Abstract
The pol and gag gene fragments of small ruminant lentivirus field isolates collected in the last decade in Italy were amplified, sequenced, and analyzed. Phylogenetic analysis revealed that the majority of ovine isolates form a distinct cluster more similar to caprine lentivirus prototypes than to the visna virus prototype. These findings confirm and extend those reported by Leroux et al. (Arch. Virol., 142:1125-1137, 1997). Moreover, we observed that a variable region of Gag, included in the fragment analyzed, corresponded to one of the three major capsid antigen epitopes, which suggests that the antibody response to this epitope may be type specific. To test this hypothesis, two recombinant peptides, derived from the Icelandic prototype K1514 and this novel genotype, were expressed and used in an enzyme-linked immunosorbent assay to screen a panel of ovine and caprine sera collected from different geographical locations in Italy. Several sera reacted in a type-specific manner, indicating that in a diagnostic setting the combination of at least these two type-specific peptides is necessary to cover a wide range of infections. Additionally, these results support the hypothesis of cross-species transmission based on the phylogenetic analysis described above. This has implications for the control and eradication of small ruminant lentivirus infections.
To date, maedi-visna virus (MVV) and caprine arthritis encephalitis virus (CAEV) are considered to be two antigenically related and genetically distinct lentiviruses of the Retroviridae family (3). Since the cross-reactivity between MVV and CAEV involves the major structural proteins (7), a number of serological tests have been proposed, based on ovine strains, to detect specific antibodies in both species. Ovine lentiviruses are usually easier to grow in tissue culture, and well-adapted strains have been extensively used to develop native-based immunoassays (9, 19). Furthermore, since sequence information was first produced from ovine strains, recombinant antigens have been largely employed and characterized from ovine isolates (12, 13, 14, 25, 33). The development of a diagnostic test capable of detecting the widest range of infection is obvious from a practical point of view. The current concept of the universality of a single-strain-based immunoassay is based on the finding that the gag-encoded capsid antigen (CA) and the env-encoded transmembrane (TM) protein are conserved among small ruminant lentiviruses (7, 21), despite the variability of the env-encoded surface antigen (1, 31). However, further characterization of the immunodominant epitopes of the major CA has shown that these epitopes are at least in part quite variable, questioning the use of single-strain-based immunoassays for diagnostic purposes. In particular, the immunodominant region of the CA involved in the cross-reaction between ovine and caprine infection was recently identified (22). Partial mapping studies suggested that at least two consecutive linear epitopes, located in the N-terminal half of the CA, and a third epitope located in the C terminus of the same protein are important to preserve a spectrum of cross-reactivity. Unfortunately, few sequences are available to date which cover the region of interest. These are from the full-length visna virus genome of the Icelandic strain K1514 (28), the British strain EV1 (26), the South African strain SAOVV (17), the partial sequence of the Dutch strain ZZV1050 (33), and the full-length genome of CAEV strain CO (24).
Although both MVV and CAEV infections are widely distributed in Italy, no information is currently available on the genetic background of the Italian field isolates. The aim of this work was to characterize small ruminant lentivirus strains isolated in Italy and to evaluate the variability of the immunodominant regions of the CA.
MATERIALS AND METHODS
Viruses and cells.
Sixteen small ruminant lentivirus isolates were used in the study. Seven were Italian ovine isolates obtained from lung explants (It-128, It-169, It-170, It-172, and It-2038) or udder tissue explants (It-585 and It-561) collected from animals showing typical maedi gross and histopathological lesions. One was an Italian ovine lentivirus (Pi1) isolated through explantation of synovial membranes from a sheep with severe arthritis. Four were Italian caprine lentiviruses (Siena-85, Pisa-88, TO-1/89, and AL-95) isolated by explantation of synovial membranes from goats with arthritis (isolates Pi1, Siena-85, and Pisa-88 were kindly provided by F. Tolari, University of Pisa). In addition to the Italian lentivirus isolates, two Spanish ovine lentiviruses (Sp3M and Sp4GM) were kindly provided by J. J. Badiola and B. Amorena (University of Zaragoza), one British ovine lentivirus (K-187) was kindly provided by M. Dawson (Central Veterinary Laboratory, Weybridge, United Kingdom), and one North American ovine lentivirus (Clay Center) was kindly provided by J. Kwang (U.S. Department of Agriculture, Clay Center, Nebr.). All of the isolates were adapted to ovine fetal lung or caprine fetal synovial membrane cells at a low number of passages until discrete cytopathic effect was detectable by Giemsa staining on replica 24-well microplates. Infected cells were further processed for DNA sample preparation.
DNA sample preparation.
Unintegrated proviral DNA was extracted from each infected monolayer grown in 25-cm2 flasks by a previously described modified Hirt method (8, 20). The final pellet was resuspended in 100 μl of distilled water containing RNase (20 μg/ml) and refrigerated until used for enzymatic amplification.
Primer design PCR and sequencing.
The published nucleotide sequences of the entire genomes of prototype MVV and CAEV strains K1514 (28), EV1 (26), SA-OMVV (17), and CAEV strain CO (24) were compared for conserved nucleotide sequences with the sequence comparison algorithm PILEUP from the University of Wisconsin genetics computing group software (5). PCR primer binding sites were selected on the basis of sequences conserved in all strains, length, and GC content, and primer pairs were selected on the basis of product length and 3′-end compatibility. This enabled eight primer sets to be selected, two each in the pol, gag, and env genes, respectively, and two in the long terminal repeats. Primers were synthesized for all primer sets, and these were tested against a batch of 15 representative field isolates from Italy and Spain, U.S. strain WLC-1, and from naturally infected sheep in Scotland. Three primer sets were selected that amplify PCR products from the widest range of strains and/or isolates (85 to 100%). These were in the pol, gag, and long terminal repeat regions of the genome. Two of these primer sets were used. The first set amplifies a 218-bp fragment of the pol gene (sense, 5′-ATAGTAAATGGCATCAAGATGC; antisense, 5′-TCCCGAATTTGTTTCTACCC). The second set amplifies a 600-bp fragment of the gag gene, covering nearly 95% of the entire CA (sense, 5′-TTCCAGCAACTGCAAACAGT; antisense, 5′-TCCTTCTGATCCTACATCTC). Three microliters of DNA sample was added to 46 μl of PCR mix containing 10 mM Tris-HCl, 50 mM KCl, 2.5 mM MgCl2, 200 μM (each) deoxynucleoside triphosphates, and 10 pmol of each primer. Following an initial denaturation step of 5 min at 95°C, 1 U of Taq polymerase (Life Technologies, Grand Island, N.Y.) (1-U/μl dilution in the appropriate buffer) was added at 80°C and PCR was conducted for 35 cycles (94°C for 1 min, 50°C for 1 min, and 72°C for 2 min, with a final extension step of 72°C for 5 min). PCR results were analyzed by 2% agarose gel electrophoresis.
The pol amplimers were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany), eluted in 10 mM Tris-HCl, pH 8.5, and directly sequenced with the PCR primers. The gag amplimers were cloned into pCR4-TOPO (Invitrogen, Groningen, The Netherlands) by using the TA cloning strategy and sequenced with the vector primers. Sequencing was carried out with fluorescent dye terminators on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems) according to the manufacturer's protocol.
Phylogenetic analysis.
Sequencing data were analyzed with Chromas 2.0 (Technelysium, Helensvale, Australia) and aligned with the consensus sequence of the Icelandic visna virus prototype strain K1514 with ClustalX (30).
Sequences obtained with the pol fragment were used to construct a phylogenetic tree by using the small ruminant lentivirus sequences available in the GenBank database as well as bovine, equine, feline, human, and simian lentiviruses. Genetic distances were computed by using MEGA (11) and were used to construct a neighbor-joining tree (23) with the Tamura-Nei two-parameter distance option (29). The statistical confidence of the tree's topology was assessed by using 100 bootstrap replicates (6). Sequences obtained with the gag fragments were analyzed with the available small ruminant lentivirus sequences as described above. The amino acid sequences derived were aligned with MEGA, and variability was evaluated in the immunodominant regions of the CA identified in a previous study (22).
Expression of recombinant subunits and enzyme-linked immunosorbent assay (ELISA) procedure.
Since conserved polypeptide sequence variability was observed in one of the three major epitopes of the CA, we generated two recombinant subunits of 29 residues, carrying two linear epitopes, one of which (underlined) shows the above mentioned variability. The first (K1514/B3) was generated from the Icelandic ovine strain K1514, whose sequence was GNRAQKELIQGKLNEEAERWVRQNPPGPN, and the second (It-128/B3) was generated from the Italian ovine isolate It-128, whose sequence was GNRAQKELIQGKLNEEAERWRRNNPPPPA. Briefly, the gene fragments were amplified by PCR with primers carrying the BamHI (sense) and EcoRI (antisense) restriction sites at the 5′ terminus. Amplimers were digested with the appropriate restriction enzymes and cloned into the pGex-2T expression vector in frame with glutathione S-transferase (GST) (Pharmacia, Uppsala, Sweden). Fusion proteins, as well as the GST carrier, were expressed in Escherichia coli BL21 and affinity purified (27). The purity and yield of recombinant proteins were estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and the Bradford method (2).
Fifty-five sheep and 64 goat serum samples, collected from different regions of Italy, were preliminarily tested and found positive by an agar gel immunodiffusion test (AGID) carried out by using the reference strain WLC-1 according to standard procedures (4). For the subunit ELISA, microplates were coated with 100 ng of K1514/B3, 100 ng of It-128/B3, and 60 ng of GST negative control. Samples were diluted 1/20 in phosphate-buffered saline, 1% yeast extract, and 0.05% Tween 20 and incubated (100 μl/well) against each antigen for 1 h at 37°C. Following four washes, 10 ng of peroxidase-labeled protein G (Pierce) was added (100 μl/well) and the plate was incubated as described above. After a final wash step, the reaction mixture was developed with 2,2′-azino-bis-(3-ethylbenzthiazoline-6-sulfonic acid) (Chemicon, Temecula, Calif.). Net ELISA absorbances were obtained for each serum sample by subtracting the absorbance against GST as an antigen from the absorbance for each of the two subunits.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been submitted to GenBank and given accession numbers AY044811-26 (pol amplimers) and AY044803-10 (gag amplimers).
RESULTS
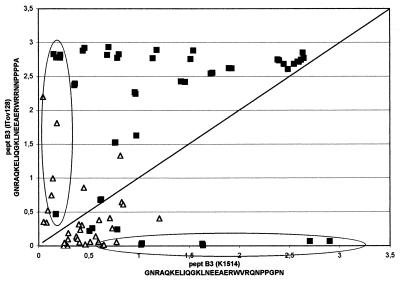
All 16 lentivirus isolates were successfully amplified with the pol primer set, giving an amplified product of the expected length. Nucleotide sequences from pol amplimers were used to construct a phylogenetic tree (Fig. 1). With the exception of isolate It-561, all Italian ovine isolates form a cluster more similar to, although distinct from, caprine isolates than to classical ovine strains. The Spanish, British, and North American isolates, as well as isolate It-561, were grouped with the latter, more heterogeneous, cluster. All Italian ovine isolates, except It-561, were also amplified with the gag primer set. Phylogenetic analysis carried out with ovine strains K1514, EV1, SAOVV, and ZZV1050 and caprine strain CO confirmed the results shown above based on pol sequences (data not shown). The deduced amino acid sequences were aligned with those of the reference strains, and the immunodominant regions, identified in a previous study, were localized (Fig. 2). In particular, a region of 17 residues carrying two epitopes (Fig. 2) showed a significant divergence in the C-terminal region between classical sheep strains and Italian sheep isolates, the latter cluster being more similar to the CAEV strain CO. Recombinant polypeptides encompassing this variable region were then generated based on the Icelandic strain K1514, the MVV prototype strain, and Italian isolate It-128. A panel of AGID-positive sheep and goat sera, collected from different Italian flocks were used to evaluate the reactivity against both peptides in an ELISA format. The majority of the samples were more reactive to the It-128 subunit, but each recombinant was able to classify as positive a number of samples which were negative to the other (Fig. 3).
FIG. 1.
Phylogenetic tree of sequence similarity between lentiviruses. Sheep (S) and goat (G) lentiviral fragments sequenced in the study were compared with sequences from small ruminant, bovine, equine, feline, human, and simian lentiviruses available in the GenBank database (accession numbers are indicated in parentheses). The neighbor-joining tree was constructed by using MEGA. The bar indicates the amount of evolution (substitutions per site) along the horizontal branches. Statistical support for the internal branches was evaluated by the bootstrap test (values greater than 50 are shown). EIAV, equine infectious anemia virus; BIV, bovine immunodeficiency virus; FIV, feline immunodeficiency virus; HIV2, human immunodeficiency virus type 2; SIV, simian immunodeficiency virus.
FIG. 2.
Amino acid sequence alignment of the gag region of eight ovine lentiviruses isolated in Italy (S) compared with reference strains. The immunodominant regions characterized in a previous report are boxed. Underlined amino acid sequences were generated by PCR primers.
FIG. 3.
Net absorbance from ELISA of sheep and goat sera against the recombinant B3 subunit from the visna virus prototype Icelandic strain K1514 and the Italian ovine isolate It-128. The samples within the circles are those which were classified as positive by one recombinant and as negative by the other. ▵, sheep; ▪, goat.
DISCUSSION
In this study, we provide further evidence that a classification of small ruminant lentiviruses based on host species origin may not be completely accurate. The existence of a new group of genetically distinct ovine isolates in France, which are more similar to the CAEV prototype, have been previously described (15, 16). Although a different pol fragment was examined in this study, it is possible that the novel genotype observed in the sheep population reared in Italy might be related to the same genetic cluster due to the extensive importation of French breeds in Italy in the last 10 to 20 years. According to the classification proposed in a previous report (32), the majority of the Italian ovine isolates belong to cluster IV, and all caprine isolates belong to cluster V, although ovine strains similar to the MVV prototype (clade I) are still circulating in Italy (i.e., It-561). The main explanation of this finding could be the cross-species transfer of goat-adapted strains to sheep, events which could have been particularly efficient in most Italian rural areas, where sheep and goats are reared in close contact. However, since ovine and caprine field isolates segregate into two different clusters, cross-species infections likely occurred in the past. Interestingly, most ovine isolates of cluster IV were associated with gross and histopathological lesions indistinguishable from typical maedi, whereas only the ovine isolate Pi1 was associated with arthritis in sheep. Among these isolates, some were highly cytopathic in vitro while others were nonlytic; thus, ovine isolates may not necessarily be differentiated in their pathogenic properties in vivo and in vitro.
Since the majority of ovine isolates belonging to cluster IV were also successfully amplified with the gag primers, the antigenic variability (if any) could be verified in the major diagnostically relevant antigen. The amino acid sequence of a subset of ovine isolates covering nearly 95% of the CA showed significant homology to the CAEV strain CO while divergent sequences from the sheep lentivirus prototype were detected in one of the three major linear epitopes. To assess whether this variability could affect the sensitivity of the CA-based serological test, we expressed two recombinant polypeptide subunits whose amino acid sequences enabled us to classify small ruminant lentiviruses in two main groups: group A, which includes the classical MVV strains whose sequences were largely employed to express diagnostically utilized recombinant antigens (Icelandic strain K1514 [13, 14], South African strain SAOVV [P. Pourquier, personal communication], British strain EV1 [25], and Dutch strain ZZV1050 [33]), and group B, which includes CAEV strains and the Italian ovine isolates.
The two subunits share a common N-terminal epitope (sequence LNEEAERW) and have distinct C-terminal epitopes (group A, VRQNPPGPN; group B, RRNNPPPPA). As shown in Fig. 3, when field AGID-positive sera showed a similar net absorbance for each antigen, it was assumed that the reactivity was mainly directed against the common N-terminal epitope, whereas when absorbance against one antigen was significantly higher with respect to the other, it is assumed that the reactivity against the C-terminal epitope was predominant. Surprisingly, some sera from either sheep or goats were classified as positive to the group A antigen and negative to the group B antigen and vice versa, leading us to suppose that goats might also have been infected with a sheep-like lentivirus. Therefore, when the two recombinant subunits are employed together, an increased number of seropositive animals could be detected in both species. It may be argued that reactivity against other highly conserved epitopes in most diagnostic tests may compensate for this lack of sensitivity. We then examined 103 goat sera collected from five CAEV-infected flocks, 75 of which were positive for a multiple-epitope antigen (recombinant CA-TM fusion protein) derived from the Icelandic strain K1514 (group A). Seven of 28 negative samples were reactive against It-128/B3 (group B), confirming the combined effect of the group B epitope on the group A antigen (Table 1).
TABLE 1.
Antibody reactivity of 103 goat serum samples collected from five CAEV-infected flocks to strain K1514-derived recombinant antigens (CA-TM and B3 subunits) and the strain It-128-derived recombinant B3 subunit
| Subunit (status) | No. of serum samples reacting to subunit (status):
|
|||||
|---|---|---|---|---|---|---|
| CA-TM (+) | CA-TM (−) | K1514/B3 (+) | K1514/B3 (−) | It-128/B3 (+) | It-128/B3 (−) | |
| CA-TM (+) | 75 | |||||
| CA-TM (−) | 28 | |||||
| K1514/B3 (+) | 26 | 1 | 27 | |||
| K1514/B3 (−) | 49 | 27 | 76 | |||
| It-128/B3 (+) | 61 | 7 | 26 | 42 | 68 | |
| It-128/B3 (−) | 14 | 21 | 1 | 34 | 35 | |
To date, several immunoassays based on a single virus strain have been described and are still widely used in eradication programs in sheep and goats. Previous reports however, have shown that the MVV antigen was less suitable than the CAEV antigen to detect CAEV diagnosis with the AGID (10, 18). The present data suggest that a double-strain-based immunoassay may increase the sensitivity of serological diagnosis of small ruminant lentivirus infections not only between sheep and goats but also within sheep or goat populations.
Acknowledgments
We thank Giuseppe Bertoni and Reto Zanoni for critical readings of the manuscript.
This work was supported by EU grant AIR3 CT94-1492 and by the Italian MURST.
REFERENCES
- 1.Bertoni, G., C. Hertig, M. L. Zahno, H. R. Vogt, S. Dufour, P. Cordano, E. Peterhans, W. P. Cheevers, P. Sonigo, and G. Pancino. 2000. B-cell epitopes of the envelope glycoprotein of caprine arthritis-encephalitis virus and antibody response in infected goats. J. Gen. Virol. 81:2929-2940. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Bulgin, M. S. 1990. Ovine progressive pneumonia, caprine arthritis-encephalitis, and related lentiviral diseases of sheep and goats. Vet. Clin. N. Am. Food Anim. Pract. 6:691-703. [DOI] [PubMed] [Google Scholar]
- 4.Cutlip, R. C., T. A. Jackson, and H. D. Lehmkuhl. 1977. Immunodiffusion test for ovine progressive pneumonia. Am. J. Vet. Res. 38:1081-1084. [PubMed] [Google Scholar]
- 5.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 7.Gogolewski, R. P., D. S. Adams, T. C. McGuire, K. L. Bauks, and W. P. Cheevers. 1985. Antigenic cross-reactivity between caprine arthritis-encephalitis, visna and progressive pneumonia viruses involves all virion-associated proteins and glycoproteins. J. Gen. Virol. 66:1233-1240. [DOI] [PubMed] [Google Scholar]
- 8.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 9.Houwers, D. J., S. L. Gielkens, Jr., and J. Schaake. 1982. An indirect enzyme-linked immunosorbent assay (ELISA) for the detection of antibodies to maedi-visna virus. Vet. Microbiol. 7:209-219. [DOI] [PubMed] [Google Scholar]
- 10.Knowles, D. P., Jr., J. F. Evermann, C. Shropshire, J. Vanerschalie, D. Bradway, H. M. Gezon, and W. P. Cheevers. 1994. Evaluation of agar gel immunodiffusion serology using caprine and ovine lentiviral antigens for detection of antibody to caprine arthritis-encephalitis virus. J. Clin. Microbiol. 32:243-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar, S., K. Tamura, and M. Nei. 1994. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10:189-191. [DOI] [PubMed] [Google Scholar]
- 12.Kwang, J., and R. C. Cutlip. 1992. Analysis of antibody response to ovine lentivirus by using viral gene products expressed in a prokaryotic system. Biochem. Biophys. Res. Commun. 188:20-27. [DOI] [PubMed] [Google Scholar]
- 13.Kwang, J., J. Keen, R. C. Cutlip, and E. T. Littledike. 1993. Evaluation of an ELISA for detection of ovine progressive pneumonia antibodies using a recombinant transmembrane envelope protein. J. Vet. Diagn. Investig. 5:189-193. [DOI] [PubMed] [Google Scholar]
- 14.Kwang, J., J. Keen, R. C. Cutlip, H. S. Kim, and A. de la Concha-Bermejillo. 1995. Serological diagnosis of caprine lentivirus infection by recombinant immunoassay. Small Ruminant Res. 16:171-177. [Google Scholar]
- 15.Leroux, C., S. Vuillermoz, J. F. Mornex, and T. Greenland. 1995. Genomic heterogeneity in the pol region of ovine lentiviruses obtained from bronchoalveolar cells of infected sheep from France. J. Gen. Virol. 76:1533-1537. [DOI] [PubMed] [Google Scholar]
- 16.Leroux, C., J. Chastang, T. Greenland, and J. F. Mornex. 1997. Genomic heterogeneity of small ruminant lentiviruses: existence of heterogeneous populations in sheep and of the same lentiviral genotypes in sheep and goats. Arch. Virol. 142:1125-1137. [DOI] [PubMed] [Google Scholar]
- 17.Quèrat, G., G. Audoly, P. Sonigo, and R. Vigne. 1990. Nucleotide sequence analysis of SA-OMVV, a visna-related ovine lentivirus: phylogenetic history of lentiviruses. Virology 175:434-447. [DOI] [PubMed] [Google Scholar]
- 18.Remond, M., and A. Boutrouille. 1990. Comparison of the sensitivity of precipitation antigens in the gel immunodiffusion test for the serological detection of Visna Maedi in sheep and of caprine arthritis encephalitis virus infection. Rev. Med. Vet. 141:125-128. [Google Scholar]
- 19.Rosati, S., J. Kwang, F. Tolari, and J. Keen. 1994. A comparison of whole virus and recombinant transmembrane ELISA and immunodiffusion for detection of ovine lentivirus antibodies in Italian sheep flocks. Vet. Res. Commun. 18:73-80. [DOI] [PubMed] [Google Scholar]
- 20.Rosati, S., J. Kwang, and J. Keen. 1995. Genome analysis of North American small ruminant lentiviruses by polymerase chain reaction and restriction enzyme analysis. J. Vet. Diagn. Investig. 7:437-443. [DOI] [PubMed] [Google Scholar]
- 21.Rosati, S., M. Pittau, F. Tolari, G. Erre, and J. Kwang. 1995. Genetic and antigenic characterization of CAEV (caprine arthritis-encephalitis virus) recombinant transmembrane protein. Vet. Microbiol. 45:363-370. [DOI] [PubMed] [Google Scholar]
- 22.Rosati, S., A. Mannelli, T. Merlo, and N. Ponti. 1999. Characterization of the immunodominant cross-reacting epitope of visna maedi virus and caprine arthritis-encephalitis virus capsid antigen. Virus Res. 66:109-116. [DOI] [PubMed] [Google Scholar]
- 23.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 24.Saltarelli, M., G. Querat, D. A. Konings, R. Vigne, and J. E. Clements. 1990. Nucleotide sequence and transcriptional analysis of molecular clones of CAEV which generate infectious virus. Virology 179:347-364. [DOI] [PubMed] [Google Scholar]
- 25.Saman, E., G. Van Eynde, L. Lujan, B. Extramiana, G. Harkiss, F. Tolari, L. Gonzales, B. Amorena, N. Watt, and J. Badiola. 1999. A new sensitive serological assay for the detection of lentivirus infections in small ruminants. Clin. Diagn. Lab. Immunol. 6:734-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sargan, D. R., I. D. Bennet, C. Cousens, D. J. Roy, B. A. Blacklaws, R. G. Dalziel, N. J. Watt, and I. McConnell. 1991. Nucleotide sequence of EV1, a British isolate of maedi-visna virus. J. Gen. Virol. 72:1893-1903. [DOI] [PubMed] [Google Scholar]
- 27.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusion protein with glutathione-s-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 28.Sonigo, P., M. Alizon, K. Staskus, D. Klatzmann, S. Cole, O. Danos, E. Retzel, P. Tiollais, A. Haase, and S. Wain-Hobson. 1985. Nucleotide sequence of the visna lentivirus: relationship to the AIDS virus. Cell 42:369-382. [DOI] [PubMed] [Google Scholar]
- 29.Tamura, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512-526. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valas, S., C. Benoit, C. Baudry, G. Perrin, and R. Z. Mamoun. 2000. Variability and immunogenicity of caprine arthritis-encephalitis virus surface glycoprotein. J. Virol. 74:6178-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zanoni, R. G. 1998. Phylogenetic analysis of small ruminant lentiviruses. J. Gen. Virol. 79:1951-1961. [DOI] [PubMed] [Google Scholar]
- 33.Zanoni, R. G., I. Nauta, U. Pauli, and E. Peterhans. 1991. Expression in Escherichia coli and sequencing of the coding region for the capsid protein of Dutch maedi-visna virus strain ZZV 1050: application of recombinant protein in enzyme-linked immunosorbent assay for the detection of caprine and ovine lentiviruses. J. Clin. Microbiol. 29:1290-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]