Abstract
Correct docking of U3 small nucleolar RNA (snoRNA) on pre-ribosomal RNA (pre-rRNA) is essential for rRNA processing to produce 18S rRNA. In this report, we have used Xenopus oocytes to characterize the structural requirements of the U3 snoRNA 3′-hinge interaction with region E1 of the external transcribed spacer (ETS) of pre-rRNA. This interaction is crucial for docking to initiate rRNA processing. 18S rRNA production was inhibited when fewer than 6 of the 8 bp of the U3 3′–hinge complex with the ETS could form; moreover, base pairing involving the right side of the 3′-hinge was more important than the left. Increasing the length of the U3 hinge–ETS interaction by 9 bp impaired rRNA processing. Formation of 18S rRNA was also inhibited by swapping the U3 5′- and 3′-hinge interactions with the ETS or by shifting the base pairing of the U3 3′-hinge to the sequence directly adjacent to ETS region E1. However, 18S rRNA production was partially restored by a compensatory shift that allowed the sequence adjacent to the U3 3′-hinge to pair with the eight bases directly adjacent to ETS region E1. The results suggest that the geometry of the U3 snoRNA interaction with the ETS is critical for rRNA processing.
INTRODUCTION
The ribosome is a gigantic macromolecular complex composed of almost 100 proteins and several RNA components. Biogenesis of the ribosome occurs in the nucleolus [reviewed in (1–4)]; this process has been calculated to consume much of the energy of the cell (5). Maturation of the ribosomal RNA (rRNA) is assisted by ∼200 small nucleolar RNAs (snoRNAs). The vast majority of the snoRNAs direct modifications on rRNA (2′-O-ribose methylations and pseudouridylations). The function of the modifications is unknown, and the individual guide snoRNAs are not essential for cell viability.
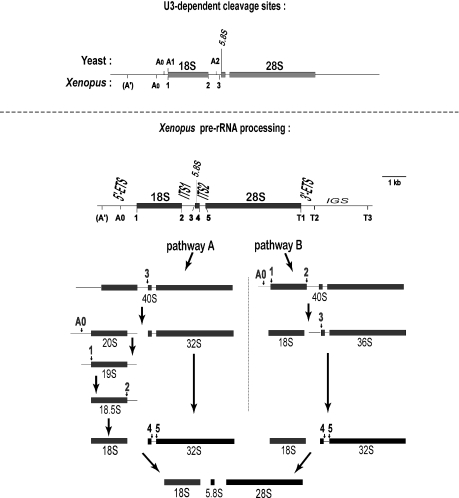
In contrast, a few snoRNAs are essential. They are required for rRNA processing to form the mature rRNAs (Figure 1). The precursor rRNA (pre-rRNA) is cleaved to remove the external and internal transcribed spacers (ETS, ITS). About half a dozen snoRNAs are needed to form 18S rRNA in eukaryotes; the most abundant of these is U3 snoRNA. The pre-rRNA cleavage sites that require U3 snoRNA (6–9) are indicated in Figure 1. Although there are some similarities between yeast and metazoa in certain U3-dependent cleavage sites (sites A0 and A1/1), there are also some differences. First, site A′ has not been found in yeast rRNA processing. Second, cleavage at the 3′ end of 18S rRNA occurs in the nucleolus at U3-dependent site 2 in higher organisms. In contrast, in yeast this cleavage occurs in the cytoplasm at site D and it requires a protein rather than U3 (10–14). Third, it is unclear if metazoan site 3 is the counterpart of site A2 in yeast. Nonetheless, despite these differences, U3 snoRNA is essential in all organisms to form 18S rRNA.
Figure 1.
Cleavage sites in rRNA processing. Upper panel: comparison of the U3-dependent cleavage sites in yeast and Xenopus, which share in common site A0, site A1/1 and perhaps site A2/3. U3-dependent cleavage at sites A′ and 2 is found in metazoa but not in yeast. Lower panel: Xenopus pre-rRNA processing pathways. Pathways A and B differ in whether site 3 or sites A0, 1 and 2 are cleaved first. Pathways A and B can co-exist in a single cell, but some frogs just use pathway A (6). Note that the 20S intermediate is diagnostic for pathway A and the 36S intermediate is diagnostic for pathway B. Conversion of 20S to 18S occurs rapidly and the transient 19S and 18.5S intermediates are not always seen.
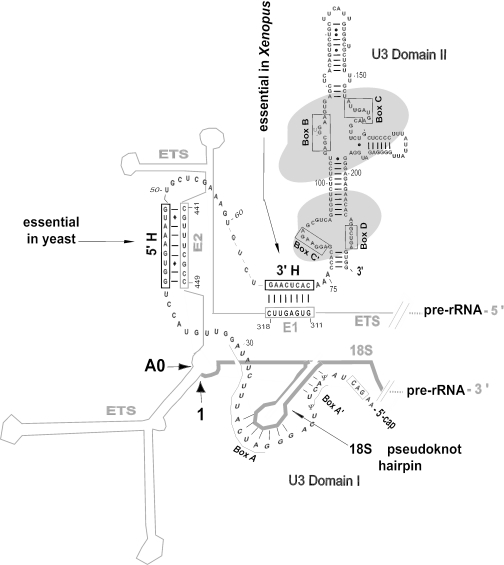
U3 snoRNA is synthesized in the nucleoplasm (15) and travels through Cajal bodies (16–18) to the nucleolus (16,19) where it functions in rRNA processing. The protein nucleolin escorts U3 snoRNP from Cajal bodies to the nucleolus (17) and is required for U3 snoRNA to dock on the ETS of pre-rRNA (20,21). U3 snoRNP can be found as a 12S ribonucleoprotein monoparticle. After docking on pre-rRNA, U3 snoRNP is part of an 80–90S ribonucleoprotein complex (22–25) named the small subunit ribosomal processing complex (SSU processome). Besides nucleolin, U3 docking on pre-rRNA requires a base-pairing interaction of the single-stranded ‘hinge’ region of U3 snoRNA with a complementary sequence in the 5′-ETS (hereafter simply called the ETS) (Figure 2). There are two hinge regions in U3 snoRNA, and they separate domains I and II of the molecule (Figure 2). Compensatory mutations in Xenopus revealed that base pairing between the 3′-hinge (3′H) of U3 snoRNA and region E1 of the ETS is essential for 18S rRNA production, whereas base pairing between the 5′-hinge (5′H) of U3 with region E2 of the ETS is helpful but not essential to form 18S rRNA (26). In contrast, it is the base pairing between the 5′-hinge of U3 and the ETS that is required in yeast (27). The role if any of the 3′-hinge in yeast U3 is unknown.
Figure 2.
U3 snoRNA interaction with the ETS. Base pairing between the 3′-hinge (3′H) of U3 snoRNA and region E1 of the pre-rRNA ETS is critical and sufficient for rRNA processing in Xenopus, whereas base pairing between the U3 5′-hinge (5′H) and region E2 of the ETS is auxiliary (26). In contrast, the 5′H–E2 interaction is essential in yeast and the 3′H–E1 interaction is not sufficient to support rRNA processing (27). Domain II of U3 snoRNA has many base-paired stems and binds several proteins (indicated by shading). Domain I of U3 is shown in an open configuration that can interact with the 5′ end of the 18S region of pre-rRNA (9,26,28). The conserved sequences comprising boxes that are conserved in U3 snoRNA from all species are indicated. Base pairing of U3 box A with the first terminal loop in 18S rRNA is thought to prevent premature pseudoknot formation (29,30). The pre-rRNA cleavage sites A0 and 1 are close to one another in the proposed secondary structure of the ETS (9,31).
Nucleolin binds to an evolutionarily conserved motif in the ETS next to the region that base pairs with the 3′-hinge of U3 snoRNA in metazoa (3,26). When the 3′-hinge is mutated, U3 snoRNA still associates with the 80S SSU processome [(32); A. Borovjagin, unpublished data], perhaps via its nucleolin-mediated association with the ETS. Nonetheless, cells with this mutation cannot direct processing to form 18S rRNA (26). Presumably, the 3′H–E1 interaction is required to properly position U3 snoRNA on the pre-rRNA substrate so that it can function in rRNA processing.
Complementary sequences exist in all eukaryotes between the U3 hinges and the ETS (33), though the actual sequences involved are not conserved. Evolutionary covariation in sequence seems to have maintained the base-pairing potential between the U3 hinges and the ETS. The species-specific sequences used for this interaction provide a potential target for a new category of antibiotics that would prevent ribosome biogenesis rather than inhibit ribosome function. RNA therapeutics (e.g. antisense oligos, ribozymes, etc.) that prevent the U3 hinge interaction with the ETS in just the pathogen but not the host would specifically prevent the formation of the pathogen's ribosomes without injurious side effects to the host. This approach has great potential to combat infections by eukaryotic pathogens. As a first step in the development of this approach, we describe here the requirements for base pairing as well as positioning of the U3 docking interaction on the ETS.
In the present report, we examine the structural basis for the functional interaction between the hinge region of metazoan U3 snoRNA with the ETS. We use a novel approach of compensatory base changes to alter the position of U3 snoRNA base pairing with the ETS. This strategy provides insight into the topographic requirements for base pairing between the partner molecules. Our results provide a first step toward understanding the principles of spatial organization of the complex formed during U3 snoRNA docking on pre-rRNA. The data suggest reasons for the differences in the U3 docking interaction between yeast and higher organisms acquired in the course of evolution.
MATERIALS AND METHODS
U3 snoRNA depletion-rescue and rRNA analysis
For experiments with endogenous rather than tagged rRNA, Xenopus laevis stages 5 and 6 oocytes were isolated and injected with antisense oligonucleotide 39–54 to disrupt endogenous U3 snoRNA as described previously (8). Subsequently, in vitro transcripts of U3 were injected at a concentration of 5 ng/oocyte. When two different concentrations of U3 were injected, the 1× concentration was 2.5 ng/oocyte and the 3× concentration was 7.5 ng/oocyte. The procedures for in vivo labeling of rRNA synthesized after U3 snoRNA depletion, RNA isolation and analysis by gel electrophoresis were as described previously (8). Each experiment shown here was repeated two or more times, confirming the results. Moreover, two different approaches confirmed the conclusions of Figures 3 and 4.
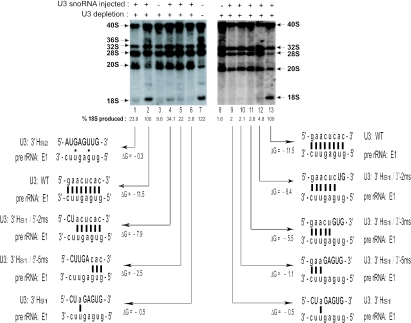
Figure 3.
Base-pairing disruption of the U3 3′-hinge with ETS region E1. Intact endogenous U3 snoRNA was depleted by antisense oligonucleotide injection into oocyte nuclei. Subsequently, synthetic wild-type or mutated U3 was injected, and the restoration of 18S rRNA production was assayed by in vivo labeling with 32P-UTP, gel electrophoresis and autoradiography. The amount of 18S rRNA produced was quantified (see Materials and Methods) to normalize for any loading differences between gel lanes. Left panel: increasing disruption of base pairing at the left side of the 3′H–E1 interaction; right panel: increasing disruption of base pairing at the right side of the 3′H–E1 interaction.
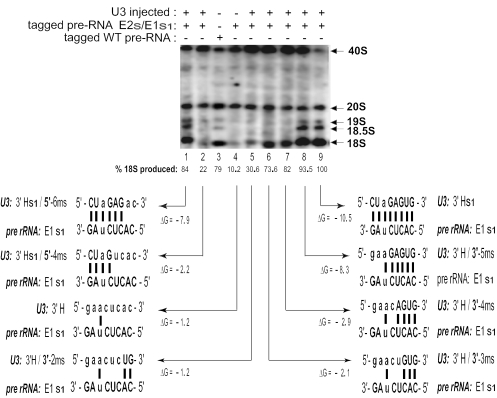
Figure 4.
Restoration of base pairing of the U3 3′-hinge with ETS region E1. A plasmid with tagged pre-rRNA carrying mutations in ETS regions E1 and E2 was co-injected into Xenopus oocyte nuclei with an increasing number of substituted bases in the 3′-hinge (3′H) of U3 snoRNA. The number of base pairs between the U3 3′H and ETS region E1 required for the production of 18S rRNA was analyzed by Northern blots using a probe against the sequence tag in the 18S coding region of the injected rRNA plasmid. Less 40S pre-rRNA is seen in lanes 3 and 9 where there was complete base pairing between the U3 hinge and the ETS than in the mutations that shortened the base-pair interaction, suggesting that 40S pre-rRNA was processed more rapidly in the wild-type situation.
The gel lanes were scanned and quantified using Scion Image release Beta 4.0.2 (2000) software. All peaks (bands) within each lane were plotted and the intensities (areas under each peak less background) were integrated. The amount of 18S rRNA produced was calculated as its percent of the total intensity of all bands in the lane. The value for 18S rRNA produced in samples where injection restored complete base pairing between the U3 3′-hinge and E1 regions was set at 100% and the values of 18S rRNA production in the other lanes were calculated as their percentage relative to the control.
U3 snoRNA mutagenesis and synthesis
U3 snoRNA mutants were created by multi-step PCR as described previously (33). The PCR DNA products of all the U3 snoRNA mutants were gel-purified and cloned into the pT7 blue-R cloning vector (Invitrogen). DNA sequencing was used to confirm all mutations. DNA of the plasmid constructs was used to produce PCR templates for in vitro T7 transcription as described previously (9). Sequences of mutagenic oligonucleotides used to introduce sequence substitution mutations in U3 snoRNA can be provided upon request.
Pre-rRNA mutagenesis
Construction of a plasmid with Xenopus pre-rRNA containing a tag in the 18S region has been described previously (26). Briefly, plasmid pXlr101, kindly provided by Dr Ronald Reeder, containing an entire genomic repeat of X.laevis rDNA was the starting material. An 8 nt tag sequence 5′-CCUCGAGU-3′ was introduced into an expansion segment of 18S rRNA (nt 284–291) by standard PCR mutagenesis. Mutations in E1 and E2 of the ETS were made by standard two-step PCR. Owing to the high GC content of the X.laevis ETS, PCR was performed with Failsafe PCR PreMix Selection Kit (Epicentre Technologies, Madison, WI), using the reaction mixtures G or K.
Pre-rRNA expression and Northern blot analysis of rRNA
Plasmid DNA carrying full size wild-type or mutated rDNA with a Pol I promoter and the 18S-tag was injected into stage 5 oocytes at a concentration of 2.5–7.5 ng/oocyte with U3 transcripts as described above. RNA from 25–50 manually isolated nuclei was analyzed by Northern blots using a probe complementary to the 18S-tag. Short wave ultraviolet light was used to visualize endogenous rRNA on the filters to confirm equal loading of RNA in the gel lanes.
The free energy (ΔG) of the hinge base pairing with the ETS in the wild type and various mutations was calculated using the ‘two state hybridization server’ application of Mfold with the default RNA and salt concentrations and temperature of 25°C [(34); www.bioinfo.rpi.edu/applications/mfold/old/rna/form6.cgi].
RESULTS
Base-pairing requirements of the 3′H–E1 duplex for 18S rRNA formation
Xenopus oocytes were injected with 32P-UTP; nuclear RNA was isolated, and the 40S pre-rRNA and the processing intermediates that give rise to the mature 18S and 28S rRNAs were visualized by gel electrophoresis and autoradiography (Figure 3, lane 7). The amount of 40S pre-rRNA varies between oocytes, perhaps reflecting differences in the rate of cleavage. The endogenous U3 snoRNA can be disrupted by injection into the oocyte of an antisense oligonucleotide targeted to U3 snoRNA (6,8). The RNA–DNA duplex is then cleaved by endogenous RNase H. In the absence of intact U3 snoRNA, mature 18S rRNA is not formed (Figure 3, lanes 3 and 8). The injected antisense oligo is not stable in the oocyte. Thus, synthetic wild-type U3 RNA can be injected subsequently to restore 18S rRNA production (Figure 3, lanes 2 and 13).
We have used mutagenesis to test the functional importance of U3 hinge–ETS base pairing. Mutation of the U3 3′-hinge inhibits 18S rRNA formation, which can be restored by compensatory mutation in region E1 of the ETS (26). This restoration indicates that it is the base pairing and not the sequences per se that is important. Moreover, the U3 hinge mutations do not seem to create an altered (non-functional) secondary structure of U3 snoRNA because the ETS compensatory mutations can restore 18S rRNA formation. When the 3′-hinge of U3 snoRNA is replaced by substitution s1 (U3: 3′H-s1) or by substitution s2 (U3: 3′H-s2), the ability of U3 to base pair with the ETS is abrogated and production of 18S rRNA is inhibited (Figure 3, lanes 1, 6 and 9). These substitutions remove all but 1 or 2 bp between the 3′-hinge of U3 snoRNA and region E1 of the ETS (Figure 3). A series of substitutions were used to elucidate the detailed base-pairing requirements for the 3′H–E1 functional interaction. The substitutions in the 3′-hinge sequence of U3 snoRNA were introduced from either its 5′ or 3′ end, replacing the wild-type nucleotides with those from the U3 3′Hs1 mutation. The negative free energy (ΔG) decreased concurrent with the decrease in the number of base pairs in the U3 hinge–ETS interaction (Figure 3).
When the first 2 bp on the left side of the duplex were disrupted by U3 snoRNA mutation 3′Hs1/5′-2ms, only 34.1% of 18S rRNA accumulated (Figure 3, lane 4) as compared with rescue with wild type U3 (Figure 3, lane 2). The inhibition was even more pronounced for a 2 nt mismatch on the right side of the duplex that resulted in 4.8% of 18S rRNA production (Figure 3, lanes 12). Since the ΔG values for pairing with ETS region E1 are basically the same for each of the two nucleotide U3 mutants (−8.4 and −7.9 for the right and left side substitutions, respectively), it appears that the right side of the duplex is more important than the left. Further disruption of base pairing on the right side of the duplex almost completely blocked 18S production (∼2% remaining; Figure 3, lanes 8–11). In contrast, disruption of up to 5 bp on the left side of the 3′H–E1 duplex by mutations 3′Hs1/5′-3ms, 3′Hs1/5′-4ms (data not shown) or 3′Hs1/5′-5ms (Figure 3, lane 5) was less inhibitory (Figure 3, lane 5: 22% of 18S rRNA was produced) than disruption of only 3 bp on the right side (Figure 3, lane 11: 2.6% of 18S rRNA produced), despite the formation of a potentially less stable duplex (Figure 3, compare lane 5 ΔG = −2.5 and lane 11 ΔG = −5.5). This reinforces the conclusion of the greater importance of the right side of the 3′H–E1 duplex.
Therefore, in vivo labeling with 32P demonstrated that as the length of the mutation in the U3 3′-hinge increased (decreasing the number of base pairs with E1 of the endogenous pre-rRNA), there was a gradual reduction in 18S rRNA production. This was more pronounced when the right side of the duplex was disrupted.
This conclusion was further supported by an alternate experimental approach where the substituted nucleotides of the same set of U3 3′-hinge mutants were now used to restore base pairing with E1 from the left or right side instead of disrupting it. This was achieved by co-injection into Xenopus oocytes of an expression vector carrying tagged pre-rRNA with an E1 sequence substitution (E1s1) in the ETS together with a set of U3 3′H substitution mutants that could base pair with E1s1. The 8 nt sequence substitution (‘tag’), introduced into an expansion segment [evolutionarily variable region (35)] in the 18S portion of pre-rRNA, was hybridized to a probe for Northern blots to monitor the production of tagged 18S rRNA and its precursors. The 18S tag did not interfere with rRNA processing (26).
In contrast to the previous in vivo labeling experiment (Figure 3), the base-pairing requirement for the 3′H–E1 interaction in the U3/plasmid co-injection experiment was studied in the absence of 5′H–E2 base pairing, which is non-essential but auxiliary in Xenopus (26).This was achieved by co-injecting the E2s/E1s1 double mutant rDNA with U3 3′H mutants. In this set of experiments, as the number of mutated nucleotides in the 3′-hinge of U3 snoRNA increased, the length of the duplex with the E1s1 region in the plasmid-encoded pre-rRNA also increased. In line with previous U3 depletion-rescue experiments, restoration of three or more base pairs on the right side of the duplex by U3 mutations 3′H/3′-3ms, 3′H/3′-4ms and 3′H/3′-5ms resulted in gradual restoration of tagged 18S rRNA production up to almost full rescue levels when 6 of the 8 bp were formed (Figure 4, lanes 6–8). On the other hand, restoration of 4 bp on the left side of the duplex was not as effective in rescuing production of tagged 18S rRNA as its counterpart mutation on the right side [Figure 4, compare lane 2 (22%) with lane 6 (73.6%)], although the duplexes have almost the same negative free energy (ΔG = −2.2 and −2.1, respectively). Similarly, restoration of 6 bp on the left side was somewhat less effective than restoration of 6 bp on the right side of the 3′H–E1 duplex [Figure 4, compare lane 1 (84%) with lane 8 (93.5%)]. Accumulation of 19S and 18.5S pre-rRNA intermediates also appeared in parallel with 18S rRNA restoration. The 19S and 18S rRNA intermediates are rapidly processed and are not always visible, but sometimes they can be seen even for the full length 3′H–E1 interaction.
The data obtained in both types of compensatory base change experiments are consistent. However, the absolute amount of 18S rRNA produced for similar base pairing and ΔG values of the U3 hinge–ETS duplex was greater for the co-injection/Northern blot experiment (Figure 4, lanes 1, 6 and 8) than for the in vivo labeling assay (Figure 3, lanes 12, 5 and 4, respectively), perhaps reflecting the different experimental designs. The sensitivity of the in vivo labeling approach (Figure 3) is lower than that of the tagged rDNA plasmid/U3 co-injection approach (Figure 4). In the former, 18S rRNA is likely only made from the 40S pre-rRNA that was transcribed after U3 was injected (4 h after the antisense oligonucleotide and 32P label injection), as inferred from previous work (36). Thus, the 40S and 20S pre-rRNAs that were labeled during 0–4 h will not be processed to 18S rRNA by U3 transcripts that were injected subsequently. Nonetheless, the trends in the results are the same in both approaches. They reinforce the conclusion that the ability to support a functional interaction between U3 snoRNA and pre-rRNA is determined by the number of base pairs in the 3′H–E1 duplex (its negative free energy) and, in addition, on which side of the duplex those base pairs form. The right side of the 3′-hinge of U3 (proximal to U3 domain II) that forms a duplex with region E1 of the ETS appears to be more important than the left side (distal to U3 domain II).
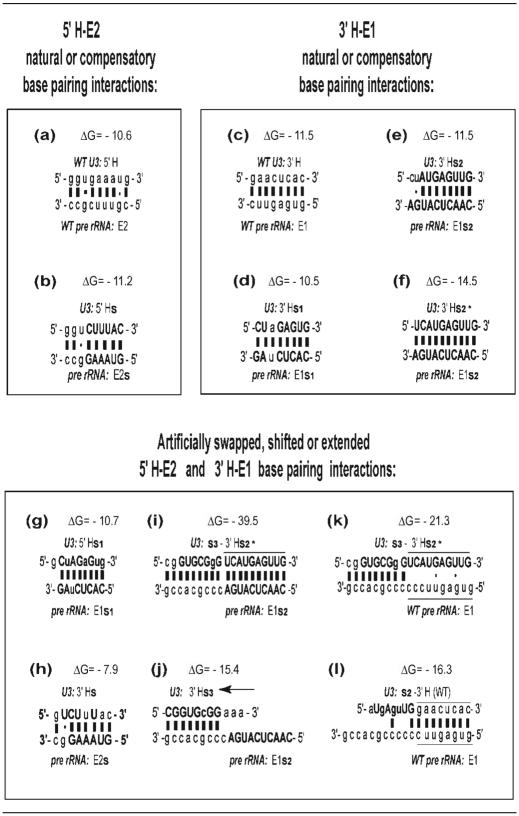
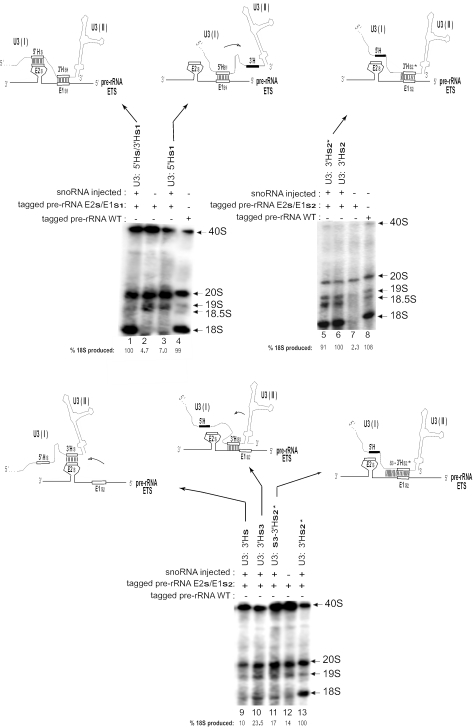
Having found that destabilization of the 3′H–E1 duplex impaired rRNA processing, we next analyzed the consequence of increasing the duplex stability. Mutations used for this and for other experiments are summarized in Table 1. Generally, the negative free energy (ΔG) increases concurrent with an increase in the number of base pairs between U3 and the ETS. The duplex potentially formed between the 3′-hinge of U3 and region E1 of the ETS rRNA is extended by 2 bp at its left boundary when U3 carries the 3′Hs2* substitution, which can base pair with E1s2 of pre-rRNA (Table 1, panel f). When plasmid encoding the pre-rRNA double mutant E2s/E1s2 (to prevent the 5′H–E2 interaction) was co-injected with transcripts from the single mutant 3′Hs2* of U3 snoRNA, 18S rRNA production was still mostly restored (Figure 5, compare lane 5 with lane 6). This suggests that the increase in the length of the 3′H–E1 duplex by 2 bp (Table 1, compare panel f with panel e) is not critical.
Table 1.
U3 hinge–ETS base pairing
The sequences involved in the wild-type or mutated interaction between the 5′-hinge (5′H) of U3 and region E2 of the ETS or between the 3′-hinge (3′H) of U3 and region E1 of the ETS are shown. Lower case letters depict the wild-type sequence, and capital letters indicate mutated bases. When the interaction was extended to involve flanking sequences, the positions of the U3 3′-hinge and of ETS region E1 are marked by horizontal lines. The arrow in panel (j) indicates the 8 bp shift in the U3–ETS interaction.
Figure 5.
18S rRNA production requires precise positioning of base pairing between the U3 hinge and the ETS. U3 snoRNA containing sequence substitutions in the hinges or flanking regions was co-injected with a plasmid containing tagged 18S rRNA and substitutions in ETS E1 or E2 (see Table 1 for the sequences used). The mutated U3 hinges and mutated ETS regions E1 and E2 are indicated by open boxes, whereas the wild-type sequence for these areas is shown by a blackened box. Domain I and domain II of U3 snoRNA is marked. Shifts in the relative position of U3 relative to the pre-rRNA are denoted by an arrow. The production of tagged 18S rRNA after these treatments was assayed by Northern blots using a probe complementary to the 18S tag sequence. Internal controls are used for each panel because the rate of 40S pre-rRNA processing can vary between frogs (e.g. compare lanes 5 and 13).
In contrast, an increase in the 3′H–E1 duplex by 9 bp compromises 18S rRNA production to 17% of its level with wild-type U3 (Figure 5, lane 11). In this case, 19 contiguous base pairs (Table 1, panel i) were created by substitution of seven bases (s3) directly upstream of the 3′H in U3 snoRNA. The sequence in this area of U3 snoRNA is not essential for its function in rRNA processing (33). Substitution s3 in this non-essential region was coupled with the 3′Hs2* substitution in U3 and tested for its ability to restore tagged rRNA processing when co-injected with the rDNA plasmid carrying mutation E1s2. This double substitution increased the length of potential interaction by 9 bp and significantly stabilized the duplex (ΔG = −39.5), resulting in less 18S rRNA produced (Figure 5, compare lane 11 with lane 13). The data suggest that not only shortening, but also an excessive increase in length of the 3′H–E1 duplex is detrimental for rRNA processing.
18S rRNA production requires precise positioning of base pairing between the U3
We next investigated whether the position of that base-pairing interaction relative to other U3 or pre-rRNA sequences is critical or not. To address this question, we created mutations that would allow the 5′-hinge of U3 snoRNA to artificially base pair with the E1 region of the ETS (instead of its normal E2 pairing partner), thus shifting the relative position of U3 base pairing with pre-rRNA toward the 5′-hinge end of the precursor (Figure 5, lane 3 schematic). To induce this artificial interaction, we injected 18S-tagged rDNA plasmid carrying the ETS mutations E2s and E1s1, neither of which can base pair with the endogenous U3 snoRNA or support rRNA processing. This plasmid was co-injected with transcripts of U3 carrying the mutation 5′Hs1, making the 5′-hinge complementary to E1s1 on the rDNA plasmid (Table 1, panel g). In this situation, the 5′-hinge of U3 snoRNA could base pair with region E1 rather than with its natural partner, region E2 of the ETS. As can be seen from Figure 5 (lane 3), hardly any tagged 18S rRNA was formed despite the potential complementarity. Moreover, the negative free energy (ΔG = −10.7) was also the same as for the functional 3′Hs1/E1s1 (control) interaction (ΔG = −10.5), as listed in Table 1. This result suggests that the E1 region of the ETS needs to base pair with a specific region of U3 snoRNA. Therefore, shifting the interaction from the 3′-hinge to the 5′-hinge of U3 is not adequate to support rRNA processing.
We also carried out the converse experiment where mutations resulted in complementarity of the 3′-hinge of U3 with region E2 rather than E1 of the ETS. 18S-tagged rDNA plasmid carrying the ETS mutation E2s/E1s2 was injected into oocytes together with U3 transcripts containing the mutation 3′Hs. In this situation, 3′Hs of U3 can only base pair with E2s of the ETS, rather than with its natural E1s1 partner on mutant pre-rRNA (Table 1, panel h). In this artificial situation, the relative position of U3 base pairing on pre-rRNA is shifted toward the 3′ end of the precursor (Figure 5, lane 9 schematic). Despite the potential complementarity between U3 and the ETS of pre-rRNA, again hardly any tagged 18S rRNA was produced (Figure 5, lane 9). This result and the one above suggest that positional shifts that move the U3 interaction toward the 5′ or 3′ end of the ETS greatly impair rRNA processing and 18S rRNA formation.
We next inquired whether a less drastic change in the position of U3 base pairing with the ETS would permit rRNA processing. Since the 3′H–E1 interaction is most important and sufficient in Xenopus, a situation was created by mutagenesis in which the 3′H–E1 interaction was replaced by base pairing of the U3 3′-hinge with the sequences directly adjacent to region E1. For this case, the 18S-tagged rDNA plasmid containing E2s and E1s2 was co-injected with transcripts of U3 containing the mutation 3′Hs3 that is complementary to ETS sequences directly adjacent to 3′ side of E1 (Table 1, panel j). Interestingly, even this slight shift in the base-pairing regions reduced the production of tagged 18S rRNA detected by Northern blot hybridization to 23.5% (Figure 5, lane 10).
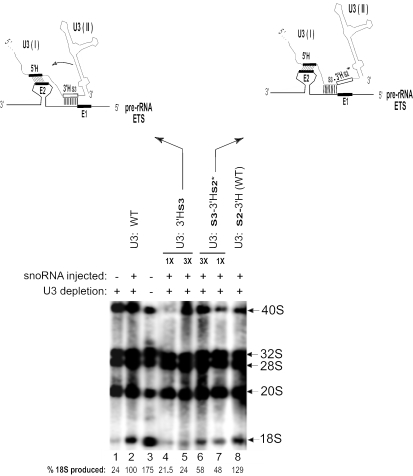
A similar result was also obtained in an in vivo labeling experiment where the same base-pairing configuration was recapitulated by depleting endogenous U3 snoRNA and replacing it with the U3: 3′Hs3 mutant. Even though base pairing is possible between 3′Hs3 of U3 and sequences directly adjacent to region E1 of endogenous pre-rRNA, there was hardly any rescue of 18S rRNA production (Figure 6, lanes 4 and 5). In this case, unlike the plasmid co-injection/Northern blot above, the 5′H–E2 interaction can occur, but, as noted previously (26), it is not sufficient to restore 18S rRNA production. Both types of experiments demonstrate that even this slight shift in base pairing between the 3′H of U3 and sequences in the ETS has some deleterious effects.
Figure 6.
A compensatory shift in base pairing between U3 snoRNA and the ETS restores the relative position of U3 on pre-rRNA and 18S rRNA can be produced. After antisense oligonucleotide-mediated depletion of endogenous U3 snoRNA, Xenopus oocyte nuclei were injected with 1× or 3× amounts of synthetic transcripts of U3 carrying a sequence substitution in the 3′-hinge or the region immediately upstream to pair with the ETS of endogenous wild-type pre-rRNA. Open boxes show regions of sequence substitution, and blackened boxes show regions of wild-type sequence for the U3 hinges and for ETS region E1 and E2. The production of 18S rRNA was assayed by gel electrophoresis and autoradiography of 32P-UTP in vivo labeled RNA.
Consequently, we investigated whether a shift in the U3 base-pairing site could be compensated by a concomitant shift in its ETS base-pairing partner. This would approximate the proper geometry of the interaction between U3 and pre-rRNA, and therefore might better rescue rRNA processing. To accomplish this, U3 transcripts carrying the s3 substitution immediately upstream of the 3′-hinge were injected into oocytes after depletion of endogenous U3 snoRNA. The mutant U3 transcripts also carried the 3′Hs2* substitution which cannot base pair with wild-type E1 of the endogenous pre-rRNA ETS. However, as noted above, the s3 sequence can base pair with the sequence directly adjacent to E1 (Table 1, panel k). Thus, although there is a shift in the U3–ETS base-pairing region, the relative position of U3 snoRNA with respect to pre-rRNA remains the same as for wild-type molecules. As a result of U3s3-3′Hs2* injection into U3-depleted oocytes, we observed a partial restoration of 18S rRNA production (Figure 6, lanes 6 and 7). In addition to the wild-type base pairing (Figure 6, lane 2), we used another control to demonstrate that the sequence directly upstream of the U3 3′-hinge that was replaced by s3 is not important. For this purpose, we utilized U3 RNA that had wild-type sequences for both hinge–ETS interactions, but had substitution s2 in the same position (Table 1, panel l) as substitution s3 (directly upstream of the U3 3′-hinge). Although the s2 sequence cannot pair significantly with the ETS (unlike the s3 sequence), 18S rRNA is still produced (Figure 6, lane 8), because the wild-type 3′H–E1 interaction can occur, consistent with our earlier findings (33).
This study demonstrates that precise positioning of U3 snoRNA relative to pre-rRNA by means of base pairing with sequences in the ETS is critical for its ability to function in rRNA processing to produce 18S rRNA in Xenopus.
DISCUSSION
U3 snoRNA docking on pre-rRNA
Docking of U3 snoRNP on the ETS of pre-rRNA seems to promote the formation of the SSU processome. Electron microscopy of Miller spreads of transcribing rDNA suggests that the processome may be the terminal balls on nascent pre-rRNA. Mutation or depletion that prevents the U3 hinge interaction with the ETS (22,37) or depletion of U3-associated proteins (38,39) obliterates the terminal balls.
U3 snoRNA docking on pre-rRNA is assisted by nucleolin (20), but functional docking requires base pairing between U3 and the ETS (26). The 3′H–E1 base-pairing interaction is essential and sufficient for U3 snoRNA function in rRNA processing in Xenopus, whereas the 5′H–E2 interaction is auxiliary (26). The sequences involved in the 3′H–E1 interaction are not evolutionarily conserved, and base pairing is maintained in diverse organisms through sequence covariation (8). Therefore, this interaction could serve as a target for a new category of drugs to block 18S rRNA formation, thus preventing ribosome biogenesis in eukaryotic pathogens. With this in mind, we have presented data here to characterize the requirements of the 3′H–E1 interaction. There are 8 bp in the 3′H–E1 interaction in Xenopus, and those on the right side of the duplex appear to be more important than those on the left side. A decrease to <6 bp impedes 18S rRNA formation, suggesting that at least 6 bp are required for stability of the duplex. We found that an increase in the length of the 3′H–E1 duplex to 10 bp can be tolerated but further increase to 19 bp hampers 18S rRNA production (Figure 5, lane 11), suggesting that the U3–ETS docking interaction (3′H–E1 base pairing) may be a transient interaction that needs to be displaced at some later point in the 18S rRNA processing pathway. Thus, increasing stability of the duplex over a certain threshold may result in slowing down its dissociation and freeze the U3 docking complex with pre-rRNA, thereby preventing subsequent U3–pre-rRNA interactions necessary for pre-rRNA cleavages and 18S formation.
After U3 snoRNA docking on the ETS, the proteins Imp3p and Imp4p stabilize the U3–hinge interaction (40). These proteins associate with Mpp10p (41). They are not found in the 12S monoparticle of U3 snoRNP, but associate with U3 snoRNP in the 80S SSU processome (32,42).
An evolutionary shift in which U3 hinge–ETS interaction is more important
In yeast, the interaction between ETS region E2 with the U3 5′-hinge is essential for rRNA processing (27). In contrast, in metazoa, such as Xenopus, the essential functional interaction occurs between ETS region E1 and the U3 3′-hinge (26). Therefore, it can be deduced that the site of preferred interaction between U3 snoRNA and the ETS of pre-rRNA may have shifted during evolution. Our data suggest that correct positioning of U3 snoRNA relative to pre-rRNA is absolutely critical for U3 function in rRNA processing and may be the basis for selective evolutionary pressure favoring this shift, as discussed below. Evidence is presented here that rRNA processing cannot occur if the 5′-hinge of U3 snoRNA is forced to pair with E1 instead of E2 of the ETS, or if the 3′-hinge is artificially paired with E2 instead of E1. The interaction of the U3 hinge regions with the ETS seems important to align U3 correctly on the pre-rRNA for function in rRNA processing.
Previously, we found that insertions of 10 or 16 nt between the 3′-hinge and domain II of U3 snoRNA diminished or prevented 18S rRNA production, respectively (33). This finding suggested the importance of either the distance in U3 snoRNA between its 3′-hinge tether on the ETS (i.e. the site of 3′H–E1 base pairing) and U3 domain II and/or the position of U3 domain II relative to the pre-rRNA complex. To distinguish between these two possibilities, we designed a new set of compensatory mutations that increased the distance of U3 domain II from its base-pairing tether on pre-rRNA. The results reported here indicate that when the normally occurring interaction of the U3 3′-hinge with region E1 of the ETS was shifted to the immediately adjacent sequence by the mutation U3s3-3′Hs2*, partial rescue of rRNA processing occurred (Figure 6, lanes 6 and 7). These results suggest that the distance between the tether and domain II is not absolutely essential (since some 18S rRNA processing was restored), but it may play some role (since there was only partial rescue of rRNA processing). Moreover, these results document the importance for rRNA processing of the position of U3 domain II relative to other elements of the pre-rRNA complex.
This conclusion is further supported by the result with the mutation U3:3′Hs3 which hindered 18S rRNA processing (Figure 6, lanes 4 and 5). This mutation should alter the spatial positioning of U3 domain II with respect to sequence (E1) and structural elements of the ETS but keep constant the distance from the site of 3′H–E1 base pairing (tether) to domain II. Even though the latter is preserved, little 18S rRNA was produced, showing that this feature alone is not sufficient for rRNA processing. Thus, we conclude that the number of nucleotides separating U3 domain II from its base-paired tether is not absolutely essential, though it may have some impact on the functional activity of U3 snoRNA in rRNA processing. These results may also explain why the right side (closest to U3 domain II) is more important than the left in the 3′H–E1 interaction. Our observations suggest that as the length and structure of the ETS changed during evolution, selective pressure may have acted to favor novel U3 base pairing with the ETS to maintain an optimal geometry of U3 on pre-rRNA to allow its efficient function in rRNA processing.
In addition to U3 domain II, U3 domain I also needs to be correctly positioned on the pre-rRNA by the 3′H–E1 interaction. Within domain I, certain conserved sequences (GAC element, box A′ and box A) are essential for metazoan rRNA processing (9). Our previous observations (33) suggest that the U3 hinge–ETS interaction may be maintained, while domain I engages in other interactions with pre-rRNA for 18S rRNA production (see Figure 2). It has been proposed that the evolutionarily conserved complementarity between boxes A′ and A of U3 with the 5′-most stem and loop in 18S rRNA may prevent premature pseudoknot formation in 18S rRNA (29). This chaperone-like function of U3 has been supported by results of chemical modification (28) and compensatory base changes (30) in yeast. Our data from Xenopus suggest that the base pairing between the U3 hinges and the ETS may be required to position U3 boxes A′ and A for base pairing with the 18S stem–loop in pre-rRNA (Figure 2).
The essential base pairing between the U3 3′-hinge and ETS E1 would allow boxes A and A′ of U3 domain I to bind to the hairpin near the 5′ end of 18S (Figure 2), as required for 18S rRNA formation (9). According to the secondary structure model of this area of Xenopus pre-rRNA, the 5′-most hairpin of 18S that base pairs with U3 appears to be juxtaposed with the site of U3–pre-rRNA (3′H–E1) interaction (Figure 2). In the case of the secondary structure model proposed for this area of yeast pre-rRNA (31), the 5′-most hairpin of 18S is juxtaposed instead with the 5′H–E2 interaction. This may explain the reason for the observed change in the U3 preferred binding site between lower and higher eukaryotes. Thus, the physical length of the ETS and the geometry of its folded structure may determine which of the possible sites of base pairing between the ETS and U3 snoRNA will be used as a preferred site of functional association.
Moreover, the secondary structure models for the ETS from Xenopus (9) and yeast (31) both show processing sites A0 and 1 (= site A1 in yeast) directly opposite one another at the base of a folded stem (Figure 2). The mechanism of site A1 selection in yeast involves measuring the distance from the hairpin at the 5′ end of 18S to the A1 cleavage site (43,44). We propose that a similar measuring process may occur for the functional action of U3 snoRNA in rRNA processing. Specifically, the 3′H–E1 base pairing may position U3 at the correct distance relative to cleavage sites A0, 1 and 2 in the tertiary folded structure of pre-rRNA.
The present study of U3 hinge base pairing with the ETS may help to explain evolution of this molecular interaction and to elucidate the mechanism for U3 snoRNA-dependent formation of 18S rRNA. Moreover, understanding the details of the U3 hinge–ETS interaction serves as a platform for future drug design targeted to prevent this complex. Our data suggest that disruption of two or more base pairs of the complex, especially on the right side, should hamper ribosome production in pathogenic eukaryotes.
Acknowledgments
The authors thank Thilo Sascha Lange, Stephen Doris and Elliot Lieberman for helpful comments on this paper, and Elaine Butler and Michael Ezrokhi for help with the figures. The authors gratefully acknowledge grant support from NIH GM61945. Funding to pay the Open Access publication charges for this article was provided by NIH GM61945.
Conflict of interest statement. None declared.
REFERENCES
- 1.Venema J., Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Ann. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 2.Gerbi S.A., Borovjagin A.V., Ezrokhi M., Lange T.S. Ribosome biogenesis: role of small nucleolar RNA in maturation of eukaryotic rRNA. Cold Spring Harbor Symp. Quant. Biol. 2001;LXVI:575–590. doi: 10.1101/sqb.2001.66.575. [DOI] [PubMed] [Google Scholar]
- 3.Gerbi S.A., Borovjagin A.V. Pre-ribosomal RNA processing in vertebrates. In: Olson M.O.J., editor. The Nucleolus. New York: Kluwer Academic/Plenum Publishers; 2004. pp. 170–198. [Google Scholar]
- 4.Raué H.A. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevisiae: the machine that makes the machine. In: Olson M.O.J., editor. The Nucleolus. New York: Kluwer Academic/Plenum Publishers; 2004. pp. 199–222. [Google Scholar]
- 5.Warner J.R. The economics of biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 6.Savino R., Gerbi S.A. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990;9:2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes J.M.X., Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borovjagin A.V., Gerbi S.A. U3 small nucleolar RNA is essential for cleavage at sites 1, 2 and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J. Mol. Biol. 1999;286:1347–1363. doi: 10.1006/jmbi.1999.2527. [DOI] [PubMed] [Google Scholar]
- 9.Borovjagin A.V., Gerbi S.A. Xenopus U3 snoRNA GAC-Box A' and Box A sequences play distinct functional roles in rRNA processing. Mol. Cell. Biol. 2001;21:6210–6221. doi: 10.1128/MCB.21.18.6210-6221.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanrobays E., Gleizes P.E., Bousquet-Antonelli C., Noaillac-Depeyre J., Caizergues-Ferrer M., Gelugne J.P. Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J. 2001;20:4204–4213. doi: 10.1093/emboj/20.15.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanrobays E., Gelugne J.P., Gleizes P.E., Caizergues-Ferrer M. Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:2083–2095. doi: 10.1128/MCB.23.6.2083-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tabb-Massey A., Caffrey J.M., Logsden P., Taylor S., Trent J.O., Ellis S.R. Ribosomal proteins RpsO and Rps21 of Saccharomyces cerevisiae have overlapping functions in the maturation of the 3′ end of 18S rRNA. Nucleic Acids Res. 2003;31:6798–6805. doi: 10.1093/nar/gkg899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fatica A., Oeffinger M., Dlakic M., Tollervey D. Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol. Cell. Biol. 2003;23:1798–1807. doi: 10.1128/MCB.23.5.1798-1807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fatica A., Tollervey D., Dlakic M. PIN domain of Nob1 is required for D-site cleavage in 20S pre-rRNA. RNA. 2004;10:1698–1701. doi: 10.1261/rna.7123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao L., Frey M.R., Matera A.G. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted structure. Nucleic Acids Res. 1997;25:4740–4747. doi: 10.1093/nar/25.23.4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narayanan A., Speckmann W., Terns R., Terns M.P. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell. 1999;10:2131–2147. doi: 10.1091/mbc.10.7.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verheggen C., Mouaikel J., Thiry M., Blanchard J.M., Tollervey D., Bordonné R., LaFontaine D.L., Bertrand E. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J. 2001;20:5480–5490. doi: 10.1093/emboj/20.19.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verheggen C., LaFontaine D.L., Samarksy D., Mouaikel J., Blanchard J.M., Bordonné R., Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002;21:2736–2745. doi: 10.1093/emboj/21.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lange T.S., Ezrokhi M., Borovjagin A.V., Rivera-León R., North M.T., Gerbi S.A. Nucleolar localization elements of Xenopus laevis U3 small nucleolar RNA. Mol. Biol. Cell. 1998;9:2973–2985. doi: 10.1091/mbc.9.10.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginisty H., Amalric F., Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ginisty H., Amalric F., Bouvet P. Two different combinations of RNA-binding domains determine the RNA binding specificity of nucleolin. J. Biol. Chem. 2001;276:14338–14343. doi: 10.1074/jbc.M011120200. [DOI] [PubMed] [Google Scholar]
- 22.Dragon F., Gallagher J.E., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y., et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schäfer T., Kuster B., Tschochner H., Tollervey D., et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 24.Schäfer T., Strauss D., Petfalski E., Tollervey D., Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein K.A., Gallagher J.E., Mitchell B.M., Granneman S., Baserga S.J. The small-subunit processome is a ribosome assembly intermediate. Eukaryotic Cell. 2004;3:1619–1626. doi: 10.1128/EC.3.6.1619-1626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borovjagin A.V., Gerbi S.A. Xenopus U3 snoRNA docks on pre-rRNA through a novel base-pairing interaction. RNA. 2004;10:942–953. doi: 10.1261/rna.5256704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beltrame M., Tollervey D. Base-pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Méreau A., Fournier A., Gregoire A., Mougin P., Fabrizio R., Lührmann R., Branlant C. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNA:protein–RNA contacts and base-pair interactions with pre-ribosomal RNA. J. Mol. Biol. 1997;273:552–571. doi: 10.1006/jmbi.1997.1320. [DOI] [PubMed] [Google Scholar]
- 29.Hughes J.M.X. Functional base-pairing interaction between highly conserved elements of U3 small nuclear RNA and the small ribosomal subunit. J. Mol. Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- 30.Sharma K., Tollervey D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol. 1999;19:6012–6019. doi: 10.1128/mcb.19.9.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Intine R.V., Good L., Nazar R.N. Essential structural features in the Schizosaccharomyces pombe pre-rRNA 5′ external transcribed spacer. J. Mol. Biol. 1999;286:695–708. doi: 10.1006/jmbi.1998.2502. [DOI] [PubMed] [Google Scholar]
- 32.Granneman S., Vogelzangs J., Lührmann R., van Venrooij W.J., Pruijn G.J., Watkins N.J. Role of pre-rRNA base pairing and 80S complex formation in subnucleolar localization of the U3 snoRNP. Mol. Cell. Biol. 2004;24:8600–8610. doi: 10.1128/MCB.24.19.8600-8610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borovjagin A.V., Gerbi S.A. The spacing between functional cis-elements of U3 snoRNA is critical for rRNA processing. J. Mol. Biol. 2000;300:57–74. doi: 10.1006/jmbi.2000.3798. [DOI] [PubMed] [Google Scholar]
- 34.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerbi S.A. Expansion segments: Regions of variable size that interrupt the universal core secondary structure of ribosomal RNA. In: Zimmermann R.A., Dahlberg A.E., editors. Ribosomal RNA Structure, Evolution, Processing and Function in Protein Synthesis. Boca Raton, FL: CRC Press; 1996. pp. 71–87. [Google Scholar]
- 36.Peculis B.A. snoRNA nuclear import and potential for cotranscriptional function in pre-rRNA processing. RNA. 2001;7:207–219. doi: 10.1017/s1355838201001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mougey E.B., O'Reilly M., Osheim Y., Miller O.L., Jr, Beyer A., Sollner-Webb B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 1993;7:1609–1619. doi: 10.1101/gad.7.8.1609. [DOI] [PubMed] [Google Scholar]
- 38.Gallagher J.E., Dunbar D.A., Granneman S., Mitchell B.M., Osheim Y., Beyer A.L., Baserga S.J. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–25217. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osheim Y.N., French S.L., Keck K.M., Champion E.A., Spasov K., Dragon F., Baserga S.J., Beyer A.L. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 40.Gérczei T., Correll C.C. Imp3p and Imp4p mediate formation of essential U3-precursor rRNA (pre-rRNA) duplexes, possibly to recruit the small subunit processome to the pre-rRNA. Proc. Natl Acad. Sci. USA. 2004;101:15. doi: 10.1073/pnas.0406819101. 301–315, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S.J., Baserga S.J. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol. 1999;19:5441–5452. doi: 10.1128/mcb.19.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Granneman S., Gallagher J.E., Vogelzangs J., Horstman W., van Venrooij W.J., Baserga S.J., Pruijn G.J. The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60–80S ribonucleoprotein complexes. Nucleic Acids Res. 2003;31:1877–1887. doi: 10.1093/nar/gkg300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venema J., Henry Y., Tollervey D. Two distinct recognition signals define the site of endonucleolytic cleavage at the 5′-end of yeast 18S rRNA. EMBO J. 1995;14:4883–4892. doi: 10.1002/j.1460-2075.1995.tb00169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma K., Venema J., Tollervey D. The 5′ end of the 18S rRNA can be positioned from within the mature rRNA. RNA. 1999;5:678–686. doi: 10.1017/s1355838299990052. [DOI] [PMC free article] [PubMed] [Google Scholar]