Abstract
Many studies have shown that an amphipathic structure and a threshold of hydrophobicity of the peptidic chain are crucial for the biological function of AMPs (antimicrobial peptides). However, the factors that dictate their cell selectivity are not yet clear. In the present study, we show that the attachment of aliphatic acids with different lengths (10, 12, 14 or 16 carbon atoms) to the N-terminus of a biologically inactive cationic peptide is sufficient to endow the resulting lipopeptides with lytic activity against different cells. Mode-of-action studies were performed with model phospholipid membranes mimicking those of bacterial, mammalian and fungal cells. These include determination of the structure in solution and membranes by using CD and ATR-FTIR (attenuated total reflectance Fourier-transform infrared) spectroscopy, membrane leakage experiments and by visualizing bacterial and fungal damage via transmission electron microscopy. The results obtained reveal that: (i) the short lipopeptides (10 and 12 carbons atoms) are non-haemolytic, active towards both bacteria and fungi and monomeric in solution. (ii) The long lipopeptides (14 and 16 carbons atoms) are highly antifungal, haemolytic only at concentrations above their MIC (minimal inhibitory concentration) values and aggregate in solution. (iii) All the lipopeptides adopt a partial α-helical structure in 1% lysophosphatidylcholine and bacterial and mammalian model membranes. However, the two short lipopeptides contain a significant fraction of random coil in fungal membranes, in agreement with their reduced antifungal activity. (iv) All the lipopeptides have a membranolytic effect on all types of cells assayed. Overall, the results reveal that the length of the aliphatic chain is sufficient to control the pathogen specificity of the lipopeptides, most probably by controlling both the overall hydrophobicity and the oligomeric state of the lipopeptides in solution. Besides providing us with basic important information, these new lipopeptides are potential candidates that can target bacteria and/or fungi, especially in cases where the bacterial flora should not be harmed.
Keywords: antifungal, antimicrobial peptide, diastereomeric peptide, fatty acid, inactive peptide, lipopeptide
Abbreviations: AMP, antimicrobial peptide; ATR-FTIR, attenuated total reflectance Fourier-transform infrared; CFU, colony-forming units; DA, decanoic acid; DDA, dodecanoic acid; diS-C3-5, 3-3′-dipropylthiadicarbocyanine-iodide; DMF, dimethylformamide; Fmoc, fluoren-9-ylmethoxycarbonyl; hRBC, human red blood cell; LB, Luria–Bertani; LPC, lysophosphatidylcholine; MA, myristic acid; MIC, minimal inhibitory concentration; PA, palmitic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; RP, reverse phase; SUV, small unilamellar vesicle; TFA, trifluoroacetic acid
INTRODUCTION
During the last two decades, the vast repertoire of a novel type of natural AMPs (antimicrobial peptides) has been identified, sequenced and studied both functionally and structurally [1–4]. The target of many of these peptides is most probably the cytoplasmatic membrane [5–7]. AMPs are employed as a protective device by most of the known organisms, from prokaryotes, such as bacteria, to arthropods, vertebrates and mammals, including humans. They serve as a non-specific defensive mechanism, which complements the highly specific, and relatively slow, adaptive immune system. Shortly after microbial infection, the AMPs are released and rapidly mobilized to neutralize a broad range of microorganisms. The growing resistance of both bacteria and fungi to existing drugs has made AMPs attractive potential drug candidates with a new mode of action.
So far, hundreds of different AMPs from different sources have been isolated and many have been designed de novo [8–15]. These peptides vary considerably in their length, sequence and structure. Most of them, however, share two common features: their net charge is positive and they are moderately hydrophobic and typically membrane-active. Moreover, when in contact with membranes, most of these peptides adopt an amphipathic arrangement that contains α-helical, β-sheet, or both structures, with hydrophobic and positively charged faces at opposite directions. AMPs can be classified according to their spectrum of activity into two general subgroups: (i) peptides that selectively act against microbial cells and (ii) non-cell-selective peptides that are toxic both to microorganisms and to normal mammalian cells. Since fungi and mammalian cells are both eukaryotic, peptides that are active on the membrane of one cell type usually have the potential to damage the membrane of the other as well [7].
Many studies have shown that the amphipathic structure is a key element in promoting the cytolytic action, rather than precisely defined structures such as an α-helix or a β-sheet [5,13,16,17]. Other studies showed that a threshold of hydrophobicity and a net positive charge are sufficient to endow a peptide with antimicrobial activity [15]. It has also been shown that substitutions of amino acids that increase the hydrophobicity of cationic AMPs result in increased antifungal activity [18–20]. Usually such changes in the peptide sequence cause undesired changes in other activity-related aspects of the peptides, such as the secondary structure and amphipathicity. Previous studies have shown that the attachment of aliphatic acids to AMPs can increase [21] their antibacterial activity or endow them with antifungal activity [22]. It has also been shown recently that the attachment of a palmitoyl group, but not shorter fatty acids, to a membrane-inactive cationic peptide makes it membrane-active [23]. However, cell specificity was determined by the sequence of the peptidic chain.
Collectively, these studies revealed that the factors that determine antimicrobial potency and the ability to target specific cells of AMPs have not yet been elucidated. To shed light on these factors, we used a biologically inactive cationic peptide containing both D- and L-amino acids (diastereomer), and attached aliphatic acids with different lengths to its N-terminal. These include: DA (decanoic acid, 10 carbons), DDA (dodecanoic acid, 12 carbons), MA (myristic acid, 14 carbons) and PA (palmitic acid, 16 carbons). This enabled us to increase the hydrophobicity of the resulting lipopeptides without altering the properties of their peptidic chain. The activity of the lipopeptides was examined against different species of pathogenic fungi and bacteria and against hRBCs (human red blood cells). Their structure in solution and in membranes and their plausible mode of action was studied by using a variety of biophysical methods including fluorescence, CD, ATR-FTIR (attenuated total reflectance Fourier-transform infrared) spectroscopy, as well as negative staining electron microscopy. The data revealed that the length of the aliphatic chain is sufficient to control the pathogen specificity of the lipopeptides, most probably by controlling both the overall hydrophobicity and the oligomeric state of the lipopeptides in solution.
EXPERIMENTAL
Materials
Rink amide MBHA (4-methyl benzhydrylamine resin) resin and Fmoc (fluoren-9-ylmethoxycarbonyl) amino acids were obtained from Calbiochem–Novabiochem AG (Läufelfingen, Switzerland). Other reagents used for peptide synthesis included TFA (trifluoroacetic acid; Sigma), piperidine (Merck, Gibbstown, NJ, U.S.A.), N,N-di-isopropylethylamine (Sigma), N-methylmorpholine (Fluka, Rolconkoma, NY, U.S.A.), DMF (dimethylformamide, peptide synthesis grade; Bio Lab, Jerusalem, Israel) and dichloromethane (peptide synthesis grade; Bio Lab). Fatty acids, namely capric acid (n-decanoic acid), lauric acid (DDA), MA (tetradecanoic acid) and PA (hexadecanoic acid), PC (phosphatidylcholine), egg PG (phosphatidylglycerol), PE (phosphatidylethanolamine from Escherichia coli), PI (phosphatidylinositol; from bovine liver), LPC (lysophosphatidylcholine), Triton, PBS, ergosterol and cholesterol, were purchased from Sigma. diS-C3-5 (3-3′-dipropylthiadicarbocyanine-iodide) was purchased from Molecular Probes (Eugene, OR, U.S.A.). All other reagents were of analytical grade. Buffers were prepared in double-distilled water. Amphotericin B was purchased from Sigma (Rehovot, Israel). RPMI 1640 was purchased from Biological Industries (Kibbutz Beit Haemek, Israel).
Peptide synthesis, acylation and purification
Peptides were synthesized by a solid-phase method using standard Fmoc chemistry on rink amide MBHA resin. The fatty acids were attached to the N-terminus of a resin-bound peptide by the same Fmoc chemistry. Briefly, after removal of the Fmoc from the N-terminus of the peptide with a solution of 20% (v/v) piperidine in DMF, the fatty acid (7 mol, 1 M in DMF) was coupled with the resin under conditions similar to those used for the coupling of amino acids. The peptides were cleaved from the resin by 95% (v/v) TFA. Cleavage of the peptides resulted in C-terminus amidated peptides. The peptides were then purified by RP (reverse phase)-HPLC on a C4 (for lipopeptides) or C18 (for the parental peptide) Bio-Rad semi-preparative column [250 mm×10 mm; 300 Å pore size (1 Å=0.1 nm), 5 μm particle size]. The purified peptides were shown to be homogeneous (>98%) by analytical RP-HPLC. The column was eluted in 80 min, using a linear gradient of 0–80% (v/v) acetonitrile in water, both containing 0.05% TFA, at a flow rate of 1.8 ml/min. Electrospray MS was used to confirm their molecular masses.
Antifungal, antibacterial and haemolytic activities of the lipopeptides
Antifungal activities of the lipopeptides were performed according to the conditions of NCCLS (National Committee for Clinical Laboratory Standards, PA, U.S.A.) document M27-A. The lipopeptides were examined in sterile 96-well plates (Nunc F96 microtitre plates) in a final volume of 200 μl as follows: 100 μl of a suspension containing fungi at a concentration of 2×103 CFU/ml (where CFU stands for colony-forming units) in a culture medium (RPMI 1640, 0.165 M Mops, pH 7.0, with L-glutamine, without NaHCO3 medium) was added to 100 μl of RPMI 1640 medium containing the peptide in serial 2-fold dilutions. Amphotericin B served as the control. The fungi were incubated for 24 h in the case of Aspergillus fumigatus or 48–72 h for Candida albicans and Cryptococcus neoformans at 35 °C in a Binder KB115 incubator. Growth inhibition was determined by measuring the absorbance at 620 nm in a microplate autoreader El309 (BioTek Instruments, Winooski, VT, U.S.A.). Antifungal activity was expressed as MIC (minimal inhibitory concentration). The antibacterial activity of the lipopeptides was examined in sterile 96-well plates (Nunc F96 microtitre plates) in a final volume of 100 μl as follows. Aliquots (50 μl) of a suspension containing bacteria, at a concentration of 106 CFU/ml in culture medium [LB (Luria–Bertani) medium], were added to 50 μl of LB containing the lipopeptide in serial 2-fold dilutions. Gentamicin served as the control. Inhibition of growth was determined by measuring the absorbance at 492 nm with a Microplate autoreader El309 (BioTek Instruments), after an incubation of 18–20 h at 37 °C. Haemolytic activity was tested as described previously using fresh hRBCs (4%, v/v) with EDTA [23]. The release of haemoglobin after peptide addition was monitored by measuring the absorbance of the supernatant at 540 nm. Levels of haemolysis were determined relative to Triton (1%), which represents 100% haemolysis.
Preparation of liposomes
SUVs (small unilamellar vesicles) were prepared by sonication as described earlier [24]. Briefly, dry lipids were dissolved in chloroform/methanol (2:1, v/v). The solvents were then evaporated under a stream of nitrogen and the lipids were freeze-dried overnight. The lipids were resuspended in the appropriate buffer (50 mM K2SO4 and 25 mM Hepes-sulphate, pH 6.8; 7 mg/ml) with vortex-mixing, and the resulting lipid dispersions were sonicated (10–30 min) in a bath-type sonicator (G1125SP1 sonicator, Laboratory Supplies Company, Hicksville, NY, U.S.A.) until the turbidity had cleared. Vesicles were visualized using a JEOL JEM 100B electron microscope (Japan Electron Optics Laboratory, Tokyo, Japan). Three different compositions of lipid films were prepared: PC/PE/PI/ergosterol (5:2.5:2.5:1, by weight), PE/PG (7:3, w/w) and PC/cholesterol (10:1, w/w), which mimic the outer leaflet of the plasma membrane of Ca. albicans [25], E. coli [26] and hRBCs [27] respectively.
Peptide-induced membrane permeation measured by a diffusion potential assay
Membrane destabilization, which results in the collapse of transmembrane potential, was conducted on model phospholipid vesicles and detected fluorimetrically using a fluorescence dye, as described previously [28,29]. Briefly, a liposome suspension, prepared in K+ buffer (50 mM K2SO4 and 25 mM Hepes-sulphate, pH 6.8), was added to an isotonic K+-free buffer (50 mM Na2SO4 and 25 mM Hepes-sulphate, pH 6.8). Then the dye diS-C3-5 was added. The subsequent addition of valinomycin created a negative diffusion potential inside the vesicles, which resulted in quenching of its fluorescence. Peptide-induced membrane permeation resulted in an increase in fluorescence. The wavelengths of excitation and emission were 650 and 675 nm respectively. The percentage of fluorescence recovery Ft was defined by
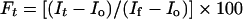 |
where It is the fluorescence observed after the addition of a peptide at time t, Io is the fluorescence after addition of valinomycin and If is the maximum fluorescence obtained by 2% (v/v) Triton.
CD spectroscopy
The CD spectra of the peptides were measured with an Aviv 202 spectropolarimeter, in a quartz optical cell with a path length of 1 mm at 25 °C. Each spectrum was recorded at wavelengths in the range of 260–190 nm (0.2 nm steps, 3 s averaging). The peptides were scanned at a concentration of 100 μM in PBS and in a membrane mimetic environment of 1% LPC in PBS. Fractional helicities were calculated as follows [30,31]:
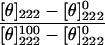 |
where [θ]222 is the experimentally observed mean residue ellipticity at 222 nm, and the values for [θ]0222 and [θ]100222, which correspond to 0 and 100% helix content respectively at 222 nm are estimated to be 2000 and 32000 deg·cm2/dmol respectively [31].
ATR-FTIR measurements
Spectra were obtained with a Bruker equinox 55 FTIR spectrometer equipped with a deuterated triacylglycerol sulphate detector and coupled with an ATR device. For each spectrum, 150 scans were collected, with a resolution of 4 cm−1. Samples were prepared as described previously [32,33]. Briefly, a mixture of lipids (0.5 mg) alone or with a peptide (∼30 μg) was deposited on a ZnSe horizontal ATR prism (80 mm×7 mm). Before sample preparations, the trifluoroacetate (CF3COO−) counterions, which strongly associate with the peptides, were replaced with chloride ions by several freeze-drying steps of the peptides in 0.1 M HCl to eliminate an absorption band near 1673 cm−1 [34]. Lipid/peptide mixtures were prepared by dissolving them together in a methanol/chloroform (1:2) mixture and spread with a Teflon bar on the ZnSe prism. The solvents were eliminated by drying under vacuum for 30 min. Spectra were recorded and the respective pure phospholipids spectra were subtracted to yield the difference spectra. The background for each spectrum was a clean ZnSe prism. Hydration of the sample was achieved by introducing excess 2H2O into a chamber placed on top of the ZnSe prism in the ATR casting, and incubating for 7 min before acquisition of the spectra. The H/2H exchange was considered complete after total shift of the amide II band. Any contribution of 2H2O vapour to the absorbance spectra near the amide I peak region was eliminated by subtracting the spectra of pure lipids equilibrated with 2H2O under the same conditions.
ATR-FTIR data analysis
Before curve fitting, a straight baseline passing through the co-ordinates at 1700 and 1600 cm−1 was subtracted. To resolve overlapping bands, the spectra were processed using PEAKFIT™ (Jandel Scientific, San Rafael, CA, U.S.A.) software. Second-derivative spectra accompanied by 13-data point Savitsky–Golay smoothing were calculated to identify the positions of the component bands in the spectra. These wave numbers were used as initial parameters for curve fitting with Gaussian component peaks. Positions, bandwidths and amplitudes of the peaks were varied until (i) the resulting bands shifted by not more than 2 cm−1 from the initial parameters, (ii) all of the peaks had reasonable half-widths (<20–25 cm−1) and (iii) good agreement between the calculated sum of all the components and the experimental spectra was achieved (r2>0.99). The relative contents of different secondary structure elements were estimated by dividing the areas of individual peaks assigned to a specific secondary structure by the whole area of the amide I band.
Examination of peptides-induced bacterial and fungal damage by TEM (transmission electron microscopy)
(i) Samples containing E. coli ATCC 25922 (1×106 CFU/ml) in LB medium were incubated with the lipopeptides at concentrations corresponding to their MIC values or 50 μM for the parental peptide, for 15 min. A drop containing the bacteria was placed on to a gold-coated grid and negatively stained with 2% (w/v) phosphotungstic acid (pH 6.8). The grids were examined using a JEOL JEM 100B electron microscope (Japan Electron Optics Laboratory). (ii) Samples containing Ca. albicans ATCC 10231 (3.5×107 CFU/ml) were incubated with or without (control) the lipopeptides dissolved in PBS at their MIC for 15 min. The fungi were fixed by incubation with 1% glutaraldehyde in PBS for 20 min. A drop containing the fungi was deposited on to a carbon-coated grid and negatively stained with 1% uranyl acetate and visualized as described above.
RESULTS
Design of peptides
The parental peptide comprises both D- and L-amino acids, and was designed to create an amphipathic α-helical structure if assumed to be in all L-form, based on Schiffer and Edmundson's wheel projection [35], but its hydrophobicity is insufficient for strong membrane binding and antimicrobial activity. Four lipopeptides were prepared by attaching fatty acids of various lengths to the N-terminus of the peptide; DA (10 carbons), DDA (12 carbons), MA (14 carbons) and PA (16 carbons). All the lipopeptides were amidated at their C-terminus and had a net +6 charge (see Table 1). There is a gradual increase in the hydrophobicity of the lipopeptides as a function of the fatty acid length.
Table 1. Characterization of the investigated lipopeptides.
| Peptide | Sequence* | Molecular mass (g/mol) | RP-HPLC retention time (min)† |
|---|---|---|---|
| [D]-L6K6 | LKKLLKKLLKKL-NH2 | 1466 | 16.3 |
| [D]-L6K6-DA | CH3(CH2)8CO-LKKLLKKLLKKL-NH2 | 1620.3 | 29.9 |
| [D]-L6K6-DDA | CH3(CH2)10CO-LKKLLKKLLKKL-NH2 | 1648.3 | 32.9 |
| [D]-L6K6-MA | CH3(CH2)12CO-LKKLLKKLLKKL-NH2 | 1676.3 | 36.1 |
| [D]-L6K6-PA | CH3(CH2)14CO-LKKLLKKLLKKL-NH2 | 1704.3 | 41.5 |
* Underlined and boldface amino acids are D-enantiomers. All the peptides are amidated at their C-terminus.
† A C4 RP analytical column was used. The peptides were eluted in 80 min, using a linear gradient of 0–80% acetonitrile in water containing 0.05% TFA.
Antifungal, antibacterial and haemolytic activities of the lipopeptides
The MIC values of the lipopeptides are shown in Tables 2 and 3 for fungi and bacteria respectively. Amphotericin B and gentamicin were used as controls for fungi, and bacteria respectively. The parental peptide is inactive toward all the cells tested. Importantly, there is a direct correlation between the length of the acyl chain attached to the peptide and its antifungal activity. In contrast, the antibacterial activity of the lipopeptides generally decreased with an increase in the fatty acid chain.
Table 2. MIC of the peptides towards fungi.
Results are the means for three independent experiments, each performed in duplicate.
| MIC (μM) | |||
|---|---|---|---|
| Peptide | Ca. albicans (ATCC 10231) | Cr. neoformans (ATCC 26430) | A. fumigatus (ATCC MY A-422) |
| [D]-L6K6 | >50 | >50 | 50 |
| [D]-L6K6-DA | 25 | 1.56 | 12.5 |
| [D]-L6K6-DDA | 3.12 | 0.78 | 6.25 |
| [D]-L6K6-MA | 1.56 | 0.78 | 1.56 |
| [D]-L6K6-PA | 1.56 | 0.78 | 1.56 |
Table 3. MIC of the peptides towards bacteria.
Results are the means for three independent experiments, each performed in duplicate.
| MIC (μM) | |||
|---|---|---|---|
| Peptide | Staph. aureus (ATCC 6538P) | Ps. aeruginosa (ATCC 27853) | E. coli (ATCC 35218) |
| [D]-L6K6 | >50 | >50 | >50 |
| [D]-L6K6-DA | 6.25 | 12.5 | 12.5 |
| [D]-L6K6-DDA | 12.5 | 25 | 25 |
| [D]-L6K6-MA | 50 | 50 | 50 |
| [D]-L6K6-PA | 50 | >50 | 50 |
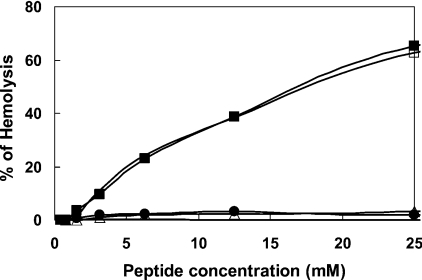
The haemolytic activity of the lipopeptides towards 4% hRBCs is shown in Figure 1. The two short DA and DDA lipopeptides were non-haemolytic, whereas the longer MA and PA lipopeptides were haemolytic, but only above their MICs. Nevertheless, they have significantly reduced haemolytic activity compared with well-known lytic peptides such as melittin, which induces 100% haemolysis already at a concentration of 1–3 μM under similar conditions.
Figure 1. Dose-dependent haemolytic activity of the peptides towards hRBCs.
The assay was performed using a final concentration of 4% red blood cells in PBS that were incubated with serial 2-fold dilutions of the different peptides for 1 h. The parental peptide [D]-L6K6 was non-haemolytic at all tested concentrations. Designations: upper curves are for [D]-L6K6-MA and [D]-L6K6-PA, and lower curves are for [D]-L6K6, [D]-L6K6-DA and [D]-L6K6-DDA.
Membrane destabilization induced by the lipopeptides
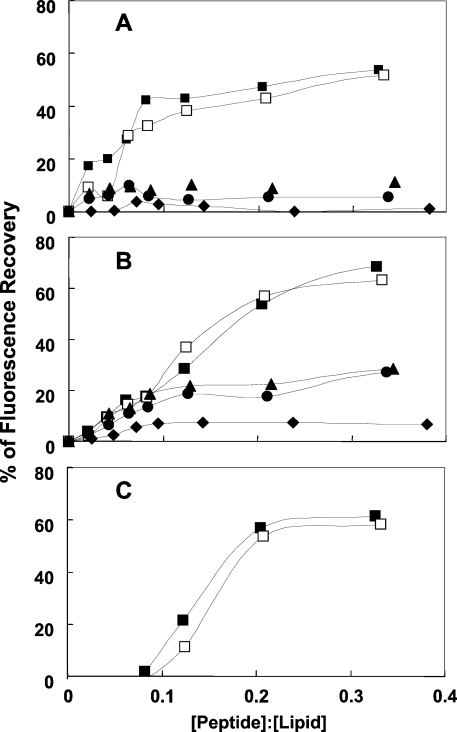
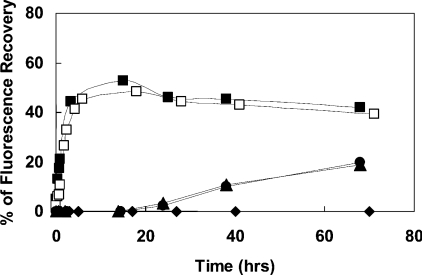
The potency of the lipopeptides to destabilize membranes, measured by using the diffusion potential assay, are shown in Figures 2(A)–2(C) for PC/cholesterol, PE/PG and PC/PE/PI/ergosterol SUVs respectively. The results can be summarized as follows: (i) in all three types of vesicles, the MA- and PA-lipopeptides were equally active and possessed a potency significantly higher than that of the two shorter lipopeptides. Their higher activity towards PC/cholesterol and PC/PE/PI/ergosterol compared with the DA and DDA lipopeptides is in agreement with their higher haemolytic and antifungal activities respectively. However, their activity towards PE/PG is in contrast with their very low antimicrobial activity. This is thought to result from the high oligomeric state of the two longer lipopeptides in aqueous solution based on their CD spectroscopy (see the following section). The enhanced oligomerization of these peptides endows them with a large volume, which makes it difficult for them to penetrate through the bacterial cell wall into the cytoplasmic membrane via a specific uptake pathway, termed self-promoted uptake [36]. Self-association of AMPs has been shown to be an important parameter, which affects their selectivity toward different cells. Studies have shown that dermaseptin [18,20], LL-37 [37,38] and model AMPs [14] that self-associate in solution have increased antifungal and haemolytic activities. Other studies have shown that cyclization or incorporation of D-amino acids into melittin abolished its ability to assemble in solution and as a result increased activity towards Gram-negative bacteria, but reduced the haemolytic activity [39]. The shorter lipopeptides, which have a lesser tendency to oligomerize, should traverse the bacterial cell wall more efficiently, and act on the phospholipid membranes. (ii) The parental peptide was only slightly active on PE/PG vesicles. (iii) The DA and DDA lipopeptides were active on PE/PG displaying a rapid kinetics, in agreement with their antibacterial activity, but they were not active on PC/cholesterol, in agreement with their inability to lyse red blood cells. (iv) In contrast with the two longer lipopeptides, the two shorter ones were active on PC/PE/PI/ergosterol SUVs only after a long incubation time (Figure 3).
Figure 2. Dose-dependent dissipation of the diffusion potential in vesicles, induced by the lipopeptides.
The lipopeptides were added to isotonic K+-free buffer containing LUVs pre-equilibrated with the fluorescent dyes diS-C3-5 and valinomycin. Fluorescence recovery was measured for 60 min after the peptides were mixed with the vesicles. Lipid concentration was 30 μM and peptide concentration was in the range 0.3–12 μM. (A) PC/cholesterol vesicles, (B) PE/PG vesicles and (C) PC/PE/PI/ergosterol vesicles. Peptide designations: ◆, [D]-L6K6; ▲, [D]-L6K6DA; ●, [D]-L6K6-DDA; □, [D]-L6K6-MA; and ■, [D]-L6K6-PA.
Figure 3. Time-dependent dissipation of the diffusion potential in PC/PE/PI/ergosterol vesicles, induced by the lipopeptides.
The lipopeptides were added to isotonic K+-free buffer containing LUVs pre-equilibrated with the fluorescent dyes diS-C3-5 and valinomycin. Fluorescence recovery was measured as a function of time after the peptides were mixed with the vesicles. Peptide designations: ◆, [D]-L6K6; ▲, [D]-L6K6-DA; ●, [D]-L6K6-DDA; □, [D]-L6K6-MA; and ■, [D]-L6K6-PA.
A final interesting point is the similar activities of [D]-L6K6-MA and [D]-L6K6-PA (Tables 1 and 2 and Figures 3 and 4). This suggests a maximal threshold of effective hydrophobicity, which in our case is the conjugation of MA. Crossing that level did not improve the potency of [D]-L6K6-PA compared with [D]-L6K6-MA in any of the assays.
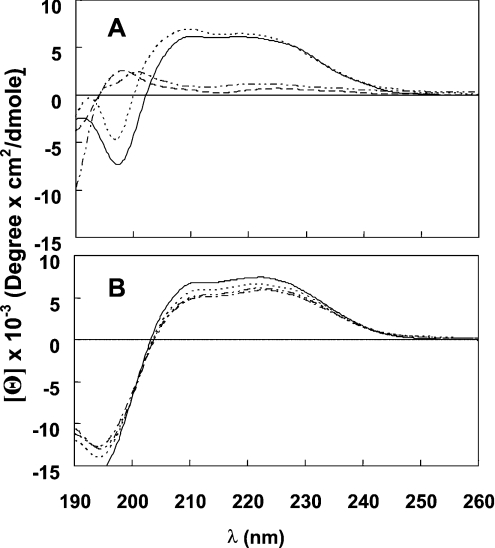
Figure 4. CD spectra of the peptides in solution and LPC.
(A) CD spectra of the lipopeptides in PBS. Spectra were measured at a peptide concentration of 100 μM. Peptide designations: [D]-L6k6-PA (———), [D]-L6k6-MA (·······), [D]-L6k6-DDA (----) and [D]-L6k6-DA (·–·–·–). (B) CD spectra of the lipopeptides in 1% LPC. Spectra were measured at a peptide concentration of 100 μM. Symbols are as in (A).
Secondary structure of the lipopeptides in micelles and solution and their ability to assemble in solution determined by CD spectroscopy
A short amphipathic peptide will not adopt a stable secondary structure in solution unless it exists as an oligomer. Figures 4(A) and 4(B) show the spectra of the lipopeptides at 100 μM in PBS and 1% LPC solutions respectively. The parental peptide displayed no structure as expected (results not shown). Note that the featured spectra are mirror images of those corresponding to all L-amino acid α-helical peptides, because these peptides are diastereomers, thus manifesting altered chirality. In LPC solution, all the lipopeptides adopted a partial α-helical structure. In PBS, however, only [D]-L6K6-MA and [D]-L6K6-PA formed a helical structure, indicating that they form oligomers in aqueous solution. The micellization of the lipophilic acid moiety probably has a pivotal role in this process.
Note that CD in proteins is a phenomenon that results when chromophores (mainly non-bonding electrons of the carbonyl oxygens) in an asymmetrical environment interact with polarized light. A diastereomer, as opposed to all-L- or all-D-amino acid peptides, is composed of both D- and L-amino acids that have opposite magnetic dipole moment and optical activity. The opposite circularly polarized absorption components of the D- and L-amino acid can decrease or even eliminate the overall absorbance of the right-handed helix. Thus the analysed results collected from the CD spectroscopy, regarding the diastereomers, should not be considered quantitatively, but rather qualitatively, as reflecting a helical secondary structure.
Secondary structure of the lipopeptides in phospholipid membranes as determined by FTIR spectroscopy
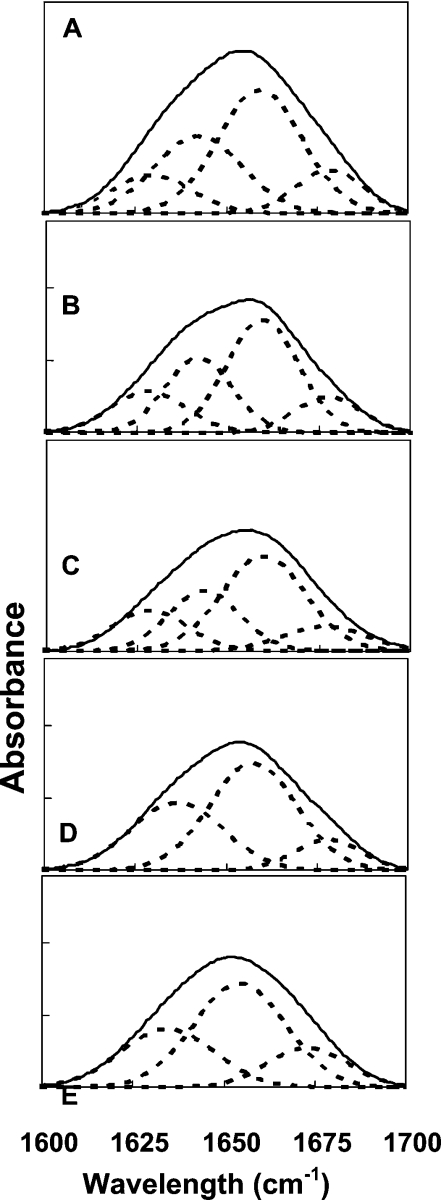
The lipopeptides were assayed on the three types of multibilayers, i.e. PC/PE/PI/ergosterol, PC/cholesterol and PE/PG, at a lipid/peptide molar ratio of 30:1 after complete deuteration as described before [38]. Assignment of the different secondary structures to the various amide I regions was calculated according to the values taken from Jackson and co-workers [40,41] and our previous results [39,42,43]. Deconvoluted amide I region spectra of the investigated peptides, when incorporated into PC/PE/PI/ergosterol, are shown as representative spectra in Figure 5 for the parental peptide and all the lipopeptides. The relative areas that were assigned to the corresponding structures for all the lipopeptides when bound to all types of lipids are summarized in Table 4.
Figure 5. Deconvolution of the amide I spectra of the fully deuterated peptides incorporated into the PC/PE/PI/ergosterol (5:2.5:2.5:1, by weight) multibilayers as detected by FTIR spectroscopy.
The component peaks are the result of curve fitting using a Gaussian line shape. The sums of the fitted components superimpose on the experimental amide I region spectra. A molar ratio of approx. 1:30 was used. ———, deconvoluted amide I region spectra; -----, fitted structural components. (A) [D]-L6K6, (B) [D]-L6K6-DA, (C) [D]-L6K6-DDA, (D) [D]-L6K6-MA and (E) [D]-L6K6-PA.
Table 4. Composition of the peptide secondary structures as determined by ATR-FTIR spectroscopy from the deconvolution of the amide I bands of the peptides incorporated into the lipid multibilayers.
A 30:1 lipid/peptide molar ratio was used. cho, cholesterol; erg, ergosterol.
| Peptide | Lipid composition | Helical structure* (%) | β-Sheet structure† (%) | Random coil (%) |
|---|---|---|---|---|
| [D]-L6K6 | PC/cho | 57 | 43 | – |
| PE/PG | 54 | 46 | – | |
| PE/PC/PI/erg | 45 | 25 | 30 | |
| [D]-L6K6-DA | PC/cho | 62 | 38 | – |
| PE/PG | 47 | 53 | – | |
| PE/PC/PI/erg | 45 | 28 | 27 | |
| [D]-L6K6-DDA | PC/cho | 70 | 30 | – |
| PE/PG | 58 | 42 | – | |
| PE/PC/PI/erg | 46 | 29 | 25 | |
| [D]-L6K6-MA | PC/cho | 64 | 36 | – |
| PE/PG | 58 | 42 | – | |
| PE/PC/PI/erg | 53 | 47 | – | |
| [D]-L6K6-PA | PC/cho | 78 | 22 | – |
| PE/PG | 65 | 35 | – | |
| PE/PC/PI/erg | 53 | 47 | – |
* The sum of both α-helical and 310-helix/dynamic helix regions.
† β-Sheet structure refers to both low-frequency β-sheet and high-frequency β-turns component regions.
The data reveal similar and mainly helical (both α-helical and 310/dynamic helix) and β-sheet (both low and high frequency) structures for all the peptides in PC/cholesterol and PE/PG multibilayers. Interestingly, however, the parental peptide, the DA and the DDA peptides adopt a significant random coil in PC/PE/PI/ergosterol multibilayers, which correlates with their reduced activity against fungi, and for the slow kinetics of these three peptides in dissipating the membrane potential of PC/PE/PI/ergosterol liposomes. Note, however, that the same peptides are similarly less active towards hRBCs and PC/cholesterol phospholipid membranes without the same random-coil component being observed. This indicates that additional factors besides the structure are important for achieving antifungal activity.
The effect of the lipopeptides on the phospholipid acyl-chain order
The orientation of the three types of phospholipid multibilayers with or without a bound peptide was determined by polarized ATR-FTIR spectroscopy. The symmetric [υsym(CH2)≈2853 cm−1] and the antisymmetric [υantisym(CH2)≈2922 cm−1] vibrations of lipid methylene C–H bonds are perpendicular to the molecular axis of a fully extended hydrocarbon chain. Thus measurements of the dichroism of IR light absorbance can reveal the order and orientation of the membrane sample relative to the prism surface. Antisymmetric and symmetric peaks indicate that the membranes are predominantly in a liquid–crystalline phase similar to biological cell membranes [44,45]. Assessing the influence of individual peptides on the orders of the acyl-chains in the multibilayers results from comparing the CH2 stretching dichroic ratios of phospholipid multibilayers alone and in a peptide-bound state. Since the intensity values of the antisymmetric CH2 vibration exceeded the value of 1, our calculations were based only on dichroic ratio (R) values taken from the symmetric vibration. The calculated values of the dichroic ratios are presented in Table 5. Contrary to the diffusion potential assay results, these results revealed a similar pronounced effect for all the peptides on each type of tested multibilayers. This supports the notion that the peptides can affect the packing of the phospholipid membranes, which is probably their major target. Once bound to the membrane, they all cause a similar disturbance to it.
Table 5. Dichroic ratios of the phospholipid multibilayers.
A 1:30 peptide/lipid molar ratio was used.
| Dichroic ratio of υsym (CH2) | |||
|---|---|---|---|
| Peptide | PC/cholesterol | PC/PE/PI/ergosterol | PE/PG |
| Phospholipid | 1.33 | 1.47 | 1.38 |
| [D]-L6K6 | 1.43 | 1.61 | 1.62 |
| [D]-L6K6-DA | 1.46 | 1.64 | 1.57 |
| [D]-L6K6-DDA | 1.47 | 1.63 | 1.59 |
| [D]-L6K6-MA | 1.44 | 1.64 | 1.57 |
| [D]-L6K6-PA | 1.46 | 1.66 | 1.62 |
Visualization of morphological changes by TEM
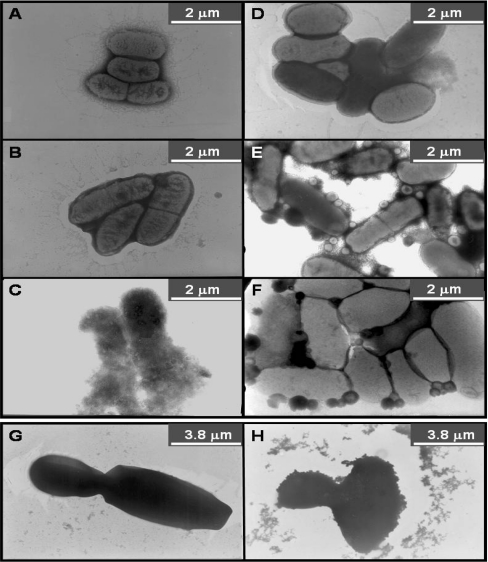
The effects of the lipopeptides on bacteria are shown in Figures 6(A)–6(F). The data shown correlate with the MICs shown in Table 3 for the short lipopeptides, and both endowed many of the cells with a ghost-like appearance. Very interestingly, the longer lipopeptides induced a unique effect on the cell, which might reflect the formation of spheroplast-like structures. These could be the result of a partial vesiculation of some of the bacterial membranes into smaller vesicles, or a complete breakdown of some of the cells, while others survive. The appearance of a number of intact bacteria could be explained by the shorter incubation time (15 min) compared with the antibacterial assay (18 h).
Figure 6. Electron micrographs of negatively stained E. coli ATCC 35218 (A–F) and Ca. albicans ATCC 10231 (G and H) before and after treatment with the lipopeptides.
(A) Untreated bacteria, (B) [D]-L6K6, (C) [D]-L6K6-DA, (D) [D]-L6K6-DDA, (E) [D]-L6K6-MA, (F) [D]-L6K6-PA, (G) untreated Ca. albicans and (H) [D]-L6K6-DDA.
All the lipopeptides at their MICs caused also a massive disruption of a large portion of the Candida cell wall after treatment for 15 min, as shown for example for [D]-L6K6-DDA (Figures 6G and 6H, for untreated and treated Candida respectively).
DISCUSSION
Many studies have shown that antimicrobial activity is reduced upon alteration of the amphipathicity of AMPs [5,46,47]. In the present study, we isolated the hydrophobicity element without changing the peptidic chain. The starting compound was a non-active short cationic diastereomeric peptide, to which fatty acids with different lengths were then attached. The main findings are: (i) the attachment of the fatty acid at the N-terminus can compensate for the hydrophobicity of the peptidic chain. (ii) The length of the fatty acid controls cell selectivity (Tables 2 and 3 and Figure 1). (iii) A specific secondary structure does not play a central role in controlling the biological activity, since all the lipopeptides were active, although with different cell specificities. (iv) When examined against fungal biomimetic membrane vesicles, [D]-L6K6-DA and [D]-L6K6-DDA displayed slow kinetics (Figure 3). (v) All the active lipopeptides cause physical damage to the bacterial and fungal cells (Figure 6).
Despite differences in the hydrophobicities of the lipopeptides, their secondary structures in the membrane are similar to the parental non-acylated peptide (Figure 4B and Table 4 respectively). An exception was found in the case of the PC/PE/PI/ergosterol multibilayers, in which [D]-L6K6, [D]-L6K6-DA and [D]-L6K6-DDA exhibited a fraction of the random coil structure. However, the effect of bound peptides on the acyl-chain order of the phospholipid multibilayers was similar and did not reveal any distinguishable effect for the random-coil structure component. This may indicate that all the lipopeptides disrupt the membrane similarly post binding. The additional random coil structural element may provide a structurally based correlation for the slow kinetics of these two lipopeptides against fungal model membranes.
CD spectra analysis of the lipopeptides in PBS solution indicated the assembly of [D]-L6K6-MA and [D]-L6K6-PA in aqueous solution, most probably as a consequence of the micellization of the long aliphatic chains. The stronger tendency of these two lipopeptides to self-associate in solution and to form oligomers may explain their microbicidal activity on cells that do not have highly negatively charged outer membranes. Indeed, with the exception of the antibacterial assay, there is a direct correlation between hydrophobicity and activity. The longer lipopeptides are more potent on the model membranes (Figure 2) as well as towards fungi (Table 2) and red blood cells (Figure 1). Note that whereas the lipopeptides with the longer aliphatic chains have low activity towards bacteria, they are highly active against PE/PG vesicles, which mimic bacterial membranes. This is probably directly connected to the inter-relationship between the oligomeric state and the composition of the cell surface. Oligomerization of the lipopeptides makes it difficult for them to traverse the bacterial cell wall and to reach and perturb the cell membrane. However, it is not clear what factors constitute the critical elements in a cell surface that interact with the peptidic oligomers to determine their potency against that cell type. Two possible factors are the polarity and density of the external layer of the cell surface. Conversely, the lipopeptides with the shorter attached fatty acids can easily traverse the bacterial cell wall as evident from their potent biological activities. It is therefore reasonable to assume that the relatively lower activity of the shorter lipopeptides towards fungi originate from their diminished ability to permeate their membranes owing to insufficient hydrophobicity-derived oligomerization, rather than difficulty to reach the membrane.
The data reveal that [D]-L6K6-DA and [D]-L6K6-DDA consistently had a weaker membrane-lytic activity towards vesicles compared with [D]-L6K6-MA and [D]-L6K6-PA. This reduced activity probably stems from the lower hydrophobicity of the shorter chains, which is associated with the less ordered structure of the shorter lipopeptides in solution. Furthermore, in the presence of fungal membranes, which are less negatively charged than those of bacteria, the less ordered structure of the two shorter lipopeptides may cause electrostatic repulsive forces owing to the positive charges of the peptides, thus amplifying the unordered structure. This in turn might reduce their activity.
In summary, the attachment of aliphatic acids to cationic peptides can be a key factor in controlling their self-assembly and in determining their oligomeric state; the shorter and less hydrophobic lipopeptides have a broad spectrum of antibacterial and fungal activities, without harming hRBCs, whereas the more hydrophobic lipopeptides are selectively active toward fungi, but also possess haemolytic activity at concentrations higher than their MICs. The overall correlation between biological activity and the ability to disrupt model membranes indicates that membrane perturbation is at least one of the targets of these lipopeptides.
Besides giving us basic important information, the potential resistance of diastereomeric peptides to proteolytic degradation and to inactivation in the serum [15] serves as another incentive for continuing research on them as potential templates for antifungal and antibacterial peptides.
Acknowledgments
This study was supported by the Israel Science Foundation. Y.S. holds the Harold S. and Harriet B. Brady Professorial Chair in Cancer Research. We thank Dr Y. Marikovsk for his assistance with the electron microscopy.
References
- 1.Boman H. G., Nilsson I., Rasmuson B. Inducible antibacterial defence system in Drosophila. Nature (London) 1972;237:232–235. doi: 10.1038/237232a0. [DOI] [PubMed] [Google Scholar]
- 2.Steiner H., Hultmark D., Engstrom A., Bennich H., Boman H. G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature (London) 1981;292:246–248. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- 3.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. U.S.A. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mor A., Nguyen V. H., Delfour A., Migliore-Samour D., Nicolas P. Isolation, amino acid sequence, and synthesis of dermaseptin, a novel antimicrobial peptide of amphibian skin. Biochemistry. 1991;30:8824–8830. doi: 10.1021/bi00100a014. [DOI] [PubMed] [Google Scholar]
- 5.Lehrer R. I., Ganz T. Antimicrobial peptides in mammalian and insect host defence. Curr. Opin. Immunol. 1999;11:23–27. doi: 10.1016/s0952-7915(99)80005-3. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann J. A., Kafatos F. C., Janeway C. A., Ezekowitz R. A. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 7.Shai Y. Mode of action of membrane active antimicrobial peptides. Biopolymers. 2002;66:236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- 8.Blondelle S. E., Takahashi E., Dinh K. T., Houghten R. A. The antimicrobial activity of hexapeptides derived from synthetic combinatorial libraries. J. Appl. Bacteriol. 1995;78:39–46. doi: 10.1111/j.1365-2672.1995.tb01671.x. [DOI] [PubMed] [Google Scholar]
- 9.Hancock R. E., Lehrer R. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 1998;16:82–88. doi: 10.1016/s0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- 10.Boman H. G. Gene-encoded peptide antibiotics and the concept of innate immunity: an update review. Scand. J. Immunol. 1998;48:15–25. doi: 10.1046/j.1365-3083.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 11.Nakajima Y., Alvarez-Bravo J., Cho J., Homma K., Kanegasaki S., Natori S. Chemotherapeutic activity of synthetic antimicrobial peptides: correlation between chemotherapeutic activity and neutrophil-activating activity. FEBS Lett. 1997;415:64–66. doi: 10.1016/s0014-5793(97)01101-0. [DOI] [PubMed] [Google Scholar]
- 12.Alvarez-Bravo J., Kurata S., Natori S. Mode of action of an antibacterial peptide, KLKLLLLLKLK-NH2. J. Biochem. (Tokyo) 1995;117:1312–1316. doi: 10.1093/oxfordjournals.jbchem.a124860. [DOI] [PubMed] [Google Scholar]
- 13.Tossi A., Sandri L., Giangaspero A. Amphipathic, α-helical antimicrobial peptides. Biopolymers. 2000;55:4–30. doi: 10.1002/1097-0282(2000)55:1<4::AID-BIP30>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 14.Avrahami D., Oren Z., Shai Y. Effect of multiple aliphatic amino acids substitutions on the structure, function, and mode of action of diastereomeric membrane active peptides. Biochemistry. 2001;40:12591–12603. doi: 10.1021/bi0105330. [DOI] [PubMed] [Google Scholar]
- 15.Papo N., Oren Z., Pag U., Sahl H. G., Shai Y. The consequence of sequence alteration of an amphipathic α-helical antimicrobial peptide and its diastereomers. J. Biol. Chem. 2002;277:33913–33921. doi: 10.1074/jbc.M204928200. [DOI] [PubMed] [Google Scholar]
- 16.Segrest J. P., De L. H., Dohlman J. G., Brouillette C. G., Anantharamaiah G. M. Amphipathic helix motif: classes and properties. Proteins. 1990;8:103–117. doi: 10.1002/prot.340080202. [DOI] [PubMed] [Google Scholar]
- 17.Oren Z., Shai Y. Mode of action of linear amphipathic α-helical antimicrobial peptides. Biopolymers. 1998;47:451–463. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 18.Strahilevitz J., Mor A., Nicolas P., Shai Y. Spectrum of antimicrobial activity and assembly of dermaseptin-b and its precursor form in phospholipid membranes. Biochemistry. 1994;33:10951–10960. doi: 10.1021/bi00202a014. [DOI] [PubMed] [Google Scholar]
- 19.Lee D. G., Kim P. I., Park Y., Woo E. R., Choi J. S., Choi C. H., Hahm K. S. Design of novel peptide analogs with potent fungicidal activity, based on PMAP-23 antimicrobial peptide isolated from porcine myeloid. Biochem. Biophys. Res. Commun. 2002;293:231–238. doi: 10.1016/S0006-291X(02)00222-X. [DOI] [PubMed] [Google Scholar]
- 20.Kustanovich I., Shalev D. E., Mikhlin M., Gaidukov L., Mor A. Structural requirements for potent versus selective cytotoxicity for antimicrobial dermaseptin S4 derivatives. J. Biol. Chem. 2002;277:16941–16951. doi: 10.1074/jbc.M111071200. [DOI] [PubMed] [Google Scholar]
- 21.Majerle A., Kidric J., Jerala R. Enhancement of antibacterial and lipopolysaccharide binding activities of a human lactoferrin peptide fragment by the addition of acyl chain. J. Antimicrob. Chemother. 2003;51:1159–1165. doi: 10.1093/jac/dkg219. [DOI] [PubMed] [Google Scholar]
- 22.Avrahami D., Shai Y. Conjugation of a magainin analogue with lipophilic acids controls hydrophobicity, solution assembly, and cell selectivity. Biochemistry. 2002;41:2254–2263. doi: 10.1021/bi011549t. [DOI] [PubMed] [Google Scholar]
- 23.Avrahami D., Shai Y. A new group of antifungal and antibacterial lipopeptides derived from non-membrane active peptides conjugated to palmitic acid. J. Biol. Chem. 2004;279:12277–12285. doi: 10.1074/jbc.M312260200. [DOI] [PubMed] [Google Scholar]
- 24.Shai Y., Bach D., Yanovsky A. Channel formation properties of synthetic pardaxin and analogues. J. Biol. Chem. 1990;265:20202–20209. [PubMed] [Google Scholar]
- 25.Schneiter R., Brugger B., Sandhoff R., Zellnig G., Leber A., Lampl M., Athenstaedt K., Hrastnik C., Eder S., Daum G., et al. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw N. Lipid composition as a guide to the classification of bacteria. Adv. Appl. Microbiol. 1974;17:63–108. doi: 10.1016/s0065-2164(08)70555-0. [DOI] [PubMed] [Google Scholar]
- 27.Verkleij A. J., Zwaal R. F., Roelofsen B., Comfurius P., Kastelijn D., Deenen L. V. The asymmetric distribution of phospholipids in the human red cell membrane. A combined study using phospholipases and freeze-etch electron microscopy. Biochim. Biophys. Acta. 1973;323:178–193. doi: 10.1016/0005-2736(73)90143-0. [DOI] [PubMed] [Google Scholar]
- 28.Sims P. J., Waggoner A. S., Wang C. H., Hoffmann J. R. Studies on the mechanism by cyanine dyes measure membrane potential in red blood cells and phosphatidylcholine vesicles. Biochemistry. 1974;13:3315–3330. doi: 10.1021/bi00713a022. [DOI] [PubMed] [Google Scholar]
- 29.Loew L. M., Rosenberg I., Bridge M., Gitler C. Diffusion potential cascade: conventional detection of transferable membrane pores. Biochemistry. 1983;22:837–844. doi: 10.1021/bi00273a020. [DOI] [PubMed] [Google Scholar]
- 30.Greenfield N., Fasman G. D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 31.Wu C. S., Ikeda K., Yang J. T. Ordered conformation of polypeptides and proteins in acidic dodecyl sulfate solution. Biochemistry. 1981;20:566–570. doi: 10.1021/bi00506a019. [DOI] [PubMed] [Google Scholar]
- 32.Gazit E., Miller I. R., Biggin P. C., Sansom M. S. P., Shai Y. Structure and orientation of the mammalian antibacterial peptide cecropin P1 within phospholipid membranes. J. Mol. Biol. 1996;258:860–870. doi: 10.1006/jmbi.1996.0293. [DOI] [PubMed] [Google Scholar]
- 33.Oren Z., Shai Y. Cyclization of a cytolytic amphipathic α-helical peptide and its diastereomer: effect on structure, interaction with model membranes, and biological function. Biochemistry. 2000;39:6103–6114. doi: 10.1021/bi992408i. [DOI] [PubMed] [Google Scholar]
- 34.Surewicz W. K., Mantsch H. H., Chapman D. Determination of protein secondary structure by Fourier transform infrared spectroscopy: a critical assessment. Biochemistry. 1993;32:389–394. doi: 10.1021/bi00053a001. [DOI] [PubMed] [Google Scholar]
- 35.Schiffer M., Edmundson A. B. Use of helical wheels to represent the structures of protein and to identify segments with helical potential. Biophys. J. 1967;7:121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawyer J. G., Martin N. L., Hancock R. E. Interaction of macrophage cationic proteins with the outer membrane of Pseudomonas aeruginosa. Infect. Immun. 1988;56:693–698. doi: 10.1128/iai.56.3.693-698.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansson J., Gudmundsson G. H., Rottenberg M. E., Berndt K. D., Agerberth B. Conformation-dependent antibacterial activity of the naturally-occurring human peptide LL-37. J. Biol. Chem. 1998;273:3718–3724. doi: 10.1074/jbc.273.6.3718. [DOI] [PubMed] [Google Scholar]
- 38.Oren Z., Lerman J. C., Gudmundsson G. H., Agerberth B., Shai Y. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell selective activity. Biochem. J. 1999;341:501–513. [PMC free article] [PubMed] [Google Scholar]
- 39.Sharon M., Oren Z., Shai Y., Anglister J. 2D-NMR and ATR-FTIR study of the structure of a cell-selective diastereomer of melittin and its orientation in phospholipids. Biochemistry. 1999;38:15305–15316. doi: 10.1021/bi991225t. [DOI] [PubMed] [Google Scholar]
- 40.Jackson M., Mantsch H. H. The use and misuse of FTIR spectroscopy in the determination of protein structure. Crit. Rev. Biochem. Mol. Biol. 1995;30:95–120. doi: 10.3109/10409239509085140. [DOI] [PubMed] [Google Scholar]
- 41.Frey S., Tamm L. K. Orientation of melittin in phospholipid bilayers. A polarized attenuated total reflection infrared study. Biophys. J. 1991;60:922–930. doi: 10.1016/S0006-3495(91)82126-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oren Z., Hong J., Shai Y. A comparative study on the structure and function of a cytolytic alpha-helical peptide and its antimicrobial β-sheet diastereomer. Eur. J. Biochem. 1999;259:360–369. doi: 10.1046/j.1432-1327.1999.00047.x. [DOI] [PubMed] [Google Scholar]
- 43.Hong J., Oren Z., Shai Y. Structure and organization of hemolytic and nonhemolytic diastereomers of antimicrobial peptides in membranes. Biochemistry. 1999;38:16963–16973. doi: 10.1021/bi991850y. [DOI] [PubMed] [Google Scholar]
- 44.Ishiguro R., Kimura N., Takahashi S. Orientation of fusion-active synthetic peptides in phospholipid bilayers: determination by Fourier transform infrared spectroscopy. Biochemistry. 1993;32:9792–9797. doi: 10.1021/bi00088a034. [DOI] [PubMed] [Google Scholar]
- 45.Cameron D. G., Casal H. L., Gudgin E. F., Mantsch H. H. The gel phase of dipalmitoyl phosphatidylcholine. An infrared characterization of the acyl chain packing. Biochim. Biophys. Acta. 1980;596:463–467. doi: 10.1016/0005-2736(80)90135-2. [DOI] [PubMed] [Google Scholar]
- 46.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 47.Hancock R. E., Diamond G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000;8:402–410. doi: 10.1016/s0966-842x(00)01823-0. [DOI] [PubMed] [Google Scholar]