Abstract
Two PCR protocols targeting the 18S rRNA gene of Cryptococcus neoformans were established, compared, and evaluated in murine cryptococcal meningitis. One protocol was designed as a nested PCR to be performed in conventional block thermal cyclers. The other protocol was designed as a quantitative single-round PCR adapted to LightCycler technology. One hundred brain homogenates and dilutions originating from 20 ICR mice treated with different azoles were examined. A fungal burden of 3 × 101 to 2.9 × 104 CFU per mg of brain tissue was determined by quantitative culture. Specific PCR products were amplified by the conventional and the LightCycler methods in 86 and 87 samples, respectively, with products identified by DNA sequencing and real-time fluorescence detection. An analytical sensitivity of 1 CFU of C. neoformans per mg of brain tissue and less than 10 CFU per volume used for extraction was observed for both PCR protocols, while homogenates of 70 organs from mice infected with other fungi were PCR negative. Specificity testing was performed with genomic DNA from 31 hymenomycetous fungal species and from the ustilaginomycetous yeast Malassezia furfur, which are phylogenetically related to C. neoformans. Twenty-four strains, including species of human skin flora like M. furfur and Trichosporon spp., were PCR negative. Amplification was observed with Cryptococcus amylolentus, Filobasidiella depauperata, Cryptococcus laurentii, and five species unrelated to clinical specimens. LightCycler PCR products from F. depauperata and Trichosporon faecale could be clearly discriminated by melting curve analysis. The sensitive and specific nested PCR assay as well as the rapid and quantitative LightCycler PCR assay might be useful for the diagnosis and monitoring of human cryptococcal infections.
Cryptococcal disease, once rare, has become a major fungal infection in Europe, Northern America, and Africa in immunocompromised, mainly human immunodeficiency virus (HIV)-infected, patients (13). Diagnosis is established easily for cryptococcal meningoencephalitis by staining cerebrospinal fluid with india ink, by antigen detection, or by culture, but it can be more difficult for other clinical specimens such as bronchoalveolar lavage fluids, lung biopsies, or blood (7). While antigen detection is a sensitive and specific diagnostic tool, false-positive as well as false-negative assays have been described, and it is unreliable for monitoring the efficacy of antifungal treatment (3, 13, 18). Recently, molecular biological tools like PCR assays have been introduced successfully to diagnose cryptococcal disease (5, 9, 11, 12, 17). The LightCycler (Roche Molecular Biochemicals, Mannheim, Germany) technology is capable of detecting and quantifying specific fungal DNA simultaneously (8).
The LightCycler uses hot and cool air for rapid temperature cycling in the range of 20°C/s, and the PCR process is monitored either by fluorescence quantification of the DNA-binding dye SYBR Green I for the general detection of double-stranded DNA or by hybridization probes. In contrast to other real-time PCR platforms, LightCycler hybridization probes consist of a pair of oligonucleotides annealing next to each other on a given nucleic acid sequence and labeled with two different fluorescent dyes on their 3′ and 5′ ends, respectively. In the presence of the specific target sequence, the probes hybridize head-to-tail, bringing the two fluorescent dyes into close physical proximity. After each primer-annealing step, the fluorescein dye at the 3′ end of probe 1 is excited. According to the principle of fluorescence resonance energy transfer, this emitted energy is transferred to the acceptor fluorophore located on the 5′ end of probe 2. The fluorescence intensity, which is directly proportional to the amount of specific target sequence present in the amplification mixture, is quantitatively measured during each PCR cycle. Moreover, this concept is capable of providing sequence confirmation for the amplified product by a simple melting curve analysis. After the PCR process is completed, melting curve analysis starts at 40°C and the temperature in the thermal chamber is slowly increased. When one of the probes melts off and the two fluorescent dyes are no longer in close proximity, fluorescence decreases and a characteristic melting point (Tm) is observed for the given target sequence. Every point mutation present in the hybridization probe annealing region will lower the observed Tm. Since 18S ribosomal DNA (rDNA) sequences of different fungal species share a relatively high degree of homology, melting curve analysis could be a major advantage for the design of species-specific assays.
Compared to block cycler technology, a rapid cycling real-time PCR protocol could simplify the diagnostic laboratory workflow and could reduce the possibility of product contaminations frequently associated with amplicon manipulations. However, every modified or new diagnostic protocol has to be carefully examined before it is introduced into routine testing. Therefore, we evaluated and compared the sensitivities of a nested block cycler PCR protocol and the single-round LightCycler PCR protocol using tissue samples of known Cryptococcus neoformans burden. Specificity testing was performed with cultured strains of phylogenetically related fungal species, some of which might occur in clinical specimens either as pathogens or as contaminants.
MATERIALS AND METHODS
Animal model.
Two strains of C. neoformans (strains 8107 and 8108; M. G. Rinaldi, Fungus Testing Laboratory, University of Texas Health Science Center at San Antonio) isolated from an HIV-infected patient before and during fluconazole treatment were tested with ICR mice. The mice were infected intracranially according to a protocol described by Nguyen et al. (10). After treatment with one of two doses of either fluconazole or posaconazole or without treatment, mice were sacrificed at day 11 postinfection. The complete brains were removed aseptically, weighed, and homogenized in 2 ml of sterile 0.9% saline supplemented with piperacillin and amikacin. Then, the homogenates were serially diluted 10-fold up to 10−4, and 2 × 100 μl of each dilution was plated on Sabouraud dextrose agar followed by incubation at 37°C for 72 h. The CFU on each plate were counted, and the number of CFU/milligram of brain tissue was calculated. The remaining homogenates and dilutions from the brains of 20 mice, in total 100 specimens representing a broad range of CFU/milliliter, as well as brain homogenates of 2 uninfected mice, were stored frozen until DNA extraction was done.
Template DNA preparation.
After thawing, 200 μl of each sample was processed with the QIAamp tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions, with minor modifications. Briefly, incubation with 180 μl of ATL-buffer and proteinase K at a final concentration of 1 mg/ml was performed in screw-cap reaction tubes for 3 h or overnight at 55°C. Then, three cycles of freezing in liquid nitrogen for 1 min and boiling for 2 min were applied to inactivate the proteolytic enzyme and to disrupt the rigid cell walls of the fungal organisms. After cooling to room temperature, AL-buffer and ethanol were added and the mixture was applied to the spin column. Following the centrifugation and washing steps, total DNA was eluted with 100 μl of AE-buffer, and 5- or 10-μl aliquots (depending on the amplification platform used) were directly transferred to a PCR mixture. The remainder was stored at −20°C for further experiments.
Primer and LightCycler hybridization probe design.
Oligonucleotide primers Fungus I and Fungus II are complementary to highly conserved regions within the nuclear gene coding for a small subunit of rRNA (18S rDNA) of several pathogenic fungi, including C. neoformans, and generate a 429-bp amplicon (2).
Oligonucleotides Cryp I and Cryp II are complementary to C. neoformans-selective regions within the 18S rDNA target, spanning a 278-bp region, and can serve as nested primers for the Fungus I/II amplicon. The relatively high melting temperatures of Cryp I and Cryp II allowed for a two-temperature amplification profile, associated with an increased specificity and shorter effective cycle times. For the real-time detection of C. neoformans-specific amplicons, a pair of LightCycler (Roche Diagnostics, Mannheim, Germany) hybridization probes was designed. The fluorescence-labeled oligonucleotides Cryp-HP-1 and Cryp-HP-2 were designed to hybridize adjacent to each other in a C. neoformans-selective region within the 278-bp amplicon, generated by primers Cryp I and Cryp II. The nucleotide sequences of PCR primers and LightCycler hybridization probes are listed in Table 1.
TABLE 1.
Oligonucleotide primers and LightCycler hybridization probes used in the PCR assays
| Oligonucleotide | Sequence (5′-3′)a | Target gene | Nucleotide position | GenBank accession no. | Reference |
|---|---|---|---|---|---|
| Fungus I | GTT AAA AAG CTC GTA GTT G | 18S rDNA | 617-635 | L05427 | 2 |
| Fungus II | TCC CTA GTC GGC ATA GTT TA | 18S rDNA | 1045-1026 | L05427 | 2 |
| Cryp I | TCC TCA CGG AGT GCA CTG TCT TG | 18S rDNA | 661-683 | L05427 | This study |
| Cryp II | CAG TTG TTG GTC TTC CGT CAA TCT A | 18S rDNA | 938-914 | L05427 | This study |
| Cryp-HP-1 | TCC TGG TTC CCC TGC ACA C-[FL] | 18S rDNA | 743-725 | L05427 | This study |
| Cryp-HP-2 | [Red640]-CAG TAA AGA GCA TAC AGG ACC ACC-Ph | 18S rDNA | 723-700 | L05427 | This study |
[FL], fluorescein; [Red 640], LightCycler-Red 640-N-hydroxy-succinimide ester; Ph, 3′-phosphate.
Block cycler PCR.
Amplification mixtures of the first-round PCR contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, and 2.5 mM MgCl2 (10× Perkin-Elmer buffer II plus MgCl2 solution [Roche Molecular Systems, Branchburg, N.J.]), 1.5 U of AmpliTaq DNA polymerase (Perkin-Elmer), 100 μM concentrations of each deoxynucleoside triphosphate (Promega, Madison, Wis.), 1 μM concentrations of each I and II primer oligonucleotides (Roth, Karlsruhe, Germany), and 10 μl of template DNA in a final volume of 50 μl.
Identical reaction mixtures were prepared for the nested PCR, except that concentrations of 1 μM for each primer oligonucleotide Cryp I and II (Roth) were used, the final concentration of deoxynucleoside triphosphates was 50 μM, and 1 μl of the first-round amplification reaction mixture was used as a template.
Following an initial denaturation at 94°C for 5 min, the 35-cycle amplification profile of the first-round PCR consisted of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min. A final elongation step was applied at 72°C for 5 min. Following initial denaturation at 94°C for 5 min, the 30-cycle two-step amplification profile of the nested PCR consisted of 94°C for 30 s and 72°C for 1 min. A final elongation step was applied at 72°C for 5 min. Amplification was performed in a Primus Tc 9600 Thermocycler (MWG Biotech, Ebersberg, Germany). The PCR products were electrophoretically analyzed on 1.5% agarose gels, stained with ethidium bromide, and visualized on a UV transilluminator.
LightCycler PCR.
Amplification mixtures contained 2 μl of 10× LightCycler FastStart DNA Master Hybridization Probes mix (Roche Diagnostics), 5 mM MgCl2 (total concentration), 0.5 μM concentrations of each Cryp I and II primer oligonucleotide (Roth), 0.2 μM concentrations of each Cryp-HP-1/-II hybridization probe oligonucleotide (TIB Molbiol, Berlin, Germany), and 5 μl of template DNA in a final volume of 20 μl. Following an initial denaturation at 95°C for 10 min to activate the FastStart Taq DNA polymerase, the 50-cycle amplification profile consisted of heating at 20°C/s to 95°C with a 10-s hold, cooling at 20°C/s to 50°C with a 10-s hold, and heating at 20°C/s to 72°C with a 20-s hold. The temperature transition rate was 20°C/s. Fluorescence values of individual LightCycler reaction capillaries were measured and recorded at 640 nm at the end of each annealing step and plotted against PCR cycle numbers. To minimize variations in positioning or geometry of individual capillaries, the absolute signal of the reporter dye (LightCycler Red 640; channel F2) divided by the signal of the donor dye (fluorescein; channel F1) is shown at the y axis of the LightCycler screen plots.
Following the amplification phase, a melting curve analysis was performed with a temperature transition rate of 0.2°C/s to determine the melting point (Tm) values for the sequences targeted by the hybridization probes. For melting curve analysis, the first negative derivative of the fluorescence (−dF/dT) was plotted versus temperature and the Tm values were manually assigned from the turning point of this curve.
Amplification controls.
Starting with a C. neoformans cell suspension adjusted to 1,000 CFU/ml, genomic DNA was extracted and 5- or 10-μl aliquots were used as positive controls in every series of PCR experiments. A water control (DNA extraction procedure performed on sterile water instead of on a clinical specimen) was applied after every 10th sample of the nested PCR to monitor for crossover contamination. A no-template control (complete amplification mixture without addition of template DNA) was included in every first-round and nested PCR experiment series to monitor for contaminants. Moreover, organ homogenates of 70 ICR or BALB/c mice infected either with Histoplasma capsulatum or Paracoccidioides brasiliensis from earlier studies (1, 2) were examined by the Cryptococcus PCR to detect nonspecific amplification or other cross-reactions.
Analytical sensitivity.
DNA extracts of C. neoformans suspensions with fungal cell concentrations defined by quantitative culture were analyzed by nested PCR and LightCycler PCR. The minimal CFU count per sample resulting in a positive nested PCR or LightCycler PCR result was determined.
Analytical specificity.
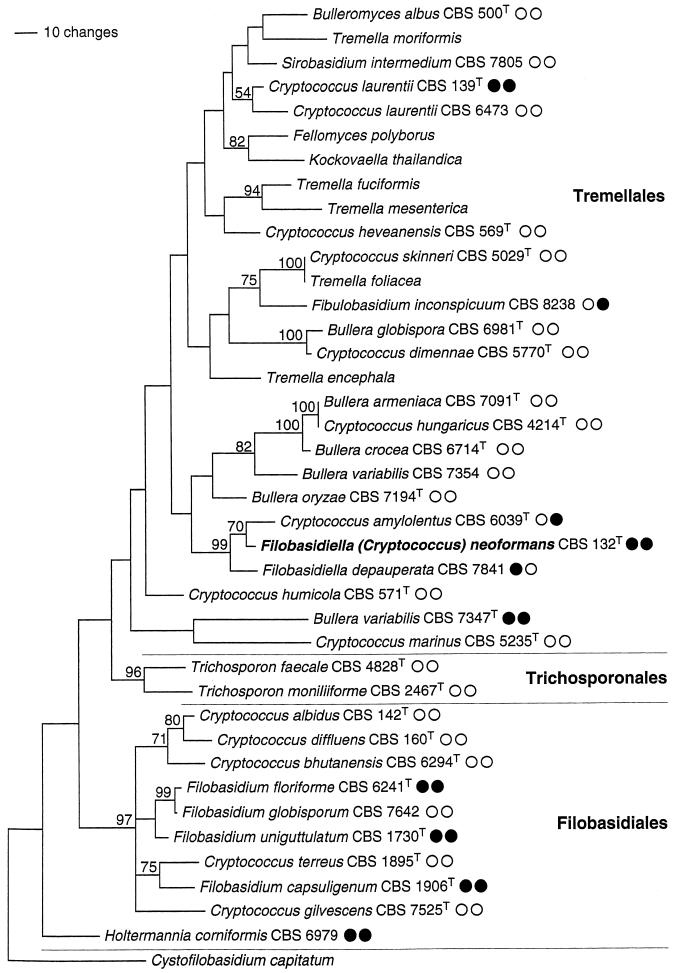
Total genomic DNA was prepared from axenic cultures of basidiomycetous yeast-like species belonging to Hymenomycetes (31 strains) and Ustilaginomycetes (Malassezia furfur) (6) using a sodium dodecyl sulfate extraction protocol, as described by Weiβ et al. (19). Species names and strain numbers of the hymenomycetous species are given in Fig. 1. All PCR assays were carried out blind: the examiner did not know the CFU concentration of the sample or the species examined for specificity testing. The numbered sample tubes were correlated after results were obtained.
FIG. 1.
Estimate of phylogenetic relationships in the Tremellomycetidae. Maximum parsimony analysis of an alignment of nuclear DNA sequences coding for the D1/D2 region of the ribosomal large subunit. Strict consensus of the three most parsimonious trees found in 1,000 rounds of heuristic search. Numbers on branches are bootstrap values from 1,000 replicates. The topology was rooted with Cystofilobasidium capitatum. Strain numbers are given for the species used in the PCR experiments of this study. CBS, Centraalbureau voor Schimmelcultures; T superscript, type strain; open and solid circles, negative and positive reaction results, respectively, of nested-PCR and LightCycler hybridization experiments.
DNA sequencing.
Nested PCR amplification products were purified with the QIAquick PCR purification kit (Qiagen), based on size-selective DNA binding to a silica membrane. Sequencing reactions were performed with the BigDye terminator cycle sequencing kit (Perkin-Elmer) using the set of amplification primers and analyzed on an ABI 373 automated DNA sequencer (Applied Biosystems Division, Perkin-Elmer Biosystems, Foster City, Calif.). Sequences of both strands were determined, edited, and aligned with the Sequence Navigator software (Applied Biosystems) and used in a BLAST search of the GenBank database (National Center for Biotechnology Information, Washington, D.C.).
Quantification of LightCycler PCR products.
The quantitative interpretation of LightCycler results was assisted by the fit point method algorithm with the minimize error option (LightCycler software version 3.5; Roche Diagnostics). The LightCycler software performs all additional calculation steps necessary for generation of a standard curve (Absolute quantification with external standards, Technical Note LC 11/2000, p. 1-18, Roche Molecular Biochemicals). Briefly, a noise band (threshold) is automatically set at a fluorescence level at which the fluorescence signal development reflects that the PCR is in the log-linear phase. The software then calculates the logarithmic values by interpolating a straight line through three data points above the threshold value, and the points of intersection (crossing points) with the noise band are determined. Finally, the crossing points are plotted against the logarithm of the concentration. By using standard concentrations, a curve is generated to determine unknown concentrations of samples by comparison of their crossing points with the standard curve.
Phylogenetic analysis.
To estimate the phylogenetic relationships of the hymenomycetous species used in the tests for analytical specificity, a DNA alignment was constructed with sequences of the nuclear coded large ribosomal subunit (D1/D2 region) including the strains used in this study and also some sequences of additional species. PAUP* version 4.0b8 (15) was used to perform a bootstrapped maximum parsimony analysis (16) on this data set (1,000 rounds of heuristic search; TBR branch swapping on starting trees obtained by random addition; MULTREES and steepest descent options in effect, gaps treated as missing data; bootstrap analysis with 1,000 replicates, each employing 10 rounds of heuristic search with parameters as indicated above).
RESULTS
Tissue burden.
The quantification by culture determined a concentration range of 3 × 101 to 2.9 × 104 CFU of C. neoformans per mg of brain tissue, depending on the fungal isolate and treatment of the 20 mice investigated in this study. The dilutions used for template DNA preparation had calculated concentrations of 1.1 × 10−1 to 1.1 × 106 CFU per 200-μl sample volume.
PCR results.
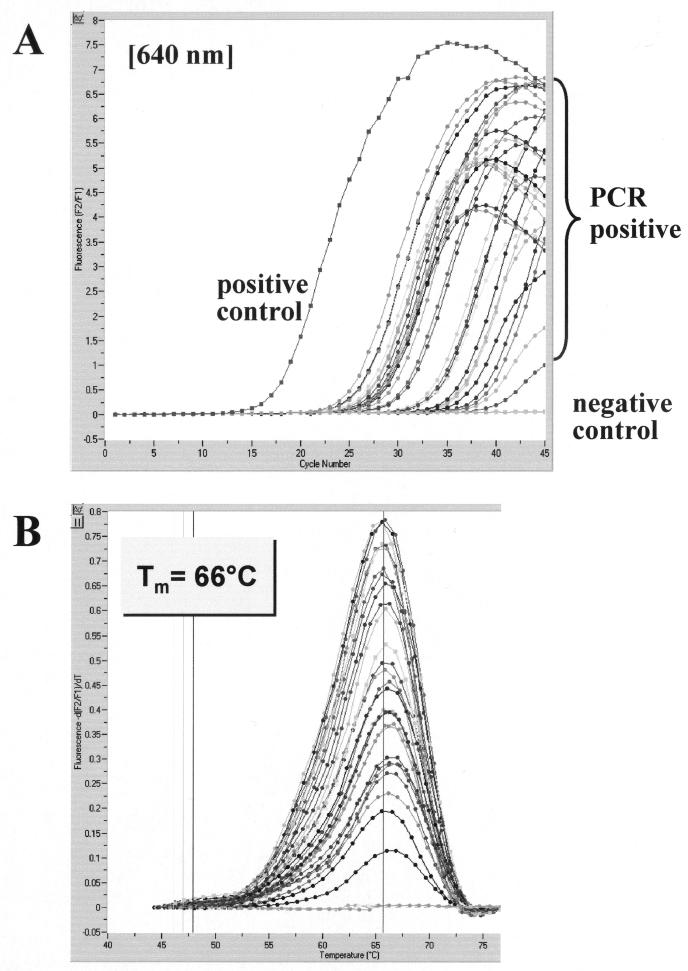
Amplification products were obtained with all 20 brain homogenates by both PCR protocols but not with 2 brain homogenates from uninfected ICR mice kept under identical conditions. Both PCR assays gave rise to positive results in 63 samples containing more than 102 CFU of C. neoformans per processed sample volume of 200 μl, i.e., before DNA was extracted. Of 37 samples with C. neoformans concentrations of less than 102 CFU/200 μl, amplification products were detected in 23 samples by nested PCR and in 24 samples by LightCycler PCR. Thus, of the 100 samples (total), 14 and 13 samples from 10 infected mice were found negative by nested PCR and by LightCycler PCR, respectively. Whereas 10 samples were concordantly negative in the two PCR assays, 3 were negative by the LightCycler PCR assay alone and 4 were negative by the nested PCR assay alone. Since all water and no-template controls gave negative results, crossover or external-contamination events were ruled out. Visual examination of ethidium bromide-stained agarose gels or amplification screen plots generated by the LightCycler software (cycle number versus fluorescence) allowed for a clear discrimination between positive and negative samples. The LightCycler results of a representative panel of brain homogenates are depicted in Fig. 2A. Post-PCR melting curve analysis demonstrated a consistent hybridization probe melting point of 66°C for all the C. neoformans-positive samples investigated (Fig. 2B). Specificity of nested PCR was further verified by sequencing of characteristic PCR products. The amplicon sequences of the 86 positive samples were found to be identical to the C. neoformans 18S rDNA sequence in GenBank (accession no. L05427) (data not shown).
FIG. 2.
Evaluation of the LightCycler PCR assay with brain tissue sections of C. neoformans-infected mice. A representative set of 28 samples with various C. neoformans concentrations was tested for the presence of C. neoformans 18S rDNA. Amplicon curves representing C. neoformans PCR-positive isolates are indicated by braces (A), and the corresponding results in melting curve analysis are depicted (B).
Analytical sensitivity.
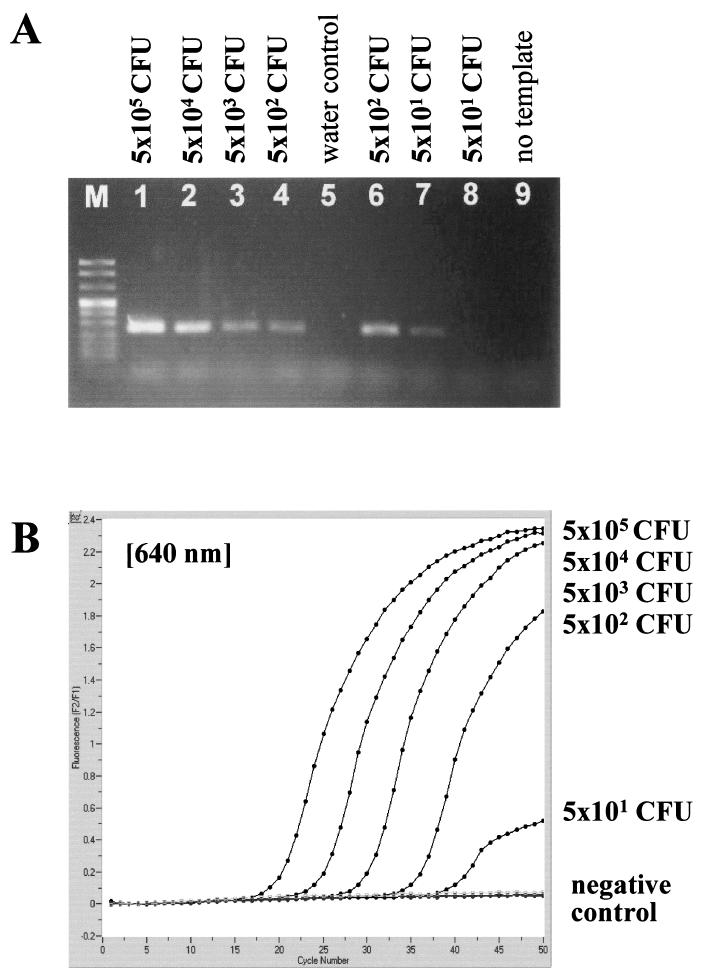
When nested PCR or LightCycler PCR was applied to dilution series of cultured C. neoformans organisms, a detection limit of 1 to 10 CFU per processed sample volume, i.e., in 200 μl used for DNA extraction, was observed for both amplification protocols (Fig. 3).
FIG. 3.
Analytical sensitivity of nested PCR (A) and LightCycler PCR (B) for the C. neoformans small-subunit rRNA gene (18S rDNA), determined with serial dilutions of a cultured C. neoformans strain. Detection limits in CFU per milliliter, determined by standard plating procedure, are given at the corresponding lanes in agarose gel electrophoresis or next to the LightCycler amplicon curves.
With mouse brain homogenates and dilutions, 11 of 14 samples negative by the nested PCR assay and 10 of 13 samples negative by the LightCycler PCR had a calculated concentration of less than 4 CFU of C. neoformans in 200-μl samples used for DNA extraction. The remaining negative samples had concentrations of 10 to 30 CFU per processed sample volume. Thus, the detection limit determined by defined culture suspensions is in good correlation with the PCR results from tissue samples. Neither the fungal isolate used for infection nor the treatment regimen had significant influence on the PCR results (data not shown).
Specificities of the nested PCR and LightCycler PCR assays.
For specificity testing, strains of 32 fungal species, most of them belonging to the Tremellomycetidae (Hymenomycetes), and 70 homogenates of H. capsulatum- or P. brasiliensis-infected organs were examined. Of the Tremellomycetidae strains, 12 revealed amplification products by the nested PCR. DNA sequencing identified the nested PCR products from one of two strains of Cryptococcus laurentii, from Filobasidium capsuligenum, Filobasidium floriforme, Filobasidium uniguttulatum, Holtermannia corniformis, and from one of two strains of Bullera variabilis, in total six products, as being identical with the 278-bp segment of C. neoformans 18S rDNA (Fig. 1). Additionally, the sequence gained from Filobasidiella depauperata showed two different nucleotide positions but was identified as being 99% homologous to C. neoformans. It could not be identified to the species level, because the 18S rDNA sequence of F. depauperata has not yet been deposited in GenBank. Amplicons from the remaining five strains, like B. variabilis, Bullera globispora, or Cryptococcus amylolentus, were correctly differentiated down to species level by their nucleotide sequences.
Template DNA from these seven cross-reacting strains, from Trichosporon faecale and Fibulobasidium inconspicuum as well as from C. amylolentus, were positive by LightCycler PCR. However, T. faecale- and F. depauperata-derived amplicons could be clearly differentiated by melting point analysis (Tm of 66°C for C. neoformans; Tm of 56°C for T. faecale; Tm of 61°C for F. depauperata) (data not shown). As expected, six strains with 100% identity of the 278-bp fragment to the sequence of C. neoformans cannot be distinguished by melting point analysis. According to the search in GenBank, the 278-bp sequence of F. depauperata is only 99% identical to C. neoformans. Since two different nucleotide positions are within the binding regions of the hybridization probes, it was recognized as being different from C. neoformans by melting point analysis.
Compared to C. neoformans sequences, the 278-bp sequences of T. faecale and F. inconspicuum are different in nine nucleotide positions within the binding sites of the primers. Therefore, the primers will not bind at the 72°C temperature used for the combined annealing and extension step in the second reaction of the nested PCR assay. Accordingly, the nested PCRs were negative. In contrast, the annealing temperature of 50°C used in the LightCycler PCR assay favors primer binding and amplification. Four nucleotide mismatch positions within the hybridization probe annealing sites revealed the product of T. faecale to be different from that of C. neoformans by subsequent melting point analysis. In contrast, the product of F. inconspicuum was not distinguished.
LightCycler PCR including discriminative melting point analysis cannot distinguish C. amylolentus and C. neoformans, because the characteristic nucleotide variations within the 278-bp PCR product are outside the annealing sites of primers and hybridization probes.
All 70 homogenates of organs infected with H. capsulatum or P. brasiliensis tested negative by both PCR protocols.
Quantification of PCR products.
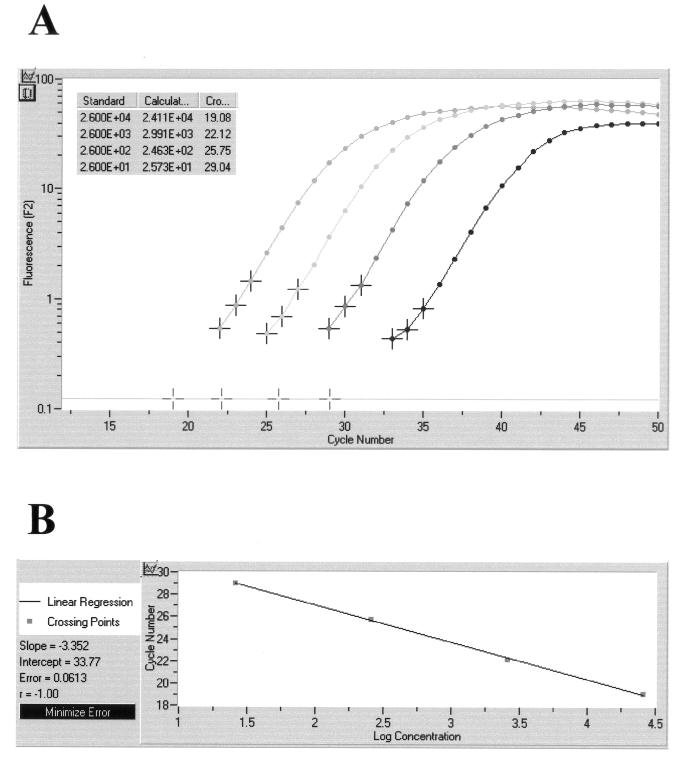
The quantitative capability of the LightCycler concept was obvious in the course of investigating defined brain samples of mice which had been infected with various doses of C. neoformans organisms. Based on the results with a series of external standards (5 × 101 to 5 × 104 CFU of C. neoformans), a standard curve was generated with the help of the LightCycler software. The calculated quantity of template DNA extracted from the tissue samples correlated well with the results of quantitative culture. The original amplification curves obtained on a representative set of four tissue sections containing 2.6 × 101 to 2.6 × 104 CFU of C. neoformans per processed sample volume are depicted in Fig. 4. This figure also shows the calculated standard curve and the individual CFU concentrations determined on the basis of crossing points.
FIG. 4.
Quantitative analysis of LightCycler results obtained on a representative set of four tissue sections containing 2.6 × 101 to 2.6 × 104 CFU of C. neoformans per processed sample volume. Based on the slope of the original amplification curves during the log-linear phase determined by three data points, artificial crossing points with the noise band (horizontal line) were determined by the LightCycler software (A). The standard curve represents the linear regression line through the data points on a plot of crossing points (threshold cycle) versus logarithm of standard sample concentration (B). Slope, y-intercept, mean squared error, and regression coefficient of the standard curve are given.
Single-round block cycler PCR.
A protocol for the block cycler PCR using the primer oligonucleotides Cryp I and Cryp II, and consisting of 45 cycles with an annealing temperature of 50°C, was unsuccessful. A significant reduction with respect to sensitivity and specificity of the PCR assay was observed. All brain samples with fewer than 102 CFU per processed sample volume became negative, and products were amplified from 24 of 32 related fungal species. As could be expected theoretically, the second round of amplification increases sensitivity, and the highly stringent annealing step at 72°C reduces nonspecific amplification significantly.
When LightCycler PCR mixtures were analyzed by agarose gel electrophoresis, amplification products of the expected sizes were detected for 30 of the 32 related fungal species investigated. This demonstrates the value of the LightCycler hybridization probe concept for total assay specificity.
Phylogenetic analysis.
Heuristic maximum parsimony analysis of the ribosomal D1/D2 DNA data set yielded three equally most parsimonious trees; a strict consensus of these is shown in Fig. 1. The phylogenetic hypothesis obtained is consistent with analyses on data sets including more species (6).
DISCUSSION
The excellent sensitivity of a novel nested PCR protocol and a LightCycler PCR protocol for the detection of C. neoformans DNA was demonstrated in a murine model of cryptococcal meningitis. When the two are compared, the LightCycler PCR assay appears more sensitive, because only 5 μl instead of 10 μl of DNA extract is used, and 45 instead of a total of 65 cycles are necessary to detect a tissue burden of less than 1 CFU of C. neoformans per mg of brain. In addition, real-time PCR using the hybridization probe detection format and the use of external standards offers quantitative results without additional work or costs. This may prove beneficial for monitoring antifungal treatment in humans in a clinical setting.
PCR assays for the detection of C. neoformans DNA in clinical specimens have been described, targeting 18S, 28S, or the ITS and 5.8S ribosomal DNA (rDNA) (5, 9, 11, 12). The detection limits of our novel PCR assays compare favorably with the reported detection limits of 1 to 10 cells ml−1 or per volume used for DNA extraction (5, 11, 12). A number of DNA extraction procedures have been published for efficient disruption of cryptococcal cells, including enzyme digestion or glass beads (17). After identifying fungal DNA contaminants in enzyme preparations, we included instead three cycles of freezing and boiling in our DNA extraction protocol for dimorphic fungi (1, 2). As demonstrated in this study, the simple freeze-and-boil procedure is sufficient to liberate cryptococcal DNA from tissue homogenates as well.
The PCR assays showed cross-reactions with some fungal species related to C. neoformans (Fig 1). Some of these can be pathogenic, and their detection is beneficial. The presence of the remaining cross-reacting species is most unlikely in clinical samples but might occur as contaminants in mycological laboratories. In general, this demonstrates the specificity limits of PCR assays targeting conserved regions of the rDNA. Targeting multicopy rDNA genes will undoubtedly raise the analytical sensitivity for a given fungal pathogen, but specificity can decrease due to possible cross-reactions with phylogenetically related fungi.
Prariyachatigul et al. showed that their primers and hybridization probe used for an 18S rDNA PCR assay for cryptococcal disease did not cross-react with phylogenetically distant pathogens like Candida spp. (11). According to GenBank, their probe will exclusively hybridize with C. neoformans 18S rDNA. However, the 18S rDNA gene sequences from most of the related fungal species used in this study have not been submitted to GenBank. Therefore, it is not surprising that although we used GenBank alignments for the selection of two unique LightCycler hybridization probes and a unique pair of nested primers for block cycler PCR within Cryptococcus-specific 18S rDNA regions, cross-reactions were observed. Whereas two of the cross-reacting species can be differentiated by LightCycler melting point analysis, others disclose a 100% sequence identity with C. neoformans in the partial sequence targeted by the PCR protocols. To minimize the event of false-positive results in the nested PCR protocol, a high annealing temperature was selected to discriminate between similar but not identical primer binding regions, and amplification products were subjected to DNA sequencing.
According to our findings, hybridization with probes complementary to segments of the rDNA may be unreliable for absolute species confirmation, and amplicons should be sequenced if an unambiguous species identification is desired.
Including a variety of other pathogens, specificity testing should include phylogenetically closely related fungal species and potential contaminants of clinical specimens, especially fungi residing on normal skin or mucous membranes. From a clinical point of view, false-positive PCR results with other fungal pathogens are acceptable, because antifungal treatment is indicated in that situation as well. But any cross-reaction of the PCR assay with harmless contaminants will falsely indicate the necessity of therapy, which might be harmful to the patient.
C. laurentii, for example, was shown to cross-react with the presented PCR assays. Since this is a potential pathogen (4), its detection is beneficial. Due to strain variation, which is known to occur (14), not all strains will be detected. Potential skin contaminants such as Trichosporon spp. or Malassezia furfur either will not be detected or will be distinguished by LightCycler melting point analysis. Bullera spp., F. depauperata, F. inconspicuum, some Filobasidium species, C. amylolentus, and H. corniformis will be incorrectly identified as C. neoformans. However, these species have not been described to cause human disease, nor is a contamination of clinical specimens likely to occur.
Since procedures for sequence-specific detection of amplicons evolved as the “gold standard” in the field of diagnostic PCR, LightCycler technology avoids the application of time-consuming and laborious postamplification procedures like Southern blotting or DNA sequencing. Enhancing the reliability of the results by sequence-specific probes and simplifying the PCR workflow by a completely automated amplification and online detection procedure, the LightCycler system proved to be a valuable tool for rapid and sensitive identification of C. neoformans. Once DNA is extracted from suitable specimens and reaction mixtures are completed, the results of sensitive and quantitative PCR are available within 60 min. A current limitation of the LightCycler system is the total reaction volume of 20 μl, which limits the input of template DNA and, as a consequence, total assay sensitivity.
In conclusion, we have successfully evaluated sensitive conventional nested PCR and LightCycler PCR protocols for the detection of C. neoformans in tissue samples. The LightCycler real-time PCR is more rapid and offers quantitative results, whereas the block cycler PCR is broadly applicable. Although we have identified some cross-reacting fungal species, the true diagnostic value of these assays will be further evaluated in prospective studies on human cryptococcal disease.
Acknowledgments
We thank Hans Wolf, Ulrike Zelck, Jürgen Knobloch, José Paulo Sampaio, and Dominik Begerow for their active support and Jeffrey Emch for critical comments. We gratefully acknowledge the technical assistance of Markus Bollwein and Birgit Leppmeier during the study.
REFERENCES
- 1.Bialek, R., A. Ibricevic, C. Aepinus, L. K. Najvar, A. W. Fothergill, J. Knobloch, and J. R. Graybill. 2000. Detection of Paracoccidioides brasiliensis in tissue samples by a nested PCR assay. J. Clin. Microbiol. 38:2940-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bialek, R., J. Fischer, A. Feucht, L. K. Najvar, K. Dietz, J. Knobloch, and J. R. Graybill. 2001. Diagnosis and monitoring of murine histoplasmosis by a nested PCR assay. J. Clin. Microbiol. 39:1506-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blevins, L. B., J. Fenn, H. Segal, P. Newcomb-Gayman, and K. C. Carroll. 1995. False-positive cryptococcal antigen latex agglutination caused by disinfectants and soaps. J. Clin. Microbiol. 33:1674-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, M.-F., C. C. Chiou, Y.-C. Liu, H.-Z. Wang, and K.-S. Hsieh. 2001. Cryptococcus laurentii fungemia in a premature neonate. J. Clin. Microbiol. 39:1608-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evertsson, U., H. J. Monstein, and A. G. Johansson. 2000. Detection and identification of fungi in blood using broad-range 28S rDNA PCR amplification and species-specific hybridisation. APMIS 108:385-392. [DOI] [PubMed] [Google Scholar]
- 6.Fell, J. W., T. Boekhout, A. Fonseca, G. Scorzetti, and A. Statzell-Tallman. 2000. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 50:1351-1371. [DOI] [PubMed] [Google Scholar]
- 7.Kralovic, S. M., and J. C. Rhodes. 1998. Utility of routine testing of bronchoalveolar lavage fluid for cryptococcal antigen. J. Clin. Microbiol. 36:3088-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeffler, J., N. Henke, H. Hebart, D. Schmidt, L. Hagmeyer, U. Schumacher, and H. Einsele. 2000. Quantification of fungal DNA by using fluorescence resonance energy transfer and the Light Cycler system. J. Clin. Microbiol. 38:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell, T. G., E. Z. Freedman, T. J. White, and J. W. Taylor. 1994. Unique oligonucleotide primers in PCR for identification of Cryptococcus neoformans. J. Clin. Microbiol. 32:253-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen, M. H., L. K. Najvar, C. Y. Yu, and J. R. Graybill. 1997. Combination therapy with fluconazole and flucytosine in the murine model of cryptococcal meningitis. Antimicrob. Agents Chemother. 41:1120-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prariyachatigul, C., A. Chaiprasert, V. Meevootisom, and S. Pattanakitsakul. 1996. Assessment of a PCR technique for the detection and identification of Cryptococcus neoformans. J. Med. Vet. Mycol. 34:251-258. [DOI] [PubMed] [Google Scholar]
- 12.Rappelli, P., R. Are, G. Casu, P. L. Fiori, P. Cappuccinelli, and A. Aceti. 1998. Development of a nested PCR for detection of Cryptococcus neoformans in cerebrospinal fluid. J. Clin. Microbiol. 36:3438-3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saag, M. S., J. R. Graybill, R. A. Larsen, P. G. Pappas, J. R. Perfect, W. G. Powderly, J. D. Sobel, and W. E. Dismukes, for the Mycoses Study Group cryptococcal subproject. 2000. Practical guidelines for the management of cryptococcal disease. Clin. Infect. Dis. 30:710-718. [DOI] [PubMed] [Google Scholar]
- 14.Sugita, T., M. Takashima, R. Ikeda, T. Nakase, and T. Shinoda. 2000. Intraspecies diversity of Cryptococcus laurentii as revealed by sequences of internal transcribed spacer regions and 28S rRNA gene and taxonomic position of C. laurentii clinical isolates. J. Clin. Microbiol. 38:1468-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, Mass.
- 16.Swofford, D. L., G. J. Olsen, P. J. Waddell, and D. M. Hillis. 1996. Phylogenetic inference, p. 407-415. In D. M. Hillis, C. Moritz, and B. K. Mable (ed.), Molecular systematics. Sinauer Associates, Inc., Sunderland, Mass.
- 17.Tanaka, K.-I., T. Miyazaki, S. Maesaki, K. Mitsutake, H. Kakeya, Y. Yamamoto, K. Yanagihara, M. A. Hossain, T. Tashiro, and S. Kohno. 1996. Detection of Cryptococcus neoformans gene in patients with pulmonary cryptococcosis. J. Clin. Microbiol. 34:2826-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tintelnot, K., S. Adler, F. Bergmann, K. Schoenherr, and M. Seibold. 2000. Case reports. Disseminated cryptococcoses without cryptococcal antigen detection. Mycoses 43:203-207. [DOI] [PubMed] [Google Scholar]
- 19.Weiβ, M., Z.-L. Yang, and F. Oberwinkler. 1998. Molecular phylogenetic studies in the genus Amanita. Can. J. Bot. 76:1170-1179. [Google Scholar]