Abstract
Genetic screens in zebrafish (Danio rerio) have identified mutations that define the roles of hundreds of essential vertebrate genes. Genetic maps can link mutant phenotype with gene sequence by providing candidate genes for mutations and polymorphic genetic markers useful in positional cloning projects. Here we report a zebrafish genetic map comprising 4073 polymorphic markers, with more than twice the number of coding sequences localized in previously reported zebrafish genetic maps. We use this map in comparative studies to identify numerous regions of synteny conserved among the genomes of zebrafish, Tetraodon, and human. In addition, we use our map to analyze gene duplication in the zebrafish and Tetraodon genomes. Current evidence suggests that a whole-genome duplication occurred in the teleost lineage after it split from the tetrapod lineage, and that only a subset of the duplicates have been retained in modern teleost genomes. It has been proposed that differential retention of duplicate genes may have facilitated the isolation of nascent species formed during the vast radiation of teleosts. We find that different duplicated genes have been retained in zebrafish and Tetraodon, although similar numbers of duplicates remain in both genomes. Finally, we use comparative mapping data to address the proposal that the common ancestor of vertebrates had a genome consisting of 12 chromosomes. In a three-way comparison between the genomes of zebrafish, Tetraodon, and human, our analysis delineates the gene content for 11 of these 12 proposed ancestral chromosomes.
Genome sequencing projects have revealed a nearly complete picture of the 20,000-30,000 genes that constitute the genomes of human and other vertebrates (Lander et al. 2001; Venter et al. 2001; Aparicio et al. 2002; Waterston et al. 2002; Kirkness et al. 2003; Gibbs et al. 2004; Jaillon et al. 2004). Although the sequences of these genes are now largely known, elucidating their functions continues to be a significant challenge. Large-scale forward genetic screens in zebrafish have uncovered thousands of mutations defining hundreds of genes essential for vertebrate embryonic development (Driever et al. 1996; Haffter et al. 1996). In addition, more recent targeted screens have identified many mutations that disrupt specific aspects of vertebrate development and physiology (e.g., Birely et al. 2005; Lyons et al. 2005). Because gene functions are often conserved in vertebrates, functional studies with zebrafish mutations provide valuable insights into the role of human genes in development and disease. There has been significant progress in identifying the molecular nature of mutations in zebrafish, but many mutated genes defined in genetic screens remain to be analyzed molecularly. Genetic maps accelerate the molecular analysis of mutations both by providing a large pool of mapped candidate genes and by localizing genetic polymorphisms that facilitate positional cloning projects.
Sequence analysis can identify orthologous genes in different species, and gene maps therefore allow comparisons among the genomes of different species. Conserved synteny (the presence of two or more orthologous gene pairs on a single chromosome in each of two different species) defines regions of ancestral chromosomes that have been maintained through evolution. By allowing the transfer of map information between species, comparative maps increase the pool of mapped genes that can be considered as candidates for mutations. Previous comparative maps (Gates et al. 1999; Barbazuk et al. 2000; Woods et al. 2000) have identified regions of conserved synteny between zebrafish and human, thereby facilitating the cloning of mutations based on analysis of human candidate genes (e.g., Karlstrom et al. 1999; Donovan et al. 2000; Miller et al. 2000; Varga et al. 2001; Lyons et al. 2005). Additional analyses using an expanded zebrafish gene map will further define syntenic regions conserved between zebrafish and other vertebrates with more extensively annotated genome sequences.
Comparative mapping can also illuminate the history of chromosome evolution. Previous analyses comparing zebrafish genetic maps with the genomes of other vertebrate species have suggested that a whole-genome duplication occurred in the teleost lineage, after its divergence from the tetrapod lineage (Amores et al. 1998; Postlethwait et al. 1998, 2000; Force et al. 1999; Gates et al. 1999; Naruse et al. 2000; Taylor et al. 2003; Woods et al. 2000). Furthermore, analysis of the patterns of duplicated genes in zebrafish and medaka suggested the hypothesis that the ancestral vertebrate karyotype comprised 12 chromosomes, which expanded in number in the teleost lineage via duplication, and in the tetrapod lineage largely by fragmentation (Postlethwait et al. 2000; Naruse et al. 2004). Support for the hypothesized ancestral karyotype number derived from analysis of the Tetraodon nigroviridis genome sequence, and extensive comparisons of gene content between human and Tetraodon chromosomes have facilitated the detailed dissection of interchromosomal rearrangements that have shaped extant chromosomes (Jaillon et al. 2004). Although these two studies proposed a similar number of chromosomes for the ancestral vertebrate karyotype, they used different approaches and data from different fish species (Jaillon et al. 2004; Naruse et al. 2004), and there has been no previous comparison to determine if the two analyses arrived at similar ancestral gene maps.
Here we report an extensive gene-based meiotic map for zebrafish, with more than double the number of genes and ESTs present on previously reported genetic maps. This map will facilitate the molecular identification of mutations, and will therefore constitute a valuable resource in elucidating gene function. Moreover, because it contains a large data set that has been extensively checked for errors, the map will provide an important framework to facilitate the assembly of the zebrafish genome sequence. We use this map in comparisons between the genomes of human and Tetraodon to define numerous regions of conserved synteny, to compare the outcome of gene duplication events in zebrafish and Tetraodon, and to present a comprehensive assessment of the possible composition of the ancestral vertebrate karyotype.
Results and Discussion
Genetic mapping of zebrafish genes and ESTs
By using a previously described homozygous diploid mapping panel (Kelly et al. 2000), we increased the number of genes and ESTs localized on the zebrafish meiotic map from 1503 (Woods et al. 2000) to 3417 (Fig. 1, enclosed poster). Assuming the zebrafish has about the same number of genes as do Tetraodon and other vertebrates (Jaillon et al. 2004), the mapped sequences probably represent >10% of the genes in the zebrafish genome. The majority (2992) of these genes and ESTs were localized via single-strand conformational polymorphism (SSCP) analysis as previously described (Kelly et al. 2000; Woods et al. 2000), whereas 425 markers were mapped by using PCR-based restriction fragment length polymorphisms (RFLPs). This work raises the total number of markers on the map to 4073, including 656 previously mapped simple sequence length polymorphisms (Shimoda et al. 1999) that allow comparison among various zebrafish meiotic and radiation hybrid maps. The data set contained a total of 165,180 genotypes, for an average of 40.6 genotypes per mapped marker. The 4073 markers occupied 902 unique map positions, and the total map length was 3192 cM, giving an average distance of 3.5 cM between groups of mapped markers. Accession numbers, primer sequences, restriction enzymes, and UniGene assignments for mapped genes and ESTs are available in Supplemental Table 1.
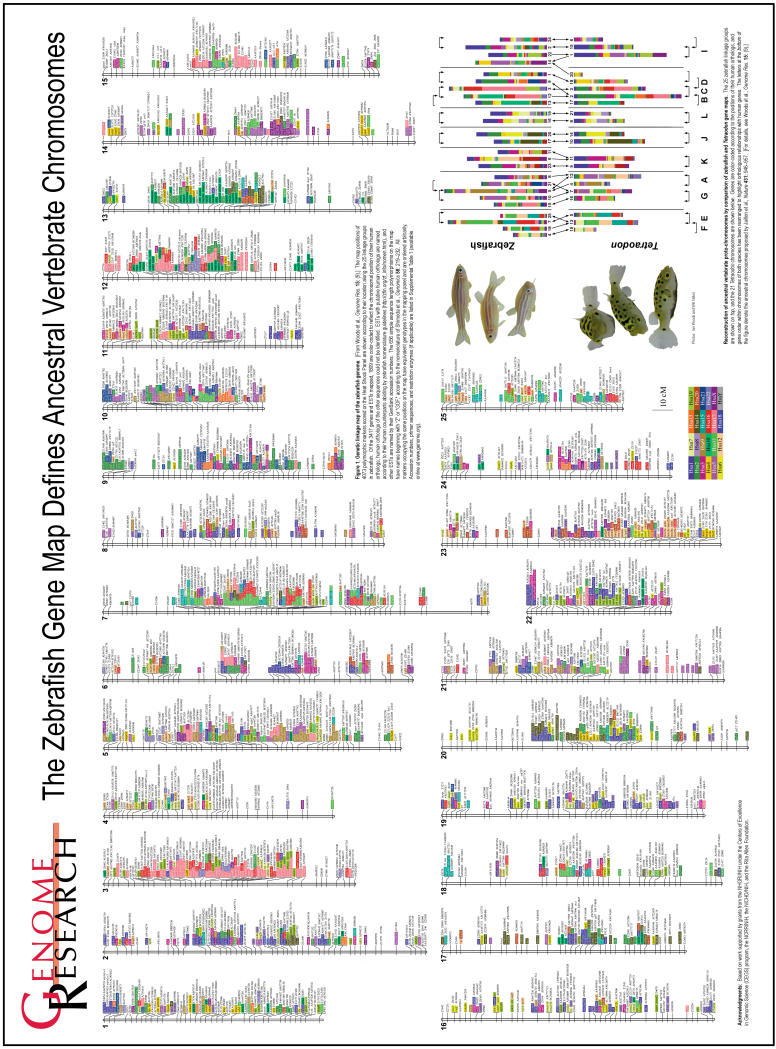
Figure 1.
The Zebrafish Gene Map Defines Ancestral Vertebrate Chromosomes
We worked to minimize the number of cases in which multiple ESTs representing the same gene were present on the map. First, we assigned 1149 ESTs to putative full-length cDNAs from the zebrafish gene collection (http://zgc.nci.nih.gov), increasing the total number of full-length cDNAs on the map to 1648. Because sequences in this collection are thought to represent full-length mRNAs, assigning matches between ESTs and these sequences decreases the possibility that two ESTs representing non-overlapping portions of the same gene are both present on the map. Second, duplicate entries for the same gene were removed when ESTs matching the same human ortholog (see below) were localized to the same position on the map. Although some of these cases may represent independent genes formed by tandem duplications, removing one member of this type of duplicate pair reduces the possibility that two non-overlapping ESTs from the same gene are retained. Third, 3208 mapped genes and ESTs were assigned to different UniGene clusters (http://www.ncbi.nlm.nih.gov/UniGene, build 83), and these are likely to represent different genes. Eight pairs of genes were assigned to the same UniGene cluster but mapped to different locations; these map positions were confirmed in duplicate genotyping analyses. Genes in the same UniGene cluster found on different chromosomes are likely to be errors in cluster assembly. Finally, the 193 genes and ESTs without UniGene assignments were analyzed via two-way BLAST comparisons with the entire set of mapped genes and ESTs, and no significant overlap was detected.
We took numerous steps to maximize the accuracy of map position assignments. First, we identified possible problematic markers by inspecting the data for markers that exhibited high numbers of genotyping failures, that caused the introduction of double recombinants into the map, or that were localized to positions discrepant with those of other zebrafish maps. The positions of markers in these categories were confirmed in additional genotyping analyses. In addition, 91 SSCP markers showing none of these potential problems were selected for random sampling of map accuracy. By using newly generated RFLPs for these markers, 90 of 91 original positions were confirmed. Furthermore, we took cases of different ESTs matching the same UniGene cluster or full-length sequence that mapped to the same region as confirmation of the map position. In total, 1522 of the 3417 genes and ESTs on the map have been confirmed in duplicate genotyping analyses.
Zebrafish-human comparison
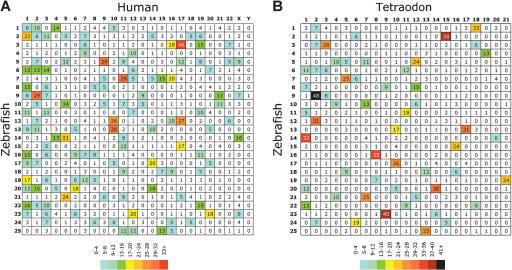
We identified putative human orthologs of the mapped zebrafish genes and ESTs by using reciprocal BLAST comparisons. By using a previously described approach (Woods et al. 2000) with increased stringency (see Methods), we identified 1809 putative human-zebrafish orthologous gene pairs. In Figure 1 (enclosed poster), mapped zebrafish genes and ESTs are color-coded according to the positions of their human orthologs. Map locations of orthologous gene pairs can be used to define regions of conserved synteny between species. These data are summarized in an Oxford grid (Fig. 2A; Edwards 1991), which shows the number of orthologous gene pairs observed on a per-chromosome basis between species. Boxes on the grid with high numbers represent conserved syntenies with many genes. The distribution of conserved syntenies is highly nonrandom (χ2 = 1673, P < 0.001; χ2 calculated according to the method of Gates et al. 1999); for example, a random distribution would predict that only two boxes would contain nine or more gene pairs, whereas 61 such boxes were observed (Fig. 2A). High degrees of conserved synteny were observed, for example, between human chromosome (Hsa) 10 and zebrafish linkage group (LG) 12 and 13, Hsa 17/LG 3, 5, 12, 15, and Hsa 9/LG 5. All major clusters reported in previous work (Gates et al. 1999; Woods et al. 2000) were supported and extended in the current analysis, with the exception of Hsa 6/LG 19, which lost three gene pairs found in the initial studies (Woods et al. 2000). In addition, a major cluster at Hsa X/LG 14 appeared in the current work that was less significant in previous analyses. These differences could result from a combination of several factors, including the enhanced detection using full-length cDNAs rather than ESTs to identify orthologous gene pairs, the increased number of human genes available for comparison, and improvements in map accuracy. The complete set of zebrafish-human orthologous gene pairs identified in this study, along with chromosome locations for each gene, is available in Supplemental Table 2.
Figure 2.
Oxford grids showing conservation of synteny among zebrafish, human, and Tetraodon. These grids plot the locations of orthologous gene pairs according to their chromosomal positions in the two compared species. The number in each square in the grids is the number of orthologous gene pairs on the indicated chromosomes in the two compared species. Boxes in the grid that contain large numbers identify conserved syntenies involving many genes. (A) Zebrafish-human comparison. (B) Zebrafish-Tetraodon comparison. Human orthologs of mapped zebrafish genes are listed in Supplemental Table 2, and Tetraodon orthologs of mapped zebrafish genes are listed in Supplemental Table 3. Both grids clearly show a nonrandom distribution of clusters (for details, see text), with particular chromosome pairs showing high degrees of conserved synteny. Map positions for zebrafish genes were derived from this work, and those of Tetraodon were obtained from http://www.ensembl.org/Tetraodon_nigroviridis/. Hox gene clusters containing multiple mapped genes are counted as a single point on each of the grids.
Zebrafish-Tetraodon comparison
By using the data obtained from the Tetraodon nigroviridis (Tetraodon) sequencing project (Jaillon et al. 2004), we identified orthologous gene pairs between zebrafish and Tetraodon. This analysis employed similar reciprocal BLAST analyses as for the zebrafish-human comparison, except that the selection criteria were more stringent (see Methods). Although we identified 1496 putative orthologs, 409 of these genes had not been mapped in Tetraodon. The distribution of conserved syntenies was extremely nonrandom (χ2 = 1226, P < 0.001); for example, no boxes containing nine or more gene pairs would be expected in a random distribution, whereas 36 of these boxes were observed in the analysis, and 20 boxes had >17 orthologous gene pairs (Fig. 2B). In many cases, the high degree of conserved synteny between individual Tetraodon chromosomes and zebrafish linkage groups suggested a 1:1 correspondence of chromosomes in the two species, for example Tetraodon chromosome (Tni) 9/LG 23, Tni 18/LG 1, Tni 15/LG 2, Tni 3/LG 3, and Tni 8/LG 16. Supplemental Table 3 contains the complete set of putative zebrafish-Tetraodon orthologous gene pairs, along with chromosome locations for each gene.
Jaillon et al. (2004) reported that the Tetraodon genome sequence sustained relatively few interchromosomal rearrangements since the divergence of the teleost and tetrapod lineages, whereas mammalian genomes have rearranged more extensively. They suggested that the large expansion of transposable elements in mammalian genomes may have facilitated rearrangements between chromosomes, and suggest that zebrafish, which has many more transposable elements than Tetraodon, might be expected to show many more interchromosomal rearrangements. The large number of chromosomes exhibiting a 1:1 correspondence in the zebrafish versus Tetraodon Oxford grid (Fig. 2B) argues against this hypothesis.
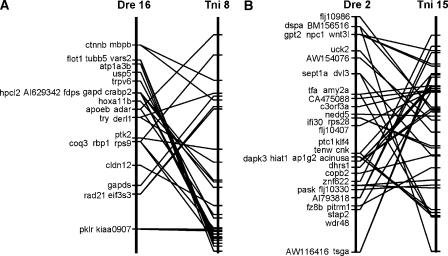
Because of the high degree of conserved synteny exhibited between the majority of zebrafish and Tetraodon chromosomes, we examined whether gene order was also conserved in regions of conserved synteny. Figure 3 shows a comparison of gene order in two regions of synteny conserved in zebrafish and Tetraodon. In both cases shown, orthologous gene pairs were distributed along the zebrafish and Tetraodon chromosomes, suggesting that a large group of genes were syntenic in their last common ancestor. Comparison of gene order along individual chromosomes indicated that numerous inversions involving large regions altered gene order. Some smaller clusters of genes have been conserved in both species through evolution, but there is also evidence of some rearrangement within these small groups of genes.
Figure 3.
Comparison of gene order between zebrafish and Tetraodon. (A) Comparison of the chromosomal locations of 28 orthologous gene pairs on zebrafish (Danio rerio, or Dre) LG 16 with Tetraodon nigroviridis (Tni) chromosome 8. (B) Comparisons of the chromosomal locations of 38 orthologous gene pairs on Dre LG 2 with Tni chromosome 15. Lines between the compared chromosomes connect positions of orthologous gene pairs in the two species. Distances between markers on a single chromosome are shown to scale, but compared chromosomes have been scaled to equivalent lengths. Map positions for zebrafish genes were derived from this work, and those of Tetraodon were obtained from http://www.ensembl.org/Tetraodon_nigroviridis/. Seven of the 35 gene pairs between Dre16/Tni8 shown in Figure 2B are not shown here, because the precise locations of these seven genes along this Tetraodon chromosome have not yet been reported.
Duplicate genes in zebrafish and Tetraodon
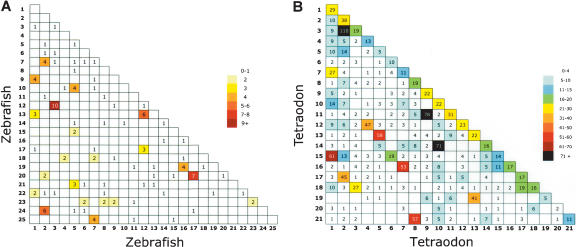
The hypothesis that a teleost ancestor experienced a whole-genome duplication event followed by widespread but incomplete loss of duplicate genes in derivative species predicts that a subset of genes throughout the zebrafish genome would be present in two copies (Amores et al. 1998; Postlethwait et al. 1998, 2000; Force et al. 1999; Gates et al. 1999; Meyer and Schartl 1999; Woods et al. 2000; Taylor et al. 2003). We used the zebrafish-human comparative analysis to identify 126 putative duplicate gene pairs in the zebrafish genome (Supplemental Table 4). Duplicate zebrafish genes were identified when (1) two zebrafish genes matched the same human protein in BLASTX searches, and (2) a reciprocal TBLASTN search returned the original zebrafish genes as one of the top two matches (which allows both members of a duplicate pair to be identified as orthologs of a single human gene). Figure 4A illustrates the locations of duplicated zebrafish genes. Clusters were evident in most linkage groups and were distinctly nonrandom (χ2 = 61, P < 0.001), indicating that duplicated genes tend to be clustered on duplicated chromosomal segments. There were 12 boxes that contained three or more duplicate gene pairs, including LG 3/LG 12, LG 7/LG 25, and LG 17/LG 20. These clusters confirmed and built upon those identified in previous analyses (Amores et al. 1998; Postlethwait et al. 1998, 1999, 2000, 2002; Woods et al. 2000); in addition, two new clusters were identified: LG 2/LG 24, and LG 5/LG 10. This analysis strongly supports the conclusion that the zebrafish genome contains many duplicated genes that were likely formed by a whole-genome duplication event.
Figure 4.
Chromosomal locations of duplicate genes in zebrafish and Tetraodon. Each box on the grids contains the number of duplicated genes (see Methods) shared between the indicated chromosomes. Boxes with large numbers denote chromosome pairs containing many duplicated genes. (A) Zebrafish-zebrafish comparison. (B) Tetraodon-Tetraodon comparison. The distribution of clusters of duplicated genes is highly nonrandom (for details, see text), indicating that most duplicated genes in these species are found on pairs of duplicated chromosomal segments. Map positions for zebrafish genes were derived from this work, and those of Tetraodon were obtained from http://www.ensembl.org/Tetraodon_nigroviridis/.
To compare complements of duplicated genes in different teleost species, we searched for duplicate gene pairs in the Tetraodon genome sequence. We identified 3327 pairs of duplicate genes in Tetraodon using criteria identical to those applied for zebrafish (Fig. 4B). Map locations of both duplicate genes were available for only 1543 gene pairs (Supplemental Table 5). Again, the distribution of clusters was highly nonrandom (χ2 = 987, P < 0.001). The locations of significant duplicate clusters—for example, Tni 2/Tni 3, Tni 10/Tni 14, and Tni 9/Tni 11—largely agree with those reported by Jaillon et al. (2004), who identified duplicate genes in the Tetraodon genome via a different approach based on all-by-all sequence comparisons within the complete set of Tetraodon genes. We identified about twice as many Tetraodon duplicates as the previous analysis (1543 mapped pairs vs. 748). A striking difference between the Tetraodon and zebrafish grids is the large number of duplicated gene pairs located on the same chromosomes in Tetraodon, as indicated by the line of filled boxes running across the diagonal of Figure 4B. These may represent fragments of incompletely assembled genes, actual tandem duplications, or a combination of both. Of the 1543 gene pairs on the Tetraodon grid, 362 (23%) are in the diagonal line denoting putative within-chromosome duplicates. In the initial analysis of the Tetraodon genome sequence, a similar fraction of annotated genes (∼5500 of 27,918 = 20%) were predicted to be incompletely assembled fragments (Jaillon et al. 2004). Most of the putative intrachromosomal duplicates observed in Figure 4B, therefore, are expected to be derived from incompletely assembled genes, rather than tandem duplicates. Even if all of the intrachromosomal duplicates result from incompletely assembled genes, our Tetraodon-human comparison has identified more putative duplicate gene pairs than the previous Tetraodon-Tetraodon comparison. In summary, the two analyses of Tetraodon duplicates independently identified a similar set of duplicated chromosomal segments, supporting the validity of both approaches. The identification of numerous duplicate chromosomes in zebrafish and Tetraodon (Fig. 4) and the finding that most zebrafish chromosomes correspond to a single orthologous chromosome in Tetraodon (Fig. 2B) support the hypothesis that a whole-genome duplication event occurred early in teleost evolution, before these two lineages diverged (Amores et al. 1998; Postlethwait et al. 1998, 2000; Gates et al. 1999; Woods et al. 2000; Taylor et al. 2003).
By analyzing extant duplicated genes in each species, rates of retention of duplicated genes through evolution can be estimated. For example, assuming that 80% of the 3327 Tetraodon duplicate gene pairs identified in our analysis are actually gene duplicates rather than misannotated gene fragments, there are ∼2660 duplicate gene pairs in the Tetraodon genome, representing ∼24% of the complete count of ∼22,400 genes in this species (Jaillon et al. 2004). A precise assessment of the rate of retention of duplicate genes in zebrafish is not feasible at present because the complete gene list is not yet available. The lower limit of retained duplicate genes, however, can be estimated with our current data. Of the 1809 zebrafish-human orthologous gene pairs identified in our analysis, 126 are present in two copies, indicating that ∼14% of the genes in our analysis are present in duplicate. The actual number of retained duplicates in zebrafish is certainly higher, because many duplicate genes have not been identified and mapped. A previous study taking particular care to identify zebrafish co-orthologs of genes located on human chromosomes 9 and 17 predicted that ≥30% of zebrafish genes are present in duplicate (Postlethwait et al. 2000). Our estimated frequency of duplicate gene retention in Tetraodon, therefore, is within the range estimated for zebrafish, suggesting that the frequency at which duplicated genes are retained may be similar between these two species.
In addition to estimating the overall frequency of retention after gene duplication, comparisons of retained duplicated genes can determine whether the same duplicates are retained in different species through evolution. For example, our analysis suggests that 60 of 126 (48%) zebrafish duplicate gene pairs are also duplicated in Tetraodon, whereas 66 appear to be represented by a single gene in Tetraodon. This proportion is similar to that observed in a previous comparison of duplicate genes between zebrafish and Takifugu rubripes (Taylor et al. 2003), who found Takifugu duplicates for 22 of 42 (52%) zebrafish duplicate gene pairs. These data demonstrate that different teleosts have retained different sets of duplicated genes. The loss of different complements of duplicated genes—a process termed divergent resolution—has been proposed to reinforce reproductive barriers between allopatric populations, therefore facilitating speciation (Lynch and Conery 2000; Lynch and Force 2000). Our analysis of retained duplicate genes in zebrafish and Tetraodon is consistent with the proposal that divergent resolution is one mechanism that has contributed to speciation in teleosts (Force et al. 1999, Postlethwait et al. 2004).
Genome-wide comparisons and the ancestral karyotype
Several previous analyses have employed comparisons between fish and human genomes, as well as between the genomes of different fish species, to attempt a reconstruction of the ancestral vertebrate karyotype. By analyzing the map positions of zebrafish-human orthologous gene pairs, Postlethwait et al. (2000) hypothesized that 12 chromosomes in a vertebrate ancestor gave rise to the current number of ∼20-30 chromosomes in Eutherian mammals via chromosome fissions, and to the current number of ∼25 chromosomes in most teleosts via chromosome duplications. A comparison of syntenic relationships among the genomes of human, zebrafish, and medaka supported this prediction of 12 ancestral chromosomes (Naruse et al. 2004). In that study, 12 sets of fish chromosomes were shown to cluster according to shared patterns of orthologous relationships with human genes. Furthermore, the analysis of the complete Tetraodon genome sequence corroborated the prediction of 12 ancestral vertebrate chromosomes, and a detailed investigation of orthologous relationships between Tetraodon and human genes uncovered the events that may have acted to mold these putative ancestral chromosomes into the chromosomes currently present in Tetraodon (Jaillon et al. 2004).
These analyses differed both in approach and in the scope of the data sets employed. In the zebrafish-medaka study, a relatively small number of mapped genes in these two fish species was analyzed in three-way comparisons with orthologous human genes to reconstruct the putative ancestral karyotype (Naruse et al. 2004), while the second study employed a two-way comparison between the complete genome sequences of Tetraodon and human to attempt a similar reconstruction (Jaillon et al. 2004). The ancestral gene maps predicted by these studies have not previously been compared, and prior to our analysis, it was not clear whether the two approaches deduced the same group of 12 ancestral chromosomes.
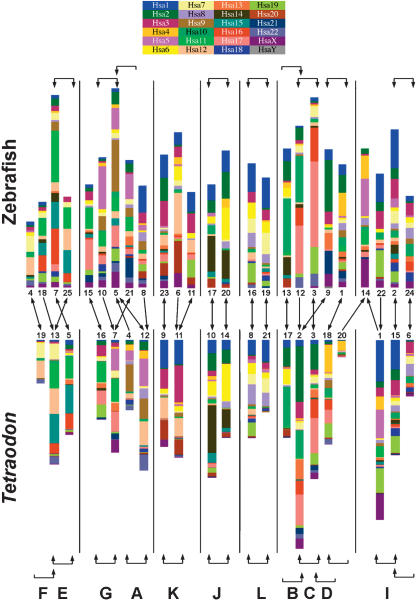
By analyzing the complete genome sequence of Tetraodon along with the most extensive zebrafish map yet produced, we undertook an independent assessment of the predicted ancestral vertebrate karyotype. The goal of this analysis was to compare gene maps of extant tetrapods (i.e., human) and teleosts to identify conserved syntenies, which represent groups of genes that were present on the same chromosome in the last common ancestor and have remained together through the vertebrate radiation. Since ancestral groups of syntenic genes may have been fragmented in one teleost lineage, the likelihood of identifying conserved syntenies is increased by comparing duplicate chromosomal segments within a species and by comparing orthologous chromosomal segments between the two species. Thus a comparative analysis using two teleost species provides a more complete picture of the ancestral gene map than does a comparison of one teleost and one tetrapod species. We used the data in the Oxford grids comparing orthologous chromosome segments between species (Fig. 2), along with paralogous chromosome segments within species (Fig. 4), to reconstruct putative relationships among extant chromosomes within and between the genomes of zebrafish and Tetraodon (Fig. 5). Chromosomes in Figure 5 are color-coded to denote the position in the human genome of fish-human orthologous gene pairs found in our analysis, and genes are rearranged according to human chromosome number, rather than map order, to enhance the clarity of the comparisons. In many cases, the pattern of human orthology clearly supports the Oxford grid data suggesting a 1:1 relationship for many chromosomes between zebrafish and Tetraodon (e.g., Tni 10/LG 17, Tni 14/LG 20, Tni 8/LG 16, Tni 21/LG 19) and between putative duplicate chromosomes within a single fish species (e.g., Tni 10/14, Tni 8/21, LG 17/20, LG 16/19).
Figure 5.
Reconstruction of ancestral vertebrate proto-chromosomes by comparison of zebrafish and Tetraodon gene maps. The 25 zebrafish linkage groups are shown on top, and the 21 Tetraodon chromosomes are shown below. Genes are color-coded according to the positions of their human orthologs, and gene order within chromosomes of both species has been rearranged to highlight orthologous relationships with human genes. Brackets at the top and the bottom of the figure indicate relationships between paralogous chromosomes in each species. Brackets with two arrowheads indicate best reciprocal two-way matches between chromosomes according to the number of duplicate genes shared between these chromosomes (derived from Fig. 4). Brackets with one arrowhead indicate cases in which one chromosome overlapped significantly with a second chromosome, yet the most significant overlap for that second chromosome was with a third chromosome. In the zebrafish-zebrafish comparison, only those chromosomes exhibiting three or more shared duplicate gene pairs are bracketed. Arrows in the middle of the figure indicate the largest conserved syntenies between orthologous chromosomes in zebrafish and Tetraodon (derived from the data in Fig. 2B). Arrowheads are shown to reflect best two-way or one-way relationships as above. The letters at the bottom of the figure denote the ancestral chromosomes proposed by Jaillon et al. (2004); their ancestral group “H” did not emerge from our analysis.
Gene content of ancestral chromosomes can be inferred from these relationships. For example, a single ancestral chromosome (J) was likely duplicated in the ancestor of both fish species and gave rise to Tni 10/14 and LG 17/20. In the lineage leading to human, this ancestral chromosome likely fragmented to form large portions of chromosomes 6 and 14, and smaller portions of other chromosomes. Because of interchromosomal rearrangements through evolution, however, not all relationships are as clear. For example, the pattern of chromosomal relationships within and between species suggests that two ancestral chromosomes (E and F) were both duplicated in the teleost lineage, and in Tetraodon the duplicate copies of Tni 5 and Tni 19 fused to form Tni 13, as was proposed in the analysis of the complete Tetraodon genome sequence (Jaillon et al. 2004). In zebrafish, duplication of ancestral chromosomes E and F, followed by fission and fusion events, likely gave rise to LGs 7, 18, 25 (Postlethwait et al. 2000), and 4. Analysis of the complete genome sequence of the zebrafish will help to clarify the interchromosomal rearrangements that may have shaped zebrafish chromosomes through evolution.
Our analysis of map locations of paralogous genes between zebrafish and Tetraodon chromosomes, combined with between-species comparisons of map locations of orthologous genes (Fig. 5), largely agreed with the ancestral karyotype reconstruction proposed by each of the previous studies (Jaillon et al. 2004; Naruse et al. 2004). In almost all cases, analysis of map positions of duplicated genes within and between zebrafish and Tetraodon chromosomes uncovered the same putative ancestral groupings (A-G, I-L) as were predicted via extensive comparisons of the complete genome sequences of human and Tetraodon. The one major exception was ancestral chromosome H, which was proposed to group Tni 1 with Tni 7 (Jaillon et al. 2004). Our analysis assigned Tni 1 and Tni 7 to different putative ancestral chromosomes. One likely explanation for this difference is that specific segments of Tni 1 and Tni 7 may have been ancestrally syntenic, as proposed in the previous study (Jaillon et al. 2004), whereas other segments of Tni 1 and Tni 7 might have derived from other ancestral chromosomes, as shown in Figure 5. In addition, the data presented in Figure 5 concurred in many cases with the prior study comparing a relatively small number (∼800) of mapped genes between zebrafish and medaka (Naruse et al. 2004). For example, both our analysis and the zebrafish-medaka study predicted the grouping of zebrafish LGs 3/12, 16/19, 17/20, 7/18/25, 11/23, and 10/21, but the previous analysis did not predict the grouping of LGs 2/24 or 5/10. The much smaller sample size of available mapped genes and ESTs employed in the medaka-zebrafish comparison likely contributes to the differences in karyotype predictions between these analyses.
Conclusions
With more than double the number of genes and ESTs present on the most recent genetic map for the zebrafish (Woods et al. 2000), the map presented here represents a valuable resource for a large community of researchers striving to elucidate gene function in vertebrates. This map is an important source of candidate genes for the hundreds of mutations that have been identified in the many previous and ongoing genetic screens in zebrafish. In addition, these genetic polymorphisms constitute a valuable source of entry points for positional cloning projects. Moreover, the map presented here will be an important framework for assembling the zebrafish genome sequence.
The analysis presented here represents the most extensive comparison to date between the genomes of zebrafish and other vertebrate species. The identification of regions of conserved synteny between the zebrafish genome and the extensively annotated genomes of other vertebrate species such as human and Tetraodon will facilitate cloning of mutations based on comparative approaches. Moreover, these comparative studies can highlight processes underlying vertebrate chromosome evolution. Our identification of numerous duplicated chromosome segments involving nearly all zebrafish chromosomes has confirmed and extended data from previous studies (Amores et al. 1998; Postlethwait et al. 1998, 2000; Force et al. 1999; Gates et al. 1999; Naruse et al. 2000; Woods et al. 2000; Taylor et al. 2003; Jaillon et al. 2004) supporting a whole-genome duplication event in an ancestor of teleosts. We also show that different duplicated genes have been lost in zebrafish and Tetraodon, although similar overall numbers of duplicated genes may have been retained in these two species. These differences in retention of duplicated genes are consistent with the proposal that divergent resolution may have facilitated the vast species radiation observed in teleosts. Finally, our three-way comparison of the zebrafish, Tetraodon, and human genomes provides an important confirmation of most elements of two previous attempts to reconstruct the ancestral vertebrate karyotype. This analysis provides a clear picture of the gene content for 11 of the 12 putative ancestral proto-chromosomes in the last common ancestor of bony vertebrates.
Methods
Linkage analysis
Primer design, PCR assays, and linkage analysis were performed as previously described (Kelly et al. 2000). In some cases, PCR primers were designed by using alignments of ESTs or genes with genomic DNA derived from the zebrafish whole-genome shotgun sequencing project (http://trace.ensembl.org). RFLPs were identified by sequencing PCR amplicons from SJD and C32 DNA. In rare cases, markers were polymorphic on only one of the two F2 families used in the mapping analysis (Kelly et al. 2000). The mapping software we used (MapManager, Manly 1993) increases the distance around these markers, leading to a slight increase in total map length, even though the total number of recombinants is highly similar between this map and previous genetic maps (Kelly et al. 2000; Woods et al. 2000). The complete genotype data set is available online at http://zebrafish.stanford.edu.
Sequence comparisons
ESTs were assigned to full-length cDNA sequences via BLASTN comparisons (Altschul et al. 1997) with the zebrafish gene collection database (http://zgc.nci.nih.gov). Matches were assigned when the sequence identity between the EST and the full-length cDNA was ≥96% over at least 200 bases. Zebrafish genes and ESTs were assigned putative human and Tetraodon orthologs via reciprocal BLAST best matches as described (Woods et al. 2000) with increased stringency, as follows. Zebrafish genes and ESTs were employed in BLASTX searches against the Sanger Institute (http://www.ensembl.org) peptide databases for human and Tetraodon; matches with expect scores >1e - 5 were excluded. Human orthologs were confirmed if a reciprocal TBLASTN search against the zebrafish sequence database identified the original gene or EST as one of the top two matches, while zebrafish-Tetraodon orthologous gene pairs were confirmed if TBLASTN searches identified the original zebrafish gene or EST as the top match. Orthologous gene pairs between human and Tetraodon were obtained using identical criteria to the zebrafish-human comparisons. Duplicate gene pairs in both fish species were identified in cases where two fish genes within a species matched the same human protein in a BLASTX search, and a TBLASTN search using that human protein returned the original fish genes as the top two matches. Map positions for human and Tetraodon genes were obtained from the Sanger Institute Web site (http://www.ensembl.org).
In some cases, phylogenetic trees were constructed to confirm or resolve orthologous relationships derived from reciprocal BLAST searches giving several matches with nearly equivalent scores (delta, msx, nodal, tenm, noggin), or when the reciprocal BLAST analysis suggested an orthology relationship that differed from published data (bmpr1) (Taylor et al. 2003). In the latter case, phylogenetic analysis of full-length cDNAs supported the BLAST data suggesting that bmpr1a and bmpr1ab are co-orthologs of human BMPR1A. Sequences were aligned by using probcons (http://probcons.stanford.edu), edited with Protein Alignment Editor (http://mendel.stanford.edu:16080/SidowLab/), and trees were constructed by using semphy (http://www.cs.huji.ac.il/~nir/SEMPHY/).
Duplicate genes in Tetraodon were identified by comparison to human, as described above. The data from the Tetraodon-Tetraodon duplicate grid (Fig. 4B) were examined to determine which chromosome pairs shared the highest number of duplicates (as indicated in Fig. 5). The data from the zebrafish-Tetraodon comparison (Fig. 2B) were used to identify orthologous chromosomes in these species. Duplicate chromosomes in zebrafish (Fig. 4A) were identified as described for Tetraodon. Collectively, these data define pairings of chromosomes both within and between species, and these pairings represent extant chromosomes likely derived from the same ancestral chromosome.
Supplementary Material
Acknowledgments
We thank members of our laboratories for helpful discussions, Tom Conlin for expert help with bioinformatics, Greg Cooper and Jon Binkley for assistance with phylogenetic analyses, and Alex Schier for critical comments on the manuscript. I.G.W. was supported by a predoctoral fellowship from the Howard Hughes Medical Institute. This work was supported by NIH grants R01RR10715 (J.H.P.), HD22486 (J.H.P.), RR12349 (W.S.T.), and HG02568 (W.S.T.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.4134305. Article published online before print in August 2005. Freely available online through the Genome Research Immediate Open Access option.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.L. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores, A., Force, A., Yan, Y.L., Joly, L., Amemiya, C., Fritz, A., Ho, R.K., Langeland, J., Prince, V., Wang, Y.L., et al. 1998. Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711-1714. [DOI] [PubMed] [Google Scholar]
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. [DOI] [PubMed] [Google Scholar]
- Barbazuk, W.B., Korf, I., Kadavi, C., Heyen, J., Tate, S., Wun, E., Bedell, J.A., McPherson, J.D., and Johnson, S.L. 2000. The syntenic relationship of the zebrafish and human genomes. Genome Res. 10: 1351-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birely, J., Schneider, V.A., Santana, E., Dosch, R., Wagner, D.S., Mullins, M.C., and Granato, M. 2005. Genetic screens for genes controlling motor nerve-muscle development and interactions. Dev. Biol. 280: 162-176. [DOI] [PubMed] [Google Scholar]
- Donovan, A., Brownlie, A., Zhou, Y., Shepard, J., Pratt, S.J., Moynihan, J., Paw, B.H., Drejer, A., Barut, B., Zapata, A., et al. 2000. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature 403: 776-781. [DOI] [PubMed] [Google Scholar]
- Driever, W., Solnica-Krezel, L., Schier, A.F., Neuhauss, S.C., Malicki, J., Stemple, D.L., Stainier, D.Y., Zwartkruis, F., Abdelilah, S., Rangini, Z., et al. 1996. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123: 37-46. [DOI] [PubMed] [Google Scholar]
- Edwards, J.H. 1991. The Oxford Grid. Ann. Hum. Genet. 55: 17-31. [DOI] [PubMed] [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.L., and Postlethwait, J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates, M.A., Kim, L., Egan, E.S., Cardozo, T., Sirotkin, H.I., Dougan, S.T., Lashkari, D., Abagyan, R., Schier, A.F., and Talbot, W.S. 1999. A genetic linkage map for zebrafish: Comparative analysis and localization of genes and expressed sequences. Genome Res. 9: 334-347. [PubMed] [Google Scholar]
- Gibbs, R.A., Weinstock, G.M., Metzker, M.L., Muzny, D.M., Sodergren, E.J., Scherer, S., Scott, G., Steffen, D., Worley, K.C., Burch, P.E., et al. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493-521. [DOI] [PubMed] [Google Scholar]
- Haffter, P., Granato, M., Brand, M., Mullins, M.C., Hammerschmidt, M., Kane, D.A., Odenthal, J., van Eeden, F.J., Jiang, Y.J., Heisenberg, C.P., et al. 1996. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123: 1-36. [DOI] [PubMed] [Google Scholar]
- Jaillon, O., Aury, J.M., Brunet, F., Petit, J.L., Stange-Thomann, N., Mauceli, E., Bouneau, L., Fischer, C., Ozouf-Costaz, C., Bernot, A., et al. 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431: 946-957. [DOI] [PubMed] [Google Scholar]
- Karlstrom, R.O., Talbot, W.S., and Schier, A.F. 1999. Comparative synteny cloning of zebrafish you-too: Mutations in the Hedgehog target gli2 affect ventral forebrain patterning. Genes & Dev. 13: 388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, P.D., Chu, F., Woods, I.G., Ngo-Hazelett, P., Cardozo, T., Huang, H., Kimm, F., Liao, L., Yan, Y.L., Zhou, Y., et al. 2000. Genetic linkage mapping of zebrafish genes and ESTs. Genome Res. 10: 558-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness, E.F., Bafna, V., Halpern, A.L., Levy, S., Remington, K., Rusch, D.B., Delcher, A.L., Pop, M., Wang, W., Fraser, C.M., et al. 2003. The dog genome: Survey sequencing and comparative analysis. Science 301: 1898-1903. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Lynch, M. and Conery, J.S. 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151-1155. [DOI] [PubMed] [Google Scholar]
- Lynch, M. and Force, A. 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons, D.A., Pogoda, H.M., Voas, M.G., Woods, I.G., Diamond, B., Nix, R., Arana, A., Jacobs, J., and Talbot, W.S. 2005. erbb3 and erbb2 are essential for schwann cell migration and myelination in zebrafish. Curr. Biol. 15: 513-524. [DOI] [PubMed] [Google Scholar]
- Manly, K.F. 1993. A Macintosh program for storage and analysis of experimental genetic mapping data. Mamm. Genome 4: 303-313. [DOI] [PubMed] [Google Scholar]
- Meyer, A. and Schartl, M. 1999. Gene and genome duplications in vertebrates: The one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 11: 699-704. [DOI] [PubMed] [Google Scholar]
- Miller, C.T., Schilling, T.F., Lee, K., Parker, J., and Kimmel, C.B. 2000. Sucker encodes a zebrafish endothelin-1 required for ventral pharyngeal arch development. Development 127: 3815-3828. [DOI] [PubMed] [Google Scholar]
- Naruse, K., Fukamachi, S., Mitani, H., Kondo, M., Matsuoka, T., Kondo, S., Hanamura, N., Morita, Y., Hasegawa, K., Nishigaki, R., et al. 2000. A detailed linkage map of medaka, Oryzias latipes: Comparative genomics and genome evolution. Genetics 154: 1773-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse, K., Tanaka, M., Mita, K., Shima, A., Postlethwait, J., and Mitani, H. 2004. A medaka gene map: The trace of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Res. 14: 820-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait, J.H., Yan, Y.L., Gates, M.A., Horne, S., Amores, A., Brownlie, A., Donovan, A., Egan, E.S., Force, A., Gong, Z., et al. 1998. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 18: 345-349. [DOI] [PubMed] [Google Scholar]
- Postlethwait, J.H., Amores, A., Force, A., and Yan, Y.L. 1999. The zebrafish genome. Methods Cell Biol. 60: 149-163. [PubMed] [Google Scholar]
- Postlethwait, J.H., Woods, I.G., Ngo-Hazelett, P., Yan, Y.L., Kelly, P.D., Chu, F., Huang, H., Hill-Force, A., and Talbot, W.S. 2000. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 10: 1890-1902. [DOI] [PubMed] [Google Scholar]
- Postlethwait, J.H., Amores, A., Yan, Y.L., and Austin, C.A. 2002. Duplication of a portion of human chromosome 20q containing topoisomerase (top1) and snail genes provides evidence on genome expansion and the radiation of teleost fish. In Aquatic genomics (eds. N. Schimizu, et al.), pp 20-31. Springer, Tokyo.
- Postlethwait, J., Amores, A., Cresko, W., Singer, A., and Yan, Y.L. 2004. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 20: 481-490. [DOI] [PubMed] [Google Scholar]
- Shimoda, N., Knapik, E.W., Ziniti, J., Sim, C., Yamada, E., Kaplan, S., Jackson, D., de Sauvage, F., Jacob, H., and Fishman, M.C. 1999. Zebrafish genetic map with 2000 microsatellite markers. Genomics 58: 219-232. [DOI] [PubMed] [Google Scholar]
- Taylor, J.S., Braasch, I., Frickey, T., Meyer, A., and Van de Peer, Y. 2003. Genome duplication: A trait shared by 22,000 species of ray-finned fish. Genome Res. 13: 382-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga, Z.M., Amores, A., Lewis, K.E., Yan, Y.L., Postlethwait, J.H., Eisen, J.S., and Westerfield, M. 2001. Zebrafish smoothened functions in ventral neural tube specification and axon tract formation. Development 128: 3497-3509. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Woods, I.G., Kelly, P.D., Chu, F., Ngo-Hazelett, P., Yan, Y.L., Huang, H., Postlethwait, J.H., and Talbot, W.S. 2000. A comparative map of the zebrafish genome. Genome Res. 10: 1903-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://zgc.nci.nih.gov; zebrafish gene collection.
- http://www.ncbi.nlm.nih.gov/UniGene, build 83; UniGene clusters.
- http://trace.ensembl.org; zebrafish whole-genome shotgun sequencing project.
- http://zebrafish.stanford.edu; the complete genotype data set.
- http://www.ensembl.org; Ensembl genome browser.
- http://probcons.stanford.edu; probcons.
- http://mendel.stanford.edu:16080/SidowLab/; Protein Alignment Editor.
- http://www.cs.huji.ac.il/~nir/SEMPHY/; semphy.
- http://zfin.org/zf_info/nomen.html; zebrafish nomenclature guidelines.
- http://www.ensembl.org/Tetraodon_nigroviridis/; map positions for Tetraodon.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.