Abstract
DNA transposons are known to frequently capture duplicated fragments of host genes. The evolutionary impact of this phenomenon depends on how frequently the fragments retain protein-coding function as opposed to becoming pseudogenes. Gene fragment duplication by Mutator-like elements (MULEs) has previously been documented in maize, Arabidopsis, and rice. Here we present a rigorous genome-wide analysis of MULEs in the model plant Oryza sativa (domesticated rice). We identify 8274 MULEs with intact termini and target-site duplications (TSDs) and show that 1337 of them contain duplicated host gene fragments. Through a detailed examination of the 5% of duplicated gene fragments that are transcribed, we demonstrate that virtually all cases contain pseudogenic features such as fragmented conserved protein domains, frameshifts, and premature stop codons. In addition, we show that the distribution of the ratio of nonsynonymous to synonymous amino acid substitution rates for the duplications agrees with the expected distribution for pseudogenes. We conclude that MULE-mediated host gene duplication results in the formation of pseudogenes, not novel functional protein-coding genes; however, the transcribed duplications possess characteristics consistent with a potential role in the regulation of host gene expression.
With their ubiquitous presence in eukaryotic genomes and their capacity to generate mutations, transposons are important players in genome evolution, influencing genome size, function, and structure (Kazazian Jr. 2004). They are involved in the duplication of parts of their host genome through several different mechanisms, such as ectopic exchange between neighboring transposons via unequal crossing over (Lim and Simmons 1994) and macrotransposition (Ralston et al. 1989). In addition to these mechanisms, which produce duplications of the genomic sequence between two transposons, fragments of host genes can be found duplicated within a single transposon. In the case of retrotransposons, this phenomenon is known as transduction and involves readthrough transcription from a retrotransposon promoter into adjacent host gene sequences and the incorporation of the host gene into the transposon sequence during reverse transcription (Bureau et al. 1994; Kazazian Jr. 2004). The mechanism of gene-fragment duplication by DNA transposons, herein referred to as “transduplication,” is unknown, but is clearly different from transduction because duplicated fragments retain introns. The potential functional impact of a duplicated gene fragment depends on whether it forms a protein-coding gene or a pseudogene; while the latter appears to be predominant for transduced gene fragments, the outcome of transduplication events is unclear.
MULEs are DNA transposons found in high-copy number in plants as well as in fungi and prokaryotes (Yu et al. 2000; Turcotte et al. 2001; Chalvet et al. 2003). They are characterized by long terminal inverted repeats (TIRs; longer than 100 bp), lengths ranging from a few hundred base pairs to more than 30 kbp, hypervariable internal sequence, and a 9-11 bp target site duplication (TSD). MULEs lacking TIRs (i.e., non-TIR MULEs) are rare in rice but predominate in Arabidopsis (Le et al. 2000; Yu et al. 2000). MULE families are typically composed of a few autonomous elements containing a transposase gene, mudrA, and tens to hundreds of nonautonomous elements that lack functional mudrA, but which may be mobilized in trans (Le et al. 2000; Yu et al. 2000; International Rice Genome Sequencing Project [IRGSP] 2005).
Gene fragment transduplication by MULEs has previously been documented in maize (Talbert and Chandler 1988), Arabidopsis (Le et al. 2000; Yu et al. 2000), and rice (Turcotte et al. 2001). Recently, it has been suggested as a mechanism for the creation of novel protein-coding genes (Jiang et al. 2004). In this report, we demonstrate that the evidence is to the converse, that widespread MULE-mediated transduplication in rice results in the formation of pseudogenes that lack protein-coding function. Even the 5.3% of the transduplicates that are transcribed have one or more features that would disable any potential translation product. Furthermore, the synonymous amino acid substitution rate is consistent with reduced purifying selection. However, the transcribed pseudogenes may function in the regulation of host gene expression.
Results and Discussion
Members of rice MULE families were identified by (1) determining the location of mudrA genes, (2) examining flanking regions for TIRs or terminal structures, (3) using the termini of complete putative autonomous elements to identify the corresponding nonautonomous elements, and (4) removing elements that lack characteristic TSDs (Supplemental Table 1). This protocol yields well-supported family classification and avoids the potential mis-annotation of elements with similar structural features to, but no evolutionary connection with, bona fide MULEs. In total, we mined 8274 MULEs from the rice genome, with an estimated 3% false positive and 17% false negative rate. The distribution of MULEs along the rice chromosomes (Supplemental Fig. 1) is similar to that of other DNA transposons (IRGSP 2005), with higher element density in euchromatic regions (23 MULEs/Mbp) than in centromeric and pericentromeric regions (16 MULEs/Mbp).
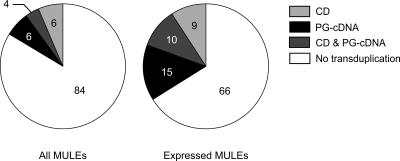
To determine the prevalence of transduplication in our data set, we identified 1968 MULEs (24%) with high sequence similarity (BLASTN E-value ≤ 1 × 10-37) to coding regions of predicted genes that are unrelated to transposons (Supplemental Table 2). Because gene prediction is relatively inaccurate, we focused on the 1337 MULEs (16%) with transduplicates for which the paralogous host gene is confirmed by a full-length cDNA (Supplemental Table 3), or which encode a conserved protein domain (Supplemental Table 4) (Fig. 1). Our methods sacrifice sensitivity for specificity, so we expect the actual number of transduplicates to be considerably larger. The size of transduplicated regions ranges from 95 to 7742 bp, with a mean of 305 bp.
Figure 1.
Distribution of supported transduplicates among rice MULEs. Percent contribution is shown for each MULE category. (PG) Predicted gene; (PG-cDNA) predicted gene supported by a cDNA; (CD) conserved domain.
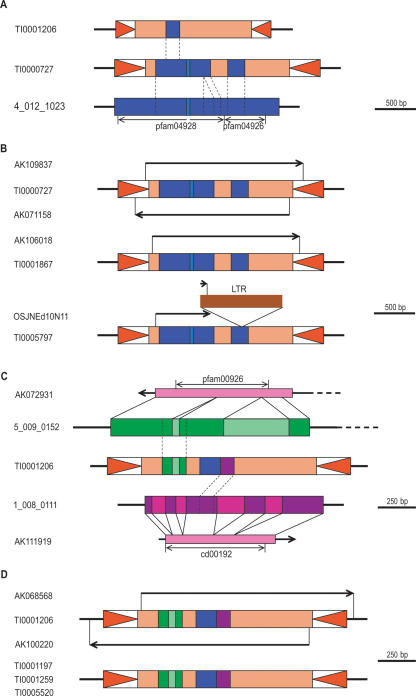
We clustered MULEs that contained the same transduplicate(s) and found that 64% are single-copy, and thus were formed by unique transduplication events. The remainder of the MULEs are multicopy, and thus presumably arose by replicative transposition of an ancestral MULE that had previously undergone transduplication. The number of MULEs per cluster followed a power-law relationship, decreasing rapidly with cluster size; 99% of the clusters had fewer than 10 members. For example, we identified seven MULEs in two families that contain a transduplicated putative poly(A) polymerase (PAP) gene fragment (Fig. 2). In addition, we found that in 25% of the cases, a single MULE contains separate transduplicates of more than one host gene fragment, presumably the result of multiple independent transduplication events. In general, the insertion and rearrangement of transduplicated sequences may account for the high level of divergence both within and between MULE families (Yu et al. 2000).
Figure 2.
Transduplication and diversification of a poly(A) polymerase (PAP) gene fragment in the rice genome. (A) The central region of a putative PAP gene on chromosome 4 (4_012_1023) was transduplicated in a MULE that later replicated and, in one branch of the group, internal deletions reduced the size of the transduplicate. (B) Three highly similar copies in the rice genome possess the larger transduplicate, each having a different pattern of expression, including TI0000727, which generates sense and antisense transcripts and TI0005797 with the transcript terminating in a retrotransposon solo long terminal repeat (LTR). (C) The MULE copy with the smaller transduplicate subsequently transduplicated regions from two additional rice genes, a ribA-like gene (5_009_0152) and a serine/threonine protein kinase (1_008_0111). (D) There are four MULEs with these three transduplicates and one of them is transcribed in both the sense and antisense direction. Mapped cDNAs and ESTs are represented by arrows, pink bars represent ORFs, and regions containing conserved protein domains are indicated by double-headed arrows. Genes are shown in blue, green, and purple, and introns are shown in a lighter shade. Inverted triangles represent TIRs.
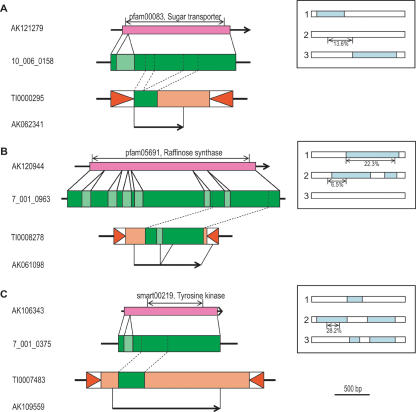
The existence of an extensive data set of rice full-length cDNAs (Kikuchi et al. 2003) allowed us to identify transcribed transduplicates by comparing the position of elements to mapped cDNAs in the rice genome. A total of 310 MULEs overlap cDNAs (Supplemental Table 5), and in 66 of these, the expressed region includes one or more transduplicates, for a total of 72 transcribed transduplicates (Fig. 1, Supplemental Table 6). All of the transcribed transduplicates have disablements preventing them from expressing a functional protein. First, six expressed transduplicates (8%) are not within a predicted ORF of the cDNA (Fig. 3A). Second, for 13 expressed transduplicates (18%) that overlap a cDNA ORF, the predicted coding region is not in the same reading frame or has accumulated frameshift mutations and premature stop codons (Fig. 3B). Third, for 22 expressed transduplicates with regions in the same reading frame as a cDNA ORF, the overlap does not contain protein domains found within the transduplicate (21%) or these domains are truncated (10%; Fig. 3C). The only transduplicate with an intact conserved domain within a possible cDNA ORF is the pentatricopeptide repeat (PPR) transduplicate found in TI0007876 (Supplemental Table 6); however, all known PPR proteins contain multiple PPRs (Lurin et al. 2004), so the functionality of a single repeat is questionable. Fourth, 23 transduplicates (32%) are expressed only in the antisense direction and therefore do not encode a protein. Finally, 10 of the expressed predicted host genes contain only truncated domains and may themselves be expressed pseudogenes. The predicted genes matching the nine remaining transduplicates do not contain any known conserved domain. In addition, a search of the SWISS-PROT and SCOP protein databases reveals that many of the transduplicates show similarity to known proteins and protein domains, but the matching regions contain frameshifts, premature stop codons, or are truncated (Supplemental Tables 6, 7). Interestingly, a recent study of CACTA-like DNA transposons with transduplicated gene fragments also revealed that they are transcribed pseudogenes (Kawasaki and Nitasaka 2004).
Figure 3.
Examples of expressed transduplicates. (A) The transduplicated conserved domain fragment is found within the cDNA, but it does not overlap with any of the possible ORFs. (Inset) White bars represent the three forward frames of MULE cDNAs, and pale blue regions indicate possible ORFs (not to scale). (B) As a result of a frameshift mutation, the transduplicated conserved domain fragment is split between two cDNA frames and two possible ORFs. (C) The transduplicate is found within one of the possible cDNA ORFs, but the conserved domain is truncated. Mapped cDNAs are represented by arrows, pink bars represent ORFs, and regions containing conserved domains are indicated by double-headed arrows. Genes are shown in green and introns are in light-green. Inverted triangles represent TIRs.
In contrast to these results, a recently published study of MULE transduplication in rice (Jiang et al. 2004), concludes that the coding sequence of over 10% of transduplicates might have been functionally constrained. Our study differed from Jiang et al. (2004) in several ways. We identified and analyzed transduplicates on all 12 rice chromosomes, whereas they limited their analysis to chromosomes 1 and 10, then extrapolated to make a genome-wide estimate. We recorded duplicated regions that are highly similar to a transcribed gene or conserved domain, whereas they used similarity to any nonhypothetical nontransposase protein, regardless of additional support. According to Jiang et al. (2004), of a sample of 100 of their transduplicates, 20 were not found to be similar to any genomic sequence outside of the MULE, seven of the remaining 80 were not similar to a predicted host coding region, and only 46 of the suspected host-coding sequences were supported by a cDNA. After taking into account these differences, many of our basic results agree with Jiang et al. (2004). For example, we identified 1337 MULEs with transduplicates, whereas Jiang et al. estimate 3200 in total, but based on their sample, fewer than 46% (1500) of these would have host genes with matching cDNAs. Additional similar results include the fraction of transduplicate-containing MULEs present in more than one copy in the genome (36%; Jiang et al. 20%); the fraction of transduplicates that are transcribed (5%; Jiang et al. 5%); and the fraction of MULEs containing multiple transduplicates (25%; Jiang et al. 23%). Despite the consistency of these basic results, our key result that virtually all transcribed transduplicates contain coding-sequence disablements contradicts the conclusion of Jiang et al. (2004) that many transduplicates show evidence of purifying selection on the coding sequence. The reason for this contradiction is that Jiang et al. (2004) based their conclusion on a misinterpreted dN/dS analysis.
The ratio of dN (nonsynonymous substitutions per nonsynonymous site) to dS (synonymous substitutions per synonymous site) is a measure of selective constraint on coding sequences (Li et al. 1981). dN and dS can be estimated using a number of substitution models and methods, and the estimates are sensitive to these choices and other complications such as the GC content of the sequences and their genomic context (Bustamante et al. 2002). A pair of sequences will have dN/dS ≪ 1 if both sequences have been under purifying selection, dN/dS < 1 if one sequence has been under purifying selection but the other drifting neutrally, dN/dS ∼ 1 if both sequences are drifting neutrally, and rarely, dN/dS > 1 at specific sites that are under adaptive selection. For example, mammalian gene-gene pairs have mean dN/dS ∼ 0.15 (Waterston et al. 2002; Jorgensen et al. 2005), while human gene-pseudogene pairs have mean dN/dS ∼ 0.5 (Torrents et al. 2003; Zhang et al. 2003).
Testing for purifying selection on gene-transduplicate pairs using pairwise dN/dS is difficult because the standard null hypothesis for gene-gene pairs, that dN/dS = 1, is invalid. In this case, we expect dN/dS < 1 due to selection on the host gene, even if the transduplicate has been drifting neutrally. Jiang et al. (2004) overlooked this critical consideration and based their conclusion that >10% of transduplicates have been under purifying selection primarily on the results of a statistical test using this invalid null hypothesis. Their conclusion is therefore unfounded.
It would be useful to construct a generally valid null hypothesis of the form dN/dS = α, where α is the expected pairwise dN/dS ratio between a neutrally drifting transduplicate and a corresponding host gene under purifying selection. Unfortunately, α would vary with the strength of selective pressure on the host gene and so would need to be estimated on a case by case basis using, for example, additional homologous functional genes. Such an approach would be difficult to implement en masse for large numbers of transduplicates.
Instead, we adopted an alternative method and compared the dN/dS distribution of all qualified transduplicates with a benchmark distribution, as follows. We calculated the dN/dS ratios of all 47 transduplicates for which the transduplicate and host gene cDNA sequences could be aligned for a minimum length of 150 bp (87%-96% identity). This gave a dN/dS distribution with a median of 0.55 and standard deviation of 0.36 (Supplemental Fig. 2; Supplemental Table 6). As a benchmark, we used the published dN/dS distribution of a large set of human processed pseudogenes (Zhang et al. 2003). If a large fraction of transduplicates had been under selection, it would have been reflected in a skewed transduplicate distribution relative to the benchmark. However, the distribution profiles are similar, both peaking near 0.5 with most dN/dS ratios in the range from 0.4 to 0.7. Although this comparison must be interpreted cautiously due to differences between human processed pseudogene benchmark population and the rice transduplicate sample, such as GC-content and sequence divergence, the similarity between the two distributions is consistent with the observed high incidence of coding-sequence disablements and our hypothesis that transduplicates are noncoding pseudogenes. This suggests that if the process of transduplication ever does result in the formation of functional protein-coding genes, it occurs only rarely. Of over 1300 transduplicates in the extant rice (japonica) genome, we were unable to identify a single candidate functional protein-coding transduplicate.
Contrary to the standard definition of pseudogenes as nonfunctional, it has been shown that an inability to encode functional proteins often does not preclude pseudogenes from performing other functional roles, such as the provision of sequence reservoirs for gene conversion, or the regulation of paralogous gene expression (Korneev et al. 1999; Balakirev and Ayala 2003; Hirotsune et al. 2003). Sense (51), antisense (23), and bidirectional (3) transcripts containing transduplicated sequences (Supplemental Table 6) could negatively affect host gene expression through RNA-mediated gene silencing (i.e., RNA interference or RNAi) by mechanisms including mRNA degradation, translational suppression, or DNA methylation of the host gene sequence (Meister and Tuschl 2004). For prokaryotic and eukaryotic organisms, endogenous noncoding transcripts are known to suppress the expression of genes with which they share short regions of high nucleotide sequence similarity (Korneev et al. 1999; Palatnik et al. 2003; Vazquez et al. 2004). Although these RNAs are often processed from a stem-loop precursor structure (i.e., miRNAs; Bartel 2004), partial complementarity is often sufficient to cause silencing (Korneev et al. 1999; Palatnik et al. 2003; Vazquez et al. 2004). Therefore, transcribed MULE transduplicates possess features that could allow them to silence host genes. Interestingly, categories of proteins that predominate among known plant miRNA targets, such as transcription factors involved in development and proteins involved in ubiquitination and sulfur assimilation (Bonnet et al. 2004), are also frequently found among sources of expressed transduplicates (Supplemental Table 6).
Transcription of MULE transduplicates is likely to be differentially regulated, since transduplicates exist in a different regulatory context from their corresponding host genes. The eukaryotic TATA box is usually found -20 to -35 bp from the transcription start site, and in most plant promoters, functionally important cis-regulatory elements reside within 300 bp upstream of the transcription start site (Guilfoyle 1997). Since the transcription start site for 153 of 235 transcripts that initiate within a MULE sequence is located <500 bp from the element terminus, the promoter is likely located within the TIR. In fact, the TIRs of the maize MuDR element harbor promoters for mudrA and mudrB (Raizada et al. 2001), and while the TIR sequences of rice and maize MULEs are divergent, the majority of rice MULE TIRs contain putative TATA boxes and other promoter elements. In addition, transposition of MULEs with transduplicates may bring them into proximity to other cis-regulatory sequences.
In summary, our study confirms that MULEs mediate the duplication of host gene fragments, but we found no evidence that such transduplicates may frequently evolve into novel functional protein-coding genes. All expressed transduplicates with conserved domains have disablements preventing them from encoding functional proteins, and their dN/dS ratios do not indicate that they have been evolving under purifying selection. However, we cannot rule out the possible existence of ancient protein-coding transduplicates that are no longer recognizable as MULEs. Of more significance may be the fact that transduplicates constitute a large source of pseudogenes with characteristics suggestive of a role in regulating host gene expression, but further analysis is needed to evaluate this possibility.
Methods
MULE detection
We used HMMER (Eddy 1998) and RepeatMasker (http://www.repeatmasker.org) to identify sequences of the rice genome [Oryza sativa, ssp. japonica; International Rice Genome Sequencing Project 2005 (IRGSP) Build 2.0 pseudomolecules; http://rgp.dna.affrc.go.jp/IRGSP/Build2/build2.html; DDBJ accessions AP006867-AP006877 and NCBI accession AE016959] similar to known MULE families with autonomous members (Juretic et al. 2004). We tentatively identified as MULEs pairs of sequences similar to MULE termini of the same family, with inverted orientation and separated by <30 kbp. We retained MULEs longer than 10 kbp only if manual inspection of their TIRs, repetitiveness, and TSDs supported their authenticity. We built custom software to identify TSDs located no more distant than 17 bp from the termini with at most two mismatches, rejecting MULEs lacking TSDs. From among overlapping MULEs, a single element was selected based on TSD quality (located nearest the predicted termini with fewest mismatches); nesting was permitted. We detected additional MULEs using BLASTN (Altschul et al. 1990) searches of 10 kbp sequences flanking either side of mudrA-like genes not associated with a MULE in our data set.
To estimate false positives, 140 MULEs were manually inspected; 136 (97%) were authentic. To estimate false negatives, we mapped 18 independently identified MULEs (Jiang et al. 2004); 15 mapped to identical positions as MULEs in our data set; 3 (17%) were absent.
To determine MULE density in euchromatic and heterochromatic regions, pericentromeric regions were defined by mapping marker positions to regions with reduced recombination surrounding the centromere of each chromosome, defined by comparing genetic to physical distances (S.I. Wright, pers. comm.).
Gene prediction
Gene models for Build 2.0 pseudomolecules were predicted using FGENESH (Salamov and Solovyev 2000) in monocot trained settings. Models were rejected if the ORFs were incomplete due to missing initiation or termination codons, if they encoded proteins shorter than 50 amino acids, or if they contained ambiguous sequence (represented by the symbol “N”) (T. Sasaki, pers. comm.).
Transposon-related predicted genes (PGs) were identified by the following four methods and rejected: (1) BLASTN against a comprehensive rice transposon nucleotide sequence database (TIGR Oryza Repeats.v2; E <10-10) (Ouyang and Buell 2004; T. Sasaki, pers. comm.; http://www.tigr.org/tdb/e2k1/osa1/blastsearch.shtml); (2) BLASTP of translated PG coding sequences (CDS) against a custom library of transposon proteins (NCBI accessions BAC15617.2, AAK27803.1, NP_920829.1, BAC15617.2, BAC15618.2, AAA70218.1, AAA70219.1, AAA70220.1, AAA66266.1, AAA66267.1, AAA66268.1, AAA21566.1, AAA21567.1, CAA25498.1, CAA25499.1, CAA26444.1, CAA26445.1, CAA27457.1, CAA27458.1, CAA27459.1, CAA29005.1, CAA29006.1, CAA36480.1); (3) overlap with a MULE or MULE fragment in our data set; (4) BLASTN of transduplicate clusters of more than 20 PGs or 40 MULEs (see Transduplicate detection, below).
cDNA mapping
We used BLAT (Kent 2002) to map rice full-length cDNAs (Kikuchi et al. 2003) to Build 2.0 pseudomolecules. We aligned 29,364 of 32,127 cDNAs (91%) by selecting the best alignment of minimum 99% sequence identity.
Transduplicate detection
Transduplicates were identified by BLASTN (E <10-37) of MULE internal sequences, trimmed by 200 bp at each end to partially remove TIRs, against nontransposon-related PG CDS (Supplemental Table 2). We then identified transduplicates corresponding to expressed PGs, defined as CDS overlapping a mapped cDNA (Supplemental Table 3).
Because some transduplicated regions shared significant similarity to more than one PG, PGs with overlapping (>30 bp) BLASTN high-scoring segment pairs (HSPs) to one or more MULEs were clustered. Eighty percent of the clusters contained only a single PG, and 99.9% were sized 10 or smaller. PG clusters occurring in only one MULE were interpreted as single, independent transduplication events, while PG clusters occurring in more than one MULE were interpreted as being descended by replicative transposition of a common transduplicate-containing ancestral MULE. In addition, individual MULEs containing multiple PG clusters were interpreted as resulting from multiple separate transduplication events.
Conserved domain detection
Transduplicates containing conserved protein domains (CD) were located by querying the NCBI Conserved Domain Database (CDD) (Marchler-Bauer et al. 2005) with 6-frame translations of each MULE (Supplemental Table 4). Five mudrA conserved domains (smart00575, pfam03108, pfam00872, pfam04434, and COG3328) were identified by querying Zea mays mudrA (NCBI accession M76978) against the CDD and excluded from consideration as transduplicates. Additional transposon-related CDs were identified by examining CD descriptions and also excluded.
Coding-sequence disablement and truncation
We identified 66 MULEs containing 72 high-stringency transduplicates (Supplemental Table 6), defined as a transduplicate that overlaps a cDNA and contains a CD or has a corresponding host PG that also overlaps a cDNA. For each high-stringency transduplicate, we verified the TSDs, TIRs, and alignments to conserved domains, PGs and cDNAs. cDNAs overlapping high-stringency transduplicates were compared with known nonfragmentary SWISS-PROT proteins (Bairoch et al. 2005) and known SCOP protein domains (Chandonia et al. 2004) to test for frameshifts and premature stop codons using a modification of procedures described previously (Harrison et al. 2002; Zhang et al. 2003) (Supplemental Tables 6, 7). We also identified transduplicate cDNAs with significant protein-domain truncations, where protein-domain mappings were shorter than 50% of the domain length (Harrison et al. 2003).
Synonymous and nonsynonymous substitution rate
We used BL2SEQ (Tatusova and Madden 1999) to align PG cDNA ORFs to corresponding high-stringency transduplicates. Forty-seven alignments longer than 150 bp (87%-96% identity) were manually curated by joining HSPs (E <10-4) on the correct strand to eliminate frameshifts and redundancies. dN/dS ratios were calculated using PAML CODONML (runmode = -2, CodonFreq = 0) (Yang 1997; Supplemental Table 6).
Supplementary Material
Acknowledgments
This study was supported by operating and genomics grants from the National Science and Engineering Research Council (NSERC) of Canada to T.E.B. and P.M.H. and a grant from the McGill University William Dawson Scholar Chair to T.E.B. The authors thank Ben Burr, Takuji Sasaki, and Takashi Matsumoto for access to the IRGSP sequence and annotation data set, Rebecca Cowan, Daniel Schoen, and Stephen Wright for critical reading of the manuscript, and Stephen Wright for analysis of pericentromeric locations.
Footnotes
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: S.I. Wright and T. Sasaki.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.4064205.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Bairoch, A., Apweiler, R., Wu, C.H., Barker, W.C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., et al. 2005. The Universal Protein Resource (UniProt). Nucleic Acids Res. 33: D154-D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakirev, E.S. and Ayala, F.J. 2003. Pseudogenes: Are they “junk” or functional DNA? Annu. Rev. Genet. 37: 123-151. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281-297. [DOI] [PubMed] [Google Scholar]
- Bonnet, E., Wuyts, J., Rouze, P., and Van de Peer, Y. 2004. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. 101: 11511-11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T.E., White, S.E., and Wessler, S.R. 1994. Transduction of a cellular gene by a plant retroelement. Cell 77: 479-480. [DOI] [PubMed] [Google Scholar]
- Bustamante, C.D., Nielsen, R., and Hartl, D.L. 2002. A maximum likelihood method for analyzing pseudogene evolution: Implications for silent site evolution in humans and rodents. Mol. Biol. Evol. 19: 110-117. [DOI] [PubMed] [Google Scholar]
- Chalvet, F., Grimaldi, C., Kaper, F., Langin, T., and Daboussi, M.J. 2003. Hop, an active Mutator-like element in the genome of the fungus Fusarium oxysporum. Mol. Biol. Evol. 20: 1362-1375. [DOI] [PubMed] [Google Scholar]
- Chandonia, J.M., Hon, G., Walker, N.S., Lo Conte, L., Koehl, P., Levitt, M., and Brenner, S.E. 2004. The ASTRAL Compendium in 2004. Nucleic Acids Res. 32: D189-D192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy, S.R. 1998. Profile hidden Markov models. Bioinformatics 14: 755-763. [DOI] [PubMed] [Google Scholar]
- Guilfoyle, T.J. 1997. In Genetic engineering (ed. J.K. Setlow), pp. 15-47. Plenum Press, New York.
- Harrison, P.M., Hegyi, H., Balasubramanian, S., Luscombe, N.M., Bertone, P., Echols, N., Johnson, T., and Gerstein, M. 2002. Molecular fossils in the human genome: Identification and analysis of the pseudogenes in chromosomes 21 and 22. Genome Res. 12: 272-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, P.M., Carriero, N., Liu, Y., and Gerstein, M. 2003. A “polyORFomic” analysis of prokaryote genomes using disabled-homology filtering reveals conserved but undiscovered short ORFs. J. Mol. Biol. 333: 885-892. [DOI] [PubMed] [Google Scholar]
- Hirotsune, S., Yoshida, N., Chen, A., Garrett, L., Sugiyama, F., Takahashi, S., Yagami, K., Wynshaw-Boris, A., and Yoshiki, A. 2003. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature 423: 91-96. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (IRGSP). 2005. The map-based sequence of the rice genome. Nature (in press) [DOI] [PubMed]
- Jiang, N., Bao, Z., Zhang, X., Eddy, S.R., and Wessler, S.R. 2004. Pack-MULE transposable elements mediate gene evolution in plants. Nature 431: 569-573. [DOI] [PubMed] [Google Scholar]
- Jorgensen, F.G., Hobolth, A., Hornshoj, H., Bendixen, C., Fredholm, M., and Schierup, M.H. 2005. Comparative analysis of protein coding sequences from human, mouse and the domesticated pig. BMC Biol. 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretic, N., Bureau, T.E., and Bruskiewich, R.M. 2004. Transposable element annotation of the rice genome. Bioinformatics 20: 155-160. [DOI] [PubMed] [Google Scholar]
- Kawasaki, S. and Nitasaka, E. 2004. Characterization of Tpn1 family in the Japanese morning glory: En/Spm-related transposable elements capturing host genes. Plant Cell Physiol. 45: 933-944. [DOI] [PubMed] [Google Scholar]
- Kazazian Jr., H.H. 2004. Mobile elements: Drivers of genome evolution. Science 303: 1626-1632. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12: 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, S., Satoh, K., Nagata, T., Kawagashira, N., Doi, K., Kishimoto, N., Yazaki, J., Ishikawa, M., Yamada, H., Ooka, H., et al. 2003. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376-379. [DOI] [PubMed] [Google Scholar]
- Korneev, S.A., Park, J.H., and O'Shea, M. 1999. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J. Neurosci. 19: 7711-7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, Q.H., Wright, S., Yu, Z., and Bureau, T. 2000. Transposon diversity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 97: 7376-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.H., Gojobori, T., and Nei, M. 1981. Pseudogenes as a paradigm of neutral evolution. Nature 292: 237-239. [DOI] [PubMed] [Google Scholar]
- Lim, J.K. and Simmons, M.J. 1994. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. Bioessays 16: 269-275. [DOI] [PubMed] [Google Scholar]
- Lurin, C., Andres, C., Aubourg, S., Bellaoui, M., Bitton, F., Bruyere, C., Caboche, M., Debast, C., Gualberto, J., Hoffmann, B., et al. 2004. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer, A., Anderson, J.B., Cherukuri, P.F., DeWeese-Scott, C., Geer, L.Y., Gwadz, M., He, S., Hurwitz, D.I., Jackson, J.D., Ke, Z., et al. 2005. CDD: A conserved domain database for protein classification. Nucleic Acids Res. 33: D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, G. and Tuschl, T. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431: 343-349. [DOI] [PubMed] [Google Scholar]
- Ouyang, S. and Buell, C.R. 2004. The TIGR Plant Repeat Databases: A collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 32: D360-D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425: 257-263. [DOI] [PubMed] [Google Scholar]
- Raizada, M.N., Benito, M.I., and Walbot, V. 2001. The MuDR transposon terminal inverted repeat contains a complex plant promoter directing distinct somatic and germinal programs. Plant J. 25: 79-91. [DOI] [PubMed] [Google Scholar]
- Ralston, E., English, J., and Dooner, H.K. 1989. Chromosome-breaking structure in maize involving a fractured Ac element. Proc. Natl. Acad. Sci. 86: 9451-9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov, A.A. and Solovyev, V.V. 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10: 516-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, L.E. and Chandler, V.L. 1988. Characterization of a highly conserved sequence related to mutator transposable elements in maize. Mol. Biol. Evol. 5: 519-529. [DOI] [PubMed] [Google Scholar]
- Tatusova, T.A. and Madden, T.L. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174: 247-250. [DOI] [PubMed] [Google Scholar]
- Torrents, D., Suyama, M., Zdobnov, E., and Bork, P. 2003. A genome-wide survey of human pseudogenes. Genome Res. 13: 2559-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte, K., Srinivasan, S., and Bureau, T. 2001. Survey of transposable elements from rice genomic sequences. Plant J. 25: 169-179. [DOI] [PubMed] [Google Scholar]
- Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A.C., Hilbert, J.L., Bartel, D.P., and Crete, P. 2004. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16: 69-79. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555-556. [DOI] [PubMed] [Google Scholar]
- Yu, Z., Wright, S.I., and Bureau, T.E. 2000. Mutator-like elements in Arabidopsis thaliana. Structure, diversity and evolution. Genetics 156: 2019-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Harrison, P.M., Liu, Y., and Gerstein, M. 2003. Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 13: 2541-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.repeatmasker.org; RepeatMasker home.
- http://rgp.dna.affrc.go.jp/IRGSP/Build2/build2.html; IRGSP Build 2.0 pseudomolecules.
- http://www.tigr.org/tdb/e2k1/osa1/blastsearch.shtml; the TIGR Oryza Repeat database.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.