Abstract
Objective:
To Validate a one-step RT-qPCR as a reliable diagnostic tool in HER-2 positive breast cancer. Further, establishing this tool as a standard procedure to quantify HER-2 expression in Breast cancer patient.
Methods:
Here we report a prospective validation study that shows the concordance of one-step RT-qPCR in assessing the HER2 levels in formalin-fixed paraffin-embedded (FFPE) tissue samples with the current paradigm of diagnosis such as Immunohistochemistry (IHC) and Fluorescence-in-situ Hybridization (FISH). We collected 275 FFPE samples from the Department of Pathology, Ibn Rochd University Hospital Center. IHC was carried out in all the samples and those with a score of 2+ were also analyzed using FISH. We extracted mRNA from FFPE samples and performed RT-qPCR, and the results were compared with those obtained using IHC/FISH. HER2 mRNA levels were quantified and normalized using the reference genes RPL30 and RPL37, based on our earlier reports.
Results:
HER2 cut-off value was fixed at 11.954 corresponding to the combination of the best sensitivity and specificity (93.4% and 100%) respectively, and with positive predictive values PPV that reached 100% and a negative predictive value NPV that reached 89.4%. The results showed 100% concordance with FISH. The Kappa coefficient was 0.863 which indicated concordance with IHC. The area under the curve (AUC) which is an important parameter that determines the diagnostic accuracy of the test was calculated as AUC=0.955. The results were comparable to some of the recent studies published in a similar direction.
Conclusion:
Our study shows that one-step RT-qPCR-based quantitation is an accurate and reproducible test to record HER2 gene expression for a better treatment orientation focusing on maximal therapeutic and survival outcome.
Key Words: Breast cancer, HER2, one-step RT, qPCR, immunohistochemistry, fluorescence in situ hybridization
Introduction
Despite the theragnostic advances made so far, breast cancer remains the second most common cause of mortality among women after lung cancer and the most common cancer affecting women worldwide [1]. It is a highly heterogeneous cancer in terms of histology, presentation, molecular profile, disease progression, etc., which makes the treatment options and clinical outcomes different for each subtype [2]. The National Cancer Institute (NCI) estimates that more than 297,000 women in the United States will be diagnosed positive for breast cancer by the end of the year 2023, and more than 43,000 deaths will be registered [3]. Most importantly, although recent reports indicate that the rate of breast cancer in developing countries is less compared to that of developed ones, there has been an alarming increase in the mortality rate in low- and middle-income countries (LMIC), which may be largely attributed to the lack of accurate screening and diagnostic procedures [4].
Invasive breast cancer is categorized into five different molecular subtypes based on the expression of certain genes in the cancer cells, which determines their clinical behavior [5]: (i) Luminal A breast cancer cells express estrogen (ER) and progesterone receptors (PR). Since they have low levels of Ki-67, and do not express Human epidermal growth factor 2 receptor (HER2), the cells tend to grow at a slower rate compared to other subtypes and have a good prognosis, (ii) Luminal B breast cancer is positive for ER and negative for HER2, PR and the cells have high expression of Ki-67, (iii) Luminal B-like breast cancer cells express both ER and HER2 and may be either PR positive or negative. The cells also have different Ki-67 cells (either high or low), and hence this type of cancer has a slightly worse prognosis compared to luminal A cancers, (iv) HER2-enriched subtype is ER and PR negative. Although the prognosis is worse compared to the luminal A subtype, timely treatment with HER2-targeted therapies has shown successful treatment outcomes, (v) Triple-negative or basal-like subtype breast cancer cells do not express ER, PR, or HER2. The incidence of this subtype is mostly reported among young women who have a BRCA1 mutation, and it is more aggressive compared to luminal subtypes [6].
Approximately 20 to 30% of breast cancer cases have HER2 gene amplification and hence HER2 receptor overexpression [7]. HER2 (185kD) is a 1255 amino acid long transmembrane tyrosine kinase that belongs to the family of epidermal growth factor (EGF) encoded by the ERBB2 gene (also named HER2, neu, or p185) and was discovered in the year 1984. Being a tyrosine kinase, HER2 overexpression is implicated in rapid cell division which drives the oncogenesis process. HER2 gene is located on the long arm of chromosome 17 (17q12). The clinical diagnosis of HER2-positive breast cancer is most often performed by immunohistochemistry (IHC) or fluorescence in situ hybridization (FISH). The discovery of trastuzumab (Herceptin) almost 25 years ago revolutionized the treatment of HER2-positive breast cancer [8]. It is the first-line treatment option and is also used as an adjuvant for combined therapy with either paclitaxel, docetaxel, or carboplatin [9].
American Cancer Society recommends that all invasive breast cancers should be tested for the presence of HER2. Currently FISH remains the gold standard for HER2 diagnosis, whereas IHC is more popular in preclinical settings since it is easy and cost-effective compared to that of FISH [10]. However, a definitive diagnosis is not currently possible by IHC. Although FISH is a highly sensitive and accurate method for diagnosing HER2 amplification, the prohibitive cost, and the requirement of specialized laboratory facilities, tools, and expertise makes it less attractive for low-resource settings [11]. Moreover, using FISH, the optimization of tumor morphology is difficult, and it is also challenging to differentiate between the diverse types of carcinomas. The rapid quenching of the fluorescent probes at room temperature is also a potential disadvantage of FISH. Furthermore, using the current techniques, it is not possible to stratify patients with lower ranges of HER2 expression [12]. The false negative results may lead to trastuzumab-induced cardiotoxicity and unnecessary financial burden, whereas false negative results may cause loss of quality-of-life years (QALY), metastasis, disease recurrence, and burden of additional treatment costs [13]. It is also impossible to evaluate the tumor progression from the results of diagnosis using either FISH or IHC, which is a crucial factor before the administration of the treatment [14]. There is also a discrepancy reported in the HER2 expression in the tumor and the affected lymph node [15]. These limitations urge the need for more valid, quantitative, and reliable standardized assays for the definitive diagnosis of HER2-positive breast cancer. Since HER2 gene amplification is correlated with the over-expression of mRNA and protein levels, it is possible to measure the HER2 at DNA, mRNA, and protein levels. Quantitative real-time reverse transcription-PCR (RT-qPCR) is an advanced PCR technology that allows reliable and rapid detection and quantification of gene products at each PCR cycle, which enables the quantification of HER2 gene products [16]. Apart from absolute and relative quantification of gene expression it is also widely used for identifying circulating tumor cells, validating DNA microarray results, single nucleotide polymorphism (SNP) discovery and validation, assessing viral, bacterial, and fungal loads, etc [17–20]. Previous analysis performed in a retrospective study has shown that RT-qPCR could be used as a possible technique for accurate quantification of HER2 using breast cancer biopsy samples [21]. The results indicate that RT-qPCR-based tests could bring a paradigm shift in the current diagnosis workflow of HER2-positive breast cancer. Moreover, in contrast to the current dosing regimen which is primarily based on the weight of the patient, the new dosing regimen advocated by several research groups worldwide based on pharmacokinetics, quantitative HER2 expression, etc. necessitates new strategies for HER2 quantification such as RT-qPCR in the clinics [22]. It will also channel the intelligent use of the drug to reduce both the financial and clinical toxicities associated with the current use of trastuzumab.
Materials and Methods
Samples
Altogether 275 tumor samples were collected from breast cancer patients who reported/diagnosed at the Ibn Rochd University Hospital Center (Casablanca, Morocco), after their informed consent. The tumor samples obtained from the Pathology Department of the Ibn Rochd University Hospital Center were formalin-fixed and paraffin-embedded (FFPE) and collected prospectively from the patients over a period of one year. The sample collection and the study outlined in this article were approved by The Institutional Ethics Committee.
Cell lines and cell culture
Breast cancer cell lines used in this study (HER2 over-expressing SKBR-3, ATCC® HTB-30™, and HER2 low MCF7, ATCC® HTB-22™) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). SKBR3 cells were cultured in ATCC-formulated Modified McCoy’s 5a medium and MCF7 cells in ATCC-formulated Eagle’s Minimum Essential Medium respectively. The media were supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/ streptomycin antibiotic solution as directed by ATCC, and the cells were cultured at standard culture conditions (37°C, 5% CO2) and sub-cultured when the cells reached ~80% confluence.
Immunohistochemistry (IHC)
IHC was done at the Pathology Department of the Ibn Rochd University Hospital Center. VENTANA anti-HER2/neu (4B5) Rabbit Monoclonal Primary Antibody was used for the IHC studies on a fully automated BenchMark IHC automat. The results were reported in accordance with the 2018 ASCO/CAP recommendations [23].
Fluorescence in situ hybridization – FISH
Tissue samples which were scored 2+ were validated using FISH. 3 μm paraffin tissue sections were used for FISH analysis. HER2 FISH pharmDx™ (Dako Denmark A/S, Glostrup, Denmark) probe was used to perform the FISH, and the images were captured using a fluorescence microscope with a suitable filter. HER2 amplification was reported as per 2018 ASCO/CAP recommendations [23].
RNA extraction from cell lines
RNeasy Mini kit from QIAGEN was used for extracting RNA from SKBR3 (HER2high) and MCF-7 cells (HER2low) as per instructions from the manufacturer. Approximately 30µl of RNA was eluted in RNase free water and the samples were stored at -80°C, until used. The quality and concentration of RNA were recorded in a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
Deparaffinization of FFPE tissue and total RNA extraction
PureLink™ FFPE RNA Isolation Kit (Invitrogen, Carlsbad, California) was used to remove paraffin and extract total RNA from tissue samples. Two to three 10-μm tissue sections were used for RNA extraction according to the instructions from the manufacturer. Approximately 30-50µl RNA was eluted in RNase free water (according to the quantity of the tissues) and stored at −80°C until used. The quality and concentration of RNA were recorded in a NanoDrop ND-2000 spectrophotometer as mentioned earlier.
Reverse transcription-quantitative realtime- PCR
Primers and probes design
Primer 3 plus software version 2.0 was used to design the primers and probes sequences. The probes were manufactured by Eurofins (Germany) with a 5ʹ6- carboxyfluorescein (FAM) for HER2 and the two control genes RPL30 (FAM) and RPL37 (VIC) as reporter dyes, and a 3ʹBlack Hole Quencher-1 (BHQ-1) as a quencher dye for both the probes. The primers were designed to exclude the amplification of genomic DNA. The specificity of the primers and probes was validated using the HER2 mRNA levels in MCF-7 (HER2low) and SKBR3 (HER2high).
One-step RT-qPCR
HER2 mRNA levels were quantified using “one step” RT-qPCR in a QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) following the relative quantification method, and the data analysis was done using QuantStudio 6 Flex software V1.0. The results obtained from one-step RT-qPCR were expressed as relative levels of HER2 mRNA with respect to the calibrator sample obtained from the HER2low MCF-7 cell line and normalized as 1X expression of HER2. HER2high SKBR- 3 cell line was used as a positive control. TaqMan one-step Master Mix (Applied Biosystems) was used to perform one-step RT-qPCR as per the manufacturer’s instructions. The Master Mix contained Amplitaq Gold hot start fast DNA polymerase and M-MLV reverse transcriptase. 10 μL RT-qPCR reaction mix contained Taqman 1-step master mix (4X, 2.5μL volume), primers (300 nM each), and probe (200 nM). A total of 6.5 μL of the total purified RNA (200ng/µL) was added to the 3.5μL PCR Mix. RT-qPCR reactions were performed in duplicates. The cycle conditions were maintained the same for the 3 transcripts at 50°C for 5min (reverse transcription reaction), 95°C for 20s (initial denaturation), and 50 cycles at 95°C for 15s. The annealing and extension steps were combined and performed at 60°C for a duration of 60s. For the relative quantification of HER2, the reference genes reported in our previous study were used, and the same protocol was used for quantifying HER2 relative to the geometric means of the reference genes with respect to the calibrator sample and the errors were calculated as per the rules of error propagation [24]. The risk of contamination was eliminated by performing the experiments in a pre-PCR, a PCR, and a post-PCR room, wherein the sample transportation was performed via airlock windows. To rule out contamination, a negative control containing RT-qPCR reaction mix added to pure water was also kept.
Relative quantification = 2–ΔΔCt
2−ΔΔCt method was used for the relative quantification of HER2 gene expression. The algorithm considered the calibrator gene expression (in this case HERlow MCF7) represented as 1X expression of HER2. HER2 gene expression was calculated as per the following equation: fold induction = 2− [ΔΔCt], where ΔΔCt = [Ct HER2 (Tumor sample) − Ct endogenous control (tumor sample)] − [CtHER2 (MCF7) − Ct endogenous control (MCF7)]. The ROC curve method was used for differentiating between HER2 positive and negative samples. The RT-qPCR data obtained in this study were classified into two expression levels. Overexpression of HER2 was categorized as positive cases, whereas normal expression of HER2 was categorized as negative cases. A normalized ratio was used to represent the result and if the value was above the predetermined threshold, HER2 was considered overexpressed in the samples.
Results
HER2 expression in breast cancer patients
The expression of HER2 was assessed in all breast cancer patients using IHC, FISH, and qRT-PCR. The studies were performed in the Department of Pathological Anatomy, CHU Ibn Rochd, Casablanca. The baseline characteristics of the patients (275 patients) included in this study have been shown in Table 1. IHC was performed using paraffin-embedded tissue sections, and the representative image is shown in Figure 1 panel 1. HER2 overexpression can be clearly visualized in amplified HER2 infiltrating breast carcinoma (Figure 1a) as against minimal HER2 expression in a non-amplified HER2 infiltrating breast carcinoma (Figure 1b). HER2 gene amplification was also confirmed using FISH (Figure 1, panel 2), wherein the red signals indicate HER2 amplification and green signals indicate centromeres of chromosomes. Figure 1c shows a sample with low/basal expression of HER2 as indicated by the scattered red clusters and the presence of two green signals from the centromere of chromosome 17. In sharp contrast, a high level of HER2-infiltrating breast carcinoma sample (Figure 1d) showed HER2 amplification as indicated by the clustering of red signals and the polysomy of centromere of chromosome 17 (green signals). Later, HER2 mRNA expression was performed using “one-step” qRT-PCR after extracting the total RNA from all 275 FFPE tissues. The standard curve was plotted for HER2 and the reference genes RPL30 and RPL37, and the R2 value and slope indicated the efficacy of the RT-qPCR reaction, and also the primers and probes of the target genes. Figure 1 Panel 3 shows the amplification curves of HER2 mRNA in two different samples using RT-qPCR, wherein Figure 1e is the negative control which had normal expression of HER-2, and Figure 1d is the sample with over-expression of HER2. The assay reproducibility was determined on 3 pools of HER2high patient samples (3 levels of mRNA overexpression: high, medium, and low) and one pool of HER2low patient samples with a 10-fold passage number. Repeatability was within the set analytical objectives (CV<5%).
Table 1.
Baseline Characteristics of Patients Included in This Study
| Characteristics | Number of patients | Percentage | |
|---|---|---|---|
| Age | 20-30 | 4 | 1.45 |
| 31-40 | 33 | 13.82 | |
| 41-50 | 107 | 38.91 | |
| 51-60 | 65 | 23.64 | |
| >60 | 61 | 22.18 | |
| SBR Grade | Grade 1 | 17 | 6.18 |
| Grade 2 | 152 | 55.27 | |
| Grade 3 | 106 | 38.55 | |
| Presence of | Oui | 130 | 47.27 |
| emboli | Non | 145 | 52.73 |
| Tumor size | <1cm | 36 | 13.09 |
| 1-5cm | 221 | 80.36 | |
| 5-10cm | 2 | 0.73 | |
| >10cm | 16 | 5.82 | |
| RE% | Negative (<1%) | 62 | 22.55 |
| Positive (>1%) | 213 | 77.45 | |
| RE intensity | Intense | 123 | 44.73 |
| Moderate | 67 | 24.36 | |
| Negative | 85 | 30.91 | |
| RP% | Negative (<1%) | 75 | 27.27 |
| Positive (>1%) | 200 | 72.73 | |
| PR intensity | Intense | 95 | 34.55 |
| Moderate | 72 | 26.18 | |
| Negative | 108 | 39.27 | |
| HER2 SCORE | 0+ | 133 | 48.36 |
| 1+ | 62 | 22.55 | |
| 2+ | 25 | 9.09 | |
| 3+ | 55 | 20.00 | |
| KI-67% | High (>10%) | 241 | 87.64 |
| Low (<10%) | 34 | 12.36 | |
| T,n | pT1 | 174 | 63.27 |
| pT2 | 99 | 34.91 | |
| pT3 | 2 | 0.73 | |
| N, n | pN0 | 154 | 56 |
| pN1 | 78 | 28.36 | |
| pN2 | 31 | 11.27 | |
| pN3 | 12 | 4.26 | |
| M, n | 0 | 275 | 100 |
| pStage | Stade I | 117 | 42.55 |
| Stade II | 114 | 41.46 | |
| Stade III | 43 | 15.63 | |
| Stade IV | 1 | 0.36 |
Figure 1.
Examples of HER2 protein Overexpression (IHC), HER2 gene amplification (FISH) and mRNA quantification by RT-qPCR. Panel 1. Immunohistochemistry a. Sample showing no HER2 overexpression in non-amplified HER2 infiltrating breast carcinoma. b. Sample showing strong HER2 overexpression in an amplified HER2 infiltrating breast carcinoma. Panel 2. HER2 gene amplification detected by FISH (red signals: HER2, green signals: centromeres of chromosome 17) c. Sample showing low level of HER2 gene amplification, without clusters and two green signals from centromere of chromosome 17. d. Sample showing high level of HER2 amplification in HER2-infiltrating breast carcinoma which shows clusters of red signals, indicating HER2 amplification in presence of polysomy of centromere of chromosome 17 (green signals). Panel 3. Amplification curves using RT-qPCR of HER2 mRNA in two different samples. e. negative sample with normal HER2 expression. d. positive sample with HER2 overexpression.
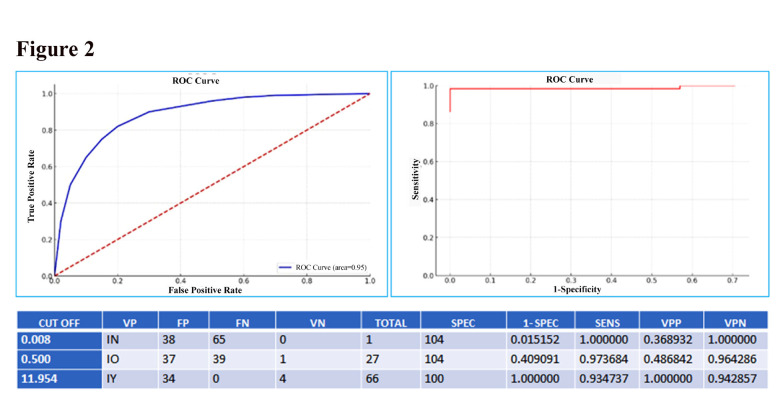
Sensitivity and specificity of the test by ROC analysis
The ROC curve analysis which is the receiver operating characteristic curve indicates the true positive cases against false positive cases. ROC curve was plotted for the samples using the HER2 mRNA levels obtained using RT-qPCR and IHC score to determine the optimal diagnostic cut-off value with minimized false-negative and false-positive (Figure 2). The area under the curve (AUC) which is an important parameter that determines the diagnostic value of the test was calculated as AUC=0.955. A test with a strong discriminative power must have an AUC value close to 1 and far from 0.5, and our results were close to 1, which indicates the diagnostic accuracy of RT-qPCR. As per the ROC analysis, the optimal cut-off value in our context was 11.954 corresponding to the combination of the best sensitivity and specificity (93.4% and 100%) respectively, and with positive predictive values PPV that reached 100% and a negative predictive value NPV that reached 89.4%.
Figure 2.
Sensitivity and Specificity of the Test by ROC Analysis: The ROC curve is used to determine both sensitivity and specificity of the test. The optimal cut-off value in our context is the value of 11.954 corresponding to the combination of the best sensitivity and specificity (93.4% and 100%). The area under curve which is a parameter to determine the diagnostic value of the test was also calculated (AUC=0.955).
The concordance between IHC vs RT-qPCR and FISH vs RT-qPCR is shown in Table 2. The results show that among the 211 samples that were IHC negative, RT-qPCR was negative for 209 samples and positive for 2 samples. Similarly, among the 53 samples that were positive as per IHC, RT-qPCR was negative for 9 samples and was positive only for 44 samples. The Cohen’s Kappa coefficient was around 0.863 which indicates a very good concordance between IHC and RT-qPCR tests. FISH vs RT-qPCR results showed 100% concordance since 11 samples that were negative in FISH were also RT-qPCR negative. Hence the results indicate that IHC and RT-qPCR were consistent in assessing the molecular status of the patients in the study. High concordance also indicates that depending on the availability and feasibility, one technique may be used instead of the other, and the treatment decisions will generally be similar based on the results, although the patient’s overall clinical context is also a crucial factor in determining the treatment.
Table 2.
Concordance between IHC vs RT-qPCR / and FISH vs RT-qPCR in Our Study
| RT-qPCR Negative | RT-qPCR Positive | |
|---|---|---|
| IHC negative | 209 | 2 |
| IHC positive | 9 | 44 |
| Kappa coefficient | 0.863 | |
| FISH negative | 11 | 11 |
| FISH positive | 0 | 0 |
The HER2 expression studies showed that larger tumors were associated with higher HER2 scores (Figure 3), most likely due to the higher expression of HER2, which in turn might have fueled an aggressive tumor growth. However, we did observe a certain degree of variability in this study, since some of the medium-sized tumors showed higher HER2 scores, whereas certain other tumors with approximately same size showed low HER2 scores.
Figure 3.
Relationship between Tumor Size and HER2 Score. The graph illustrates the relationship between tumor size and HER2 score in 269 patients.
Figure 4 shows the relationship between HER2 score and SBR grade. HER2 score and SBR grade are two key parameters for characterizing breast tumors. The results indicate that there is an association between HER2 score and SBR grade, two key diagnostic criteria in breast cancer. The observation of an association between these two parameters, although not perfectly linear, could suggest that patients with HER2 overexpression also tend to have histologically more aggressive tumors.
Figure 4.
Relationship between HER2 Score and SBR Grade
Discussion
HER2 amplification status is a crucial factor in the response to targeted anti-HER2 therapy, and hence there is a pressing need to streamline the therapy based on the differential expression of HER2 and the underlying signaling mechanisms. There is ample evidence that indicates HER2 levels can predict the response to anti-HER2 therapy, and survival outcomes [25]. Although FISH is a gold standard technique to assess the amplification of HER2, IHC is more frequently used, the latter being a comparatively inexpensive technique [10]. However, IHC has a high interoperator variability and a high rate of false positives and false negatives [26]. Despite the specificty imparted by the antibodies, the assay positivity in IHC is largely dependent on the enzymatic activity of Horseradish peroxidase (HRP) which is conjugated to the antibodies. The staining intensity is significantly influenced by the quality and concentration of HRP and substrate, and also storage conditions, and duration of incubation [27].
The main focus of this study was to evaluate the feasibility of one-step RT-qPCR to be used as a test for quantifying the HER2 gene expression in breast cancer and validate the results with those obtained using FISH and IHC. HER2 overexpression in the breast cancer biopsy samples was confirmed using IHC, and classified as negative (score 0/1+), or positive (score 3+), as per the standard guidelines [23, 28]. If the score obtained was 2+ another method of reference i.e FISH was also used. However, FISH has its own limitations, apart from being an expensive technique, the fluorescence quenching happens fast, and hence the slides cannot be preserved for long [29]. Moreover, the facilities to carry out FISH may not be available at all laboratories. This necessitates a sensitive, quantitative, rapid, and accurate test to standardize and automate the diagnosis of HER2+ve breast cancer.
Previous studies on the use of RT-qPCR for quantifying HER2 were contradictory, and even certain studies do not recommend its use [30, 31]. We presume that this is largely due to the mRNA degradation that might have occurred while its extraction from formalin fixed paraffin embedded (FFPE) tissues, or the absence of suitable reference genes. However, in the current study, we could overcome the discrepancies in the results by using appropriate endogenous controls/reference genes, thereby making the results more accurate. We used RPL30 and RPL37 as endogenous controls which we selected based on our previous results [24]. We carried out clinical validation of one-step RT-qPCR using FFPE samples and compared the results with those of IHC and FISH. The first step was to identify an optimal diagnostic cut-off value using ROC-based analysis and using the IHC reference score. We found that an optimal value of 11.954 represented the best specificity (100%) and sensitivity (89.4%), and an AUC value of 0.955. The optimal cut-off value thus indicates that if the HER2 expression is above 11.954, it is considered positive and if the expression is below 11.954, the sample is HER2 negative. Our study showed a high concordance of RT-qPCR and IHC results. Cohen’s Kappa coefficient of 0.863 (Table 2) indicates very good concordance between IHC and RT-qPCR tests, which in turn suggests that the two methods are largely consistent in their assessment of the clinical status of patients in the study. Furtermore, a 100% concordance obtained with FISH further validates the use of RT-qPCR. A high concordance also reinforces the reliability of both diagnostic methods in the clinical context, which indicates that the healthcare professionals can rely on either of these tests to provide accurate information about patients’ disease state, depending on the availability and/or feasibility. However, we did observe a discordance for a few cases (11 patients in our study), which necessitates re-evaluation of the results and consideration of other pathophysiological factors. So far there are no elaborate studies which show a similar trend, and we believe that further studies in this direction may have profound clinical implications especially in personalizing the treatment approach.
Our results are in conjunction with several other studies published in this direction which show concordance of RT-qPCR with IHC/FISH. In a very recent study conducted by Li et al. using 323 patient samples which included different molecular subtypes, a high concordance (89.4%) was found to exist between IHC and RT-qPCR in the case of HER2 expression [32]. Previously, Chen et al. conducted studies using 397 breast cancer patient samples of different subtypes and found that for HER2-positive cases there was 81.6% concordance between RT-qPCR and IHC. Their study recommends the use of RT-qPCR as a complementary method to molecular subtyping under circumstances of ambiguous IHC results [33]. Caselli et al. have conducted studies using different subtypes of breast cancer and showed that the mRNA level expression of breast cancer biomarkers such as HER2, ER, PR, and Ki67 highly correlated with that of IHC. Most importantly, RT-qPCR results were reproducible, and offered a higher degree of standardization for Ki67, and also solved HER2 cases that were uncertain as per IHC/FISH assessment [34]. Similarly, a high degree of concordance i.e., 94% was observed by Gheni and Westenberg in their studies using 54 paired tissue samples (FFPE tissues). Their study concluded that IHC can be used only for initial screening and a more accurate, quantitative and a reproducible result may be obtained by using RT-qPCR in routine clinical practice [35]. Al Banyahyati also reported that one-step RT-qPCR method was quite accurate in giving correlated results with those of FISH/IHC methods, and even recommended using the former as the initial choice of diagnosis for HER2 amplification and also to predict the response to trastuzumab therapy [36]. Some of the earlier studies also show that in terms of the quality of results RT-qPCR may outperform IHC/FISH [37, 38]. At the same time, it is also important to discuss some of the studies that demonstrated discordance of RT-qPCR with IHC/FISH. Gupta et al. performed studies using 63 patients and found that the results obtained using RT-qPCR and IHC/immunofluorescence were not correlating [31]. Another recent study showed that RT-qPCR may not be a reliable technique for evaluating the HER2 amplification [30]. Even in the scenario that a few studies show discordance of RT-qPCR results with that of IHC/FISH, most of them stress the need for reliable quantitative techniques to assess HER2 amplification, especially when the IHC results are ambiguous.
Although our study showed that the larger tumors had higher HER2 scores (Figure 3), compared to the smaller ones, there was some variability in the data, which might be attributed to the inherent complexity and hetergoneity of cancer [39]. Other factors such as genetics, environment, and other molecular features, can influence HER2 expression, and this warrants further research, since this could be significant for personalizing the treatment approach. It is a well-established fact that HER2 amplification is often associated with aggressive tumor growth and poor prognosis [40]. In our samples, the majority of tumours with a high HER2 score (3+) also had a high SBR grade (grade 3), which is based on three histological features: tubular differentiation, nuclear pleomorphism, and mitotic proliferation index (Figure 4). A high SBR score generally indicates more aggressive tumor behavior and an increased risk of recurrence [41]. Although not linear, the association suggests that tumours with HER2 overexpression may also have more aggressive histological features, which is crucial for therapeutic decision-making, particularly in identifying patients with optimal response to anti-HER2 targeted therapy.
In conclusion, the current study shows that one-step RT-qPCR is an accurate, reliable, and fast technique for quantifying HER2 mRNA. Moreover, compared to the gold standard techniques, it is less expensive, rapid, and offers reproducibility of results. Current studies indicate that the therapy strategy should not be solely designed based on IHC results, under circumstances that IHC offers only a limited dynamic range of analysis. Apart from designing the therapy, quantitative HER2 expression is significant in tracking the progress of therapeutic response. Our results are in conjunction with several other recently published studies which show that RT-qPCR is a reliable technique and may be easily adapted to all clinical settings. However, further studies at multiple locations are warranted in this direction, recruiting patients with different levels of HER2 amplification, and at different stages of the disease. It should be noted that the quality of mRNA and the choice of reference genes play a significant role in the results and as laid out by previous studies, the FFPE samples should contain more than 50% of tumor cells for reliable results. Taken together, our studies shed light to the significance of a more standardized quantification of HER2 expression for better treatment orientation.
Author Contribution Statement
BA carried out most of the experiments. HE helped in performing experiments. HE, AM participated in the design of the study, performed the statistical analysis and helped in the drafted the manuscript. BY participated in the design of the study and were involved with revising the manuscript critically. MK and AB provided and managed the patient’s samples and revised the manuscript critically and conducted the external validation of the test. AM conceived, designed, coordinated the study and drafted the paper. All authors read and approved the final manuscript.
Acknowledgements
We thank all patients that have kindly agreed to provide us with the samples analyzed here. We are also very grateful to all members of the medical biotechnology laboratory in MAScIR for their help in performing this research. We are thankful to the technical staff of Pathology Department in Hospital Ibn Rochd Casablanca and National Institute of Oncology Rabat. We are indebted to MAScIR administration staff for their support.
Ethical approval for the use of patient’s samples has been obtained from the Casablanca medical school ethic committee
Funding statement
This work was funded by both the Moroccan foundation for advanced science, innovation and research (MAScIR) and the UM6P.
Any conflict of interest
The authors declare that they have no competing interests.
References
- 1.Li J, Goh ELK, He J, Li Y, Fan Z, Yu Z, et al. Emerging Intrinsic Therapeutic Targets for Metastatic Breast Cancer. Biology. 2023;12 doi: 10.3390/biology12050697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finkelman BS, Zhang H, Hicks DG, Turner BM. The Evolution of Ki-67 and Breast Carcinoma: Past Observations, Present Directions, and Future Considerations. Cancers. 2023;15:808. doi: 10.3390/cancers15030808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaurasia M, Singh R, Sur S, Flora SJS. A review of FDA approved drugs and their formulations for the treatment of breast cancer. Front Pharmacol. 2023;14:1184472. doi: 10.3389/fphar.2023.1184472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manson EN, Achel DG. Fighting breast cancer in low-and-middle-income countries – What must we do to get every woman screened on regular basis? Sci Afr. 2023;21:e01848. [Google Scholar]
- 5.Mohammed AA. The clinical behavior of different molecular subtypes of breast cancer. Cancer Treat Res Commun. 2021;29:100469. doi: 10.1016/j.ctarc.2021.100469. [DOI] [PubMed] [Google Scholar]
- 6.Orrantia-Borunda E, Anchondo-Nuñez P, Acuña-Aguilar LE, Gómez-Valles FO, Ramírez-Valdespino CA. Subtypes of Breast Cancer. In: Mayrovitz HN, editor. Breast Cancer. Brisbane (AU): Exon Publications; 2022. [PubMed] [Google Scholar]
- 7.Najjar MK, Manore SG, Regua AT, Lo HW. Antibody-Drug Conjugates for the Treatment of HER2-Positive Breast Cancer. Genes. 2022;13:2065. doi: 10.3390/genes13112065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swain SM, Shastry M, Hamilton E. Targeting HER2-positive breast cancer: advances and future directions. Nat Rev Drug Discov. 2023;22:101–26. doi: 10.1038/s41573-022-00579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becerra-Chauca N, Nieto-Gutierrez W, Taype-Rondan A. 6-month or 12-month adjuvant trastuzumab regimen for HER2-positive breast cancer? Decision-making in a resource-limited setting. Lancet Oncol. 2022;23:e100. doi: 10.1016/S1470-2045(22)00060-2. [DOI] [PubMed] [Google Scholar]
- 10.Aznab M, Izadi B, Amirian F, Khazaei S, Madani SH, Ramezani M. Comparison of Immunohistochemical Methods (IHC) and Fluorescent in Situ Hybridization (FISH) in the Detection of HER 2 /Neu Gene in Kurdish Patients with Breast Cancer in Western Iran. Int J Hematol-Oncol Stem Cell Res. 2022;16:217. doi: 10.18502/ijhoscr.v16i4.10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chrzanowska NM, Kowalewski J, Lewandowska MA. Use of Fluorescence In Situ Hybridization (FISH) in Diagnosis and Tailored Therapies in Solid Tumors. Molecules. 2020;25:1864. doi: 10.3390/molecules25081864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moutafi M, Robbins CJ, Yaghoobi V, Fernandez AI, Martinez-Morilla S, Xirou V, et al. Quantitative measurement of HER2 expression to subclassify ERBB2 unamplified breast cancer. Lab Investig J Tech Methods Pathol. 2022;102:1101–8. doi: 10.1038/s41374-022-00804-9. [DOI] [PubMed] [Google Scholar]
- 13.Esposito A, Ablah E, Okut H, Tenofsky PL. Characteristics, treatment and outcomes of HER2 positive male breast cancer. Am J Surg. 2023;225:489–93. doi: 10.1016/j.amjsurg.2022.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Kong H, Bai Q, Li A, Zhou X, Yang W. Characteristics of HER2-negative breast cancers with FISH-equivocal status according to 2018 ASCO/CAP guideline. Diagn Pathol. 2022;17:5. doi: 10.1186/s13000-021-01187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Englander K, Chintapally N, Gallagher J, Elleson K, Sun W, Whiting J, et al. Factors Influencing Lymph Node Positivity in HER2/neu+ Breast Cancer Patients. Curr Oncol. 2023;30:2825–33. doi: 10.3390/curroncol30030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendoza G, Portillo A, Olmos-Soto J. Accurate breast cancer diagnosis through real-time PCR her-2 gene quantification using immunohistochemically-identified biopsies. Oncol Lett. 2013;5:295. doi: 10.3892/ol.2012.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andergassen U, Zebisch M, Kölbl AC, König A, Heublein S, Schröder L, et al. Real-Time qPCR-Based Detection of Circulating Tumor Cells from Blood Samples of Adjuvant Breast Cancer Patients: A Preliminary Study. Breast Care. 2016;11:194–8. doi: 10.1159/000447041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galazzo G, van Best N, Benedikter BJ, Janssen K, Bervoets L, Driessen C, et al. How to Count Our Microbes? The Effect of Different Quantitative Microbiome Profiling Approaches. Front Cell Infect Microbiol. 2020:10. doi: 10.3389/fcimb.2020.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehrotra M, Luthra R, Abraham R, Mishra BM, Virani S, Chen H, et al. Validation of quantitative PCR-based assays for detection of gene copy number aberrations in formalin-fixed, paraffin embedded solid tumor samples. Cancer Genet. 2017;212–213:24–31. doi: 10.1016/j.cancergen.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Porter WT, Kelley EJ, Bowers JR, Engelthaler DM. Normalization of SARS-CoV-2 viral load via RT-qPCR provides higher-resolution data for comparison across time and between patients. Virus Res. 2021;306:198604. doi: 10.1016/j.virusres.2021.198604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noske A, Loibl S, Darb-Esfahani S, Roller M, Kronenwett R, Müller BM, et al. Comparison of different approaches for assessment of HER2 expression on protein and mRNA level: prediction of chemotherapy response in the neoadjuvant GeparTrio trial ( NCT00544765) Breast Cancer Res Treat. 2011;126:109–17. doi: 10.1007/s10549-010-1316-y. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh PH, Kacew AJ, Dreyer M, Serritella AV, Knoebel RW, Strohbehn GW, et al. Alternative trastuzumab dosing strategies in HER2-positive early breast cancer are associated with patient out-of-pocket savings. NPJ Breast Cancer. 2022;8:32. doi: 10.1038/s41523-022-00393-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zavalishina LE, Andreeva YY, Olyushina EM, Moskvina LV, Frank GA. [Updated 2018 ASCO/CAP recommendations on the definition of human epidermal growth factor receptor type 2 (HER2) in breast cancer] Arkh Patol. 2019;81:82–5. doi: 10.17116/patol20198106182. [DOI] [PubMed] [Google Scholar]
- 24.El Hadi H, Abdellaoui-Maane I, Kottwitz D, El Amrani M, Bouchoutrouch N, Qmichou Z, et al. Development and evaluation of a novel RT-qPCR based test for the quantification of HER2 gene expression in breast cancer. Gene. 2017;605:114–22. doi: 10.1016/j.gene.2016.12.027. [DOI] [PubMed] [Google Scholar]
- 25.Atallah NM, Alsaleem M, Toss MS, Mongan NP, Rakha E. Differential response of HER2-positive breast cancer to anti-HER2 therapy based on HER2 protein expression level. Br J Cancer. 2023;129:1692–705. doi: 10.1038/s41416-023-02426-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casterá Redal C, Bernet Vegué L. “HER2 immunohistochemistry inter-observer reproducibility in 205 cases of invasive breast carcinoma additionally tested by ISH” Answer to the statistical issue to avoid misinterpretation. Ann Diagn Pathol. 2020;48:151566. doi: 10.1016/j.anndiagpath.2020.151566. [DOI] [PubMed] [Google Scholar]
- 27.Ivanova IA, Ershova MO, Shumov ID, Valueva AA, Ivanov YD, Pleshakova TO. Atomic Force Microscopy Study of the Temperature and Storage Duration Dependencies of Horseradish Peroxidase Oligomeric State. Biomedicines. 2022;10:2645. doi: 10.3390/biomedicines10102645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolff AC, Hammond MEH, Allison KH, Harvey BE, Mangu PB, Bartlett JMS, et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J Clin Oncol Off J Am Soc Clin Oncol. 2018;36:2105–22. doi: 10.1200/JCO.2018.77.8738. [DOI] [PubMed] [Google Scholar]
- 29.Logt EMJ van der, Kuperus DAJ, Setten JW van, Heuvel MC van den, Boers JE, Schuuring E, et al. Fully Automated Fluorescent in situ Hybridization (FISH) Staining and Digital Analysis of HER2 in Breast Cancer: A Validation Study. PLOS ONE. 2015;10:e0123201. doi: 10.1371/journal.pone.0123201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carretero-Barrio I, Caniego-Casas T, Rosas M, Sánchez MC, Martínez-Jáñez N, Chiva M, et al. Evaluation of ERBB2 mRNA Expression in HER2-Equivocal (2+) Immunohistochemistry Cases. Cancers. 2023;15:1688. doi: 10.3390/cancers15061688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta S, Neumeister V, McGuire J, Song YS, Acs B, Ho K, et al. Quantitative assessments and clinical outcomes in HER2 equivocal 2018 ASCO/CAP ISH group 4 breast cancer. Npj Breast Cancer. 2019;5:1–8. doi: 10.1038/s41523-019-0122-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Chen T, Du F, Wang H, Ma L. Concordance of RT-qPCR with immunohistochemistry and its beneficial role in breast cancer subtyping. Medicine (Baltimore). 2023;102:e35272. doi: 10.1097/MD.0000000000035272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen CJ, Chen TH, Lei J, Liang JA, Yang PS, Huang CS, et al. Correlation of ER, PR, and HER2 at the protein and mRNA levels in Asian patients with operable breast cancer. Biosci Rep. 2022;42:BSR20211706. doi: 10.1042/BSR20211706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caselli E, Pelliccia C, Teti V, Bellezza G, Mandarano M, Ferri I, et al. Looking for more reliable biomarkers in breast cancer: Comparison between routine methods and RT-qPCR. PLOS ONE. 2021;16:e0255580. doi: 10.1371/journal.pone.0255580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gheni N, Westenberg D. Quantitative real-time PCR assay with immunohistochemical evaluation of HER2/neu oncogene in breast cancer patients and its correlation with clinicopathological findings. Indian J Pathol Microbiol. 2020;63:123. doi: 10.4103/IJPM.IJPM_136_19. [DOI] [PubMed] [Google Scholar]
- 36.Al Banyahyati B. 164P A prospective approach for the evaluation of a one-step RT-qPCR based test for the quantification of HER2 protein in formalin-fixed and paraffin-embedded breast cancer tissues. Ann Oncol. 2020;31:S75. [Google Scholar]
- 37.Wasserman BE, Carvajal-Hausdorf DE, Ho K, Wong W, Wu N, Chu VC, et al. High Concordance of a Closed-System, RT-qPCR Breast Cancer Assay for HER2 mRNA, Compared to Clinically Determined Immunohistochemistry, Fluorescence in situ Hybridization, and Quantitative Immunofluorescence. Lab Investig J Tech Methods Pathol. 2017;97:1521–6. doi: 10.1038/labinvest.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zoppoli G, Garuti A, Cirmena G, di Cantogno LV, Botta C, Gallo M, et al. Her2 assessment using quantitative reverse transcriptase polymerase chain reaction reliably identifies Her2 overexpression without amplification in breast cancer cases. J Transl Med. 2017;15:91. doi: 10.1186/s12967-017-1195-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gunnarsson C, Olsson H. Methods for evaluating HER2 status in breast cancer: comparison of IHC, FISH, and real-time PCR analysis of formalin-fixed paraffin-embedded tissue. Pathol Lab Med Int. 2013:31. [Google Scholar]
- 40.Aman NA, Doukoure B, Koffi KD, Koui BS, Traore ZC, Kouyate M, et al. HER2 overexpression and correlation with other significant clinicopathologic parameters in Ivorian breast cancer women. BMC Clin Pathol. 2019;19:1. doi: 10.1186/s12907-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boudy A-S, Ferrier C, Selleret L, Zilberman S, Arfi A, Sussfeld J, et al. Prognosis of HER2-positive pregnancy-associated breast cancer: Analysis from the French CALG (Cancer Associé à La Grossesse) network. Breast. 2020;54:311–8. doi: 10.1016/j.breast.2020.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]