Abstract
We report here that microarrays comprised of several thousand peptoids (oligo-N-substituted glycines) are useful tools for the identification of proteins via a “fingerprinting” approach. By using maltose-binding protein, glutathione S-transferase, and ubiquitin, a specific and highly reproducible pattern of binding was observed when fluorescently labeled protein was hybridized to the array. A similar pattern was obtained when binding of an unlabeled protein to the array was visualized by secondary hybridization of a labeled antibody against that protein, showing that native proteins can be identified without the requirement for prior chemical labeling. This work suggests that small-molecule microarrays might be used for more complex fingerprinting assays of potential diagnostic value.
Keywords: profiling, proteomics, small molecule microarrays
Small-molecule microarrays (SMMs) (1–3) are becoming increasingly important tools in combinatorial chemistry. These arrays are generally produced by first synthesizing a combinatorial library on a suitable bead resin, separating the beads into the wells of microtiter plates, and then releasing the compounds from the beads (4, 5). The resulting solutions then are spotted robotically onto a chemically modified glass slide such that the library-derived molecule is attached covalently to the surface. Alternatively, methods exist for the synthesis of certain classes of compounds in situ on the array surface (6–10).
By far the most common application of SMMs has been as a versatile platform for library screening, usually with the goal of identifying small-molecule ligands for a given protein of interest (1, 11–13). However, little work has focused on the development of SMMs as analytical tools for biological research (14–16), with the notable exception of peptide arrays as tools for determining the substrate preferences for proteases and other protein-modifying enzymes (12, 17–21) and the binding preferences of antibodies (22–24). We hypothesized that hybridization of any particular protein to a SMM with thousands of features is likely to provide a unique pattern of binding to the array, allowing that factor to be identified by virtue of this “molecular fingerprint.” Indeed, screening experiments have shown qualitatively that different high-affinity ligands are usually, although not always, identified when different proteins are incubated with a combinatorial library (1). The fingerprinting idea simply extends this concept to the quantitative measurement of protein binding at most or all features on a SMM. The expectation is that whereas only a few molecules in a library will be high-affinity ligands for a given protein, a larger number would exhibit above-background binding with a broad spectrum of affinities. Quantitation therefore would provide a fingerprint unique to that protein, because it seems exceedingly unlikely that any two proteins would bind to thousands of different compounds with similar affinities.
Although this approach to protein identification seems reasonable, a number of important technical issues must be addressed to determine its feasibility. For example, libraries that are either large and/or rich in general protein-binding compounds must be made and arrayed, because the fingerprinting concept would not work if only a few compounds on an array bound the target protein above background. Another critical issue is the reproducibility of this sort of experiment. Derivation of a fingerprint will be far more demanding in this regard than identifying a few of the brightest spots on the array, as is the case in ligand discovery screens.
We show here that arrays comprised of several thousand peptoids (oligo-N-substituted glycines) (25–28) can indeed be used to measure protein fingerprints in a highly reproducible fashion. We also demonstrate that this approach can be combined with an antibody-based “sandwich assay” to identify proteins in complex mixtures without chemical labeling of the sample. These studies suggest that peptoid-based SMMs could be developed as useful tools for protein identification in biological milieu.
Materials and Methods
General Remarks. All chemicals and solvents were purchased from commercial suppliers and used without further purification. Ubiquitin (Ub) was obtained from Sigma–Aldrich, maltose-binding protein (MBP) was from New England Biolabs, goat anti-mouse IgG and Alexa Fluor 488 were obtained from Molecular Probes, and anti-glutathione S-transferase (GST) antibody was from Santa Cruz Biotechnology. All proteins were labeled by using standard protocols (27, 29). The slides were scanned by using a ScanArray ExpressHT Microarray Scanner (PerkinElmer) at 10-μm resolution with 488- and 532-nm excitation lasers.
Peptoid Library. The library used in this study was synthesized by using the “submonomer” method (30) as described in ref. 27, except that microwave irradiation (31) was used to accelerate all of the synthetic steps.
Preparation of Peptoid Microarrays. Peptoid stock solutions were printed onto a chemically functionalized (maleimide) glass slides by using SpotArray 72 Microarray Printing System (PerkinElmer). The slides then were allowed to stand for 15 h on the printer platform and washed 1 h each with DMSO, dimethylformamide, tetrahydrofuran, and isopropanol. Slides were dried by centrifugation and stored under argon at room temperature. Full details of slide preparation are available from the authors on request.
Microarray Hybridization and Image Analysis. Peptoid microarrays were scanned and hybridized with various proteins as described in ref. 29. Slides were equilibrated with 1× TBST (50 mM Tris/150 mM NaCl/0.1% Tween 20, pH 7.4) for 15 min and blocked with Escherichia coli lysate for 1 h at 4°C. After each step, microarray slides were rinsed in 1× TBST before applying the next protein. Each protein was diluted (see text for concentrations) with 1× TBST containing 100-fold excess of E. coli lysate and applied to the slides. Microarray slides then were incubated for 2 h at 4°C with gentle shaking. The slides were rinsed once and washed with 1× TBST (3 × 4 min), then dried by centrifugation.
Microarrays were scanned with a ScanArray ExpressHT Microarray Scanner using blue 488-nm (fluorescein-labeled proteins) and green 543-nm (Cy3-labeled proteins) lasers at 100% power and 70% photomultiplier tube gain. All of the scanned images were analyzed by using the genepix pro 5.0 software (Axon Instruments, Union City, CA). Local background subtracted mean spot intensities were used for further analysis. All of the spot intensities of a slide before protein hybridization were subtracted from the same slide after protein hybridization in excel to get the true signal intensity due to protein binding to each feature. These true signal intensities were used for downstream analysis with genespring software (Silicon Genetics, Palo Alto, CA). Only features that gave positive signal intensity values on both experiments were used to determine reproducibility and standard correlation values. We used these intensities rather than a ratio in which the intensity at any given spot is referenced to that obtained at a control feature. This approach, which is used routinely in DNA microarray analysis, is problematic in this case. In a DNA microarray, or any array in which each feature is designed to capture a specific analyte, one can design controls that are not expected to bind anything specifically. In a protein-fingerprinting experiment, it is not possible to do so.
Results
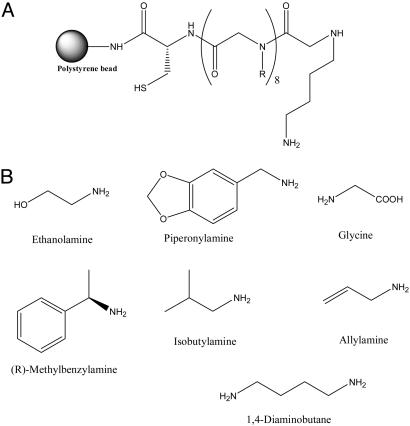
Different Proteins Exhibit Unique and Reproducible Fingerprints When Hybridized to a Peptoid Microarray. We constructed microarrays consisting of 7,680 different octameric peptoids spotted covalently on a maleimide-functionalized glass microscope slide by using a robotic pin spotter. The peptoid library was created by split and pool synthesis on 500-μm polystyrene macrobeads (Rapp Polymere, Tübingen, Germany) by using the amines shown in Fig. 1. A C-terminal cysteine residue was included in each molecule to facilitate coupling to the array surface.
Fig. 1.
The peptoid library used in this study. (A) General structure of the peptoid library synthesized on polystyrene macrobeads. (B) Amines used in the synthesis of the library.
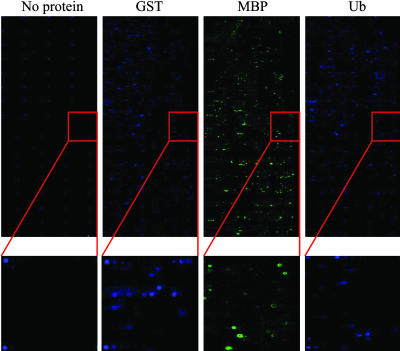
To these arrays was hybridized either fluorescein-labeled Ub, fluorescein-labeled GST, or Cy3-labeled MBP in the presence of a 100-fold excess of unlabeled proteins derived from a crude E. coli extract (to mimic a moderately abundant protein in a crude extract). The concentration of the labeled protein was 500 nM in each case. After washing, the pattern of binding of the labeled protein to the array was visualized by using a standard commercial microarray scanner. Each experiment was done twice in a completely independent fashion. The raw array images from the first set of hybridizations are shown in Fig. 2, along with an image of an array taken before protein hybridization (far left). The visible spots on this control array represent fluorescein-containing marker peptoids spotted as navigation aids. Hundreds of features on each array captured labeled protein at a level detectable above the background under these conditions, as can be better seen in the expanded regions shown in Fig. 2. These data confirm that a library of 7,680 peptoids is sufficiently rich in protein ligands to support a fingerprinting application.
Fig. 2.
Protein profiling using peptoid microarrays. Images were obtained by incubating fluorescently labeled GST, MBP, or Ub to a peptoid microarray containing 7,680 different compounds. For comparison, a fourth array to which no protein has been hybridized is shown on the far left. In this image, the fluorescent spots are labeled peptoids that have been attached to the slide as navigation aids. These images were obtained by scanning the arrays with a standard commercial array scanner used for DNA microarray analysis after hybridization and washing. The Insets provide a magnified view to illustrate the signal-to-noise ratio obtained in the hybridization of 500 nM labeled protein to the array.
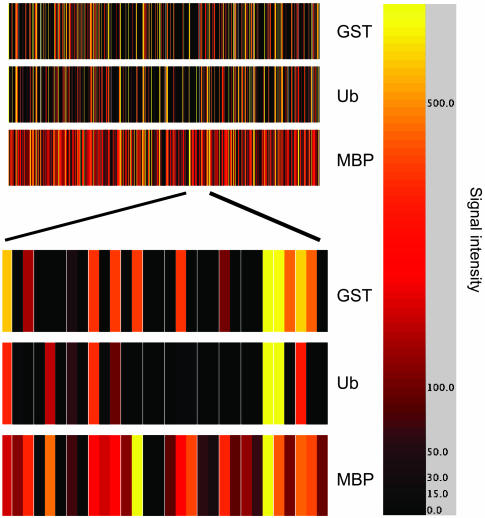
Even with the naked eye, it can be seen that the binding patterns are distinct on each array (Fig. 2). To better illustrate and quantify this distinction, the fluorescent intensity at each feature (from 1 to 7,680) was quantified and, after subtraction of the background intensity (see Materials and Methods), was displayed as a color-coded bar graph with each line representing the background-subtracted intensity of a single feature using the genespring software package (Fig. 3). Examination of these bar codes revealed that each protein provided a unique pattern of binding to the array, although, surprisingly, the Ub and GST data sets superficially resembled one another, whereas that produced in the MBP hybridization was quite different. Focusing on almost any smaller set of features clearly illustrates the differences between the Ub and GST binding patterns (Fig. 3, bottom).
Fig. 3.
A protein fingerprint. The intensities at each of the 7,680 features on the arrays shown in Fig. 2 (and the replicate experiments) were assigned a color code (shown on the right) and displayed as a bar code to allow visualization of the binding patterns. A portion of this bar code is expanded to show clearly the differences between the binding patterns.
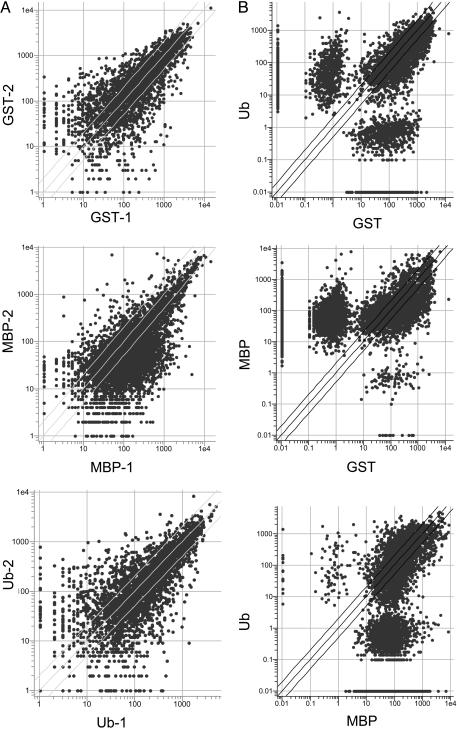
To address the critical issue of reproducibility and the uniqueness of the fingerprint in another way, all of the data were visualized as a series of scatter plots. When the two independent hybridizations for a given protein were compared, a high degree of correlation was obtained (Fig. 4A)(R = 0.97 for GST, 0.96 for MBP, and 0.97 for Ub). An even better correlation was obtained if only the higher-intensity features were considered, as one would expect, because the data closest to the background tend to be the “noisiest” (see Fig. 6, which is published as supporting information on the PNAS web site) (R = 0.99 for all proteins). To better display how many peptoids bind promiscuously to all of the proteins and how many are specific, all of the features that display an intensity value of ≥10-fold above background were incorporated into a Venn diagram (see Fig. 7, which is published as supporting information on the PNAS web site); 191, 126, and 61 peptoids were found to be quite specific for GST, MBP, and Ub, respectively. We conclude that the technique is sufficiently reproducible and has a high enough degree of specificity to serve as a platform for protein fingerprinting.
Fig. 4.
Reproducibility and the degree of uniqueness of the protein fingerprint. (A) Scatter plots comparing the data obtained in two completely independent experiments employing the same protein. High correlation coefficients were obtained (see text). (B) Scatter plots comparing the data obtained in the three different protein hybridization experiments, showing the large number of off-diagonal spots, which represent features that discriminate between the two proteins. Much lower correlation coefficients were obtained (see text).
When the data sets obtained from two different protein hybridizations were compared, the correlation was far lower (see Fig. 4B) (R = 0.56 for GST vs. Ub, 0.28 for GST vs. MBP, and 0.25 for Ub vs. MBP). Based on these data, we conclude that the three different proteins used in this study exhibit highly reproducible and distinctive patterns when hybridized to the peptoid microarray.
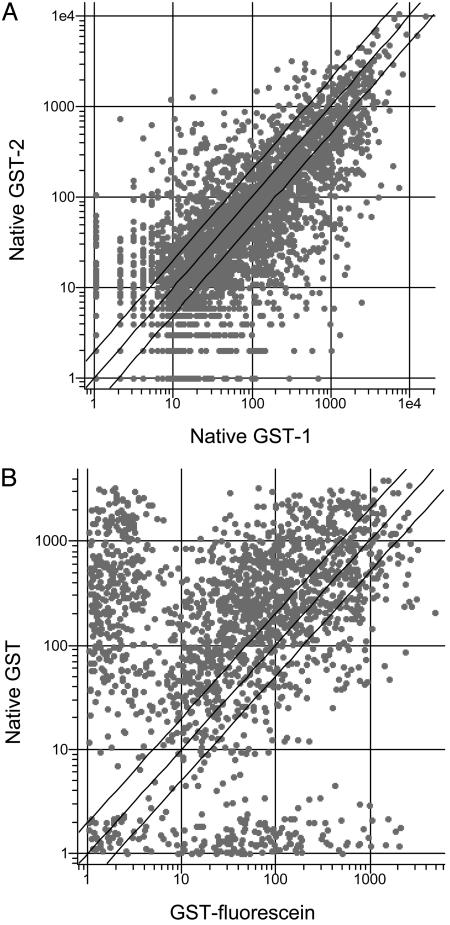
An Antibody Sandwich Assay Allows Detection of the Binding Pattern of an Unlabeled Protein. The proof of principle experiments presented above used purified proteins that had been labeled covalently and then mixed with the bacterial extract. Obviously, this type of protocol could not have been used to detect a native protein. An alternative method that would achieve this goal would be to hybridize to the array a mixture of native proteins, then probe the chip with an antibody raised against a particular protein of interest, followed by a labeled secondary antibody (13). Therefore, we performed an experiment in which unlabeled GST was doped into a 100-fold excess of bacterial proteins and hybridized to the array. To visualize the GST binding pattern selectively, the array was then probed with an anti-GST antibody followed by a secondary antibody labeled with Alexa Fluor 488. As a control, a second array was treated in the same way, except that the GST was omitted. The fluorescent intensities at each feature then were measured, and the values obtained in the “–GST” experiment were subtracted from those in the “+GST” experiment to provide a corrected data set from which the binding patterns of the antibodies had been subtracted. Again, two completely independent experiments were conducted, and these experiments were highly reproducible (Fig. 5A)(R = 0.96). A comparison of this data set with that obtained with the chemically labeled GST (Fig. 5B) revealed a similar, but not identical, protein fingerprint. The correlation coefficient was R = 0.84. Although errors introduced in the subtraction of the “antibody-only” control data could play some role in this reduced correlation, the similarity of this scatter plot to those generated by using data sets from the hybridization of different proteins (Fig. 4B) suggests an alternative explanation. The dramatically “off-diagonal” features clearly cluster into two groups, one of which registers a much higher signal intensity in the antibody-using experiment whereas the other provides a much higher signal in the experiment by using chemically labeled GST. This result suggests that the features in these two groups discriminate significantly between the labeled and native forms of the GST. In other words, the simplest explanation of the data are that native and fluorescein-labeled GST appear to the array as related, but different, proteins, although detailed binding studies of some of these peptoids to each form of the protein will be required to substantiate this hypothesis. In any case, this experiment shows clearly that a distinctive fingerprint of a native protein can be detected on the peptoid microarray by using an antibody-based sandwich assay.
Fig. 5.
Fingerprinting native GST by using an antibody sandwich assay (see text for details). (A) Scatter plot comparing the two completely independent experiments using native GST. (B) Scatter plot comparing the data obtained with native GST and fluorescein-labeled GST (Fig. 2).
Sensitivity of the Peptoid Microarrays. The experiments discussed above used the target protein at a concentration of 500 nM in the presence of a 100-fold excess of bacterial proteins. To determine whether more dilute proteins could be detected and finger-printed by using the peptoid microarray, fluorescein-labeled GST solutions of 100 and 10 nM, again in a solution containing a 100-fold excess of bacterial proteins, were prepared and hybridized to the peptoid arrays. After washing, the arrays were scanned, and the intensity observed at each feature was quantified as described above. The data then were compared. This comparison revealed an excellent correlation (see Fig. 8, which is published as supporting information on the PNAS web site) between the data sets obtained at the different GST concentrations, with correlation coefficients of R = 0.95, R = 0.97, and R = 0.95 for comparison of the 500 vs. 100 nM, 100 vs. 10 nM, and 500 vs. 10 nM data sets, respectively. In other words, very similar protein fingerprints could be discerned at each of the protein concentrations examined. In another experiment, GST (500 nM) was serially diluted into an E. coli lysate to create solutions with a 500-fold (GST = 0.5% of total protein) or 1,000-fold (GST = 0.1% of total protein) excess of bacterial proteins. After hybridization to the arrays and detection as described above, the data sets were compared on a scatter plot (see Fig. 9, which is published as supporting information on the PNAS web site). The R values derived from these scatter plots were 0.96 for 100-fold vs. 500-fold, 0.97 for 100-fold vs. 1,000-fold, and 0.97 for 500 vs. 1,000-fold. In other words, these data show that a recognizable and consistent fingerprint is observed for a protein present in an extract at different levels and is not sensitive to the abundance of that protein. Another issue that can be addressed with these 8data sets is whether the signal intensities on the arrays accurately reflected the absolute concentration of the protein. To address this point, the signal intensities for all of the features that displayed above-background binding in the 500 and 100 nM GST data sets were placed on a scatter plot (see Fig. 8). The slope of the best-fit line through these data was ≈5.7. When only the 100 most intense spots were considered, the average intensity difference between the two experiments was 5.2-fold (data not shown). Thus, the array results reflect the protein concentration in these two experiments. However, when the same type of analyses were repeated for the 100- and 10-nM GST data sets, the average difference was only ≈3-fold rather than the 10-fold expected ideally. In other words, the intensities in the 10-nM data set compared with 100-nM data set did not drop off as much as one would expect based on the absolute protein concentration. These data suggest that we are approaching the sensitivity limit of the array somewhere between 100 and 10 nM fluorescein-labeled GST.
Discussion
Both biological (32) and artificial (33, 34) sensors containing a large number of low to modest affinity receptors can differentiate between different molecules by distinguishing quantitative patterns of binding of an analyte to the different receptors. We have combined this concept with peptoid microarrays to create a powerful platform for the identification of proteins. Proof of principle experiments using chemically labeled proteins (Figs. 2, 3, 4) demonstrated that different proteins exhibit a different pattern of binding to the thousands of peptoids on the array. We also demonstrated that a protein fingerprint could be discerned when a native protein (GST) in a solution containing a large excess of bacterial proteins was hybridized to the array (Fig. 5). Selective visualization of the GST binding pattern even in the presence of a large excess of other proteins was achieved by conducting a second hybridization with an anti-GST antibody. The pattern due to antibody binding was determined in a control experiment and subtracted from these data. This finding is critical in further development of these arrays as bioanalytical tools, because selective chemical labeling of a protein of interest in a complex mixture can rarely be achieved. A small amount of analytical work has been done to distinguish purified proteins with simpler arrays in other configurations (14–16). However, this work provides a previously undescribed demonstration that a glass slide-based SMM can “fingerprint” a native protein in a complex mixture. Because the proteins used in this study, GST, MBP, and Ub, are not related to one another and because the molecules in the library used to make the array (Fig. 1) were not biased in any way, we presume that the peptoid microarray is capable of fingerprinting almost any protein, although substantiation of this hypothesis obviously will require further study.
This array-based fingerprinting technique proved to be fairly sensitive, with a recognizable fingerprint produced by using a GST solution of only 10 nM in the presence of a 100-fold excess of bacterial proteins (Fig. 8). This issue was a major question to be addressed going into this study, because none of the peptoids were anticipated to be high-affinity ligands for GST or any other protein. Our experience with peptoid library screening suggests that even the best ligands in an unbiased library are likely to form complexes with KD values in the low-micromolar range (27). However, the linearity of the signal intensities deviated from the expected values between 100 and 10 nM for labeled GST, indicating that the sensitivity limit for truly quantitative work was somewhere between 100 and 10 nM GST in this particular case. Nonetheless, this limit would be sufficient to fingerprint many highly and modestly abundant proteins in serum or cell extracts. Furthermore, this limit is not necessarily a general sensitivity limit of the array, because this limit will depend on the affinities of the protein analyzed for the peptoids on the array. For example, we have evidence that antibodies and certain other proteins bind with unusually high affinity to many peptoids (M.M.R. and T.K., unpublished results) on the microarray. It is also possible that much higher sensitivity could be achieved by using more intensely fluorescent dyes such as quantum dots or with detection schemes that employ some sort of signal amplification.
The experiments reported here are model studies and break no new biological ground. The presence and levels of a protein in a complex mixture could have been determined with SDS/PAGE and Western blotting. Nonetheless, these experiments demonstrate two critical points that were far from obvious before these studies. The first is that the patterns obtained are highly reproducible (Fig. 4) in completely independent experiments. The second is that even an array with a relatively modest number of features (<8,000) is capable of a high level of discrimination between two proteins. In other words, the collection of peptoids is sufficiently rich in specific ligands for any given protein that a unique pattern is produced. Indeed, it was quite interesting that a comparison between the binding patterns obtained with native GST and fluorescein-labeled GST were noticeably different. This finding suggests that the distinguishing power of the array may be sufficient to derive different patterns for the same protein with different posttranslational modifications. This application would be an attractive use of these arrays, because antibodies that recognize specific forms of a given protein are often difficult to obtain. To read the array pattern, only a standard antibody that recognized any form of the protein would be necessary.
Another potential application of this array technology is protein profiling for diagnostic purposes. By using other analytical platforms, particularly surface-enhanced laser desorption ionization mass spectrometry, several investigators have argued that protein profiling of complex samples can be used to diagnose cancers and other disease states (for reviews, see refs. 35–37). This application is a different kind of fingerprint or profiling experiment than we have reported here using SMM array technology. In Protein Chip/surface-enhanced laser desorption ionization experiments, the subset of the serum proteome that binds to the surfaces on the chip and that are ionized efficiently in the mass spectrometer is analyzed to produce a pattern of peaks that is taken as a potential fingerprint of a disease state. This technique thus looks at a large number of different proteins whose identities are unknown. Although we have focused on binding patterns of single proteins in this study, one could easily imagine examination of binding “superpatterns” comprised of the superposition of many individual protein-binding events. Such an experiment would use a visualization reagent that would recognize many different proteins, such as an anti-phosphotyrosine antibody that would “light up” any protein with this modification. Another possibility would be to label certain classes of enzymes, such as serine proteases, with activity-based labeling reagents before hybridization (38). The high level of reproducibility observed in the simpler experiments reported here suggests that the peptoid array technology would be highly competitive with surface-enhanced laser desorption ionization for more complex profiling applications.
Supplementary Material
Acknowledgments
We thank Jonathan Lawson, Rhonda Friedberg, Prof. Harold Garner (University of Texas Southwestern), and Srujana Chandi for technical assistance with the genespring software program. We also thank Prof. Stephen Albert Johnston (University of Texas Southwestern), Prof. Harold Garner, and Dr. Robert Carlson (Receptors, LLC, Chaska, MN) for helpful discussions and comments on the manuscript. This work was supported by National Heart, Lung, and Blood Institute Contract NO1-HV-28185 for University of Texas Southwestern Proteomics Research.
Author contributions: T.K. designed research; M.M.R. performed research and contributed new reagents/analytic tools; and M.M.R. and T.K. analyzed data and wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MBP, maltose-binding protein; SMM, small-molecule microarray; Ub, ubiquitin.
References
- 1.Uttamchandani, M., Walsh, D. P., Yao, S. Q. & Chang, Y.-T. (2005) Curr. Opin. Chem. Biol. 9, 4–13. [DOI] [PubMed] [Google Scholar]
- 2.MacBeath, G., Koehler, A. N. & Schreiber, S. L. (1999) J. Am. Chem. Soc. 121, 7967–7968. [Google Scholar]
- 3.Lam, K. S. & Renil, M. (2002) Curr. Opin. Chem. Biol. 6, 353–358. [DOI] [PubMed] [Google Scholar]
- 4.Blackwell, H. E., Perez, L., Stavenger, R. A., Tallarico, J. A., Eatough, E. C., Foley, M. A. & Schreiber, S. L. (2001) Chem. Biol. 8, 1167–1182. [DOI] [PubMed] [Google Scholar]
- 5.Clemons, P. A., Koehler, A. N., Wagner, B. K., Sprinings, T. G., Spring, D. R., King, R. W., Schreiber, S. L. & Foley, M. A. (2001) Chem. Biol. 8, 1183–1195. [DOI] [PubMed] [Google Scholar]
- 6.Fodor, S. P., Read, J. L., Pirrung, M. C., Stryer, L., Lu, A. T. & Solas, D. (1991) Science 251, 767–773. [DOI] [PubMed] [Google Scholar]
- 7.Frank, R. (2002) J. Immunol. Methods 267, 13–26. [DOI] [PubMed] [Google Scholar]
- 8.Li, S., Bowerman, D. J., Marthandan, N., Garner, H. R. & Kodadek, T. (2004) J. Am. Chem. Soc. 126, 4088–4089. [DOI] [PubMed] [Google Scholar]
- 9.Shaginian, A., Patel, M., Li, M.-H., Flickinger, S. T., Kim, C., Cerrina, F. & Belshaw, P. J. (2004) J. Am. Chem. Soc. 126, 16704–16705. [DOI] [PubMed] [Google Scholar]
- 10.Li, S., Marthandan, N., Bowerman, D., Garner, H. R. & Kodadek, T. (2005) Chem. Commun., 581–583. [DOI] [PubMed]
- 11.Kuruvilla, F. G., Shamji, A. F., Sternson, S. M., Hergenrother, P. J. & Schreiber, S. L. (2002) Nature 416, 653–657. [DOI] [PubMed] [Google Scholar]
- 12.Winssinger, N., Ficarro, S., Schultz, P. G. & Harris, J. L. (2002) Proc. Natl. Acad. Sci. USA 99, 11139–11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koehler, A. N., Shamji, A. F. & Schreiber, S. L. (2003) J. Am. Chem. Soc. 125, 8420–8421. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi, M., Nokihara, K. & Mihara, H. (2003) Chem. Biol. 10, 53–60. [DOI] [PubMed] [Google Scholar]
- 15.Usui, K., Ojima, T., Takahashi, M., Nokihara, K. & Mihara, H. (2004) Biopolymers (Pept. Sci.) 76, 129–139. [DOI] [PubMed] [Google Scholar]
- 16.Baldini, L., Wilson, A. J. & Hamilton, A. D. (2004) J. Am. Chem. Soc. 126, 5656–5657. [DOI] [PubMed] [Google Scholar]
- 17.Harris, J. L., Alper, P. B., Li, J., Rechsteiner, M. & Backes, B. J. (2001) Chem. Biol. 8, 1131–1141. [DOI] [PubMed] [Google Scholar]
- 18.Mason, D. E., Ek, J., Peters, E. C. & Harris, J. L. (2004) Biochemistry 43, 6535–6544. [DOI] [PubMed] [Google Scholar]
- 19.Martin, K., Steinberg, T. H., Cooley, L. A., Gee, K. R., Beechem, J. M. & Patton, W. F. (2004) Proteomics 3, 1244–1255. [DOI] [PubMed] [Google Scholar]
- 20.Falsey, J. R., Renil, M., Park, S.-H., Li, S. & Lam, K. S. (2001) Bioconj. Chem. 12, 346–353. [DOI] [PubMed] [Google Scholar]
- 21.Lesaicherre, M.-L., Mahesh, U., Chen, G. Y. J. & Yao, S. Q. (2002) Bioorg. Med. Chem. Lett. 12, 2085–2088. [DOI] [PubMed] [Google Scholar]
- 22.Jellis, C. L., Cradick, T. J., Rennert, P., Salinas, P., Boyd, J., Amirault, T. & Gray, G. S. (1993) Gene 137, 63–68. [DOI] [PubMed] [Google Scholar]
- 23.Beattie, J., Shand, J. H. & Flint, D. J. (1996) Eur. J. Biochem. 239, 479–486. [DOI] [PubMed] [Google Scholar]
- 24.Reineke, U., Ivascu, C., Schlief, M., Landgraf, C., Gericke, S., Zahn, G., Herzel, H., Volkmer-Engert, R. & Schneider-Mergener, J. (2002) J. Immun. Methods 267, 37–51. [DOI] [PubMed] [Google Scholar]
- 25.Simon, R. J., Kania, R. S., Zuckermann, R. N., Huebner, V. D., Jewell, D. A., Banville, S., Ng, S., Wang, L., Rosenberg, S., Marlowe, C. K., et al. (1992) Proc. Natl. Acad. Sci. USA 89, 9367–9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Figliozzi, G. M., Goldsmith, R., Ng, S. C., Banville, S. C. & Zuckermann, R. N. (1996) Methods Enzymol. 267, 437–447. [DOI] [PubMed] [Google Scholar]
- 27.Alluri, P. G., Reddy, M. M., Bacchawat-Sikder, K., Olivos, H. J. & Kodadek, T. (2003) J. Am. Chem. Soc. 125, 13995–14004. [DOI] [PubMed] [Google Scholar]
- 28.Heine, N., Ast, T., Schneider-Mergener, J., Reineke, U., Germeroth, L. & Wenschuh, H. (2003) Tetrahedron 59, 9919–9930. [Google Scholar]
- 29.Reddy, M. M., Bachhawatt-Sikder, K. & Kodadek, T. (2004) Chem. Biol. 11, 1127–1137. [DOI] [PubMed] [Google Scholar]
- 30.Zuckermann, R. N., Kerr, J. M., Kent, S. B. H. & Moos, W. H. (1992) J. Am. Chem. Soc. 114, 10646–10647. [Google Scholar]
- 31.Olivos, H. J., Alluri, P. G., Reddy, M. M., Saloney, D. & Kodadek, T. (2002) Org. Lett. 4, 4057–4059. [DOI] [PubMed] [Google Scholar]
- 32.Axel, R. (1996) Cold Spring Harbor Symp. Quant. Biol. 61, 135–145. [PubMed] [Google Scholar]
- 33.Rakow, N. A. & Suslick, K. S. (2000) Nature 406, 710–713. [DOI] [PubMed] [Google Scholar]
- 34.Goodey, A., Lavigne, J. J., Savoy, S. M., Rodriguez, M. D., Curey, T., Tsao, A., Simmons, G., Wright, J., Yoo, S.-J., Sohn, Y., et al. (2001) J. Am. Chem. Soc. 123, 2559–2570. [DOI] [PubMed] [Google Scholar]
- 35.Wulfkuhle, J. D., Liotta, L. A. & Petricoin, E. F. (2003) Nat. Rev. Cancer 3, 267–276. [DOI] [PubMed] [Google Scholar]
- 36.Pweletz, C. P., Gillespie, J. W., Ornstein, D. K., Simone, N. L., Brown, M. R., Cole, K. A., Wang, Q.-H., Huang, J., Hu, N., Yip, T.-T., et al. (2000) Drug. Dev. Res. 49, 34–42. [Google Scholar]
- 37.Petricoin, E. F., Wulfkuhle, J., Espina, V. & Liotta, L. A. (2004) J. Proteome Res. 3, 209–217. [DOI] [PubMed] [Google Scholar]
- 38.Barglow, K. T. & Cravatt, B. F. (2004) Chem. Biol. 11, 1523–1531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.