chromosome 3 (Cheng et al. 2001), suggesting that the chromosome arm nomenclature should be reversed. In addition, at 36.1 Mb, chromosome 3 is the second longest chromosome in rice. A total of 339 genetic loci from 595 sequences could be located on chromosome 3, leaving 45 genetically mapped sequenced markers representing 34 genetic loci absent from the virtual contig.
Table 1.
Statistics of rice chromosome 3
| Feature | Statistic |
|---|---|
| Total number of BACs/PACs | 323 |
| Total BAC length (Mb) | 44.6 |
| Total nonoverlapping sequence (Mb) | 36.1 |
| Short arm (Mb) | 19.4 |
| Long arm (Mb) | 16.7 |
| Integrated genetic markers | 339 (loci) |
| G+C content | |
| Overall | 43.7 |
| Exons | 54.4 |
| Introns | 38.5 |
| Intergenic regions | 41.5 |
| Total number of genesa | 5915 (5002) |
| Known/putative genes | 3064 (51.8%) |
| Expressed genes | 757 (12.8%) |
| Hypothetical genes | 2094 (35.4%) |
| Gene density (kb)b | 6.1 (7.2) |
| Average gene length (bp)c | 2567 |
| Total number of gene models | 6232 |
| Average exon size (bp) | 281 |
| Average intron size (bp) | 369 |
| Average number exons per gene model | 4.5 |
Total number of genes includes the 913 TE-related genes and the 5002 non-TE-related genes.
Gene density is reported for all genes and non-TE-related genes (in parentheses).
Average length of the genes is determined using the length from the start codon to the stop codon of the longest isoform for all gene models.
Figure 1.
Distribution of features on japonica chromosome 3. The pseudomolecule was annotated for features including gene models, transcript evidence, repetitive sequences, tRNA genes, genetic markers, organellar insertions, and flanking sequence tags (FSTs). These are identified on the pseudomolecule using a false color display. Genetic markers are positioned at 10-cM intervals (approximately). Physical gaps are noted separately (black ticks) and include the gap at the centromere (red). Gene model density (GD) is noted in two separate tiles, all gene models and non-TE-related gene models. Expression density reflects gene models with transcript evidence. FST denotes the presence of a sequence-tagged insertion among tagged populations. Repetitive sequences are represented by the TE DNA and MITE tiles. tRNA genes and organellar insertions (CP/MT) are also denoted.
The total amount of repetitive sequence present on rice chromosome 3 is 21.4%, with transposable elements (TE) accounting for >90% of the repetitive sequences (Supplemental Table 1). The TEs are predominated with retrotransposons and miniature inverted repeat transposable elements (MITEs). Segments of retrotransposons (2745 segments in total) are present on the chromosome, totaling 3.38 Mb. In comparison, 11,865 MITEs totaling 2.29 Mb are present on chromosome 3. Another feature of chromosome 3 includes the insertions of organellar sequences. In total, 57 (37.8 kb) and 70 (27.6 kb) sequences with significant similarity to the rice chloroplast and mitochondrion, respectively, are present.
Functional features of japonica chromosome 3
A total of 5915 genes could be identified on chromosome 3 (Table 1; Fig. 1). Of these, 913 were annotated as transposable element related (TE-related), leaving 5002 non-TE genes. There are 267 genes with alternative splice forms, some of which have multiple forms, resulting in a total of 6232 gene models. Of the 5915 total genes, a function could be assigned to 3064 (51.8%), while 757 (12.8%) were annotated as encoding an expressed protein, and 2094 (35.4%) were annotated as encoding a hypothetical protein. The distribution of known versus expressed/hypothetical genes is consistent with annotation of the other completed japonica chromosomes 1, 4, and 10 (Feng et al. 2002; Sasaki et al. 2002; Rice Chromosome 10 Sequencing Consortium 2003). If TE-related genes were excluded, one gene was present every 7.2 kb, consistent with the gene density reported by Yu et al. (2005) for the entire japonica genome. The distribution of non-TE genes was not even along the chromosome as a reduced frequency of non-TE genes was observed near the centromere with an increased frequency at the telomeres (Fig. 1). The average gene model had 4.5 exons and was 2567 bp in length (Table 1), consistent with reports on other finished chromosomes and the whole japonica genome (Feng et al. 2002; Rice Chromosome 10 Sequencing Consortium 2003; Yu et al. 2005). In addition to these protein-coding genes, 78 transfer RNA genes were identified.
Using EST frequency to assess transcription levels, a clear reduction of expression was apparent near the centromere (Fig. 1). A detailed examination of expression in six tissues did not reveal a tissue-specific pattern of expression along the chromosome (Supplemental Fig. 1). In contrast to the reduced frequency of expressed genes at the centromere, there was a clear enrichment of transposable elements, with the exception of MITEs, present near the centromere (Fig. 1). This general pattern has also been observed in Arabidopsis (CSHL/WUGSC/PE Biosystems Arabidopsis Sequencing Consortium 2000; Lippman et al. 2004). Consistent with previous reports of association with genes, MITEs were preferentially present in the euchromatic arms (Feng et al. 2002; Sasaki et al. 2002; Rice Chromosome 10 Sequencing Consortium 2003).
The predicted proteome of japonica chromosome 3 is similar to that of other published rice chromosomes. Searches against the predicted proteome of Arabidopsis thaliana with the deduced protein sequences from the 6232 gene models revealed 4201 rice proteins (67%) with a putative ortholog/paralog in the Arabidopsis proteome as defined by a BLASTP E-value of <10-5. A reduced frequency of homology, 12%-41% of the rice proteins with a putative ortholog, was apparent between japonica chromosome 3 and other model organisms such as Escherichia coli, Synechocystis, yeast, fly, worm, and human (Supplemental Table 2). In addition to searches against model organisms, we ascertained the presence of Pfam domains in the predicted rice chromosome 3 proteome. Excluding TE-related proteins, a total of 2462 proteins contained at least one Pfam domain, with 929 Pfam domains represented within the rice chromosome 3 proteome. The most prevalent non-TE related Pfam domain is protein kinase with 127 members. The most prevalent Pfam domains on rice chromosome 3 are presented in Supplemental Table 3. Construction of paralogous families using a combination of Pfam domains and BLASTP similarity revealed 662 paralogous families representing 2807 proteins. The distribution of proteins in paralogous families is shown in Supplemental Figure 2 with a majority of the proteins belonging to a paralogous family that contained only two members. The largest paralogous family, with 85 members, encodes protein kinase.
To ascertain the number of proteins that could be potentially novel to rice, we assessed the predicted japonica chromosome 3 proteome for similarity to predicted proteins from other genomes and/or the presence of a Pfam domain (Supplemental Fig. 3). We first excluded all TE-related proteins from our analysis. Of the 5317 non-TE-related proteins, only 1377 did not exhibit sequence similarity to an Arabidopsis protein or a non-rice sequence present in GenBank (BLASTP E-value cutoff of 10-5) or contain a Pfam domain above the trusted cutoff. Of these remaining 1377 proteins, 67 had an EST match to rice (>97% identity), suggesting these are expressed genes. Another 42 and 38 proteins matched a monocot or dicot EST, respectively. A search of assembled gene-enriched sequences from maize (Whitelaw et al. 2003) and sorghum (A. Chan and P. Rabinowicz, unpubl.) revealed 377 proteins with similarity to one or both of these related cereals, leaving 853 proteins without similarity to an entry in the public databases and, therefore, potentially novel rice genes or artifacts of the annotation methods. Another possibility is that these are rice-specific transposon sequences that we have failed to identify in our filtering processes. Interestingly, 426 of the 853 rice proteins from chromosome 3 have matches to other proteins present in the rice genome.
Further annotation of the predicted proteome was obtained through assignment of Plant GOSlim ontologies. A total of 2250 of the 6232 predicted proteins (36.1%) could be assigned a minimum of one GO term with 8897 GO terms assigned in total, of which 3982 were biological process, 3737 were molecular function, and 1178 were cellular component terms. Molecular function Plant GOSlim terms with at least 20 assignments are shown in Supplemental Figure 4. A search of databases that contain genomic DNA sequences flanking tagged insertion sites (FSTs; Tos17, Ac/Ds, T-DNA) revealed a total of 3977 insertions in chromosome 3, of which 2798 could be aligned only with rice chromosome 3. Of the 3977 FST insertions, 2787 were either in a gene or within 500 bp of a gene providing a candidate insertion line for 1146 genes (19.4 % of the 5915 genes) on rice chromosome 3.
Comparison with O. nivara
Overall alignment
A total of 3163 paired O. nivara BES were mapped on the japonica chromosome 3 pseudomolecule with an average of ≥98% sequence similarity. Sequence similarity in transcribed regions was 98.8% ± 1.1%, and that in nontranscribed regions was 97.7% ± 1.6%. These paired end sequences were initially found to be associated with 27 FPC contigs that could be further reduced to 16 FPC contigs based on sequence alignment to the japonica chromosome 3 pseudomolecule. The alignment between BES in the FPC contigs and japonica chromosome 3 is graphically represented in Supplemental Figure 5. A tremendous amount of collinearity between two species was detected across the chromosome, and no major rearrangements were identified based on this alignment.
Reconstructed contigs and coverage
Based on the sequence alignment, ∼35 Mb (∼97%) of the japonica chromosome 3 was covered by the 16 O. nivara FPC contigs. FPC contig size ranged from 0.9 to 5.7 Mb with an average of 2.2 Mb (Supplemental Table 4). In the reconstructed O. nivara chromosome 3, 14 gaps, excluding a centromere gap, represented ∼1.2 Mb of sequence. Most gaps were small and ranged from 2.5 to 53 kb with an average of 20 kb in size. However, two large gaps (420 and 490 kb, respectively) were detected near 12.5 Mb and 24.7 Mb on the japonica pseudomolecule. We expect that those regions may contain sequences unique to both japonica and O. nivara or that our analysis failed to detect positive paired BES due to higher sequence divergence than in other regions of the chromosome.
Insertion/deletion analysis
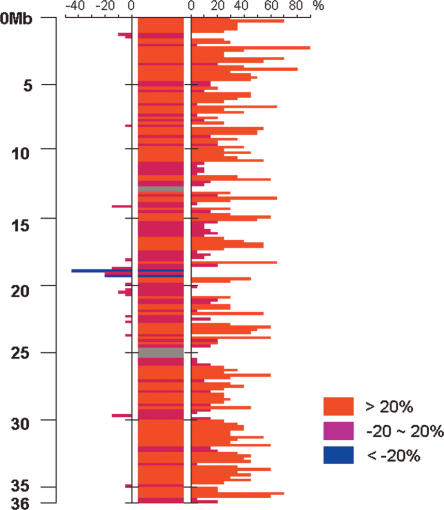
To determine the level of insertions or deletions between japonica chromosome 3 and the reconstructed O. nivara chromosome, we compared distances between 2414 paired O. nivara BES, relative to the japonica genome, with empirically determined insert sizes of these same BACs. O. nivara paired BAC ends align to the japonica pseudomolecule with an average distance of 165 kb apart, whereas the average insert size of these same clones was 136 kb. This suggested that japonica chromosome 3 is ∼21% larger than that of O. nivara. Size difference between the two chromosomes was further analyzed in a 200-kb window across the pseudomolecule, and we identified 34 insertion blocks (representing 106 200-kb windows), two deletion blocks (two 200-kb windows), and 36 invariable size blocks (67 200-kb windows) using a 20% size difference cutoff parameter on japonica chromosome 3 (Fig. 2; Supplemental Table 5). Insertion blocks accounted for 5.2 Mb of sequence increase in japonica or a 5.2-Mb decrease in O. nivara. Moreover, ∼70% of the insertion blocks (74 of 106 200-kb windows) were localized within 10 Mb from the ends of the chromosome, which suggests that insertion or deletion may occur unequally across the chromosome. Even though repetitive sequences and paralogous gene contents were slightly higher than average (20% for repetitive sequences and 45% for paralogs per genes) within the insertion blocks (Supplemental Table 5), we could not detect a sufficient correlation to account for the 5.8-Mb expected sequence insertion into japonica or deletion within O. nivara. These data are consistent with prior reports by Bennetzen and colleagues that rice intergenic regions are highly variable (Ma and Bennetzen 2004; Ma et al. 2004).
Figure 2.
Plot of insertion/deletion/null blocks relative to the O. nivara chromosome 3 FPC map using a 200-kb sliding window across the japonica chromosome 3 pseudomolecule. Percent difference (%D) between sizes from O. nivara paired BAC ends and high-resolution HindIII fingerprints was plotted using a 200-kb window across the japonica chromosome 3 pseudomolecule, where a >20% difference is red (insertion), -20%∼20% is purple (null), and <-20% is blue (deletion). Percentage difference was calculated using the following formula: {[(paired BES size) - (FP size)]/(FP size)} × 100. A percentage decrease (left) or increase (right) was plotted on the bar graph on either side of the consensus. Regions of the japonica chromosome 3 pseudomolecule that were not covered by O. nivara paired BAC end sequences are represented in gray.
Polymorphic SSRs
Among molecular marker systems, simple sequence repeats (SSRs) have been used successfully not only to detect genetic variation within or between species but also to develop molecular markers tightly linked to agronomically important traits in breeding programs. Since O. nivara is an important source of new genes and allelic variation for the improvement of cultivated rice, we can use the O. nivara chromosome 3 contig/BES map described above to construct a virtual SSR map between japonica and O. nivara that can be used for high-resolution marker-assisted selection in breeding populations.
Our analysis of japonica chromosome 3 revealed a total of 6489 SSRs using RepeatMasker with default parameters. Among those, the trinucleotide repeat was the most abundant repeat class and accounts for ∼57% of the total amount of SSRs, followed by di- (21%), penta- (12%), tetra- (8%), and hexanucleotide repeats (3%). The frequency of chromosome 3 SSRs was very similar to the SSRs found in the entire japonica genome (Supplemental Table 6). Further repeat motif analysis showed that CGG/CCG was the predominant repeat motif in both the japonica genome (42%) and japonica chromosome 3 (41%), and that TA was the second most numerous motif (11% in genome and 11% in chromosome 3). Previous work reported that monocot plants are more enriched in GC-rich trinucleotide repeats, mainly CGG/CCG repeats, than dicots because of their higher overall GC content (Kantety et al. 2002; Morgante et al. 2002), which is in agreement with our results.
The O. nivara BES showed similar SSR composition with that of japonica chromosome 3 except for a 2% increase in dinucleotide repeats and a 2% decrease in tri- and pentanucleotide repeats (Supplemental Table 6). We detected an ∼7% reduction in the CGG/CCG in O. nivara sequences; however, other GC-rich motifs increased by 1%-2% each in trinucleotide repeats.
To detect polymorphic SSRs on chromosome 3 between japonica and O. nivara, we compared the SSR content of 326 mapped O. nivara BES containing SSRs to their corresponding regions in japonica. Of those, 206 SSRs showed the same motif and length (length difference ± 2 bp considered the same length), 118 SSRs representing 87 unique loci showed different SSR length, and two SSRs were present only in the O. nivara sequence. Trinucleotide and dinucleotide repeats were the predominant polymorphic SSRs with 49 and 32 SSRs, respectively. CGG/CCG was the most abundant polymorphic SSR type (25 loci), and GA/TC and TA motifs represented 15 loci each in the chromosome. The base pair positions and SSR compositions of these 118 polymorphic SSRs are shown in detail in Supplemental Table 7.
Synteny between rice and other major cereals
The genome sizes of the grass family (Poaceae) vary dramatically from ∼400 Mb in rice to 16,000 Mb in wheat. Despite a 35-fold difference in genome size and 50 million years of evolution, a remarkable degree of collinearity has been discovered among these species by comparative genetic mapping (Ahn et al. 1993; Moore et al. 1995; Gale and Devos 1998; Feuillet and Keller 1999). This collinearity greatly facilitates the isolation of agronomically important genes and our understanding of genome evolution. With the finished sequence of the rice genome, we can obtain greater insight into the syntenic relationship between rice and the major cereals.
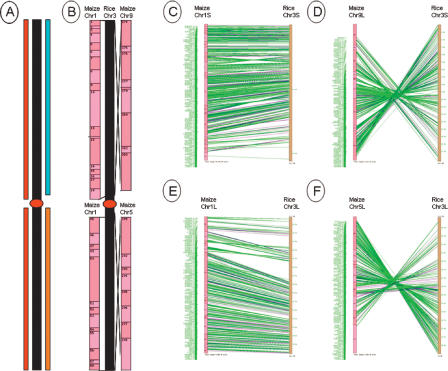
Although Goff et al. (2002) published a general idea of macrosynteny between rice and maize, and maize itself, their results were preliminary and contained errors due to the low-resolution genetic map of maize and their partially unanchored rice draft sequence. Recently Yu et al. (2005) performed a similar analysis with improved japonica and indica whole genome draft sequence and a 1063-marker maize genetic map. We substantially improved the fidelity of this analysis by using our high-resolution FPC physical and genetic map of the maize genome, which is comprised of >15,000 sequenced markers (Coe et al. 2002; http://www.genome.arizona.edu/fpc/maize/) and the finished sequence of japonica chromosome 3. Except for the centromere region, rice chromosome 3 is highly collinear with the maize genome. The short arm of japonica chromosome 3 is highly collinear with the short arm of maize chromosome 1 (1S) and the inverted long arm of maize chromosome 9 (9L), while japonica 3L is highly conserved with maize 1L and the inverted maize 5S (Fig. 3). These highly syntenic chromosomal regions of maize aligned with the japonica chromosome 3 pseudomolecule, clearly demonstrating the ancient tetraploid origin of the maize genome. Our results corroborate and refine those found by Goff et al. (2002) and Yu et al. (2005) but to a much higher resolution and provide contig and BAC tile information that can be used for localized draft sequence analysis and positional cloning.
Figure 3.
Syntenic relationship between japonica chromosome 3 and maize chromosomes 1, 5, and 9. We used maize markers, such as anchored markers and EST-derived overgoes and BAC end sequences to construct this syntenic map with the rice chromosome 3 pseudomolecule. Graphics were generated using SyMAP (www.agcol.arizona.edu/symap). (A) Overall picture of the rice chromosome 3-maize synteny. Rice chr3: the filled black bar; maize chr1 (red), chr5 (orange), and chr9 (blue). (B) Synteny of rice chromosome 3 pseudomolecule to the maize FPC map. (C,D) Synteny of the short arm of rice chromosome 3 with maize chromosomes 1 and 9 in detail. (E,F) Synteny of the long arm of rice chromosome 3 with maize chromosomes 1 and 5 in detail. (Green line) Marker and overgoes-originated EST sequences; (purple line) maize BAC end sequences.
The hexaploid nature of bread wheat can tolerate the large deletion of a chromosome fragment. A collection of deletion lines has allowed for the establishment of an overlapping cytogenetically based physical map of wheat (Werner et al. 1992). Wheat ESTs can be mapped to physical bins with these deletion lines. Recently, Sorrells et al. (2003) established a japonica-wheat synteny map by comparing 4485 bin-mapped wheat ESTs to the ordered japonica BAC/PAC sequence. With more bin-mapped ESTs (Qi et al. 2004) and our nonoverlapping japonica chromosome 3 pseudomolecule, a wheat-japonica chromosome 3 syntenic map was constructed (Supplemental Fig. 6). We can clearly observe collinearity between japonica chromosome 3S and wheat 4BL/4DL, or with wheat 5AL and 4AS. In addition, japonica 3L is conserved with wheat 5BL/5DL and 4DS or with wheat 4AL and 5AL. The wheat B and D genomes are more similar to each other than to the A genome. The two arms of the 4A genome are switched in comparison to the arms of 4B and 4D. The japonica 3S-wheat 5L synteny, therefore, appears unique to the A genome of wheat. Overall, the sequence of japonica chromosome 3 is highly preserved among the cereals and will be an ideal model chromosome for comparative evolutionary and functional genomics studies in the future.
Conclusions
The availability of sequence and annotation for japonica chromosome 3 provides further insight into the dynamics of the rice genome. The availability of partial genomic sequences for O. nivara, maize, and wheat allowed us to further demonstrate the utility of rice as an “anchor” species for cereal comparative genomics. The completion of japonica chromosome 3 along with the entire rice genome will provide the foundation for trait identification and genome evolutionary studies in the most important family of flowering plants.
Methods
Chromosome 3 was sequenced using a clone-by-clone approach in which large insert BAC and PAC clones were selected for sequencing from multiple libraries (Baba et al. 2000; Chen et al. 2002) using a combination of hybridization, BAC end sequence alignment, and/or FPC. Each BAC/PAC was sequenced to ∼8-10-fold sequence redundancy, and gaps were closed using a combination of resequencing, alternative chemistries, microlibraries, and/or transposon-mediated sequencing. The pseudomolecule, or virtual contig for the chromosome, was constructed by aligning the BAC/PAC clones and trimming overlapping regions. The complete sequence of the pseudomolecule is available in Gen-Bank (accession no. DP000009).
Genes and gene models were identified through computational methods using the ab initio gene prediction program FGENESH (Salamov and Solovyev 2000), or through manual curation efforts as described previously (Yuan et al. 2002). Genes are distinct loci on the chromosome and are represented by at least one gene model, which is the structure of the transcribed mRNA. Some genes are represented by more than one gene model because of the presence of alternative splice forms. tRNA genes were identified using tRNAScan (Lowe and Eddy 1997). Gene models were searched against a nonredundant amino acid database, and domains were identified using the HMMer program with the Pfam database (Bateman et al. 2004). The fine structure of the gene models was improved using EST and full-length cDNA evidence using the Program to Assemble Spliced Alignments (Haas et al. 2003). Genes were annotated as known or putative if the protein had significant similarity to proteins or Pfam domains in the database. Genes with alignments only to ESTs or other transcripts that lack a known function were annotated as encoding expressed proteins. Genes that were determined solely by ab initio gene finders were annotated as encoding hypothetical proteins. Genes encoding transposable elements were identified and transitively annotated by searching against the TIGR Oryza Repeat Database (Ouyang and Buell 2004) using TBLASTN (Altschul et al. 1990) with an E-value cutoff of 10-10. Transmembrane domains were identified using TMHMM V2.0 (http://www.cbs.dtu.dk/services/TMHMM/), while signal peptides were identified using SignalP V2.0 (http://www.cbs.dtu.dk/services/SignalP-2.0/). The predicted proteome was searched against other model organisms using BLASTP with an E-value cutoff of 10-5, and these matches were used to identify putative orthologs and paralogs. Paralogous families were identified using methods described previously (Wortman et al. 2003) involving an algorithmic approach combining Pfam domains (Bateman et al. 2004) and novel domains as identified by BLASTP. Plant GOSlim ontologies were assigned by searching the japonica chromosome 3 proteome against the TIGR Arabidopsis thaliana Release 5 proteome (http://www.tigr.org/tdb/e2k1/ath1) using BLASTP. Chromosome 3 proteins were transitively annotated with Plant GOSlim terms derived from Arabidopsis using BLASTP with an E-value cutoff of 10-10. Proteins annotated as hypothetical, expressed, TE-related, and proteins assigned with GO ids with “unknown” definition were not included. Sequence and annotation features of the japonica chromosome 3 pseudomolecule can be downloaded at ftp://ftp.tigr.org/pub/data/rice/rice.chr03.v2.1/.
The pseudomolecule was further annotated for features such as repetitive sequences, transcription activity, and distribution of FSTs. Repetitive sequences were identified on japonica chromosome 3 using RepeatMasker (http://www.repeatmasker.org) with the TIGR Oryza Repeat Database and quantitated using a 100-kb window. To identify transcriptional activity, the pseudomolecule was searched against the TIGR Rice Gene Index (Quackenbush et al. 2001; http://www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=rice), in which the ESTs and Expressed Transcripts (ETs) were binned into six tissue categories (leaf, shoot, flower, seed, root, callus). Using a cutoff criterion of 95% identity over 80% of the EST/FL-cDNA sequence length, the frequency of transcript evidence was determined at 100-kb windows along the chromosome. To identify genes that have a tagged insertion or FST, the pseudomolecule was searched against a collection of 26,725 sequence tags generated from several insertional mutagenesis projects (Greco et al. 2001; Miyao et al. 2003; Kim et al. 2004; Sallaud et al. 2004).
For synteny analysis between maize and rice, we used the integrated physical and genetic maps of maize. The maize FPC physical map covers ∼94% of the maize genome with 1931 genetically anchored markers and 13,634 EST-derived overgo markers (http://www.genome.arizona.edu). Using the Synteny Mapping and Analysis Program (SyMAP, http://www.agcol.arizona.edu), sequences of these genetic markers and overgo-originated ESTs were BLASTN-searched against the rice chromosome 3 pseudomolecule using an E-value cutoff of 10-10, and the synteny blocks were computed. The results can be interactively browsed at http://www.agcol.arizona.edu/symap/maize. The rice-wheat synteny analysis was performed using BLASTN (E-value cutoff of 10-10) using the wheat genetic markers as the query against the japonica pseudomolecule. The wheat genetic markers were derived from wheat ESTs and bin-mapped with wheat deletion lines (Qi et al. 2004; Sorrells et al. 2003).
For reconstruction of O. nivara chromosome 3, we used the O. nivara FPC map and BES (Wing et al. 2005). The initial FPC map was built by assembling 51,056 successful SNaPshot (Luo et al. 2003) fingerprints with a cutoff e-50 and tolerance 4 using FPC. The FPC map was further refined by iterative reassembling with lower stringent cutoff values followed by removal of q-clones and automatic contig merging of contigs. The final FPC map assembled at cutoff e-21 was manually merged using evidence from the SyMAP results of the O. nivara FPC/BESs aligned to the IRGSP assembled pseudomolecules (http://www.omap.org). O. nivara paired BES were mapped onto the 36.1-Mb japonica chromosome 3 pseudomolecule using MegaBLAST with an E-value of 10-200 and ≥95% similarity and then associating the sequences with their corresponding FPC contigs. We applied the high-resolution agarose fingerprinting method with HindIII restriction digestion followed by band calling using IMAGE software in order to size 3163 O. nivara chromosome 3 BAC clones (Chen et al. 2002). RepeatMasker with default parameters was used to identify polymorphic SSRs between the japonica chromosome 3 pseudomolecule and O. nivara BAC end sequences. We considered an SSR polymorphic when the repeat length differed by 2 bp or more.
Complete list of authors
The Institute for Genomic Research (TIGR)
C. Robin Buell,2,3 Qiaoping Yuan,3 Shu Ouyang,3 Jia Liu,3 Wei Zhu,3 Aihui Wang,3 Rama Maiti,3 Brian Haas,3 Jennifer Wortman,3 Mihaela Pertea,3 Kristine M. Jones,3 Mary Kim,3 Larry Overton,3 Tamara Tsitrin,3 Douglas Fadrosh,3 Jayati Bera,3 Bruce Weaver,3 Shaohua Jin,3 Shivani Johri,3 Matt Reardon,3 Kristen Webb,3 Jessica Hill,3 Kelly Moffat,3 Luke Tallon,3 Susan Van Aken,3 Matthew Lewis,3 Teresa Utterback,3 Tamara Feldblyum,3 Victoria Zismann,3 Stacey Iobst,3 Joseph Hsiao,3 Aymeric R. de Vazeille,3 Steven L. Salzberg,3 Owen White,3 and Claire Fraser3
Arizona Genomics Institute (AGI)
Yeisoo Yu,4,5 HeyRan Kim,4,5 Teri Rambo,4,5 Jennifer Currie,4,5 Kristi Collura,4,5 Shelly Kernodle-Thompson,4,5 Fusheng Wei,4 David Kudrna,4 Jetty Siva S. Ammiraju,4 Meizhong Luo,4,5 Jose Luis Goicoechea,4,5 and Rod A. Wing2,4,5
Clemson University Genomics Institute (CUGI)
Rod A. Wing,2,4,5 David Henry,5 Ryan Oates,5 Michael Palmer,5 Gina Pries,5 Christopher Saski,5 Jessica Simmons,5 and Carol Soderlund5,6
Arizona Genomics Computational Laboratory (AGCoL)
Cold Spring Harbor Laboratory (CSHL)
Melissa de la Bastide,7 Lori Spiegel,7 Lidia Nascimento,7 Emily Huang,7 Raymond Preston,7 Theresa Zutavern,7 Lance Palmer,7 Andrew O'Shaughnessy,7 Sujit Dike,7 and W. Richard McCombie2,7
Washington University School of Medicine Genome Sequencing Center (WUGSC)
University of Wisconsin
Purdue University
Scott Jackson10
Supplementary Material
Acknowledgments
Funding for work on rice chromosome 3 was provided by grants from the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (Grants 99-35317-8275, 2003-35317-13173 to C.R.B.; Grants 99-35317-8505, 2002-35317-12414 to R.A.W.); the National Science Foundation (Grants DBI998282, DBI0321538 to C.R.B.; Grant DBI0241181 to R.A.W.); and the U.S. Department of Energy (Grant DE-FG02-99ER20357 to C.R.B.; Grant DE-FG03-02ER15363 to R.A.W.). C.R.B. acknowledges the assistance of the TIGR Sequencing Facility, the TIGR Informatics Department, the TIGR IT Group, and the J. Craig Venter Joint Technology Center. R.A.W. acknowledges the assistance of the AGI and CUGI Sequencing, Physical Mapping, BAC/EST Resource Centers, and the Arizona Genomics Computational Laboratory. W.R.M. acknowledges the assistance of the CSHL sequencing and informatics groups. We thank the Japanese Ministry of Agriculture, Forestry and Fishery (MAFF) for genetic markers used to develop initial seed BACs for sequencing.
Footnotes
[Supplemental material is available online at www.genome.org. The chromosome 3 pseudomolecule sequence data from this study has been submitted to GenBank under accession no. DP000009.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3869505. Article published online before print in August 2005.
The Institute for Genomic Research, Rockville, Maryland 20850, USA.
Arizona Genomics Institute (AGI), Department of Plant Sciences and BIO5 Institute, The University of Arizona, Tucson, Arizona 85721, USA.
Clemson University Genomics Institute (CUGI), Clemson University, Clemson, South Carolina 29634, USA.
Arizona Genomics Computational Laboratory (AGCoL), Department of Plant Sciences and BIO5 Institute, The University of Arizona, Tucson, Arizona 85721, USA.
Cold Spring Harbor Laboratory (CSHL), Cold Spring Harbor, New York 11723, USA.
Washington University School of Medicine Genome Sequencing Center (WUGSC), St. Louis, Missouri 63108, USA.
University of Wisconsin, Department of Horticulture, Madison, Wisconsin 53706, USA.
Purdue University, Department of Agronomy, West Lafayette, Indiana 47907, USA.
References
- Ahn, S., Anderson, J.A., Sorrells, M.E., and Tanksley, S.D. 1993. Homoeologous relationships of rice, wheat and maize chromosomes. Mol. Gen. Genet. 241: 483-490. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Meyers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Baba, T., Katagiri, S., Tanoue, H., Tanaka, R., Chiden, Y., Saji, S., Hamada, M., Nakashima, M., Okamoto, M., Hayashi, M., et al. 2000. Construction and characterization of rice genomic libraries: PAC library of japonica variety, Nipponbare and BAC library of indica variety, Kasalath. Bull. NIAR 14: 41-49. [Google Scholar]
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E.L., et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32: D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., Presting, G., Barbazuk, W.B., Goicoechea, J.L., Blackmon, B., Fang, G., Kim, H., Frisch, D., Yu, Y., Sun, S., et al. 2002. An integrated physical and genetic map of the rice genome. Plant Cell 14: 537-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., Buell, C.R., Wing, R.A., Gu, M., and Jiang, J. 2001. Toward a cytological characterization of the rice genome. Genome Res. 11: 2133-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., Dong, F., Langdon, T., Ouyang, S., Buell, C.R., Gu, M., Blattner, F.R., and Jiang, J. 2002. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14: 1691-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe, E., Cone, K., McMullen, M., Chen, S.S., Davis, G., Gardiner, J., Liscum, E., Polacco, M., Paterson, A., Sanchez-Villeda, H., et al. 2002. Access to the maize genome: An integrated physical and genetic map. Plant Physiol. 128: 9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cold Spring Harbor Laboratory, Washington University Genome Sequencing Center, and PE Biosystems Arabidopsis Sequencing Consortium. 2000. The complete sequence of a heterochromatic island from a higher eukaryote. Cell 100: 377-386. [PubMed] [Google Scholar]
- Feng, Q., Zhang, Y., Hao, P., Wang, S., Fu, G., Huang, Y., Li, Y., Zhu, J., Liu, Y., Hu, X., et al. 2002. Sequence and analysis of rice chromosome 4. Nature 420: 316-320. [DOI] [PubMed] [Google Scholar]
- Feuillet, C. and Keller, B. 1999. High gene density is conserved at syntenic loci of small and large grass genomes. Proc. Natl. Acad. Sci. 96: 8265-8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M.D. and Devos, K.M. 1998. Comparative genetics in the grasses. Proc. Natl. Acad. Sci. 95: 1971-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S.A., Ricke, D., Lan, T.H., Presting, G., Wang, R., Dunn, M., Glazebrook, J., Sessions, A., Oeller, P., Varma, H., et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92-100. [DOI] [PubMed] [Google Scholar]
- Greco, R., Ouwerkerk, P.B., Taal, A.J., Favalli, C., Beguiristain, T., Puigdomenech, P., Colombo, L., Hoge, J.H., and Pereira, A. 2001. Early and multiple Ac transpositions in rice suitable for efficient insertional mutagenesis. Plant Mol. Biol. 46: 215-227. [DOI] [PubMed] [Google Scholar]
- Haas, B.J., Delcher, A.L., Mount, S.M., Wortman, J.R., Smith Jr., R.K., Hannick, L.I., Maiti, R., Ronning, C.M., Rusch, D.B., Town, C.D., et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31: 5654-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima, Y., Yano, M., Shomura, A., Sato, M., Shimano, T., Kuboki, Y., Yamamoto, T., Lin, S.Y., Antonio, B.A., Parco, A., et al. 1998. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148: 479-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantety, R.V., La Rota, M., Matthews, D.E., and Sorrells, M.E. 2002. Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol. Biol. 48: 501-510. [DOI] [PubMed] [Google Scholar]
- Kim, C.M., Piao, H.L., Park, S.J., Chon, N.S., Je, B.I., Sun, B., Park, S.H., Park, J.Y., Lee, E.J., Kim, M.J., et al. 2004. Rapid, large-scale generation of Ds transposant lines and analysis of the Ds insertion sites in rice. Plant J. 39: 252-263. [DOI] [PubMed] [Google Scholar]
- Lippman, Z., Gendrel, A.V., Black, M., Vaughn, M.W., Dedhia, N., McCombie, W.R., Lavine, K., Mittal, V., May, B., Kasschau, K.D., et al. 2004. Role of transposable elements in heterochromatin and epigenetic control. Nature 430: 471-476. [DOI] [PubMed] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M.C., Thomas, C., You, F.M., Hsiao, J., Ouyang, S., Buell, C.R., Malandro, M., McGuire, P.E., Anderson, O.D., and Dvorak, J. 2003. High-throughput fingerprinting of bacterial artificial chromosomes using the snapshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics 82: 378-389. [DOI] [PubMed] [Google Scholar]
- Ma, J. and Bennetzen, J.L. 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. 101: 12404-12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., Devos, K.M., and Bennetzen, J.L. 2004. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14: 860-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao, A., Tanaka, K., Murata, K., Sawaki, H., Takeda, S., Abe, K., Shinozuka, Y., Onosato, K., and Hirochika, H. 2003. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15: 1771-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, G., Devos, K.M., Wang, Z., and Gale, M.D. 1995. Cereal genome evolution: Grasses, line up and form a circle. Curr. Biol. 5: 737-739. [DOI] [PubMed] [Google Scholar]
- Morgante, M., Hanafey, M., and Powell, W. 2002. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 30: 194-200. [DOI] [PubMed] [Google Scholar]
- Ouyang, S. and Buell, C.R. 2004. The TIGR Plant Repeat Databases: A collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 32: D360-D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, L.L., Echalier, B., Chao, S., Lazo, G.R., Butler, G.E., Anderson, O.D., Akhunov, E.D., Dvorak, J., Linkiewicz, A.M., Ratnasiri, A., et al. 2004. A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168: 701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush, J., Cho, J., Lee, D., Liang, F., Holt, I., Karamycheva, S., Parvizi, B., Pertea, G., Sultana, R., and White, J. 2001. The TIGR Gene Indices: Analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 29: 159-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Rice Chromosome 10 Sequencing Consortium. 2003. In-depth view of structure, activity, and evolution of rice Chromosome 10. Science 300: 1566-1569. [DOI] [PubMed] [Google Scholar]
- Salamov, A.A. and Solovyev, V.V. 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10: 516-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud, C., Gay, C., Larmande, P., Bes, M., Piffanelli, P., Piegu, B., Droc, G., Regad, F., Bourgeois, E., Meynard, D., et al. 2004. High throughput T-DNA insertion mutagenesis in rice: A first step towards in silico reverse genetics. Plant J. 39: 450-464. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., Matsumoto, T., Yamamoto, K., Sakata, K., Baba, T., Katayose, Y., Wu, J., Niimura, Y., Cheng, Z., Nagamura, Y., et al. 2002. The genome sequence and structure of rice chromosome 1. Nature 420: 312-316. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., Humphray, S., Dunham, A., and French, L. 2002. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 10: 1772-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrells, M.E., La Rota, M., Bermudez-Kandianis, C.E., Greene, R.A., Kantety, R., Munkvold, J.D., Miftahudin, M.A., Ma, X., Gustafson, P.J., Qi, L.L., et al. 2003. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 13: 1818-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, J.E., Endo, T.R., and Gill, B.S. 1992. Toward a cytogenetically based physical map of the wheat genome. Proc. Natl. Acad. Sci. 89: 11307-11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw, C.A., Barbazuk, W.B., Pertea, G., Chan, A.P., Cheung, F., Lee, Y., Zheng, L., van Heeringen, S., Karamycheva, S., Bennetzen, J.L., et al. 2003. Enrichment of gene-coding sequences in maize by genome filtration. Science 302: 2118-2120. [DOI] [PubMed] [Google Scholar]
- Wing, R.A., Ammiraju, J.S.S., Luo, M., Kim, H.R., Yu, Y., Kudrna, D., Goicoechea, J.L., Wang, W., Nelson, W., Rao, K., et al. 2005. The Oryza Map Alignment Project: The golden path to unlocking the genetic potential of wild rice species. Plant Mol. Biol. (in press). [DOI] [PubMed]
- Wortman, J.R., Haas, B.J., Hannick, L.I., Smith Jr., R.K., Maiti, R., Ronning, C.M., Chan, A.P., Yu, C., Ayele, M., Whitelaw, C.A., et al. 2003. Annotation of the Arabidopsis genome. Plant Physiol. 132: 461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Maehara, T., Shimokawa, T., Yamamoto, S., Harada, C., Takazaki, Y., Ono, N., Mukai, Y., Koike, K., Yazaki, J., et al. 2002. A comprehensive rice transcript map containing 6591 expressed sequence tag sites. Plant Cell 14: 525-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Wang, J., Lin, W., Li, S., Li, H., Zhou, J., Ni, P., Dong, W., Hu, S., Zeng, C., et al. 2005. The genomes of Oryza sativa: A history of duplications. PLoS Biol. 3: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Q., Hill, J., Hsiao, J., Moffat, K., Ouyang, S., Cheng, Z., Jiang, J., and Buell, C.R. 2002. Genome sequencing of a 239 kb region of rice chromosome 10L reveals a high frequency of gene duplication and a large chloroplast insertion. Mol. Genet. Genomics 267: 713-720. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.tigr.org/pub/data/rice/rice.chr03.v2.1/; annotation data for chromosome 3.
- http://www.agcol.arizona.edu; Arizona Computational Genomics Lab.
- http://www.agcol.arizona.edu/symap; SyMAP.
- http://www.cbs.dtu.dk/services/SignalP-2.0/; SignalP Server.
- http://www.cbs.dtu.dk/services/TMHMM/; TMHMM Server v 2.0.
- http://www.genome.arizona.edu; AGI/AGCoL.
- http://www.omap.org; Oryza Map Alignment Project.
- http://www.repeatmasker.org;: RepeatMasker.
- http://www.shigen.nig.ac.jp/rice/oryzabase/top/top.jsp; Oryzabase Database.
- http://www.tigr.org/tdb/e2k1/ath1; TIGR Arabidopsis.
- http://www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=rice; TIGR Rice Gene Index.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.