Abstract
During early zebrafish (Danio rerio) development zygotic transcription does not begin until the mid-blastula transition (MBT) ∼3 h after fertilization. MBT demarcates transition from synchronous short cell cycles of S and M phases exclusively to full cycles encompassing G1 and G2 phases. Transcriptional profiling and RT–PCR analyses during these phases enabled us to determine that this shift corresponds to decreased transcript levels of S/M phase cell cycle control genes (e.g. ccna2, ccnb1, ccnb2 and ccne) and increased transcript levels of ccnd1, encoding cyclin D1, and orthologs of p21 (p21-like) and retinoblastoma (Rb-like 1). To investigate the regulation of this process further, the translation of ccnd1 mRNA, a G1/S checkpoint control element, was impaired by microinjection of ccnd1-specific morpholino phosphorodiamidate (MO) 20mer or hydroxyprolyl-phosphono peptide nucleic acid (HypNA-pPNA) 16mer antisense oligonucleotides. The resulting downregulation of cyclin D1 protein resulted in microophthalmia and microcephaly, but not lethality. The phenotypes were not seen with 3-mismatch MO 20mers or 1-mismatch HypNA-pPNA 16mers, and were rescued by an exogenous ccnd1 mRNA construct with five mismatches. Collectively, these results indicate that transcription of key molecular determinants of asynchronous cell cycle control in zebrafish embryos commences at MBT and that the reduction of cyclin D1 expression compromises zebrafish eye and head development.
INTRODUCTION
Control of cell cycle progression is central to maintaining homeostasis in multicellular organisms. Loss of cell cycle control may lead to imbalances in proliferation and cell death that contribute to various disease states including neoplasia. Current knowledge of cell cycle regulation has largely been instructed by studies on cells and tissues in the adult organism and the extensive use of knockout mice (1). These studies have illuminated the complex interplay of cyclins, cyclin-dependent kinases (cdks) and regulators thereof, which control progression through the G1, S, G2 and M phases of the cell cycle. Furthermore, they highlighted the existence of cell cycle checkpoints, i.e. molecular switches controlling cell cycle progression. In malignant tumor cells, molecular determinants of checkpoint control are frequently inactivated permitting unchecked cell cycle progression and relaxed genome surveillance (2). Cell cycle checkpoints are also absent during very early stages of amphibian development as initially described in fertilized Xenopus laevis eggs (3). In Xenopus, synchronous, rapid cycling and the absence of the G1 and G2 phases of the cell cycle characterize the first rounds of DNA replication and cell division. The molecular underpinnings of this phenomenon are poorly understood.
We used the zebrafish (Danio rerio) as a simple model to study cell cycle regulation during vertebrate embryogenesis. The zebrafish has in recent years evolved as a novel and facile in vivo model to study human disease since many key genes are highly conserved between the two vertebrate species; these include cyclins, cdks and inhibitors of cdks. Importantly, zebrafish and Xenopus embryos share the absence of G1 and G2 cell cycle phases during very early development, followed by the establishment of asynchronous cell cycles within a short time frame (∼3 h) after fertilization, coincident with the mid-blastula transition (MBT) (4).
The onset of cell cycle regulation in zebrafish embryos occurs when zygotic transcription commences, raising the issue whether and how these two phenomena are linked. Here, we describe global transcription profiles in zebrafish embryos at distinct stages of embryonal development before and after establishment of cell cycle checkpoints. We observed that key cell cycle regulators involved in S to M phase transition, specifically ccnb1, encoding cyclin B1, ccnb2, encoding cyclin B2, and ccne, encoding cyclin E, were expressed at high levels in pre-MBT embryos followed by a rapid decline in post-MBT, congruent with the expected pattern of these cell cycle regulators during early zebrafish development. These observations led us to evaluate the expression profiles of other cell cycle regulatory genes, including those involved in G1 checkpoint control that is established several hours after fertilization. We identified three genes involved in the G1–S transition in vertebrates: ccnd1, encoding cyclin D1, and the orthologs of p21 and Rb, with initial low expression levels, followed by increased expression correlating with the onset of cell cycle asynchrony. Of these three, ccnd1 mRNA expression was most markedly upregulated after MBT, in accordance with an earlier study (5). This prompted us to investigate the consequences of interfering with ccnd1/cyclin D1 expression in developing zebrafish embryos.
Morpholino phosphorodiamidate (MO) oligonucleotides are used frequently for sequence-based knockdown studies in zebrafish (6). Recently, we and others have described the hybridization strength and activity of antisense oligonucleotides with alternating trans-4-hydroxy-l-proline peptide nucleic acid (PNA) and phosphono PNA residues (7,8). The hydroxyprolyl-phosphono PNA (HypNA-pPNA) oligomers are more specific than conventional MOs (9,10). The highly soluble HypNA-pPNA analog exhibited higher affinity towards DNA and RNA, and more stringent mismatch discrimination than MOs. Hence, we compared both MOs and HypNA-pPNAs for their ability to knock down ccnd1 mRNA translation to cyclin D1 protein based on the published ccnd1 cDNA sequence (4). We report that the reduction of cyclin D1 expression was associated with impaired development of the eye and the head region in zebrafish embryos, the two anatomical sites with the highest cyclin D1 expression levels during early zebrafish development.
MATERIALS AND METHODS
Embryo harvesting and maintenance
Zebrafish husbandry, embryo collection, dechorionation and embryo maintenance were performed according to the Standard Operating Procedures as described elsewhere (11) and with approval by the Institutional Animal Care and Use Committee at Thomas Jefferson University. Zebrafish were maintained in the Kimmel Cancer Center Zebrafish Facility at 28.5°C on a 14 h light/10 h dark cycle. Selected embryos with >24 h post-fertilization (hpf) were placed in embryo medium with 0.2 mM 1-phenyl-2-thio urea (Sigma, St Louis, MO) to prevent pigment formation.
Transcription profiles
Embryos were obtained from natural crosses of wild-type zebrafish at various times during development and staged as described elsewhere (12). Total RNA was isolated from groups of 100 staged embryos corresponding to 1.5, 3, 6 and 24 h post-fertilization (hpf) at 28.5°C using TriReagent (Sigma, St Louis, MO) according to the manufacturer's protocol. Gene expression in the zebrafish embryos was determined using biotin-labeled and in vitro-transcribed antisense RNA (aRNA) generated from the total RNA template. The labeled aRNA was hybridized to a microarray chip with 17 000 65mer sense oligonucleotides representing 16 399 zebrafish genes (Compugen/Sigma-Genosys, Kimmel Cancer Center Microarray Facility), with three replicate chips for each developmental time point. Each chip was scanned and quantified using a ScanArray Express laser scanner (Packard BioScience). Raw gene expression data were normalized to housekeeping gene controls on each chip. The data were exported to MS Excel for further analysis. After local background correction, each chip was normalized globally by dividing each array by its respective median signal. The different developmental time points were compared with Student's t-test. Significantly different genes were identified at P < 0.01 (3σ) and ratio >10 cutoffs. These genes were grouped using the Cluster method of Eisen et al. (13), and clusters were visualized by using TreeView. For hierarchical clustering, data were log transformed, median centered and normalized. Clusters were identified by average linkage.
RT–PCR analysis
The expression profiles of selected cell cycle control transcripts were confirmed by RT–PCR analysis of the original RNA samples using the following target-specific primer sets: cyclin A2 (ccna2, GenBank accession no. AF234784) forward 5′-GGCACGAGGTAAAAAGCAAC-3′ and reverse 5′-GGCCTCTCTCCAAAACTCC-3′; cyclin B1 (ccnb1, GenBank accession no. AB040435) forward 5′-GAGTCACAGCAATAAACCAC-3′ and reverse 5′-AGGAAGGCTCAGACACAAC-3′; cyclin B2 (ccnb2, GenBank accession no. AW422010) forward 5′-AGTTGAGTTGGACGAGAAAC-3′ and reverse 5′-GAAAGAGGCTGTTGGAAAAG-3′; cyclin D1 (ccnd1, GenBank accession no. X87581) forward 5′-ACAGCAACCTGTTGAATGAC-3′ and reverse 5′-GGCCAGATCCCACTTCAGTT-3′; cyclin E (ccne, GenBank accession no. X83594) forward 5′-GGACTGCGGAACACATCAC-3′ and reverse 5′-CGGTTCCTCGACTTCATCAG-3′; p21 ortholog (p21-like, GenBank accession no. BI887574) forward 5′-CCGTAGACCATGAGGAGC-3′ and reverse 5′-GTCTCGTCCACTTCTTTCTTTC-3′; Rb ortholog (Rb-like 1, GenBank accession no. AW281574) forward 5′-CCTTCAGCCACCCAAAGTGT-3′ and reverse 5′-GCACCTGTTCCTCTATACTG-3′; and β-actin (GenBank accession no. AF057040) forward 5′-GTTTTCCCCTCCATTGTTG-3′ and reverse 5′-GGTGTTGAAGGTCTCGAACA-3′. Reverse transcription was performed using AMV Reverse Transcriptase, and sequence amplification was accomplished using PCR Master Mix according to the manufacturer's protocol (Promega, Madison, WI). Samples were initially heated to 94°C for 2 min then run for 25 (ccne and p21-like), 30 (ccna2, ccnb1, ccnb2, ccnd1 and β-actin) or 35 (Rb-like 1) cycles in a GeneAmp System 9700 (PE Applied Biosystems, Foster City, CA) for 1 min at 94°C (denaturation), then annealed at 45°C for 1 min (ccnd1 and β-actin), at 52°C for 30 s (Rb-like 1), at 56.5°C for 1 min (ccnb2) or at 58°C for 30 s (ccne) or 1 min (ccna2, ccnb1 and p21-like), and then elongated for 1 min (ccne and Rb-like 1) or 2 min (ccna2, ccnb1, ccnb2, ccnd1, p21-like and β-actin) at 72°C. Samples were then cooled to 4°C and then analyzed by electrophoresis on 1% agarose gels.
MO and HypNA-pPNA antisense oligonucleotide treatment
We predicted that three mismatches would be necessary to inactivate ccnd1 MOs based on our earlier observations on zebrafish developmental mRNAs (9), indicating that one or two MO mismatches did not decrease knockdown activity, but four mismatches eliminated activity. In the same study, it was apparent that only one mismatch was necessary in HypNA-pPNAs. Antisense and three-mismatch 20mer MOs specific for ccnd1 RNA (Figure 1) were purchased from Gene Tools, LLC (Corvallis, OR). Antisense and single mismatch 16mer HypNA-pPNAs specific for ccnd1 mRNA (Figure 1) were provided by Active Motif (Carlsbad, CA).
Figure 1.
ccnd1 antisense sequences. The ccnd1-specific MO and HypNA-pPNA antisense and corresponding mismatch control sequences are displayed with their respective Tms. The trimer complementary to the AUG initiation codon is underlined and italicized, and the mismatched nucleotides in the control sequences are shown in lower case.
Oligomer concentrations were determined by ultraviolet absorbance spectra at room temperature on a spectrophotometer (Shimadzu UV-160), assuming molar absorptivities at 260 nm of Ade, 15.4; Gua, 11.7; Thy, 8.8 and Cyt, 7.3 × 103/M cm in 50 mM Et3N-H2CO3, pH 7.0 at 25°C. Absorbance versus temperature ramps were recorded in triplicates to determine the influence of mismatches on the hybridization efficiency of a complementary MO or HypNA-pPNA with RNA targets. Equimolar HypNA-pPNA or MO sequences (Figure 1) and a complementary RNA at 2.5 µM each in 10 mM Na2HPO4, 1.0 M NaCl, 0.5 mM EDTA, pH 7.0, were heated to 90°C for 3 min. then cooled gradually to room temperature. The absorbance of the mixtures at 260 nm was then recorded as the temperature was raised at 1°C/min from 20 to 95°C on a Varian Cary 3 spectrophotometer with Peltier temperature control. Melting temperatures (Tm) were assigned as the peak of the first derivative plot ± SD (14).
For MO or HypNA-pPNA microinjection into zebrafish, a final concentration of 0.5 mM of each oligonucleotide was prepared in 1× phosphate-buffered saline solution, containing 0.1% Phenol red dye, and ∼1 nl injected into 1–4 cell embryos using a nitrogen gas pressure injector (Harvard Apparatus, Cambridge, MA).
Sense mRNA rescue
To generate the capped mRNA, zebrafish ccnd1 cDNA (RefSeq accession no. NM_131025) was subcloned into pT3Ts plasmids (generously provided by Dr S. Ekker, University of Minnesota). The capped mRNA rescue constructs were not susceptible to the antisense oligomers because the sequence upstream of the AUG is a Kozak sequence, instead of the endogenous ccnd1 sequence, resulting in five mismatches between the antisense sequences and the rescue mRNA, as shown in Figure 2. The plasmids were linearized and transcribed with T7 RNA polymerase using the mMESSAGE mMACHINE T7 in vitro transcription kit (Ambion, Inc., Austin, TX) according to the manufacturer's instructions. Embryos were co-microinjected with ∼1 nl of antisense MO or HypNA-pPNA oligonucleotides and ∼1 nl of 1 g/l capped ccnd1 rescue mRNA, in parallel with the MO and HypNA-pPNA knockdown experiments.
Figure 2.
ccnd1 rescue mRNA construct. The capped mRNA rescue construct (bottom) includes a Kozak sequence upstream of the AUG, instead of the endogenous D.rerio sequence (top), resulting in five mismatches with the MO and HypNA-pPNA antisense sequences (middle).
Western blot analysis of cyclin D1 expression
Protein was isolated from 30 embryos per treatment condition (control, MO-injected or HypNA-pPNA-injected) by lysis using 25 mM methyl ethane sulfonate, 150 mM NaCl, 2% Triton X-100 and 3.5% octyl β-d-glucopyranoside, pH 6.4. Samples were diluted 1:1 (v/v) in 2× Tris–glycine SDS sample buffer (Invitrogen, Carlsbad, CA) and boiled for 5 min. The total lysates were resolved by 12% SDS–PAGE and transferred onto nitrocellulose membranes. For the detection of cyclin D1, the membranes were blocked with 1% powdered milk, then probed with a 1:1000 dilution of polyclonal rabbit anti-cyclin D1 antibodies (Neomarkers, Fremont, CA). The bound antibodies were visualized using a horseradish peroxidase-conjugated anti-rabbit IgG (Santa Cruz Biotechnology) and the SuperSignal West Femto Maximum Sensitivity substrate (Pierce, Rockford, IL). Samples were analyzed similarly for the expression of GAPDH as a control for sample loading using a mouse anti-rabbit polyclonal antibody (RDI, Flanders, NJ).
Zebrafish embryo retinal histology
Control uninjected, ccnd1 MO-injected and ccnd1 HypNA-pPNA-injected 5-day-old larvae were fixed, embedded in agarose arrays, sectioned (4 µm) and stained as described elsewhere (15). Slide images were acquired using a Qimaging Retiga EXi and a CRI RGB tunable filter on a Zeiss Axiophot using a ×20 0.75NA Plan-Apo objective, and processed using Adobe Photoshop and Illustrator CS.
RESULTS
Transcription profile analysis of zebrafish gene expression during embryogenesis
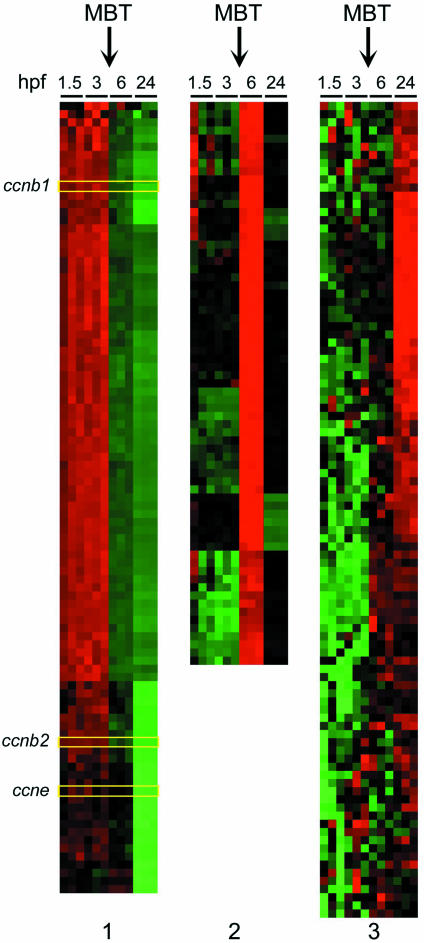
The first 10 embryonic cell cycles in zebrafish proceed in a synchronous fashion and in the absence of RNA synthesis. During this early phase, cells rapidly cycle through DNA synthesis (S phase) and mitosis (M phase) without transition through either G1 or G2 (12). After 10 divisions and ∼3 h post-fertilization (hpf) embryos enter MBT characterized by the start of zygotic transcription and the gradual lengthening of the cell cycle. The coincident start of zygotic transcription and onset of G1 phase at MBT raises the question of which early zygotic transcripts direct the establishment of cell cycle control post-MBT (5). To gain a representative view of gene expression patterns during this important transition, we analyzed transcription profiles before and after MBT. To this end, we used a 17 000-oligonucleotide microarray platform containing most of the cell cycle regulatory genes known in zebrafish to date. The analysis was performed in triplicates on RNA preparations collected in three different experiments performed independently of each other. Hierarchical cluster analysis of the results of these experiments revealed massive changes in gene expression reflecting the shift from maternal transcripts pre-MBT to zygotic transcription post-MBT. Normalized mean mRNA levels from each time point investigated were directly compared with the expression level at all other time points using pairwise analysis. Thus, a total of six possible comparisons were made for each expressed gene on the microarray (1.5 × 3 hpf; 1.5 × 6 hpf; 1.5 × 24 hpf; 3 × 6 hpf; 3 × 24 hpf; and 6 × 24 hpf). Of the 16 399 zebrafish genes represented on the array, 293 demonstrated a >10-fold difference in expression level in ≥3 of the 6 pairwise comparisons per gene (Table 1). At the >5-fold and >3-fold threshold levels, 1024 and 2464 genes were differentially expressed, respectively. Of note, the expression patterns before MBT at 1.5 and 3 hpf were virtually unchanged (Figure 3). In fact, between 1.5 and 3 hpf significant changes in transcript levels were observed only in nine cases. This result was expected based on the observation that zygotic transcription commences after MBT, i.e. after 3 hpf. In contrast, sequential waves of gene expression were apparent at subsequent stages of embryonal development after MBT. At lower threshold levels (single pairwise comparisons), more extensive changes in gene expression at 6 and 24 hpf were apparent (Table 1).
Table 1.
Change in global gene expression patterns in early zebrafish development
| Change in mRNA levels | Significant difference by pairwise comparisona | |
|---|---|---|
| ≥3 Comparisonsb | Single comparisonc | |
| >10-Fold | 293d | 945 |
| >5-Fold | 1024 | 2742 |
| >3-Fold | 2464 | 5208 |
aNormalized mean mRNA levels from each time point investigated by microarray analysis were directly compared with the expression level at all other time points for a total of six possible comparisons. Level of significance is based on P < 0.01 by Student's t-test.
bExpressed genes with three or more pairwise comparisons demonstrating the indicated fold difference (>10, >5 or >3).
cExpressed genes with only one pairwise comparison demonstrating the indicated fold difference (>10, >5 or >3).
dThis clustered dataset is shown in Figure 3.
Figure 3.
Global gene expression profiles during zebrafish embryogenesis. All transcripts with >10-fold change in expression from 1.5–24 h post-fertilization (hpf) identified by zebrafish-specific microarray analysis are clustered according to similar time-dependent expression patterns with upregulated expression in red, downregulated genes in green and unchanged expression in black. Panel 1 shows those steady-state mRNAs that are markedly downregulated when embryonal transcription commences, including ccnb1, ccnb2 and ccne (highlighted). Panels 2 and 3 present those transcripts that are markedly upregulated after MBT at 6 and 24 hpf, respectively.
Figure 3 provides a visual representation of all genes that changed >10-fold over the observation period and demonstrates a high degree of fidelity among the three sets of experimental samples for each of the time points chosen. Of note, 3 of the 96 genes with at least a 10-fold higher expression level before MBT were found to be the S to M phase cell cycle regulators, ccnb1, ccnb2 and ccne. The complete set of annotated genes in this set can be found in Supplementary Material of this report.
Stage-dependent expression of cell cycle regulatory genes in zebrafish development
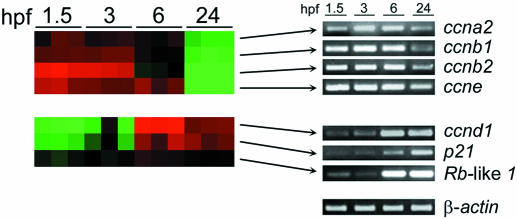
Next, we focused our analysis of the microarray results to transcripts encoding cell cycle regulators and genes involved in checkpoint control. This analysis revealed that transcripts for S/M phase cyclin genes, ccna2, ccnb1, ccnb2 and ccne, were expressed at high levels of pre-MBT and declined thereafter (Figure 4). In contrast, genes of major importance to G1/S phase transition, including orthologs of the Rb, p21 and ccnd1, were expressed at very low levels initially and increased markedly between 3 and 6 hpf (Figure 4), with the greatest increase being seen for ccnd1. We confirmed the expression patterns of these genes by RT–PCR analysis. Collectively, these results underscore that several key molecular determinants of G1/S transition, including cyclin D1, are expressed as a result of zygotic transcription consistent with the delayed establishment of cell cycle checkpoints after MBT.
Figure 4.
Cell cycle regulatory gene expression during zebrafish embryogenesis. Expression profiles of selected genes involved in cell cycle regulation (ccna2, ccnb1, ccnb2, ccne, ccnd1, p21-like and Rb-like 1) identified by zebrafish-specific microarray analysis of embryos at 1.5, 3, 6 and 24 h post-fertilization (hpf) are clustered according to similar time-dependent expression patterns with upregulated expression in red, downregulated genes in green and unchanged expression in black. The RT–PCR analyses of the corresponding mRNA levels are shown at right to test the results of the microarray analysis. Amplification of β-actin transcripts was performed as a loading control for each RT–PCR experiment with representative expression levels shown.
MO and HypNA-pPNA-mediated knockdown of cyclin D1 in zebrafish embryos
Whole mount in situ hybridization of ccnd1 mRNA demonstrated ubiquitous expression during gastrulation, and was prominently expressed in the retina and brain from the 18-somite stage to 24 hpf (16). After 48 hpf, expression remained strong in the retina and tectum of the brain (18). The distinctive expression pattern of ccnd1 mRNA in zebrafish embryos prompted us to investigate the functional contribution of cyclin D1 protein expression to embryonal development in zebrafish larvae. To this end, we used antisense oligonucleotides complementary to the ccnd1 mRNA encoding cyclin D1 in order to block translation of the targeted message; thereby, reducing the level of the target protein. To test the efficacy of MOs and HypNA-pPNAs in reducing cyclin D1 expression in zebrafish embryos, target sequences were selected as shown in Figure 1. The MO•RNA and HypNA-pPNA•RNA Tms (Figure 1) revealed that the MO 20mer was only 5°C more stable than the HypNA-pPNA 16mer. A single mismatch in the latter lowered Tm by 6°C. In the MO 20mer, three mismatches lowered Tm by 26°C.
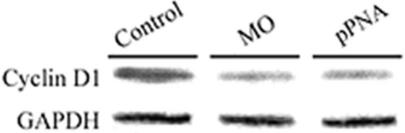
The effects of MO and HypNA-pPNA injections on cyclin D1 protein expression in the developing embryos were assessed by western blot analyses (Figure 5) showing that both treatments were equally effective in downregulating target protein expression. This analysis was performed at 24 hpf, at which time substantial expression of cyclin D1 was expected based on the transcription profiles shown in Figure 4.
Figure 5.
Western blot analysis of protein expression in antisense-treated zebrafish embryos. Western blot analysis of cyclin D1 protein in zebrafish embryo total protein extracts prepared at 24 hpf as a function of vehicle or antisense microinjection shortly after fertilization: Lane 1, Phenol red control embryo extract; lane 2, MO-treated embryo extract; lane 3, HypNA-pPNA-treated embryo extract. GAPDH expression was determined as a loading control for each sample.
Effects of cyclin D1 downregulation on zebrafish embryos by MO and HypNA-pPNA antisense strategies
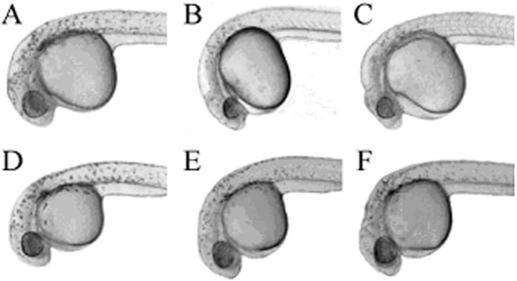
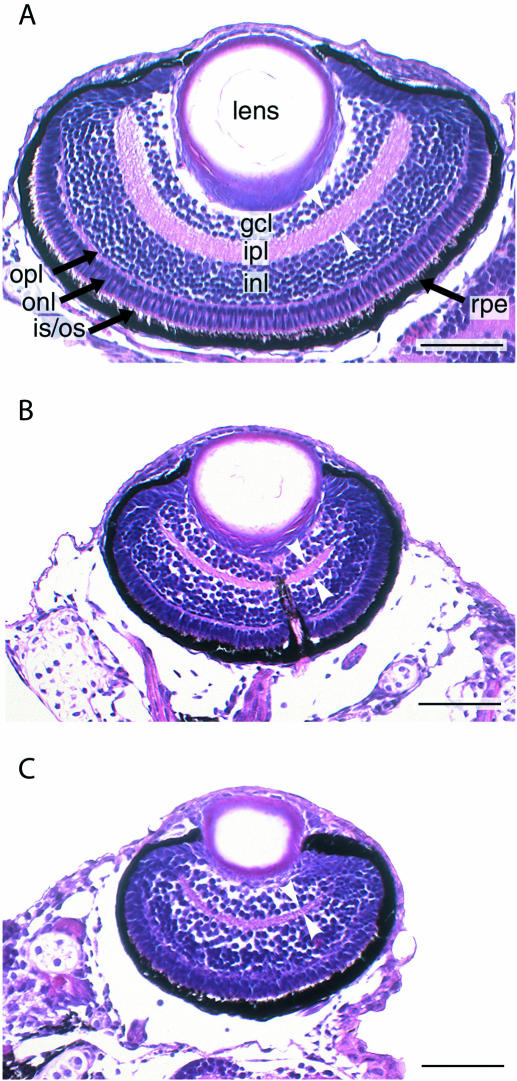
The developmental consequences of ccnd1-targeted MO and HypNA-pPNA injections were assessed first at 24 hpf (Figure 6). Cyclin D1 knockdown by MO and HypNA-pPNA injection was associated with comparable phenotypic changes confined to the head and eye regions. Knockdown of ccnd1 did not markedly affect body size, but did decrease eye and head size. Closer examination revealed that these changes consisted largely of growth inhibition as evidenced by the reduced circumference of the eye and the reduced size of the head region without overt evidence for necrosis or gross malformation. A quantitative analysis of the effects of ccnd1-targeted HypNA-pPNA and MO treatments based on measuring the diameters of the eyes clearly confirmed the visual impression of microophthalmia (Figure 7). Histological analysis of ccnd1 HypNA-pPNA-treated or MO-treated fish larvae further confirmed reduced gross size of the eyes throughout the observation period. Representative examples of control, MO-treated and HypNA-pPNA-treated larval eyes at day 5 of development are shown in Figure 8. The most striking antisense effects were a sharp reduction in the number of retinal cells and in the thickness of the inner plexiform layer (white arrowheads, Figure 8). This difference was not due to programmed cell death as only a few apoptotic cells were observed between days 2 and 5 of larval development in all experimental conditions (data not shown).
Figure 6.
Effect of ccnd1 knockdown on morphology of zebrafish embryos. Photomicrographs demonstrate the morphology of zebrafish embryos following knockdown of cyclin D1 at 24 h after microinjection with 0.1–1 mM ccnd1 antisense MO or HypNA-pPNA: (A) Uninjected control; (B) MO-treated embryo; (C) HypNA-pPNA-treated embryo; (D) Phenol Red control; (E) MO 3-base mismatch control; and (F) HypNA-pPNA 1-base mismatch control; all images were taken at ×100 magnification. The embryos injected with either antisense oligonucleotide displayed microophthalmia and microcephaly consistent with the restricted expression of cyclin D1 in 24 hpf zebrafish embryos. No gross morphologic defect was observed in the mismatch control-injected embryos. Labels: h, head; e, eye; y, yolk sac; b, body.
Figure 7.
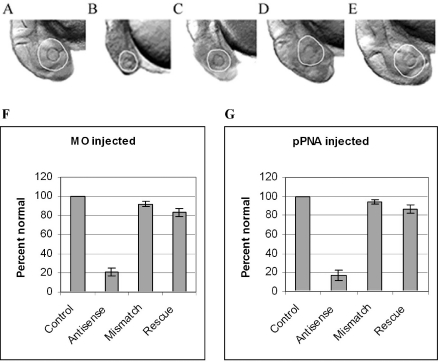
Sense mRNA rescue of ccnd1 antisense-treated zebrafish embryo morphology. Zebrafish embryos were microinjected with 0.5 mM ccnd1 antisense MO or HypNA-pPNA as described in Materials and Methods. Sense ccnd1 mRNA at 1 g/l was co-microinjected with the either MO or HypNA-pPNA into selected groups of embryos (designated ‘rescue’), and morphology recorded at 24 h after injection: (A) Phenol red-treated control; (B) MO-treated embryo; (C) HypNA-pPNA-treated embryo; (D) MO plus mRNA co-injected embryo; (E) HypNA-pPNA plus mRNA co-injected embryo; all images were taken at ×100 magnification. All embryos were treated with PTU following microinjection to inhibit pigment formation; therefore, the circumference of the eyes are delineated for better visualization. There is visual evidence of rescue of the ccnd1 antisense-induced microophthalmia and microcephaly in the ccnd1 mRNA co-injected embryos. The incidence of microophthalmia in MO- (F) or HypNA-pPNA-treated (G) embryos, with or without sense mRNA rescue, was calculated and expressed as percent (±SD) of 100 embryos per sample.
Figure 8.
Histologic appearance of zebrafish eyes following ccnd1 knockdown. Representative coronal sections of the retinas of 5-day-old larvae are shown. (A) Phenol red-treated control. (B) HypNA-pPNA-treated embryo. (C) MO-treated embryo. Layers are indicated as follows: GCL, ganglion cell layer; INL and ONL, inner and outer nuclear layer, respectively; IPL and OPL, inner and outer plexiform layer, respectively; RPE, retinal pigmented epithelium. White arrow heads indicate thickness of the inner plexiform layer. Scale bar: 50 µm.
Importantly, the developmental consequences of both MO and HypNA-pPNA cyclin D1 knockdown were sequence-specific. Even a one-base mismatch control, in the case of HypNA-pPNA injections, was sufficient to prevent the phenotypic changes produced by ccnd1-targeted HypNA-pPNA. In contrast, in the case of MOs, three-base mismatch controls were necessary to prevent inhibitory effects. To further assess the specificity of the effects observed, we performed rescue experiments by co-injection of ccnd1 mRNA and either HypNA-pPNA or MO. In either case, almost complete rescue was observed, confirming that the effects of either antisense construct were sequence-specific (Figure 7). In summary, these results revealed similar phenotypic effects produced by ccnd1 antisense oligonucleotides with quite divergent structures.
DISCUSSION
This study identifies molecular mechanisms controlling the establishment of cell cycle control during early zebrafish development. The delayed onset of G1/S transition control in this vertebrate model system allowed us to monitor expression of cell cycle regulators that contribute to the establishment of G1/S transition. The transcription profile analysis enabled simultaneous assessment of temporal changes in the expression patterns of multiple cell cycle regulators, cyclins, cdks and cdk inhibitors in early zebrafish development. Specifically, we observed high expression levels of cyclin genes ccna2, ccnb1, ccnb2 and ccne before the onset of zygotic transcription. This pattern was expected as these cyclins play prominent roles in S and M phases of the cell cycle, which are prevalent in the first hours of embryonal development (12).
A previous study demonstrated that zygotic transcription is necessary for the gradual slowing of cell cycle progression after MBT (5). In agreement with this earlier study we describe here that the expression of various molecular determinants of G1/S checkpoint control, the putative Rb and p21 gene orthologs, and ccnd1, commences after MBT. It should be noted that, as yet, zebrafish orthologs of ccnd2 and ccnd3 are not fully validated. Previous work demonstrated that, in zebrafish, ccnd1 expression occurred after MBT at the onset of G1 phase (4). Our results are consistent with this earlier report, and raised the question of what role cyclin D1 plays in zebrafish embryogenesis.
We found that downregulation of cyclin D1 by use of two antisense oligonucleotides with different backbones resulted in a phenotype restricted to the organ sites characterized by highest expression of cyclin D1 during gastrulation, i.e. the developing eye and the head region. Elimination of knockdown effects by a single mismatch in the HypNA-pPNA 16mer correlated with our previous observations of non-activity with one and two mismatches in 18mers, versus the need for four mismatches in an MO 25mer to abrogate activity (9).
The main effect of cyclin D1 downregulation on eye development was a proportional decrease in the size of the organ owing to reduced cellularity, but not overt cell death. This result is remarkably similar to results described in cyclin D1-deficient mice generated by gene targeting in embryonic stem cells (17,18). As in zebrafish (16), cnnd1 mRNA expression is very high in the developing retina and brain of mice. Furthermore, cyclin D1 deficiency in mice led to impaired development of all layers of the retina in a fashion very similar to the results described in this study. Collectively, these results show a remarkable degree of functional conservation of cyclin D1 function across species and open the door to the study of other cell cycle regulators in zebrafish.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Dr Michael Choob and Dr John Archdeacon of Active Motif for their contribution of trans-4-hydroxy-l-proline PNA/phosphono PNA oligomers. These studies were supported by the National Institutes of Health (CA81008 to U.R.; CA10663 to A.P.D.; CO27175 to E.W.), the Pennsylvania Department of Health, Tobacco Settlement Act of Pennsylvania (A.P.D.), the Mary R. Gilbert Trust (A.P.D.), and the Department of Energy (ER63055 to E.W.). Funding to pay the Open Access publication charges for this article was provided by the Department of Energy.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kiyokawa H., Koff A. Roles of cyclin-dependent kinase inhibitors: lessons from knockout mice. Curr. Top. Microbiol. Immunol. 1998;227:105–120. doi: 10.1007/978-3-642-71941-7_5. [DOI] [PubMed] [Google Scholar]
- 2.Dash B.C., El-Deiry W.S. Cell cycle checkpoint control mechanisms that can be disrupted in cancer. Methods Mol. Biol. 2004;280:99–161. doi: 10.1385/1-59259-788-2:099. [DOI] [PubMed] [Google Scholar]
- 3.Masui Y., Wang P. Cell cycle transition in early embryonic development of Xenopus laevis. Biol. Cell. 1998;90:537–548. [PubMed] [Google Scholar]
- 4.Yarden A., Salomon D., Geiger B. Zebrafish cyclin D1 is differentially expressed during early embryogenesis. Biochim. Biophys. Acta. 1995;1264:257–260. doi: 10.1016/0167-4781(95)00175-1. [DOI] [PubMed] [Google Scholar]
- 5.Zamir E., Kam Z., Yarden A. Transcription-dependent induction of G1 phase during the zebra fish midblastula transition. Mol. Cell. Biol. 1997;17:529–536. doi: 10.1128/mcb.17.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nasevicius A., Ekker S.C. Effective targeted gene ‘knockdown’ in zebrafish. Nature Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 7.Efimov V.A., Choob M.V., Buryakova A.A., Kalinkina A.L., Chakhmakhcheva O.G. Synthesis and evaluation of some properties of chimeric oligomers containing PNA and phosphono-PNA residues. Nucleic Acids Res. 1998;26:566–575. doi: 10.1093/nar/26.2.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Efimov V.A., Klykov V.N., Chakhmakhcheva O. Phosphono peptide nucleic acids with a constrained hydroxyproline-based backbone. Nucleosides Nucleotides Nucleic Acids. 2003;22:593–599. doi: 10.1081/NCN-120021962. [DOI] [PubMed] [Google Scholar]
- 9.Urtishak K.A., Choob M., Tian X., Sternheim N., Talbot W.S., Wickstrom E., Farber S.A. Targeted gene knockdown in zebrafish using negatively charged peptide nucleic acid mimics. Dev. Dyn. 2003;228:405–413. doi: 10.1002/dvdy.10394. [DOI] [PubMed] [Google Scholar]
- 10.Wickstrom E., Choob M., Urtishak K.A., Tian X., Sternheim N., Talbot S., Archdeacon J., Efimov V.A., Farber S.A. Sequence specificity of alternating hydroyprolyl/phosphono peptide nucleic acids against zebrafish embryo mRNAs. J. Drug Target. 2004;12:363–372. doi: 10.1080/10611860412331285242. [DOI] [PubMed] [Google Scholar]
- 11.Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 4th edn. Eugene, OR: University of Oregon Press; 2000. [Google Scholar]
- 12.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 13.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puglisi J.D., Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 15.Tsao-Wu G.S., Weber C.H., Budgeon L.R., Cheng K.C. Agarose-embedded tissue arrays for histologic and genetic analysis. Biotechniques. 1998;25:614–618. doi: 10.2144/98254st02. [DOI] [PubMed] [Google Scholar]
- 16.Thisse B., Pflumio S., Fürthauer M., Loppin B., Heyer V., Degrave A., Woehl R., Lux A., Steffan T., Charbonnier X.Q., et al. 2001. Expression of the zebrafish genome during embryogenesis, ZFIN Direct Data Submission. [Google Scholar]
- 17.Sicinski P., Donaher J.L., Parker S.B., Li T., Fazeli A., Gardner H., Haslam S.Z., Bronson R.T., Elledge S.J., Weinberg R.A. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 18.Fantl V., Stamp G., Andrews A., Rosewell I., Dickson C. Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes Dev. 1995;9:2364–2372. doi: 10.1101/gad.9.19.2364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.