Abstract
The mammalian chloride channel ClC-2 is a member of the CLC voltage-gated chloride channels family. This broadly expressed protein shows diverse cellular locations and despite numerous studies, its precise function is poorly understood. Disruption of ClC-2-encoding gene in mouse leads to retinal and testicular degeneration and mutations in CLC2 (gene encoding the ClC-2 channel) are associated with idiopathic generalized epilepsies. ClC-2 may also be responsible for Cl− transport in mouse salivary glands. The only CLC homologue of the yeast Saccharomyces cerevisiae, Gef1p, exhibits CLC activity. We expressed the mammalian ClC-2 protein in S. cerevisiae devoid of Gef1p in an attempt to identify yeast proteins influencing the functioning of ClC-2. The presence of such proteins in yeast could indicate the existence of their homologues in mammalian cells and would greatly aid their identification. Expression of ClC-2 in yeast required optimization of the sequence context of the AUG translation initiation codon. After obtaining an efficient translation, we found that rat ClC-2 cannot directly substitute for yeast Gef1p. Functional substitution for Gef1p was, however, achieved in the presence of an increased level of intact or C-terminally truncated yeast Kha1 protein. Based on the deduced amino acid sequence, the Kha1 protein can be classified as a Na+/H+ transporter since it has a large N-terminal domain similar to the family of NHEs (Na+/H+ exchangers). This suggests that the Kha1p may take part in the regulation of intracellular cation homoeostasis and pH control. We have established that Kha1p is localized in the same cellular compartment as Gef1p and yeast-expressed ClC-2: the Golgi apparatus. We propose that Kha1p may aid ClC-2-dependent suppression of the Δgef1-assocciated growth defects by keeping the Golgi apparatus pH in a range suitable for ClC-2 activity. The approach employed in the present study may be of general applicability to the characterization of poorly understood proteins by their functional expression in yeast.
Keywords: ClC-2, GEF1, heterologous expression, Kha1 exchanger, Kha1p, Saccharomyces cerevisiae
Abbreviations: CFTR, cystic fibrosis transmembrane conductance regulator; GFP, green fluorescent protein; Mnt1p, α-1,2-mannosyltransferase; NHE, Na+/H+ exchanger; optCLC2, optimized CLC2 gene
INTRODUCTION
The mammalian voltage-gated chloride channel ClC-2 is activated by membrane hyperpolarization, cell swelling and acidic pH. Since ClC-2 is expressed in the same epithelial tissues as the CFTR (cystic fibrosis transmembrane conductance regulator) and both channels are Cl− transporters, it has been assumed that ClC-2 expression level may modulate the severity of cystic fibrosis. Thus ClC-2 was considered as a potential target for pharmacological treatment. However, a recent study using mice with disruptions of the ClC-2 and CFTR channels has shown that ClC-2 is rather unlikely to serve as a rescue channel in cystic fibrosis [1].
Despite numerous studies, the precise function of ClC-2 is poorly understood. Disruption of the ClC-2-encoding gene in mouse leads to retinal and testicular degeneration (for a review see [2]). It has also been suggested that ClC-2 may be responsible for Cl− conductance in salivary glands [3]. On the other hand, the Caenorhabditis elegans ClC-2 orthologue ClH-3, which is present in oocytes, is activated during meiotic cell maturation, suggesting that it might play a role in the meiotic cell cycle, fertilization, and/or early development [4]. Recent studies implicate ClC-2 mutations in idiopathic generalized epilepsies [5,6].
ClC-2 has been shown to be predominantly localized in apical membranes of villi enterocytes in rat [7] and at the apical junctions of the small intestine in mouse [8]. Additionally, the protein has been detected in the basolateral membranes of rat [9] and guinea pig colonocytes [10], whereas in human colonocytes, ClC-2 appears to be expressed chiefly in an intracellular compartment [9]. Knowledge concerning the regulation of ClC-2 functioning is very limited. Recent results suggest that in human colonic epithelial cells inhibition of PI3K (phosphoinositide 3-kinase), a lipid kinase involved in the regulation of membrane traffic, diminishes the amplitude of whole cell ClC-2 current measured by the patch-clamp technique, suggesting that ClC-2 is delivered to the plasma membrane by vesicular traffic [11]. Dhani et al. [12] have shown that cell-surface expression of ClC-2 is regulated by dynein motor activity. The importance of interacting proteins in ion channel activity is further emphasized by recent results, which show that ‘a kinase-regulated mechanism controls CFTR channel gating by disrupting PDZ domain interaction’ [13]. Thus identification of proteins interacting with ClC-2, both physically and functionally, is a promising approach to further our understanding of the biological context and function of this channel protein.
The only CLC homologue of the yeast Saccharomyces cerevisiae is encoded by GEF1 (YJR040w). Deletion of this gene causes growth defects on media with non-fermentable carbon sources and a very low level of iron [14]. Moreover, the Δgef1 mutant has an increased sensitivity to Mn2+, Co2+, Zn2+ and hygromycin B [15]. The role of Gef1p is not fully understood but it is known that the protein is localized in intracellular vesicles, probably in the medial Golgi compartment and exhibits an anion channel activity [15–17].
In the present study, we expressed the mammalian ClC-2 protein in the model microorganism S. cerevisiae in an attempt to identify yeast proteins that could modulate the functioning of ClC-2. The amino acid similarity between yeast Gef1p and mammalian ClC-2 channel suggested that ClC-2 might entirely substitute for Gef1p in S. cerevisiae cells devoid of the GEF1 gene. However, it was also possible that the substitution would only be partial and yeast Δgef1 cells expressing ClC-2 cDNA would show a phenotype distinct from the wild-type one. Such a phenotype could be used to identify functional co-suppressors. This classical genetic approach would enable detection of yeast proteins interacting (directly or indirectly) with ClC-2 and permitting functional ClC-2 substitution for Gef1p in yeast. Such proteins would probably have homologues in mammalian cells, but their identification directly in mammalian cells would be, purely for technical reasons, much more difficult than in yeast. In the present study, we describe the expression of the rat ClC-2 cDNA in S. cerevisiae cells devoid of the GEF1 gene. The subcellular localization of ClC-2 protein was similar to that seen with Gef1p, but ClC-2 did not substitute directly for Gef1p. However, we identified Kha1 as a functional co-suppressor enabling the functioning of ClC-2 in Δgef1 cells of S. cerevisiae.
EXPERIMENTAL
Strains, media and microbiological techniques
Yeast strains used in the present study, isogenic with the standard strain W303, are listed in Table 1. Escherichia coli XL1-Blue MRF′ (Stratagene, Saint Quentin en Yvelines, France) was used for molecular manipulations. Yeast culture media were prepared as described in [18]. YPD contained 1% Bacto-yeast extract, 2% (w/v) Bacto-peptone and 2% (w/v) glucose. SD contained 0.67% yeast nitrogen base without amino acids (Difco, Detroit, MI, U.S.A.) and 2% glucose. For auxotrophic strains, the media contained appropriate supplements. Standard methods were used to genetically manipulate yeast cells [19].
Table 1. Yeast strains used in the present study.
| Strain | Genotype | Source or reference |
|---|---|---|
| KFY19 | Mataura3-1 leu2-3,112 trp1-1 his3-11,15 can1-100 Δgef1::LEU2 [pKF19] | [17] |
| KFY37 | Mataura3-1 leu2-3,112 trp1-1 his3-11,15::pKF37 (PMET25-optCLC2-TCYC1HIS3) can1-100 Δgef1::LEU2 | Present study |
| KFYT1-15A | Mataura3-1 leu2-3,112 trp1 his3 can1-100 Δgef1::LEU2 Δkha1(4,2491)::kanMX4 | Present study, derivative of RGY84 × KFT2-2B |
| KFYT2-2B | Matα ura3-1 leu2-3,112 trp1Δ2 his3-11 can1-100 ade2-1 Δkha1(4,2491)::kanMX4 | Present study, derivative of WBLS004-HE |
| RGY9 | Matα ura3-1 leu2-3,112 trp1-1 his3-11,15 can1-100 | [15] |
| RGY84 | Mataura3-1 leu2-3,112 trp1-1 his3-11,15 can1-100 Δgef1::LEU2 | [15] |
| WBLS004-HE | Mata/α ura3-1/ura3-1 leu2-3,112/leu2-3,112 trp1Δ2/trp1Δ2 his3-11/his3-11 can1-100/can1-100 | EUROSCARF (acc. no. 20204D) |
| ade2-1/ade2-1 KHA1/Δkha1(4,2491)::kanMX4 |
Plasmid construction
Plasmids were constructed by standard methods as described in [20] and are listed in Table 2. The original ClC-2 cDNA from Rattus norvegicus brain, clone b12-2 (GenBank® accession no. X64139) described in [21] and kindly provided by T. Jentsch (Hamburg University, Hamburg, Germany) was supplied as a 2958 bp EcoRI–EcoRI insert in pBluescript SK(+) vector. The ClC-2 encoding sequence was PCR-amplified from the original plasmid using primers: 5′-CGCGGATCCATGGCGGCGGCAACGGCC-3′ and 5′-TGCTCTAGAGAATTCCTGGCACTTGTCATCACTATC-3′. By this manipulation, the original translation STOP codon was removed, while the amino acid sequence remained unchanged, and convenient restriction sites were introduced. The resulting approx. 2.7 kb product was BamHI–EcoRI-cloned into the centromeric plasmid pUG35 (kindly provided by J. H. Hegemann, Heinrich-Heine-Universität, Düsseldorf, Germany) bearing S. cerevisiae MET25 promoter and a GFP (green fluorescent protein)-encoding sequence. The resulting plasmid pKF14 contained the CLC2 (gene encoding the ClC-2 channel) gene, 3′-fused with the GFP-encoding sequence (Table 2). To optimize CLC2 expression in yeast, a 70 nt-long double-stranded DNA was synthesized: 5′-GGATCCAAAAAAATGGCTGCCGCTACTGCCGCTGCCGCTACTGTTGCTGGTGAAGGTATGGAGCCTCGAG-3′, with the coding sequence underlined and BamHI and XhoI restriction sites double underlined. The 58 bp BamHI–XhoI fragment of pKF14 was replaced with the above synthetic DNA and the resulting plasmid was named pKF28. This plasmid, bearing an optCLC2-GFP (where optCLC2 stands for optimized CLC2 gene) fusion gene, was subsequently converted into pKF31, which carried PMET25-optCLC2-TCYC1 (no GFP-encoding sequence, the original CLC2 STOP codon reintroduced).
Table 2. Plasmids used in the present study.
| Plasmid | Description*† | Source or reference | ||
|---|---|---|---|---|
| pBluescript SK(+)/b12-2 | ClC-2 cDNA | [21] | ||
| pKF14 | CEN | URA3 | PMET25-CLC2-GFP-TCYC1 | Present study |
| pKF16 | CEN | URA3 | PMET25-GFP-CLC2-TCYC1 | Present study |
| pKF19 | CEN | URA3 | PMET25-GEF1-GFP-TCYC1 | [17] |
| pKF20 | CEN | URA3 | PMET25-CLC2-TCYC1 | Present study |
| pKF28 | CEN | URA3 | PMET25-optCLC2-GFP-TCYC1 | Present study |
| pKF31 | CEN | HIS3 | PMET25-optCLC2-TCYC1 | Present study |
| pKF37 | INT | HIS3 | PMET25-optCLC2-TCYC1 | Present study |
| pKF56 | 2μ | TRP1 | PMNT1-MNT1-3×myc | Present study |
| pKF67 | CEN | URA3 | PMET25-KHA1ΔC382-GFP-TCYC1 | Present study |
| pKF68 | CEN | URA3 | PMET25-KHA1-GFP-TCYC1 | Present study |
| pKF76 | CEN | URA3 | PMET25-8×Gly-3×myc-TCYC1 | Present study |
| pKF83 | CEN | URA3 | PMET25-KHA1-8×Gly-3×myc-TCYC1 | Present study |
| pKF86 | CEN | HIS3 | PMET25-GFP-CLC2-TCYC1 | Present study |
| pRS303 | INT | HIS3 | MCS | [22] |
| pRS424 | 2μ | TRP1 | MCS | Stratagene |
| pSM3M-414 | CEN | TRP1 | PMNT1-MNT1-3×myc | S. Munro‡ |
| pUG34 | CEN | HIS3 | PMET25-GFP-MCS-TCYC1 | J. H. Hegemann§ |
| pUG35 | CEN | URA3 | PMET25-MCS-GFP-TCYC1 | J. H. Hegemann§ |
| pUG36 | CEN | URA3 | PMET25-GFP-MCS-TCYC1 | J. H. Hegemann§ |
* Abbreviations for description of plasmids: CEN, centromeric; INT, integrative; 2μ, episomal; MCS, multiple cloning site.
† All listed plasmids convey ampicillin resistance on E. coli cells.
‡ MRC Laboratory of Molecular Biology, Cambridge, U.K.
§ Heinrich-Heine-Universität, Düsseldorf, Germany.
To obtain the GFP–ClC-2 N-terminal fusion protein the EcoRI–EcoRI insert from b12-2 was cloned into pUG36 plasmid (provided by J. H. Hegemann). From this construct, the 171 nts upstream of the CLC2 ATG sequence were removed; the resulting plasmid was named pKF16. The pKF16 plasmid was subsequently converted into pKF86 by replacing the 2982 bp PvuI–PvuI fragment of pKF16 by the respective 2530 bp PvuI–PvuI fragment of pUG34 (also provided by J. H. Hegemann). An integrative plasmid, pKF37, bearing PMET25-optClC2-TCYC1, was constructed by replacing the 3050 bp PvuI–PvuI fragment of the centromeric plasmid pKF31 by the respective 2530 bp PvuI–PvuI fragment of the integrative plasmid pRS303 [22]. The pKF19 plasmid bearing the GEF1-GFP fusion gene has been described previously [17].
The plasmid pKF68 encoding the Kha1p–GFP fusion protein was constructed by PCR amplification of the KHA1 gene using the clone pSUP7B1 (isolated as a suppressor, see the Results section) as a template and primers: 5′-CGCGGATCCATGGCAAACACTGTAGGAGGAA-3′ and 5′-GATAAGCTTTTCAGACGAAAAATGGTGCACAATAAGG-3′ (BamHI and HindIII restriction sites double underlined). The resulting approx. 2.6 kb BamHI–HindIII fragment was cloned into pUG35 (see above). The plasmid pKF67 bearing the KHA1ΔC382-GFP fusion was prepared by PCR amplification of the 5′-terminal part of the KHA1 gene using primers: 5′-CGCGGATCCATGGCAAACACTGTAGGAGGAA-3′ and 5′-GATAAGCTTGATCGCTTCTGTAGTGTTTATCACC-3′. The resulting fragment was also cloned into pUG35.
The sequence encoding Myc-tagged Kha1p (pKF83) was obtained in a multistep procedure. Initially, the GFP gene was removed from pUG36 by XbaI digestion and religation of the vector. To the resulting plasmid, double-stranded synthetic linker 5′-GAATTCAAGCTTGGTGGTGGAGGTGGTGGAGGTGGTATCGATAGATCTGACGTCGACTAATGACTCGAG-3′ was introduced using the EcoRI–XhoI restriction sites (double underlined). This sequence contained eight yeast-optimized glycine codons (underlined). In the next step, a second synthetic double-stranded DNA sequence 5′-ATCGATATGTCTGAACAAAAGTTGATTTCTGAAGAAGACTTGGGTGAACAAAAGTTGATTTCTGAAGAAGACTTGGGTGAACAAAAGTTGATTTCTGAAGAAGACTTGAGATCT-3′ (restriction sites double underlined), which encoded triple c-Myc epitope (EQKLISEEDL)3, was ClaI–BglII-cloned into the above plasmid, yielding pKF76 (for plasmid map, see Supplementary Figure 1 at http://www.BiochemJ.org/bj/390/bj390ppppadd.htm). The c-Myc encoding sequence was codon-optimized for S. cerevisiae and the deduced mRNA secondary structures and restriction sites were removed. Finally, the KHA1 gene (without the STOP codon) was BamHI–HindIII-subcloned from pKF68 into pKF76. The resulting plasmid, pKF83, encodes Kha1p triple Myc-tagged at the C-terminus.
The multi-copy plasmid pKF56 encoding triple Myc-tagged Mnt1p (α-1,2-mannosyltransferase) was constructed from the original pSM3M-414, kindly provided by S. Munro (MRC Laboratory of Molecular Biology, Cambridge, U.K.), by subcloning the approx. 1.6 kb SacI–XhoI fragment encoding Myc-tagged Mnt1p into pRS424 (Stratagene).
Suppressor isolation
The KFY37 strain, in which the original GEF1 gene was deleted and the PMET25-optCLC2-TCYC1 fusion was integrated into the HIS3 locus, exhibited phenotypes identical with those of the Δgef1 mutant. This strain was transformed with a pFL44L-based multicopy yeast genomic library [23] by high-fidelity one-step lithium acetate transformation [R. Agatep, R. D. Kirkpatrick, D. L. Parchaliuk, R. A. Woods and R. D. Gietz (1998) Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol (LiAc/ss-DNA/PEG) protocol. Technical Tips Online (http://tto.trends.com)]. After transformation cells were incubated for 4 h in SD-ura (SD medium without uracil) liquid medium and then plated. Transformants were selected for uracil prototrophy on SD medium supplemented with 2% (w/v) galactose as a carbon source and 1 mM ferrozine [3-(2-pyridyl)-5,6-bis(phenyl-sulphonic acid)-1,2,4-triazine; Sigma], an iron chelating agent. Plates were incubated for up to 7 days at 30 °C and well-growing colonies were collected and subcloned. Plasmid DNA was recovered from yeast [25] and used to transform E. coli. Plasmids that after reintroduction into KFY37 complemented the Δgef1 phenotypes were analysed further by DNA subcloning and sequencing.
Immunofluorescence and Western blotting
GFP fusion proteins were visualized in live cells or in cells fixed with 4% (v/v) formaldehyde adjusted to pH 7.5 with 0.1 M phosphate buffer. For indirect immunofluorescence, cells were prepared as described in [19] and Myc-tagged proteins were detected using anti-Myc monoclonal antibodies (9E10; Invitrogen). The ClC-2 protein was detected using polyclonal antibodies raised against a peptide corresponding to amino acids 847–862 (AIEGSVTAQGVKVRPP) of the C-terminal region of rat ClC-2, as described previously [9]. Secondary antibodies for indirect immunofluorescence were CY3-conjugated (Jackson Immunoresearch Laboratories, West Grove, PA, U.S.A.). For fluorescence, a Nikon Eclipse E800 fluorescence microscope with a 63× objective was used. Details of Kha1p–GFP and Kha1pΔC382 localization were analysed in a Zeiss Axiovert 100 M confocal microscope using yeast cells fixed in 4% formaldehyde.
For Western-blot analysis, total yeast proteins were prepared using the urea/SDS method [26], subjected to SDS/PAGE followed by blotting on to Hybond-C extra (Amersham Biosciences, Sarclay, France) and probed with polyclonal anti-GFP antibodies (Living Colors, BD Biosciences Clontech, Palo Alto, CA, U.S.A.).
RESULTS
The rat ClC-2 protein expressed in yeast has cellular localization similar to its yeast homologue Gef1p
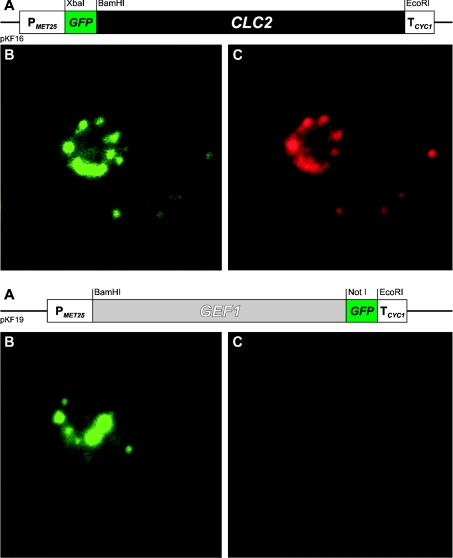
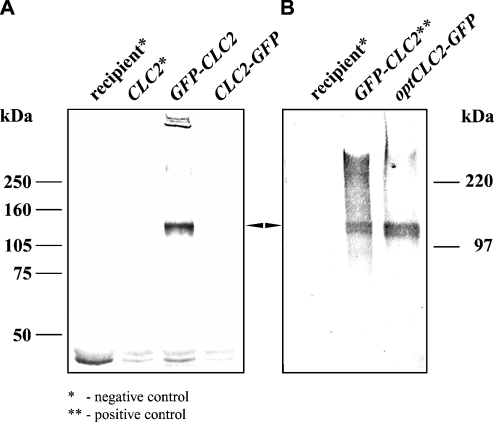
Mammalian CLC proteins have been detected in various cellular compartments (for a review see [2]). The unique S. cerevisiae CLC homologue Gef1p has probably only one localization, namely the medial Golgi compartment [16]. The GFP–ClC-2 fusion protein was expressed in yeast cells from the centromeric plasmid pKF16 and the localization of the fusion protein was examined under the fluorescence microscope. The localization based on GFP fluorescence was confirmed by detection with anti-rat ClC-2 polyclonal antibodies. Figure 1 shows the localization in yeast cells of GFP–ClC-2 (upper panel) and for comparison, that of the yeast homologue, Gef1p, tagged with GFP (lower panel). The yeast-expressed ClC-2 is predominantly intracellular and its localization pattern resembles that for Gef1p. The GFP–ClC-2 fusion protein was synthesized intact, as evidenced by the size of the corresponding band (∼126 kDa) in a Western blot developed with anti-GFP antibodies (Figure 2A).
Figure 1. Localization of the Gef1 and ClC-2 chloride channels in S. cerevisiae cells.
Genes encoding GFP-fused chloride channels were expressed in the Δgef1 strain (RGY84). Upper panel: N-terminally GFP-tagged ClC-2 channel; lower panel: C-terminally GFP-tagged yeast chloride channel Gef1p. (A) Schematic representation of the respective chimaeric genes. (B) GFP fluorescence. (C) Iodocarbocyanine (CY3) fluorescence. Yeast cells were labelled with primary rabbit anti-ClC-2 antibodies and secondary CY3-conjugated, goat anti-rabbit antibodies.
Figure 2. The effect of CLC2 gene optimization on protein expression.
Western-blot analysis of yeast expressing N- and C-terminally GFP-tagged ClC-2. Whole cell lysates were prepared using the urea/SDS method [26], analysed by SDS/PAGE (10% polyacrylamide), transferred on to Hybond-C extra and probed with anti-GFP antibodies; equal numbers of cells were analysed. Arrow indicates the expected band of 126 kDa. (A) Yeast Δgef1 strain (RGY84) was transformed with plasmid bearing the CLC2 gene that was 5′- or 3′-terminally fused to the GFP-coding sequence (pKF16 and pKF14 respectively). Only N-terminally GFP-tagged ClC-2 was detected. The recipient strain and a strain transformed with untagged version of CLC2 (pKF20) served as controls. (B) Product of the optCLC2-GFP gene (pKF28) was detected by anti-GFP antibodies. The same recipient strain as in (A) was transformed. As a positive control the strain bearing N-terminally GFP-tagged version of ClC-2 was used. The positions of molecular-mass markers are shown alongside the gels.
Expression of rat ClC-2 in S. cerevisiae requires optimization of the sequence context of the AUG translation initiation codon
For functional studies of rat ClC-2, we planned to express it in yeast without the GFP marker. To check whether the original cDNA supported efficient synthesis of ClC-2 in S. cerevisiae, we investigated expression of a C-terminal ClC-2–GFP fusion from the pKF14 construct. We could observe neither GFP fluorescence by microscopy nor an anti-GFP reactive band by Western blotting. This result taken together with the efficient expression of the N-terminal GFP–ClC-2 fusion, described in the previous section, suggested impaired translation of rat ClC-2 mRNA in yeast cells. Such impairment could be due to the presence of codons rarely used by yeast in the 5′-portion of the CLC2 ORF (open reading frame). Thus we replaced the 5′-terminal 58 bp fragment of the original CLC2 ORF with a synthetic DNA fragment encoding the same amino acid sequence, but using codons optimal for yeast. Moreover, we added six adenine nucleotides upstream of the ATG start codon to provide the yeast equivalent (5′-A/TAA/CAA/CAATGTCT/C-3′, ATG underlined; [27]) of mammalian Kozak consensus sequence [28] (for details see the Experimental section). The resulting gene was called optCLC2. It was subsequently 3′-terminally fused with GFP (plasmid pKF28, Table 2). Figure 2(B) demonstrates that the optClC-2–GFP protein of a predicted molecular mass appeared in yeast cells. Since for further experiments untagged ClC-2 (without GFP) was required, the GFP-encoding fragment was removed and the original STOP codon reintroduced into the optCLC2 gene (plasmid pKF31, Table 2).
The ClC-2 protein expressed in yeast cannot directly substitute for Gef1p
Although we achieved the expression of the mammalian ClC-2 channel in yeast and this protein had an intracellular localization similar to that of the Gef1 protein, the question remained whether it could functionally substitute for the original yeast protein. We have shown previously that Gef1p is associated with a chloride channel activity [17], but the role of this protein remains poorly understood [15,16]. When we introduced the optCLC2 gene into the Δgef1 yeast strain, we did not achieve complementation of any of the Δgef1 strain phenotypes. The strain KFY37, bearing the Δgef1 deletion and the optCLC2 gene integrated into the HIS3 locus, grew very slowly on low-iron media containing a respiratory carbon source, exactly in the same manner as the Δgef1 mutant. Furthermore, similar to Δgef1, KFY37 displayed increased sensitivity to Mn2+ and hygromycin B.
ClC-2 can functionally substitute for Gef1p in the presence of an increased level of the Kha1 protein
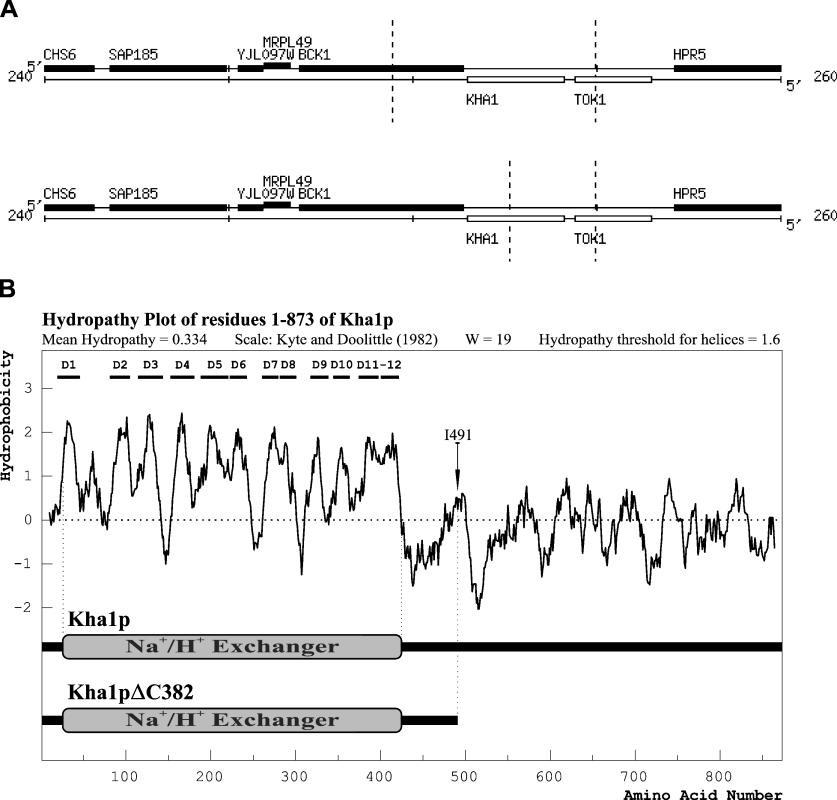
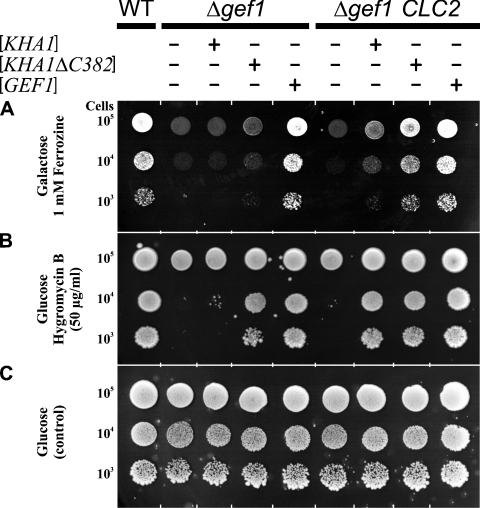
Since the presence of ClC-2 in Δgef1 mutant cells did not change the host strain phenotype, we assumed that, despite the fact that both proteins were homologous and had chloride channel activities in vitro, ClC-2 was unable to substitute for the Gef1p functions in yeast. There was, however, also a possibility that the substitution was possible, but required an additional element, interacting directly with ClC-2 or indirectly modifying its functioning. We attempted to identify such a hypothetical factor by multicopy suppressor isolation. The strain KFY37 (Δgef1 his3::PMET25-optCLC2-TCYC1) was transformed with a multicopy S. cerevisiae genomic library. We obtained approx. 650000 Ura+ transformants and ten of them complemented the defect of the KFY37 strain – the inability to grow on iron-limited minimal medium supplemented with 2% galactose. Among those ten transformants, five bore a plasmid encoding full-length GEF1, one was the gene KHA1 (YJL094c) and four were identical fragments encoding C-terminally truncated Kha1 protein (last 382 amino acids deleted, named Kha1pΔC382; Figure 3). In subsequent experiments, we confirmed that full-length KHA1 introduced into the host strain (Δgef1 bearing optCLC2) fully complemented its growth defects. The 3′-end truncated version of KHA1, encoding Kha1pΔC382, also complemented the above phenotype but additionally partially reverted the growth deficiency exhibited by the Δgef1 strain (without optCLC2). In contrast, full-length KHA1 introduced on a multicopy plasmid did not complement the Δgef1 mutation alone. Thus the ability to suppress the Δgef1 mutation by overexpression of KHA1 depends on the presence of CLC2. This dependence is partially lifted when the overexpressed Kha1p is devoid of the C-terminal 44% part of the protein. Results of all complementation tests described above are summarized in Figure 4. We conclude that an increased amount of Kha1p is a prerequisite for the functional substitution of Gef1p by the ClC-2 protein.
Figure 3. Functional co-suppressors of the Gef1p deficiency.
(A) Chromosomal location of sequences encoding the KHA1 and KHA1ΔC382 suppressors. Dashed vertical lines indicate the range of chromosomal DNA contained in suppressor plasmids. Chromosomal maps according to [51] were derived using the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org/). (B) Hydropathy profile of Kha1p deduced amino acid sequence. The Kyte–Doolittle algorithm was used with a window size of 19 amino acids. Predicted transmembrane domains are marked D1 to D12. Analysis was performed by WinPep program (Lars Hennig, University of Freiburg, Germany; http://www.biologie.uni-freiburg.de/data/schaefer/lhennig/winpep1.html). Kha1 and Kha1ΔC382 proteins are schematically presented below the plot. Area of homology to the pfam00999 domain of mammalian sodium/hydrogen exchangers is indicated according to the NCBI Conserved Domain Database [52]. Arrow indicates the position of isoleucine-491 the C-terminal amino acid of the Kha1ΔC382 protein. The N-terminal highly hydrophobic region corresponds to the exchanger domain.
Figure 4. Genetic evidence for KHA1 and KHA1ΔC382 co-suppression of Δgef1 phenotype in the presence of ClC-2.
Strains RGY84 and KFY37 are devoid of the GEF1 gene, but KFY37 additionally contains the yeast optCLC2 gene integrated into the HIS3 chromosomal locus. Both strains were transformed with multicopy plasmids isolated from yeast genomic library, as indicated. As a positive control, the wild-type RGY9 strain was used. The ability to grow on iron-limited media containing respiratory carbon sources, and to complement the hygromycin B sensitivity of Δgef1 mutant, was tested. Cells were grown overnight in SD medium and diluted to identical initial concentrations. Serial 10-fold dilutions were spotted in 5 μl portions on to SD plates supplemented as indicated. Pictures were taken after 5 days of incubation at 30 °C.
Kha1p is localized to the same cellular compartment as Gef1p and ClC-2
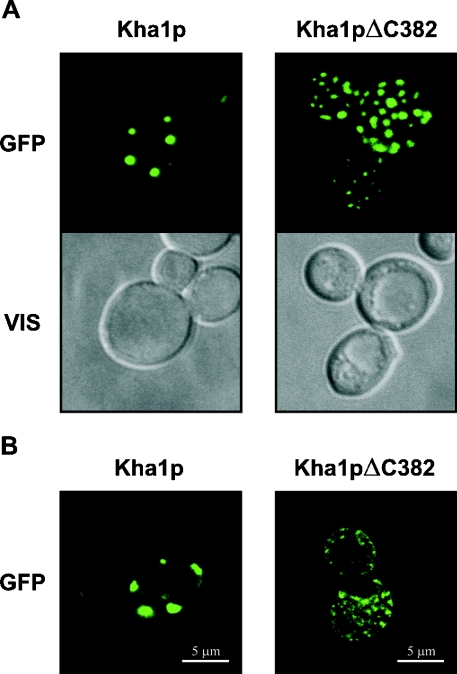
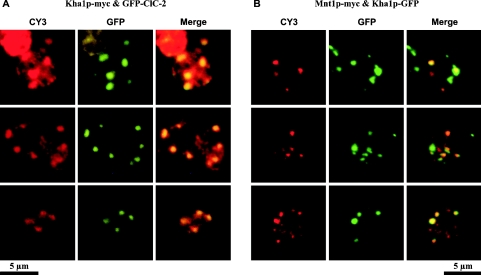
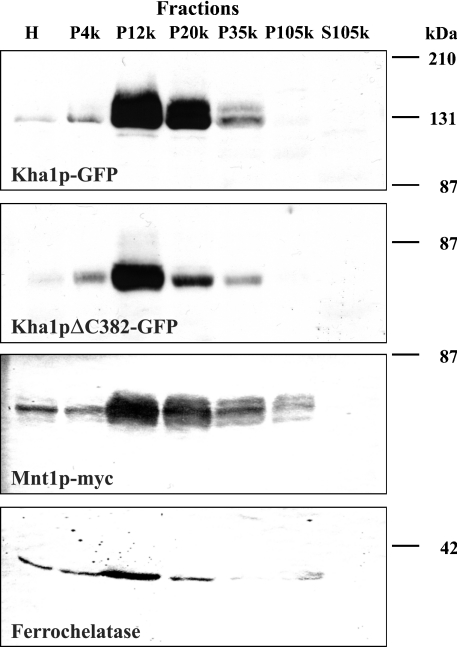
We created C-terminal GFP fusions of whole-length Kha1p and of its truncated version Kha1pΔC382. Under the fluorescence microscope (Figure 5A), we observed that Kha1p–GFP (left-hand panel) was probably intracellular and its localization resembled the localization of Gef1p or yeast-expressed rat ClC-2. C-terminally truncated Kha1p (Kha1pΔC382; right-hand panel) showed a more dispersed localization than the full-length protein. In order to confirm that the Kha1 fusion protein is indeed intracellular and not located in the cell membrane, confocal microscopy was performed. Results presented in Figure 5(B) confirmed that Kha1pΔC382 was indeed localized inside the cell, whereas the intact Kha1p localized to vesicles attached to peripheral regions of the cell, close to the plasma membrane. We also performed a co-localization study of Kha1p and ClC-2. Results presented in Figure 6(A) show that in the majority of spots Myc-tagged Kha1p co-localizes with GFP-fused ClC-2 protein. On the other hand, the Kha1p–GFP fusion has the same localization as Mnt1p, a S. cerevisiae Golgi apparatus marker protein [29] (Figure 6B). Thus we conclude that both Kha1p and ClC-2 are localized in the same compartment, i.e. Golgi vesicles. To confirm this conclusion, we examined the distribution of the Kha1p–GFP and Kha1pΔC382–GFP fusion proteins in various subcellular fractions. Figure 7 shows the results of subcellular fractionations of two strains: the RGY84 (Δgef1) strain was transformed with the pKF31 (optCLC2) and pKF56 (MNT1-myc) plasmids and additionally with a plasmid encoding one of the C-terminally GFP-tagged Kha1p variants, full-length Kha1p (pKF68) or Kha1pΔC382 (pKF67). Cells were homogenized and fractioned by differential centrifugation and the Kha1p–GFP variants were detected in the resulting subcellular fractions by Western blotting. The possibility of cross-contamination between fractions was controlled by immunostaining against marker proteins (Mnt1p, a Golgi apparatus marker; ferrochelatase, a mitochondrial inner membrane-associated protein [30]). The full-length Kha1 protein and its C-terminally truncated derivative were predominantly present in the fractions sedimenting at 12000 and 20000 g, and were also detected in the fraction sedimenting at 35000 g. A similar distribution was observed for the Mnt1 protein, but this Golgi apparatus marker was found additionally in the sediment between 35000 and 105000 g. The above results confirmed the co-localization of Kha1p and Mnt1p observed in the fluorescence microscope.
Figure 5. Cellular localization of Kha1 and Kha1ΔC382 proteins.
The Δgef1 strain (RGY84) was transformed with a plasmid bearing either KHA1 or KHA1ΔC382 3′-terminal GFP-fusion under the control of the MET25 promoter (pKF68 or pKF67 respectively). Yeast cells were grown in SD minimal medium without methionine, a repressor of PMET25. The fusion proteins were detected in the cells by monitoring GFP fluorescence. (A) Cells viewed under standard fluorescence microscopy. GFP, fluorescence of GFP fusion proteins; VIS, cells observed in Nomarski optics. (B) Fluorescence of GFP fusion proteins in fixed cells viewed under confocal microscopy (image of a 0.2 μm thick optical section).
Figure 6. Kha1 and ClC-2 proteins co-localize in the Golgi compartment.
(A) Co-localization of Kha1 and ClC-2 proteins. Both proteins were expressed in the Δgef1 strain (RGY84) transformed with pKF83 and pKF86 centromeric plasmids. Cells grown in SD medium were fixed with formaldehyde and spheroplasted. C-terminally Myc-tagged Kha1p was immunodetected using primary anti-Myc and secondary CY3-conjugated antibodies. GFP-tagged ClC-2 was detected directly by GFP fluorescence. Three representative fields are presented. Fluorescence images for CY3 (left) and GFP (middle) were merged (right). Yellow to orange colour in the merged pictures indicates co-localization. (B) Co-localization of Kha1p with Mnt1p, a Golgi apparatus marker. RGY84 strain was transformed with pKF68 and pKF56 plasmids. C-terminally Myc-tagged Mnt1p was detected in fixed cells by immunofluorescence using anti-Myc primary and CY3-conjugated secondary antibodies (CY3). Kha1p–GFP localization was followed by GFP fluorescence (GFP). Yellow to orange colour in the merged pictures (right) indicates co-localization of Kha1p and Mnt1p.
Figure 7. Localization of Kha1–GFP and Kha1ΔC382–GFP fusion proteins in subcellular fractions.
The Δgef1 strain (RGY84) was transformed with pKF31 (optCLC2) and pKF56 (MNT1-3 × myc) plasmids and additionally with a plasmid encoding full-length Kha1p (pKF68) or Kha1pΔC382 (pKF67). Cells were grown in SD medium without methionine. Subcellular fractions pelleted at 4000, 12000, 20000, 35000, 105000 g and the 105000 g supernatant were subjected to SDS/PAGE (10% polyacrylamide) and further analysed by immunoblotting. Equal amounts of protein of each fraction were loaded as indicated (H, homogenate; P, Pellet; and S, supernatant). The Kha1p variants were detected using anti-GFP antibodies. The same samples were additionally probed with anti-S. cerevisiae ferrochelatase antibodies and antibodies specific to the c-Myc epitope of Mnt1p–Myc. The positions of molecular-mass markers are shown on the right.
The KHA1 and GEF1 genes do not exhibit genetic interactions
Since the Kha1 protein was located in the same cellular compartment as Gef1p and an increased amount of Kha1p was necessary for ClC-2 to substitute for Gef1p function, we assumed that Kha1p and Gef1p could be functionally linked. To investigate these putative interactions, the double deletion mutant Δgef1 Δkha1 was constructed (strain KFYT1-15A; Table 1). We compared the growth of the Δgef1 Δkha1 double mutant and the parental Δgef1 (RGY84) and Δkha1 (KFYT2-2B) strains on SD plates supplemented with various carbon sources used instead of glucose (2% galactose, 2% raffinose, 2% melibiose and 2% glycerol) and tested for sensitivity to Mn2+ and hygromycin B. Additionally, the response to Na+ and K+ ions was checked using NaCl and KCl (0.5 and 1 M), as well as sodium and potassium acetate (1 and 2%). The usage of chloride and acetate allowed us to distinguish the sensitivity to a particular ion from a defect in pH tolerance (the acetate medium has a pH of approx. 6). We did not find a phenotype characteristic solely for the Δgef1 Δkha1 double mutant. The double mutant exhibited only the phenotypes of Δgef1: sensitivity to Mn2+ and hygromycin B and inability to grow on galactose and glycerol in the presence of low-iron concentration (results not shown). The above observations indicate that there are no genetic interactions between GEF1 and KHA1.
DISCUSSION
Numerous proteins defective in human heritable diseases show some amino acid sequence similarity to yeast proteins [31–34]. This similarity justifies further genomic study of S. cerevisiae to help identify additional genes involved in human diseases. The expression of membrane proteins in a foreign host cell is usually not as efficient as that of soluble proteins as both the translocation to the endoplasmic reticulum membrane and the transport to their final destination are more complex than the synthesis of soluble proteins in the cytosol ([35], for a review see [36]). Nevertheless, successful examples indicate that the yeast heterologous expression system can be useful in assessing precise functions of foreign, poorly characterized proteins [37–41].
Heterologous expression of chloride channels is a popular method of their investigation. Some chloride channels have already been expressed in yeast. It has been found that OmClC-3 and OmClC-5, intracellular CLC channels of the tilapia Oreochromis mosambicus, can functionally replace Gef1p [42]. Also ClC-6, an intracellular channel of mouse, can complement the Δgef1 defect of inability to grow on non-fermentable carbon sources (a short mention in [43]).
The ClC-2 channel has never been expressed in yeast, but it was expressed in Xenopus oocytes and ClC-2 associated currents were recorded [44]. We showed earlier that Gef1p has a chloride channel activity [17], thus ClC-2 should potentially substitute for it. However, despite its proper (similar to Gef1p) localization in the yeast cell, the ClC-2 protein did not substitute for Gef1p and the Δgef1 strain expressing ClC-2 retained all Δgef1-associated phenotypes. This suggested that the cellular environment in S. cerevisiae was not appropriate for ClC-2 functioning.
Literature data show that ClC-2 is activated by membrane hyperpolarization, extracellular acidification or stressors such as hypo-osmotic shock [21,44]. It is generally assumed for membrane proteins, which locate both in the plasma membrane and in intracellular membranes, that their orientation is such that the same part of the protein always faces the cytosol. In other words, from a membrane protein's ‘point of view’, the extracellular space corresponds to the lumen of cytoplasmic vesicles. We assume that in the Δgef1 yeast, the intravesicular pH is not sufficiently low to open the ClC-2 channel located in the vesicle's membrane. Since the Kha1 protein co-localizes in yeast with the ClC-2 channel, it is reasonable to assume that Kha1p may modify ClC-2 activity. Overexpression of Kha1p in the Δgef1 cells expressing ClC-2 could cause a decrease of the pH of the compartment, shared by Kha1p and ClC-2, by the electroneutral exchange of a univalent metal cation (most likely Na+ or K+), transported by Kha1p for a proton, to a level which would activate ClC-2, thus enabling its substitution for Gef1p function. Several pieces of evidence support this reasoning.
Initial characterization of the protein encoded by KHA1 has indicated that it is a putative K+/H+ antiporter [45] with strong amino acid sequence similarity to the mammalian family of NHEs (Na+/H+ exchangers) [46]. A recent study by Maresova and Sychrova [47] is in agreement with our observation that Kha1p is localized intracellularly. These authors suggest that Kha1p does not mediate potassium efflux from cells, as it was suggested before [45], but may be important for the regulation of intracellular cation homoeostasis and pH control. Since no biochemical or electrophysiological data on the Kha1 protein are available, we can only speculate on the mechanism of co-suppression described in the present study. However, the high amino acid sequence similarity of this protein to the sodium/hydrogen exchanger family suggests that it fulfils the same role as other Na+/H+ antiporters, which are key transporters in maintaining the pH of actively metabolizing cells. The shortened KHA1 version serving as a Δgef1 CLC2-dependent co-suppressor encodes a complete N-terminal region, which includes the whole putative Na+/H+ exchanger domain. This truncated version is more efficient as a co-suppressor than full-length KHA1, when checked in growth tests. Both co-suppressors require the presence of the ClC-2 channel, but it has to be noticed that the truncated variant, Kha1pΔC382, can almost entirely overcome the hygromycin B sensitivity of the Δgef1 strain without the presence of ClC-2. This indicates that Kha1p action may at least partially overlap some aspects of Gef1p functioning in yeast.
Since we noticed that under the fluorescence microscope Kha1pΔC382 was represented in the cell in more numerous spots (vesicles?) than intact Kha1p, this observation initially suggested that the mis-localization could be responsible for the higher suppressor ability of the truncated version. However, it was also possible that different locations of the full-length and truncated Kha1 proteins may reflect a dispersion of vesicles tagged with the full-length protein. The fractionation experiment confirmed that although Kha1pΔC382 was slightly less abundant in the subcellular fraction sedimenting between 12000 and 20000 g (probably containing bulkier vesicles compared with the subsequent fractions, in agreement with the microscopic data), its main proportion was in the same organellar fraction as full-length Kha1p. Thus the mis-localization could rather be excluded and another explanation was needed. Recently, Lacroix et al. [48] have shown that the NHE-1 exchanger exists in two forms: a low-affinity and a high-affinity one. Upon intracellular acidification, the low-affinity form of NHE-1 is converted into a form possessing a higher affinity for intracellular protons. They show that deletion of the C-terminal part of NHE-1 stabilizes the transporter in the low-affinity conformation. This has led the authors to propose that the conformational change of this antiporter is caused by binding of some regulatory factors to the C-terminal tail of NHE-1, as has been reported by Wakabayashi et al. [49]. We propose that, conversely, the C-terminally truncated Kha1p remains in a constitutively active state. Such a phenomenon was observed for Nha1p, a yeast plasma membrane univalent cation/proton antiporter, in which a partial truncation of the C-terminus improved the tolerance of cells to alkali metal cations compared with cells expressing the complete Nha1 protein [50]. Thus constitutive activation could be responsible for the higher suppression ability of Kha1pΔC382, but the molecular basis of its interaction with ClC-2 remains unclear.
The ClC-2 dependent co-suppression of the Δgef1 phenotype exerted by overproduction of full-length Kha1 protein seems even more complex. It is likely that excess of Kha1p overcomes the regulatory mechanisms, which control its activity, for example by changing the ratio of Kha1p to its putative negative regulator. In consequence, the full-length Kha1p may reach a permanently active state analogous to that of Kha1pΔC382. However, the lack of the C-terminus would cause activation of the whole pool of Kha1pΔC382, while the ‘over-saturation mechanism’ of Kha1p would be less efficient and only a fraction of Kha1p would become activated. This could explain the observed differences in suppression ability between the complete and truncated versions of Kha1p.
Our results also convey a more general message. Even if a foreign gene heterologously expressed in yeast has no phenotypic effects by itself, it may be possible to find co-suppressors, which aid the functioning of the foreign protein in yeast cells.
Online data
Acknowledgments
We thank J. Fritsch (INSERM U. 467, Faculté de Médecine Necker-Enfants Malades) and J. Fronk (Department of Biology, Warsaw University) for helpful comments on the manuscript. We also thank G. Fink and R. Gaxiola (Whitehead Institute for Biomedical Research, Massachusetts Institute of Technology, Cambridge, MA, U.S.A.), J. H. Hegemann, T. Jentsch and S. Munro for strains and plasmids. This work was supported by grant no. 3P05A 069 23 from the State Committee for Scientific Research, Poland. A.H. was supported by the French association ‘Vaincre la Mucoviscidose’.
References
- 1.Zdebik A. A., Cuffe J. E., Bertog M., Korbmacher C., Jentsch T. J. Additional disruption of the ClC-2 Cl(–) channel does not exacerbate the cystic fibrosis phenotype of cystic fibrosis transmembrane conductance regulator mouse models. J. Biol. Chem. 2004;279:22276–22283. doi: 10.1074/jbc.M309899200. [DOI] [PubMed] [Google Scholar]
- 2.Jentsch T. J., Stein V., Weinreich F., Zdebik A. A. Molecular structure and physiological function of chloride channels. Physiol. Rev. 2002;82:503–568. doi: 10.1152/physrev.00029.2001. [DOI] [PubMed] [Google Scholar]
- 3.Nehrke K., Arreola J., Nguyen H. V., Pilato J., Richardson L., Okunade G., Baggs R., Shull G. E., Melvin J. E. Loss of hyperpolarization-activated Cl(–) current in salivary acinar cells from Clcn2 knockout mice. J. Biol. Chem. 2002;277:23604–23611. doi: 10.1074/jbc.M202900200. [DOI] [PubMed] [Google Scholar]
- 4.Strange K. Of mice and worms: novel insights into ClC-2 anion channel physiology. News Physiol. Sci. 2002;17:11–16. doi: 10.1152/physiologyonline.2002.17.1.11. [DOI] [PubMed] [Google Scholar]
- 5.Haug K., Warnstedt M., Alekov A. K., Sander T., Ramírez A., Poser B., Maljevic S., Hebeisen S., Kubisch C., Rebstock J., et al. Mutations in CLCN2 encoding a voltage-gated chloride channel are associated with idiopathic generalized epilepsies. Nat. Genet. 2003;33:527–532. doi: 10.1038/ng1121. [DOI] [PubMed] [Google Scholar]
- 6.Niemeyer M. I., Yusef Y. R., Cornejo I., Flores C. A., Sepulveda F. V., Cid L. P. Functional evaluation of human ClC-2 chloride channel mutations associated with idiopathic generalized epilepsies. Physiol. Genomics. 2004;19:74–83. doi: 10.1152/physiolgenomics.00070.2004. [DOI] [PubMed] [Google Scholar]
- 7.Murray C. B., Chu S., Zeitlin P. L. Gestational and tissue-specific regulation of ClC-2 chloride channel expression. Am. J. Physiol. 1996;271:L829–L837. doi: 10.1152/ajplung.1996.271.5.L829. [DOI] [PubMed] [Google Scholar]
- 8.Gyomorey K., Yeger H., Ackerley C., Garami E., Bear C. E. Expression of the chloride channel ClC-2 in the murine small intestine epithelium. Am. J. Physiol. Cell Physiol. 2000;279:C1787–C1794. doi: 10.1152/ajpcell.2000.279.6.C1787. [DOI] [PubMed] [Google Scholar]
- 9.Lipecka J., Bali M., Thomas A., Fanen P., Edelman A., Fritsch J. Distribution of ClC-2 chloride channel in rat and human epithelial tissues. Am. J. Physiol. Cell Physiol. 2002;282:C805–C816. doi: 10.1152/ajpcell.00291.2001. [DOI] [PubMed] [Google Scholar]
- 10.Catalan M., Cornejo I., Figueroa C. D., Niemeyer M. I., Sepulveda F. V., Cid L. P. ClC-2 in guinea pig colon: mRNA, immunolabeling, and functional evidence for surface epithelium localization. Am. J. Physiol. Gastrointest. Liver Physiol. 2002;283:G1004–G1013. doi: 10.1152/ajpgi.00158.2002. [DOI] [PubMed] [Google Scholar]
- 11.Bali M., Lipecka J., Edelman A., Fritsch J. Regulation of ClC-2 chloride channels in T84 cells by TGF-alpha. Am. J. Physiol. Cell Physiol. 2001;280:C1588–C1598. doi: 10.1152/ajpcell.2001.280.6.C1588. [DOI] [PubMed] [Google Scholar]
- 12.Dhani S. U., Mohammad-Panah R., Ahmed N., Ackerley C., Ramjeesingh M., Bear C. E. Evidence for a functional interaction between the ClC-2 chloride channel and the retrograde motor dynein complex. J. Biol. Chem. 2003;278:16262–16270. doi: 10.1074/jbc.M209828200. [DOI] [PubMed] [Google Scholar]
- 13.Raghuram V., Hormuth H., Foskett J. K. A kinase-regulated mechanism controls CFTR channel gating by disrupting bivalent PDZ domain interactions. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9620–9625. doi: 10.1073/pnas.1633250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greene J. R., Brown N. H., DiDomenico B. J., Kaplan J., Eide D. J. The GEF1 gene of Saccharomyces cerevisiae encodes an integral membrane protein; mutations in which have effects on respiration and iron-limited growth. Mol. Gen. Genet. 1993;241:542–553. doi: 10.1007/BF00279896. [DOI] [PubMed] [Google Scholar]
- 15.Gaxiola R. A., Yuan D. S., Klausner R. D., Fink G. R. The yeast CLC chloride channel functions in cation homeostasis. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4046–4050. doi: 10.1073/pnas.95.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwappach B., Stobrawa S., Hechenberger M., Steinmeyer K., Jentsch T. J. Golgi localization and functionally important domains in the NH2 and COOH terminus of the yeast CLC putative chloride channel Gef1p. J. Biol. Chem. 1998;273:15110–15118. doi: 10.1074/jbc.273.24.15110. [DOI] [PubMed] [Google Scholar]
- 17.Flis K., Bednarczyk P., Hordejuk R., Szewczyk A., Berest V., Dolowy K., Edelman A., Kurlandzka A. The Gef1 protein of Saccharomyces cerevisiae is associated with chloride channel activity. Biochem. Biophys. Res. Commun. 2002;294:1144–1150. doi: 10.1016/S0006-291X(02)00610-1. [DOI] [PubMed] [Google Scholar]
- 18.Sherman F., Fink G. R., Hicks J. B. Plainview, NY: Cold Spring Harbor Laboratory; 1986. Methods in Yeast Genetics. [Google Scholar]
- 19.Rose M., Winston F., Hieter P. A Cold Spring Harbor Laboratory Course. Plainview, NY: Cold Spring Harbor Laboratory Press; 1990. Methods in Yeast Genetics. [Google Scholar]
- 20.Sambrook J., Fritsch E. F., Maniatis T. Plainview, NY: Cold Spring Harbor Laboratory; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 21.Thiemann A., Grunder S., Pusch M., Jentsch T. J. A chloride channel widely expressed in epithelial and non-epithelial cells. Nature (London) 1992;356:57–60. doi: 10.1038/356057a0. [DOI] [PubMed] [Google Scholar]
- 22.Sikorski R. S., Hieter P. A. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stettler S., Chiannilkulchai N., Hermann-Le Denmat S., Lalo D., Lacroute F., Sentenac A., Thuriaux P. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol. Gen. Genet. 1993;239:169–176. doi: 10.1007/BF00281615. [DOI] [PubMed] [Google Scholar]
- 24. Reference deleted.
- 25.Robzyk K., Kassir Y. A simple and highly efficient procedure for rescuing autonomous plasmids from yeast. Nucleic Acids Res. 1992;20:3790. doi: 10.1093/nar/20.14.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Printen J. A., Sprague G. F. Protein–protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamilton R., Watanabe C. K., de Boer H. A. Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res. 1987;15:3581–3593. doi: 10.1093/nar/15.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hausler A., Ballou L., Ballou C. E., Robbins P. W. Yeast glycoprotein biosynthesis: MNT1 encodes an alpha-1,2-mannosyltransferase involved in O-glycosylation. Proc. Natl. Acad. Sci. U.S.A. 1992;98:6846–6850. doi: 10.1073/pnas.89.15.6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camadro J. M., Labbe P. Purification and properties of ferrochelatase from the yeast Saccharomyces cerevisiae. Evidence for a precursor form of the protein. J. Biol. Chem. 1988;263:11675–11682. [PubMed] [Google Scholar]
- 31.Bassett D. E., Jr, Boguski M. S., Hieter P. Yeast genes and human disease. Nature (London) 1996;379:589–590. doi: 10.1038/379589a0. [DOI] [PubMed] [Google Scholar]
- 32.Steinmetz L. M., Scharfe C., Deutschbauer A. M., Mokranjac D., Herman Z. S., Jones T., Chu A. M., Giaever G., Prokisch H., Oefner P. J., et al. Systematic screen for human disease genes in yeast. Nat. Genet. 2002;31:400–404. doi: 10.1038/ng929. [DOI] [PubMed] [Google Scholar]
- 33.Barrientos A. Yeast models of human mitochondrial diseases. IUBMB Life. 2003;55:83–95. doi: 10.1002/tbmb.718540876. [DOI] [PubMed] [Google Scholar]
- 34.O'Brien K. P, Westerlund I., Sonnhammer E. L. OrthoDisease: a database of human disease orthologs. Hum. Mutat. 2004;24:112–119. doi: 10.1002/humu.20068. [DOI] [PubMed] [Google Scholar]
- 35.Shertler G. F. Overproduction of membrane proteins. Curr. Opin. Struct. Biol. 1992;2:534–544. [Google Scholar]
- 36.Bill R. M. Yeast – a panacea for the structure-function analysis of membrane proteins? Curr. Genet. 2001;40:157–171. doi: 10.1007/s002940100252. [DOI] [PubMed] [Google Scholar]
- 37.Huang P., Stroffekova K., Cuppoletti J., Mahanty S. K., Scarborough G. A. Functional expression of the cystic fibrosis transmembrane conductance regulator in yeast. Biochim. Biophys. Acta. 1996;1281:80–90. doi: 10.1016/0005-2736(96)00032-6. [DOI] [PubMed] [Google Scholar]
- 38.Kapat A., Jaakola V. P., Heimo H., Liitti S., Heikinheimo P., Glumoff T., Goldman A. Production and purification of recombinant human 2C2 adrenergic receptor using Saccharomyces cerevisiae. Bioseparation. 2000;9:167–172. doi: 10.1023/a:1008150412294. [DOI] [PubMed] [Google Scholar]
- 39.Gupta S. S., Canessa C. M. Heterologous expression of a mammalian epithelial sodium channel in yeast. FEBS Lett. 2000;481:77–80. doi: 10.1016/s0014-5793(00)01977-3. [DOI] [PubMed] [Google Scholar]
- 40.Liitti S., Matikainen M. T., Scheinin M., Glumoff T., Goldman A. Immunoaffinity purification and reconstitution of human (2)-adrenergic receptor subtype C2 into phospholipid vesicles. Protein Expr. Purif. 2001;22:1–10. doi: 10.1006/prep.2001.1410. [DOI] [PubMed] [Google Scholar]
- 41.Holz C., Prinz B., Bolotina N., Sievert V., Bussow K., Simon B., Stahl U., Lang C. Establishing the yeast Saccharomyces cerevisiae as a system for expression of human proteins on a proteome-scale. J. Struct. Funct. Genomics. 2003;4:97–108. doi: 10.1023/a:1026226429429. [DOI] [PubMed] [Google Scholar]
- 42.Miyazaki H., Uchida S., Takei Y., Hirano T., Marumo F., Sasaki S. Molecular cloning of CLC chloride channels in Oreochromis mossambicus and their functional complementation of yeast CLC gene mutant. Biochem. Biophys. Res. Commun. 1999;255:175–181. doi: 10.1006/bbrc.1999.0166. [DOI] [PubMed] [Google Scholar]
- 43.Kida Y., Uchida S., Miyazaki H., Sasaki S., Marumo F. Localization of mouse CLC-6 and CLC-7 mRNA and their functional complementation of yeast CLC gene mutant. Histochem. Cell Biol. 2001;115:189–194. doi: 10.1007/s004180000245. [DOI] [PubMed] [Google Scholar]
- 44.Jentsch T. J., Friedrich T., Schriever A., Yamada H. The CLC chloride channel family. Pflügers Arch. 1999;437:783–795. doi: 10.1007/s004240050847. [DOI] [PubMed] [Google Scholar]
- 45.Ramirez J., Ramirez O., Saldana C., Coria R., Pena A. A Saccharomyces cerevisiae mutant lacking a K+/H+ exchanger. J. Bacteriol. 1998;180:5860–5865. doi: 10.1128/jb.180.22.5860-5865.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orlowski J., Grinstein S. Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflügers Arch. 2004;447:549–565. doi: 10.1007/s00424-003-1110-3. [DOI] [PubMed] [Google Scholar]
- 47.Maresova L., Sychrova H. Physiological characterization of Saccharomyces cerevisiae kha1 deletion mutants. Mol. Microbiol. 2005;55:588–600. doi: 10.1111/j.1365-2958.2004.04410.x. [DOI] [PubMed] [Google Scholar]
- 48.Lacroix J., Poet M., Maehrel C., Counillon L. A mechanism for the activation of the Na/H exchanger NHE-1 by cytoplasmic acidification and mitogens. EMBO Rep. 2004;5:91–96. doi: 10.1038/sj.embor.7400035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wakabayashi S., Shigekawa M., Pouyssegur J. Molecular physiology of vertebrate Na+/H+ exchangers. Physiol. Rev. 1997;77:51–74. doi: 10.1152/physrev.1997.77.1.51. [DOI] [PubMed] [Google Scholar]
- 50.Kinclová O., Ramos J., Potier S., Sychrova H. Functional study of the Saccharomyces cerevisiae Nha1p C-terminus. Mol. Microbiol. 2001;40:656–668. doi: 10.1046/j.1365-2958.2001.02412.x. [DOI] [PubMed] [Google Scholar]
- 51.Cherry J. M., Ball C., Weng S., Juvik G., Schmidt R., Adler C., Dunn B., Dwight S., Riles L., Mortimer R. K., et al. Genetic and physical maps of Saccharomyces cerevisiae. Nature (London) 1997;387(6632 Suppl.):67–73. [PMC free article] [PubMed] [Google Scholar]
- 52.Marchler-Bauer A., Anderson J. B., DeWeese-Scott C., Fedorova N. D., Geer L. Y, He S., Hurwitz D. I., Jackson J. D., Jacobs A. R., Lanczycki C. J., et al. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 2003;31:383–387. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.