Abstract
Human herpesvirus 8 (HHV-8, also called Kaposi's sarcoma-associated herpes virus) has been linked to Kaposi's sarcoma and primary effusion lymphoma. HHV-8-encoded viral Fas-associated death domain-like IL-1-converting enzyme inhibitory protein (vFLIP) is one of the few viral proteins to be expressed in latently infected cells and plays a key role in the survival and proliferation of primary effusion lymphoma cells. Two main functions have been ascribed to HHV-8 vFLIP, inhibition of caspase 8/Fas-associated death domain-like IL-1-converting enzyme and activation of NF-κB. In this article, we demonstrate that vFLIP-expressing transgenic mice lack any of the features seen in mice deficient in caspase 8 or Fas-associated death domain protein and are not resistant to Fas-induced apoptosis. On the other hand, these mice display constitutive activation of classical and alternative NF-κB pathways, enhanced response to mitogenic stimuli, and increased incidence of lymphoma. Collectively, our results demonstrate that HHV-8 vFLIP is an oncogenic protein that mimics the signaling activities of caspase 8 during antigen receptor signaling and could contribute to the development of lymphoproliferative disorders via constitutive NF-κB activation independent of inhibition of Fas-induced apoptosis.
Keywords: Kaposi's sarcoma-associated herpes virus, primary effusion lymphoma
Kaposi's sarcoma (KS)-associated herpes virus, or the human herpesvirus 8 (HHV-8), was originally isolated from a patient with AIDS-associated KS and is now known to be associated with all forms of KS (classical, iatrogenic, endemic, and AIDS-related) (1). In addition to KS, HHV-8 has been linked to a number of other neoplastic disorders, such as primary effusion lymphoma (PEL), multicentric Castleman's disease, angio-immunoblastic lymphadenopathy with dysproteinemia and much rarer posttransplantation plasmacytic proliferation, and posttransplantation bone marrow failure syndromes (2-6). One of the most intriguing aspects of KS and HHV-8-linked lymphoproliferative syndromes is the involvement of several growth factors in their pathogenesis. There is increasing evidence that immune stimulation and cytokine dysregulation play a major role in the pathogenesis of these diseases (5, 7). Latency is generally assumed to be the state leading to transformed phenotype in viral-induced malignancies. Although HHV-8 encodes for homologs of cytokine, chemokines, and their receptors, none of them is expressed in latently infected PEL cell lines or KS spindle cells (5). Thus, the genes K13 (ORF71), v-cyclin, and LANA1 at the major latency locus are prime candidates for playing a causative role in the pathogenesis of HHV-8-induced malignancies (5). The K13 gene encodes a protein that resembles the prodomain of caspase 8 (Fas-associated death domain-like IL-1-converting enzyme, FLICE) in possessing two homologous copies of a death effector domain (8-10). Similar proteins have been discovered in other viruses and are believed to protect virally infected cells from death receptor-induced apoptosis by blocking the recruitment and/or activation of caspase 8/FLICE (8-10). As such, these virally encoded proteins are collectively referred to as viral FLICE inhibitory proteins (vFLIPs).
Recent in vitro results suggest that the HHV-8 vFLIP K13 may have a role beyond its ability to block caspase 8 activation. Thus, we have previously reported that HHV-8 vFLIP can also activate the NF-κB pathway, a property not shared by other vFLIPs (11). We and others have also demonstrated that vFLIP K13 plays a key role in constitutive NF-κB activity observed in PEL cells and is essential for their survival and proliferation (12-15). Finally, vFLIP K13 can transform Rat-1 and BALB/c 3T3 cells by NF-κB activation (16). Therefore, we hypothesized that K13-mediated activation of the NF-κB pathway, combined with the inhibition of death receptor-induced apoptosis, could contribute to the development of lymphoproliferative disorders seen in patients with HHV-8 infection. In addition, because the loss of caspase 8 expression is known to lead to defects in the activation of lymphocytes and immunodeficiency (17, 18), we hypothesized that expression of K13 could potentially lead to similar defects by virtue of its ability to block caspase 8 activation. To test the above hypotheses, we generated transgenic mice expressing K13. We report that transgenic expression of K13 leads to constitutive NF-κB activation and increased incidence of lymphomas but has no significant effect on Fas-induced apoptosis or the development and maturation of lymphocytes.
Materials and Methods
Production of Transgenic Mice. A cDNA encoding vFLIP K13 with three copies of a Flag tag at the carboxyl terminal was cloned under the transcriptional control of H2kb promoter in the pHSE3′ vector, which was a kind gift from Dennis Willerford (University of Washington, Seattle). The transgenic mice were developed in the transgenic core facility of the University of Texas Southwestern Medical Center in the ICR background. Heterozygous lines from the founder mice were established by backcrossing transgenic mice with K13-negative mice.
Genotyping. PCR genotyping of K13 transgenic mice was performed on DNA extracted from tail biopsies by using an upstream primer (5′-CAAGAACCAATCAGTGTCGC-3′), which corresponded to a region in the H2kb promoter, and a downstream primer (5′-CGCGGCTCGAGTGGTGTATGGCGATAGTGTTG-3′), which corresponded to the 3′ end of the K13 cDNA. PCR amplification with these primers produces a 650-bp band.
Western Blots. Splenocytes, thymocytes, and lymphocytes (inguinal and cervical lymph nodes were pooled) were washed once in ice-cold PBS and lysed on ice for 30 min in RIPA lysis buffer (50 mM Tris, pH 8/0.5% Na deoxycholate/150 mM NaCl/1% Triton X-100/0.1% SDS) supplemented with one protease inhibitor mixture tablet (Roche Applied Science, Indianapolis) per 10 ml of lysis buffer. Lysates were cleared of cellular debris by centrifugation at 20,000 × g at 4°C for 10 min. Protein concentration was determined by using the Bio-Rad Protein Assay. Thirty micrograms of protein was separated on SDS/PAGE, and Western blot was performed essentially as described (19). Antibodies were used at the following dilutions: anti-FLAG (M2) horseradish peroxidase (1:10,000; Sigma), anti-IκBα (1:2,000; Santa Cruz Biotechnology, SC-371), phospho-IκBα (1:1,000; Cell Signaling Technology, Beverly, MA), and anti-p52 (1:1,000; Upstate Biotechnology, Lake Placid, NY).
EMSA. For EMSA, nuclear extracts were prepared and analyzed for NF-κB activation as described (19).
Flow Cytometry Analysis. Single-cell suspensions prepared from thymus, spleen, and lymph nodes were stained at 4°C in PBS/5% FCS with FITC- or R-phycoerytherin-conjugated antibodies against CD4, CD8, CD45R/B220, and Thy 1.2. Events were collected on either a FACScalibur flow cytometer (Becton Dickinson) or an EPICS XL-MCL flow cytometer (Coulter). Electronic gating in forward and side scatter was used to remove spurious events, dead cells, and nonlymphocytes.
Analysis of Cellular Proliferation and Caspase Activation. B and T cells were purified from splenocytes harvested from K13 and nontransgenic littermate control (NLC) mice essentially as described (18) by using antibody-coated magnetic beads (Miltenyi Biotec, Auburn, CA). For proliferation analysis, 1.0 × 105 splenocytes, purified B cells, T cells, or thymocytes were placed in 96-well flat-bottom plates in RPMI medium 1640 containing 10% FCS and 0.1% 2-mercaptoethanol and stimulated in triplicate with soluble anti-CD3 (1 μg/ml), LPS (1 μg/ml), or phytohemagglutinin (10 μg/ml). Cells were labeled for the last 18 h with BrdUrd or [3H]thymidine, and cellular proliferation was measured 72 h after treatment as described (15, 18). Caspase activation was measured with a fluorimetric homogeneous caspase assay kit (Roche Applied Science).
Thymocyte Apoptosis. Thymocytes (1.0 × 105 cells) were treated with apoptotic stimuli: dexamethasone (5 μM) or different concentrations of an agonistic Fas antibody (Jo-2). Apoptosis was measured 24 h after treatment by flow cytometry by using an annexin/propidium iodide apoptosis detection kit (R & D Systems).
Histological Analysis. Spleens and tumors were fixed in buffered formalin, processed for paraffin-embedded sectioning at 5 μm, and stained by hematoxylin and eosin (Fisher). For immunohistochemistry, spleen sections were incubated with rat monoclonal anti-mouse B220 (Pharmingen) and rabbit polyclonal anti-mouse CD3 (DAKO) antibodies as described (18). The Liquid DAB Substrate Chromagen System (DAKO) and the Vector Red alkaline phosphatase substrate kit (Vector Laboratories) were used to reveal B220 and CD3 labeling, respectively. No background staining was observed with anti-CD3, anti-B220, or secondary antibody alone.
Statistical Analysis. Statistical analysis was performed by using a computer program for epidemiologists, pepi, version 4.0 (Sagebrush Press, Salt Lake City).
Results
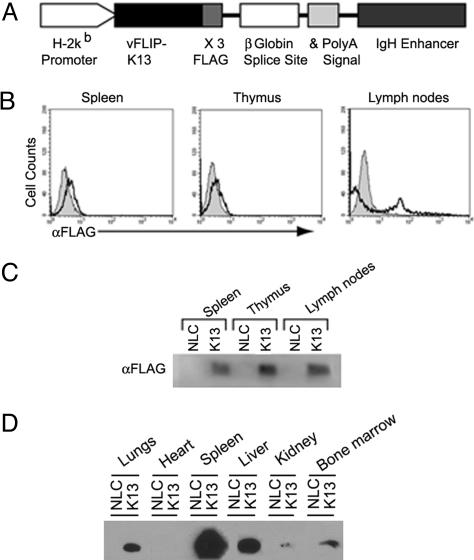
Generation of K13 Transgenic Mice. We anticipated that expression of K13 transgene under a constitutively active promoter may have a deleterious effect on normal growth and development because of constitutive NF-κB activation and interference with death receptor-induced apoptosis. Therefore, we targeted the expression of K13 transgene to tissues important for the natural history of infection by HHV-8 by using a promoter that would be switched on later in the ontogeny. For this purpose, we expressed the K13 transgene under the H2Kb promoter and Ig heavy chain enhancer (20, 21). The K13 construct was epitope-tagged with three copies of FLAG tag at its carboxyl terminus to aid in the detection of protein (Fig. 1A). We generated four independent K13 transgenic lines, designated 3586, 85, 141, and 156. As shown in Fig. 1 B and C, K13 expression was readily detected in spleen, thymus, and lymph nodes of the transgenic animals by using flow cytometry and Western blotting. Consistent with previous reports (20, 21), H2Kb promoter-driven K13 transgene expression was not limited to lymphoid organs and was also seen in liver, kidneys, lungs, and bone marrow (Fig. 1D).
Fig. 1.
Generation and characterization of transgenic mice expressing vFLIP K13. (A) Schematic representation of K13 transgene construct. IgH, Ig heavy chain. (B) Flow cytometric analyses. Intracellular staining of splenocytes, thymocytes, and lymphocytes from K13 and NLC mice with FITC-conjugated FLAG antibody. Representative histograms show FITC intensity of cells from NLC mice (gray area) and K13 (black line) mice. (C) Western blot analysis showing K13 expression. Lysates of splenocytes, thymocytes, and lymphocytes of K13 and NLC mice were probed with an antibody against the FLAG epitope tag. (D) Western blot analysis showing expression of K13 in different tissues. Cellular lysates from the indicated tissues were immunoprecipitated with FLAG antibody beads and immunoblotted with a horseradish peroxidase-conjugated FLAG antibody (Sigma).
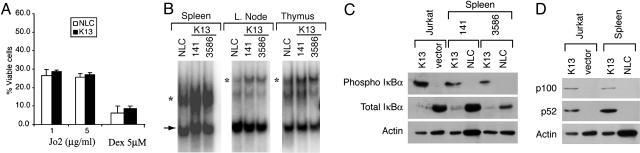
Effect of K13 Transgene on Cell Death. It has been postulated that vFLIPs block the activation of caspase 8 and thereby protect virally infected cells from death receptor-induced apoptosis (8-10). Consistent with the above hypothesis, transgenic expression of vFLIPs E8 and MC159L from equine herpes virus 2 and molluscum contagiosum virus, respectively, has been shown to protect thymocytes from Fas-induced apoptosis (22, 23). Therefore, we sought to determine whether transgenic expression of K13 would similarly block Fas-mediated apoptosis. Interestingly, K13 and NLC thymocytes exhibited similar sensitivity when apoptosis was induced by Jo2, an agonistic Fas mAb (Fig. 2A). Similarly, there was no major difference in the sensitivity of K13 and NLC thymocytes to apoptosis induced by dexamethasone, which uses the intrinsic cell death pathway (Fig. 2 A). Thus, unlike for vFLIPs E8 and MC159L, transgenic expression of vFLIP K13 fails to block either extrinsic or intrinsic cell death pathways.
Fig. 2.
Effect of K13 transgene on cell death and NF-κB pathways in lymphoid cells. (A) Effect of K13 transgene on cell death. Thymocytes from K13 and NLC mice were treated with different concentrations of Jo2 (Fas) antibody or dexamethasone (5 μM) for 24 h, and the percentage of viable cells [annexin-/propidium iodide (PI)-] was determined by annexin-FITC and PI staining. (B) EMSA showing increased NF-κB DNA binding activity in the spleens, lymph nodes, and thymi of K13 transgenic mice as compared with NLC animals. Results from transgenic lines 141 and 3586 are shown. The position of the NF-κB complex is marked by an asterisk, and an arrow indicates the position of a nonspecific band. (C) Constitutive activation of the canonical NF-κB pathway in K13 transgenic splenocytes. Western blot analysis demonstrating increased phosphorylation of IκBα (Top), a decrease in the total IκBα protein (Middle), and loading of equal amount of protein (Bottom) in transgenic K13 splenocytes versus NLC splenocytes. Results from two transgenic lines, 141 and 3586, respectively, are shown. Jurkat cells, expressing an empty vector or K13, are used as positive control. (D) Constitutive activation of the alternate NF-κB pathway in K13 transgenic mice. Up-regulation of p100 expression and its processed p52 subunit in splenocytes from K13 transgenic animals are detected by immunoblotting. Jurkat cells, expressing an empty vector or K13, are used as positive control.
Constitutive NF-κB Activation in K13 Transgenic Mice. We have previously demonstrated that vFLIP (K13) expression in a variety of cell lines leads to constitutive activation of the NF-κB pathway because of constitutive phosphorylation and degradation of IκBα (13). We used an EMSA to analyze the status of NF-κB pathway in the lymphoid compartment of K13 transgenic mice. We observed constitutive NF-κB activation in the spleens, lymph nodes, and thymi of K13 transgenic mice as compared with the NLC mice, which was associated with an increase in phosphorylation at Ser 32/36 residues of IκBα and a corresponding decline in total IκBα levels in K13 transgenic animals (Fig. 2 B and C).
An alternative (or noncanonical) pathway of NF-κB activation, which involves proteasome-mediated processing of p100/NF-κB2 into p52 subunit, has been recently described (24). The alternative NF-κB pathway is activated by a number of receptors involved in lymphocyte development and survival, such as BAFF-R, CD40, and LTβ R, and is essential for development and organization of lymphoid tissues (25-27). We have previously demonstrated that retroviral-mediated expression of vFLIP K13 in a variety of cell lines leads to the activation of the alternative NF-κB pathway that is associated with up-regulation of expression of p100/NF-κB2 subunit and its processing into p52 subunit (15). As shown in Fig. 2D, increased expression of p100 and its active processed subunit p52 was also seen in K13 splenocytes. Thus, transgenic expression of K13 leads to activation of both canonical and alternate NF-κB pathways.
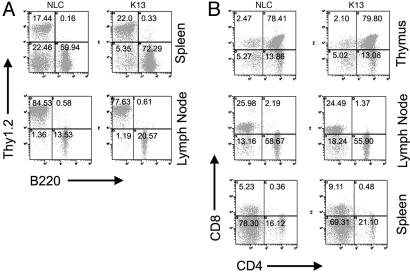
Effect of K13 Transgene on T and B Cell Homeostasis. Several recent studies suggest that, in addition to its role in activation of the apoptosis, the Fas-associated death domain (FADD)-caspase 8 pathway plays a key role in the development and proliferation of lymphocytes (17, 18). For example, FADD deficiency leads to inhibition of T cell development at the double-negative (CD4-CD8-) stage in the thymus and a reduction in the number of mature T cells (28, 29). Genetic deficiency of caspase 8 is known to lead to an immunodeficiency syndrome characterized by defects in lymphocyte apoptosis and homeostasis and defects in the activation of T and B lymphocytes and natural killer cells (17, 18). Because vFLIPs have been postulated to inhibit the FADD-caspase8 pathway, transgenic expression of vFLIP K13 would be expected to mimic the phenotype of FADD and caspase 8-deficient animals. We analyzed the cellular composition of spleen, thymus, and lymph nodes of WT and K13 transgenic mice. Although the percentage of B220+ cells was increased in the spleen and lymph nodes of K13 transgenic mice as compared with the control mice, we did not see an accumulation of an abnormal T cell population (Thy1+, B220+, CD4-, CD8-) in the periphery, which is a hallmark of Fas-mutant mice (Fig. 3). Similarly, the CD4-CD8-, CD4+CD8+, CD4+CD8-, and CD4-CD8+ subpopulations were similarly represented in the spleen, thymus, and lymph node cells of K13 transgenic and control mice (Fig. 3B).
Fig. 3.
Flow cytometric analyses of the lymphoid compartment in K13 transgenic mice. (A) Spleen and lymph node cells from 8- to 10-week-old K13 and NLC mice were stained with FITC-conjugated B220 and R-phycoerythrin-conjugated Thy 1.2. antibodies. Although the B220+ cells are increased in K13 transgenic animals, they lack the population of B220+Thy1.2+ double-positive cells known to be present in Fas-deficient animals. (B) Splenocytes, lymphocytes, and thymocytes were stained with FITC-CD4 and RPE-CD8. CD4 and CD8 T cell subsets in the spleen, thymus, and lymph nodes were similarly represented in both K13 transgenic and NLC groups. The percentages shown are representative of more than four individual experiments in three different transgenic lines.
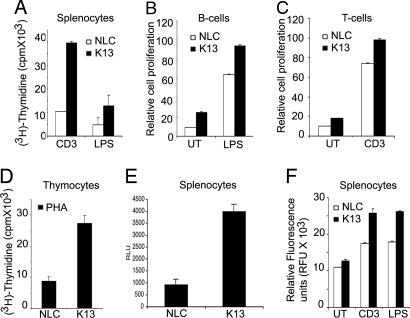
Enhanced Proliferation in K13 Transgenic Splenocytes. As the NF-κB pathway has been shown to play an essential role in proliferation of lymphocytes (30), we sought to determine whether transgenic expression of K13 would lead to increased proliferation in response to various mitogenic stimuli. Splenocytes from K13 transgenic animals demonstrated increased proliferative response to stimulation by CD3 antibody and LPS as measured by [3H] incorporation and BrdUrd incorporation assays (Fig. 4A and data not shown). The increased poliferative response was also observed in purified B and T cells after LPS and CD3 stimulation, respectively (Fig. 4 B and C). Interestingly, during the course of these studies, we observed that the basal rate of proliferation in the unstimulated B and T cells was modestly increased as compared with the NLC (Fig. 4 B and C). Moreover, the increase in proliferation was not restricted to K13-expressing splenocytes because K13 thymocytes also demonstrated a significant increase in proliferation in response to phytohemagglutinin (Fig. 4D). Consistent with an increase in the basal proliferation rate, K13 splenocytes demonstrated an increase in viable cell number as compared with the NLC splenocytes when cultured for 72 h in standard culture medium without added growth factors (i.e., RPMI medium 1640 with 10% FCS) (Fig. 4E).
Fig. 4.
Effect of K13 transgene on cellular proliferation. (A) Effect of K13 on cell proliferation. Anti-CD3- and LPS-induced proliferation of K13 and NLC splenocytes as measured by a [3H]thymidine incorporation assay. The values shown are averages (mean ± SE) of a representative of at least three independent experiments performed in triplicate. (B and C) B and T cells were purified from K13 and NLC mice essentially as described (18) and either left untreated (UT) or stimulated with LPS (1 μg/ml) and CD3 antibody (1 μg/ml), respectively. Cells were labeled with BrdUrd for the last 18 h after treatment, and cellular proliferation was measured by using BrdUrd incorporation assay 72 h after stimulation. (D) Thymocytes from K13 and NLC mice were stimulated with phytohemagglutinin (PHA; 10 μg/ml), and cellular proliferation was measured by using a [3H]thymidine incorporation assay. (E) Increased viable cell number in K13 splenocytes culture. K13 and NLC splenocytes were cultured in standard culture medium without added growth factors (RPMI medium 1640 with 10% FCS), and the number of viable cells was measured at the end of a 72-h culture with a CellTiter-Glo Luminescent cell viability assay kit (Promega). The values shown are averages (mean ± SE) of a representative of at least three independent experiments performed in duplicate or triplicate. (F) K13 transgene does not inhibit caspase activation. Caspase activation was measured in LPS-stimulated and CD3 antibody-stimulated splenocytes isolated from K13 and NLC mice with a homogeneous caspase assay kit (Roche Applied Science). The values shown are averages (mean ± SE) of one representative experiment of three experiments performed in triplicate.
K13 can potentially block caspase cascade either directly by inhibition of caspase 8 or indirectly by NF-κB-induced up-regulation of antiapoptotic genes (e.g., cFLIP, cIAP, BclxL, etc.). Therefore, we next examined whether the observed increase in the viable cell number of K13 splenocytes was secondary to inhibition of caspase activation and apoptosis. No significant difference was observed in the rate of apoptosis or caspase activation between K13 and NLC splenocytes (Fig. 4F and data not shown). In fact, we observed a modest increase in caspase activity in the culture of K13 splenocytes as compared with NLC splenocytes, which could reflect their increased proliferative rate (Fig. 4F). Collectively, the above results demonstrate that although transgenic expression of K13 leads to increased cellular proliferation, it has no significant inhibitory effect on caspase activation or apoptosis.
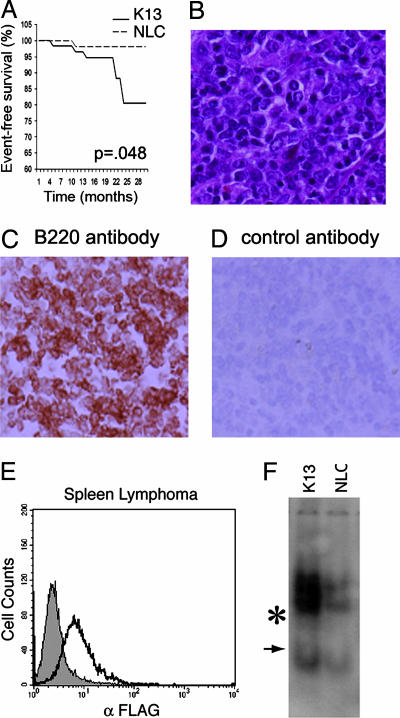
Increased Incidence of Lymphoma in K13 Transgenic Mice. A cohort of 59 animals each in the K13 and NLC groups was observed for tumor formation over 30 months. The incidence of lymphomas in the NLC was 1.8%, which is comparable to the 3.6% incidence reported in the literature (31). The incidence of lymphoma in K13 transgenic animals was increased to 11.8% (Fig. 5A). Thus, the relative risk of lymphoma development in K13 transgenic mice as compared with NLC was 6.23 (95% confidence interval, 1.54-25.22; log-rank χ2 = 3.90, 1 df, P = 0.048). Lymphomas were observed in all four lines generated, and their incidence increased after 20 months of age. A similar trend had been reported in studies with latent membrane protein 1 transgenic mice where lymphomas were found mostly in older animals (32). The mice with lymphomas usually presented with an enlarged spleen or lymph nodes (cervical, mesenteric, and inguinal). However, unlike the situation with PELs found in patients infected with HHV-8, we did not observe infiltration of body cavities with malignant cells. Histological examination of the lymphomas demonstrated a range of morphologies, including small lymphocytic, follicular, and immunoblastic subtypes (Fig. 5B and data not shown). Lymphomas in K13-transgenic animals were positive for B220 (Fig. 5 C and D), with the exception of one high-grade anaplastic lymphoma, which lacked this marker. Expression of K13 transgene was readily detected in the lymphomas by intracellular staining with FITC-conjugated M2-FLAG antibody (Fig. 5E). Furthermore, the lymphomas in the K13 transgenic animals demonstrated constitutive activation of the NF-κB pathway as measured by EMSA (Fig. 5F). K13 transgenic mice also demonstrated an increase in extramedullary hematopoieses involving liver and spleen (data not shown). Taken together, our results suggest that transgenic expression of K13 leads to increased incidence of lymphomas, which is associated with constitutive activation of the NF-κB pathway.
Fig. 5.
Development of lymphoma in K13 transgenic mice. (A) Kaplan-Meier graph showing increased incidence of lymphoma in K13 transgenic mice. Statistical analysis was done with a log-rank test. (B) Microscopic appearance of a follicular lymphoma involving the spleen in a K13-transgenic animal stained with hematoxylin-eosin. A mixture of large cleaved and noncleaved follicular center cells admixed with small cells is seen. (Original magnification: ×800.) (C and D) Immunostaining with a B220 (C) and a control (D) antibody in a splenic lymphoma, which developed in a K13 transgenic mice (Original magnifications: ×800.) (E) Expression of a K13 transgene in the lymphoma spleen as detected by intracellular staining with a FITC-conjugated FLAG antibody. Gray area indicates NLC; black line indicates K13. (F) EMSA demonstrating increased NF-κB DNA-binding activity in the spleen lymphoma of a K13 transgenic mouse as compared with the spleen of a NLC mouse. The position of the induced NF-κB complexes is marked by an asterisk, and an arrow indicates the position of a constitutive complex.
Discussion
On the basis of in vitro studies, two main biological functions have been attributed to HHV-8 vFLIP (5). Based on its homology to the prodomains of caspase 8, caspase 10, and cellular FLIP, it was initially suggested that vFLIP K13 functions to block Fas-mediated apoptosis by blocking the recruitment and/or activation of procaspase 8 (33, 34). Subsequent studies ascribed an alternative biological function to vFLIP K13, activation of the NF-κB pathway via a direct interaction with the inhibitory κB kinase (IKK) complex (11, 15, 19, 35). In this study, we used transgenic mice to determine the relative functional significance of the above two properties attributed to HHV-8 vFLIP. We demonstrate that although transgenic expression of vFLIP K13 leads to constitutive NF-κB activation it fails to block Fas-induced apoptosis. Consistent with the above results, K13 transgenic animals did not show accumulation of an abnormal T cell population (Thy1+, B220+, CD4-, CD8-), which is a hallmark of Fas-mutant mice. Furthermore, unlike the situation with caspase 8 and FADD deficiency (17, 18, 28), K13 transgenic mice did not manifest any defect in lymphocyte development or proliferation. Rather, K13-expressing splenocytes and thymocytes displayed increased proliferation in response to mitogenic stimuli. Collectively, the above results support the hypothesis that the main biological function of HHV-8 vFLIP is the activation of NF-κB pathway rather than the inhibition of caspase 8/FLICE-induced apoptosis. This conclusion is also supported by a recent study, which demonstrated that all of the soluble vFLIP in PEL cells coelutes with active IKK and coprecipitates with IKK-γ/Nemo (35).
HHV-8 vFLIP resembles vFLIP MC159 from the human molluscum contagiousum virus and vFLIP E8 from the equine herpes virus 2 in its overall structure, and all three proteins contain two death effector domains (8-10). However, unlike vFLIP K13, transgenic expression of MC159 and E8 vFLIPs has been shown to block Fas-induced apoptosis in thymocytes and impair T cell development (22, 23). Thus, despite the overall sequence similarity, vFLIPs perform distinct functions in different viruses, and the death effector domains, which are a hallmark of vFLIPs, may simply represent protein recruitment motifs that interact with different cellular proteins to activate distinct signaling pathways beneficial to different viruses. Thus, whereas vFLIPs MC159 and E8 interact with caspase 8/FLICE or FADD and protect virally infected cells from Fas-induced apoptosis, vFLIP K13 interacts with the inhibitory κB kinase complex to activate NF-κB and promote lymphocyte proliferation.
Although expression of K13 failed to protect thymocytes from Fas-induced apoptosis, it did promote their proliferation in response to mitogen stimuli. The NF-κB pathway is known to up-regulate the expression of a number of genes involved in cell cycle regulation (36, 37), and it is likely that K13-mediated constitutive NF-κB activation plays a key role in the increased proliferation of thymocytes and splenocytes observed in the current study. Consistent with the above hypothesis, we have previously demonstrated that K13 plays a pivotal role in the proliferation of PEL cells and exogenous expression of K13 using retroviral-mediated gene transfer induces increased proliferation of Rat-1 fibroblast cells by NF-κB activation (15, 16). A role of NF-κB pathway in the increased proliferation and activation of K13-transgenic splenocytes is also supported by a recent intriguing study that demonstrated that caspase 8 plays a key role in antigen receptor-induced lymphocyte proliferation by NF-κB activation (30). Thus, contrary to its perceived role as an inhibitor of caspase 8 in the cell death pathway, K13 may, in fact, function as a virally encoded structural and functional homolog of caspase 8 in the NF-κB signaling pathway and promote HHV-8-associated lymphoproliferative disorders by enhancing cellular proliferation after antigenic stimulation.
In this article, we provide in vivo evidence of the transforming ability of vFLIP K13. The lymphomas in K13 transgenic animals demonstrated constitutive NF-κB activation, thereby supporting a role of this pathway in tumorigenesis. Thus, HHV-8 vFLIP resembles Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) and human T-cell leukemia virus 1-encoded Tax in its ability to induce tumors by NF-κB activation (32, 39, 40). Interestingly, similar to the situation with the LMP1 transgenic mice (32), the incidence of lymphomas in K13 transgenic animals was relatively low, and they developed after a long period of latency. These results suggest that expression of K13 and resultant NF-κB activation may not be sufficient to induce tumors and may require cooperative interactions with other viral or cellular genes for lymphomagenesis.
Infection with HHV-8 has been associated with PEL or body cavity-based lymphoma, a type of non-Hodgkin's lymphoma that frequently lacks B cell markers and is characterized by pleural, pericardial, and peritoneal lymphomatous effusions in the absence of a solid tumor mass (41). vFLIP K13 is one of the few HHV-8-encoded proteins that are expressed in PEL cell lines and has been shown to play an essential role in the proliferation and survival of these cells by NF-κB activation (14, 15, 41). However, the lymphomas in K13 transgenic mice were primarily lymph node based and lacked the immunological or clinico-pathological features of PEL seen in the HHV-8-infected individuals. There are several possible explanations for this difference. First, previous studies have shown that the same transgene results in distinctive lymphomas on different mice backgrounds, and it is possible that the pathology of lymphomas in K13 transgenic mice observed in the current study reflects the mouse strain used (42). Second, as discussed above, although K13 may contribute to cellular transformation, it may require interaction with other HHV-8-encoded proteins for the full manifestation of the PEL phenotype. Third, PEL cells are frequently coinfected with Epstein-Barr virus (EBV), and it is conceivable that EBV-encoded proteins also contribute to the PEL phenotype (43). Finally, it is important to point out that the association of HHV-8 with lymphoproliferative diseases is not limited to PEL and multicentric Castleman's disease. Emerging evidence suggests a high incidence of HHV-8 infection in HIV-related solid immunoblastic/plasmablastic diffuse large B cell lymphomas (38, 44), which will be consistent with the immunoblastic lymphomas observed in some K13 transgenic mice in the current study. Thus, K13 transgenic mice may prove to be a useful tool for studying the cooperative interactions between various viral and cellular proteins in the pathogenesis of HHV-8-associated lymphoproliferative disorders.
Acknowledgments
We thank Dr. Dennis Willerford for providing the pHSE3′ construct. This work was supported by National Institutes of Health Grants CA85177 and AI/AR47230 and the Mario Lemieux Foundation.
Author contributions: P.M.C. designed research; P.C., H.M., S.S., S.Z., and A.K. performed research; P.C., H.M., J.A.R., A.L.S., and P.M.C. analyzed data; and P.C., H.M., and P.M.C. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HHV-8, human herpesvirus 8; KS, Kaposi's sarcoma; PEL, primary effusion lymphoma; FLICE, Fas-associated death domain-like IL-1-converting enzyme; vFLIP, viral FLICE inhibitory protein; FADD, Fas-associated death domain; NLC, nontransgenic littermate control.
References
- 1.Chang, Y., Cesarman, E., Pessin, M. S., Lee, F., Culpepper, J., Knowles, D. M. & Moore, P. S. (1994) Science 266, 1865-1869. [DOI] [PubMed] [Google Scholar]
- 2.Soulier, J., Grollet, L., Oksenhendler, E., Cacoub, P., Cazals-Hatem, D., Babinet, P., d'Agay, M. F., Clauvel, J. P., Raphael, M., Degos, L. & Sigaux, F. (1995) Blood 86, 1276-1280. [PubMed] [Google Scholar]
- 3.Gessain, A., Sudaka, A., Briere, J., Fouchard, N., Nicola, M. A., Rio, B., Arborio, M., Troussard, X., Audouin, J., Diebold, J. & de Thé, G. (1996) Blood 87, 414-416. [PubMed] [Google Scholar]
- 4.Luppi, M., Barozzi, P., Maiorana, A., Artusi, T., Trovato, R., Marasca, R., Savarino, M., Ceccherini-Nelli, L. & Torelli, G. (1996) Blood 87, 3903-3909. [PubMed] [Google Scholar]
- 5.Dourmishev, L. A., Dourmishev, A. L., Palmeri, D., Schwartz, R. A. & Lukac, D. M. (2003) Microbiol. Mol. Biol. Rev. 67, 175-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verma, S. C. & Robertson, E. S. (2003) FEMS Microbiol. Lett. 222, 155-163. [DOI] [PubMed] [Google Scholar]
- 7.Gallo, R. C. (1998) J. Natl. Cancer Inst. Monogr., 55-57. [DOI] [PubMed]
- 8.Thome, M., Schneider, P., Hofmann, K., Fickenscher, H., Meinl, E., Neipel, F., Mattmann, C., Burns, K., Bodmer, J. L., Schroter, M., et al. (1997) Nature 386, 517-521. [DOI] [PubMed] [Google Scholar]
- 9.Bertin, J., Armstrong, R. C., Ottilie, S., Martin, D. A., Wang, Y., Banks, S., Wang, G. H., Senkevich, T. G., Alnemri, E. S., Moss, B., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 1172-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu, S., Vincenz, C., Buller, M. & Dixit, V. M. (1997) J. Biol. Chem. 272, 9621-9624. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhary, P. M., Jasmin, A., Eby, M. T. & Hood, L. (1999) Oncogene 18, 5738-5746. [DOI] [PubMed] [Google Scholar]
- 12.Keller, S. A., Schattner, E. J. & Cesarman, E. (2000) Blood 96, 2537-2542. [PubMed] [Google Scholar]
- 13.Liu, H., Lo, C. R. & Czaja, M. J. (2002) Hepatology 35, 772-778. [DOI] [PubMed] [Google Scholar]
- 14.Guasparri, I., Keller, S. A. & Cesarman, E. (2004) J. Exp. Med. 199, 993-1003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Matta, H. & Chaudhary, P. M. (2004) Proc. Natl. Acad. Sci. USA 101, 9399-9404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun, Q., Zachariah, S. & Chaudhary, P. M. (2003) J. Biol. Chem. 278, 52437-52445. [DOI] [PubMed] [Google Scholar]
- 17.Chun, H. J., Zheng, L., Ahmad, M., Wang, J., Speirs, C. K., Siegel, R. M., Dale, J. K., Puck, J., Davis, J., Hall, C. G., et al. (2002) Nature 419, 395-399. [DOI] [PubMed] [Google Scholar]
- 18.Salmena, L., Lemmers, B., Hakem, A., Matysiak-Zablocki, E., Murakami, K., Au, P. Y., Berry, D. M., Tamblyn, L., Shehabeldin, A., Migon, E., et al. (2003) Genes Dev. 17, 883-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, L., Eby, M. T., Rathore, N., Sinha, S. K., Kumar, A. & Chaudhary, P. M. (2002) J. Biol. Chem. 277, 13745-13751. [DOI] [PubMed] [Google Scholar]
- 20.Pircher, H., Mak, T. W., Lang, R., Ballhausen, W., Ruedi, E., Hengartner, H., Zinkernagel, R. M. & Burki, K. (1989) EMBO J. 8, 719-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morello, D., Moore, G., Salmon, A. M., Yaniv, M. & Babinet, C. (1986) EMBO J. 5, 1877-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.OhYama, T., Tsukumo, S., Yajima, N., Sakamaki, K. & Yonehara, S. (2000) Microbiol. Immunol. 44, 289-297. [DOI] [PubMed] [Google Scholar]
- 23.Wu, Z., Roberts, M., Porter, M., Walker, F., Wherry, E. J., Kelly, J., Gadina, M., Silva, E. M., DosReis, G. A., Lopes, M. F., et al. (2004) J. Immunol. 172, 6313-6323. [DOI] [PubMed] [Google Scholar]
- 24.Pomerantz, J. L. & Baltimore, D. (2002) Mol. Cell 10, 693-695. [DOI] [PubMed] [Google Scholar]
- 25.Claudio, E., Brown, K., Park, S., Wang, H. & Siebenlist, U. (2002) Nat. Immunol. 3, 958-965. [DOI] [PubMed] [Google Scholar]
- 26.Dejardin, E., Droin, N. M., Delhase, M., Haas, E., Cao, Y., Makris, C., Li, Z. W., Karin, M., Ware, C. F. & Green, D. R. (2002) Immunity 17, 525-535. [DOI] [PubMed] [Google Scholar]
- 27.Coope, H. J., Atkinson, P. G., Huhse, B., Belich, M., Janzen, J., Holman, M. J., Klaus, G. G., Johnston, L. H. & Ley, S. C. (2002) EMBO J. 21, 5375-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, J., Cado, D., Chen, A., Kabra, N. H. & Winoto, A. (1998) Nature 392, 296-300. [DOI] [PubMed] [Google Scholar]
- 29.Kabra, N. H., Kang, C., Hsing, L. C., Zhang, J. & Winoto, A. (2001) Proc. Natl. Acad. Sci. USA 98, 6307-6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su, H., Bidere, N., Zheng, L., Cubre, A., Sakai, K., Dale, J., Salmena, L., Hakem, R., Straus, S. & Lenardo, M. (2005) Science 307, 1465-1468. [DOI] [PubMed] [Google Scholar]
- 31.Mohr, U., Dungworth, D. L., Capen, C. C., Carlton, W. W., Sundberg, J. P. & Ward, J. M. (1996) Pathobiology of the Aging Mouse (International Life Sciences Institute, Washington, DC).
- 32.Kulwichit, W., Edwards, R. H., Davenport, E. M., Baskar, J. F., Godfrey, V. & Raab-Traub, N. (1998) Proc. Natl. Acad. Sci. USA 95, 11963-11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sturzl, M., Hohenadl, C., Zietz, C., Castanos-Velez, E., Wunderlich, A., Ascherl, G., Biberfeld, P., Monini, P., Browning, P. J. & Ensoli, B. (1999) J. Natl. Cancer Inst. 91, 1725-1733. [DOI] [PubMed] [Google Scholar]
- 34.Belanger, C., Gravel, A., Tomoiu, A., Janelle, M. E., Gosselin, J., Tremblay, M. J. & Flamand, L. (2001) J. Hum. Virol. 4, 62-73. [PubMed] [Google Scholar]
- 35.Field, N., Low, W., Daniels, M., Howell, S., Daviet, L., Boshoff, C. & Collins, M. (2003) J. Cell Sci. 116, 3721-3728. [DOI] [PubMed] [Google Scholar]
- 36.Aggarwal, B. B. (2004) Cancer Cell 6, 203-208. [DOI] [PubMed] [Google Scholar]
- 37.Gilmore, T. D., Koedood, M., Piffat, K. A. & White, D. W. (1996) Oncogene 13, 1367-1378. [PubMed] [Google Scholar]
- 38.Deloose, S. T., Smit, L. A., Pals, F. T., Kersten, M. J., van Noesel, C. J. & Pals, S. T. (2005) Leukemia 19, 851-855. [DOI] [PubMed] [Google Scholar]
- 39.Devergne, O., McFarland, E. C., Mosialos, G., Izumi, K. M., Ware, C. F. & Kieff, E. (1998) J. Virol. 72, 7900-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeang, K. T. (2001) Cytokine Growth Factor Rev. 12, 207-217. [DOI] [PubMed] [Google Scholar]
- 41.Schulz, T. F. (2001) Eur. J. Cancer 37, 1217-1226. [DOI] [PubMed] [Google Scholar]
- 42.Yukawa, K., Kikutani, H., Inomoto, T., Uehira, M., Bin, S. H., Akagi, K., Yamamura, K. & Kishimoto, T. (1989) J. Exp. Med. 170, 711-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cesarman, E., Chang, Y., Moore, P. S., Said, J. W. & Knowles, D. M. (1995) N. Engl. J. Med. 332, 1186-1191. [DOI] [PubMed] [Google Scholar]
- 44.Engels, E. A., Pittaluga, S., Whitby, D., Rabkin, C., Aoki, Y., Jaffe, E. S. & Goedert, J. J. (2003) Mod. Pathol. 16, 424-429. [DOI] [PubMed] [Google Scholar]