Abstract
Apical membrane antigen 1 (AMA1) is a leading malaria vaccine candidate that possesses polymorphisms that may pose a problem for a vaccine based on this antigen. Knowledge of the distribution of the polymorphic sites on the surface of AMA1 is necessary to obtain a detailed understanding of their significance for vaccine development. For this reason we have sought to determine the three-dimensional structure of AMA1 using x-ray crystallography. The central two-thirds of AMA1 is relatively conserved among Plasmodium species as well as more distantly related apicomplexan parasites, and contains two clusters of disulfide-bonded cysteines termed domains I and II. The crystal structure of this fragment of AMA1 reported here reveals that domains I+II consists of two intimately associated PAN domains. PAN domain I contains many long loops that extend from the domain core and form a scaffold for numerous polymorphic residues. This extreme adaptation of a PAN domain reveals how malaria parasites have introduced significant flexibility and variation into AMA1 to evade protective human antibody responses. The polymorphisms on the AMA1 surface are exclusively located on one side of the molecule, presumably because this region of AMA1 is most accessible to antibodies reacting with the parasite surface. Moreover, the most highly polymorphic residues surround a conserved hydrophobic trough that is ringed by domain I and domain II loops. Precedents set by viral receptor proteins would suggest that this is likely to be the AMA1 receptor binding pocket.
Keywords: malaria, vaccine
Malaria is a major factor in maintaining the economic hardship of the world's poorest societies and is responsible for 2% of global mortalities (1, 2). Most instances of death and severe disease result from Plasmodium falciparum infections of young children. After several episodes of malaria, individuals develop resistance to the more severe forms of the disease (3), which, at least in part, is the result of the development of protective antibodies (4–6). Development of a vaccine that augments naturally acquired immunity should, therefore, be possible. Malaria parasites are highly complex, contain >5,000 genes (7), and, in the human host, develop through several antigenically distinct life cycle forms. Consequently, identifying the antigens responsible for the development of immunity has not been an easy task, although a number of experimental vaccines are now under development. The most advanced of these, the RTS,S/ASO2A vaccine based on the circumsporozoite surface protein, was shown to significantly reduce P. falciparum infections for 6 months (8). The efficacy of RTS,S/AS02A could no doubt be improved by being combined with one or more other antigens, particularly antigens capable of inducing protective immunity against asexual blood stages of P. falciparum. AMA1 is one of the most promising candidates and has recently been tested in a phase I clinical trial (9).
AMA1 is expressed in two critical life-cycle forms, the sporozoite, which invades hepatocytes, and the merozoite, which invades red blood cells, and so offers a unique opportunity as a non-stage-specific vaccine target. AMA1 is conserved in all Plasmodium species and in the phylum of apicomplexa parasites and therefore offers the potential for development of vaccines or therapeutics to a wide range of human and animal parasitic diseases (10, 11). Native and recombinant AMA1 have been shown to function as vaccines in a variety of animal models of human malaria (12–16). The protective immune effector mechanism appears to be antibodies that block merozoite invasion of red blood cells. Anti-AMA1 antibodies prevent the invasion of host cells by parasites in vitro (17, 18), but inhibition can be strain-specific (19). For example, antibodies to strain 3D7 AMA1 were noninhibitory for strain W2mef, and this was shown to be due to the AMA1 sequence differences of these two strains (20). This result is consistent with analyses of AMA1 sequences which indicate that the ama1 gene is under diversifying selection (21–23). The recent demonstration that anti-AMA1 antibodies in humans are associated with protection from malaria (24) provides support for the view that AMA1 polymorphisms are being selected in response to naturally acquired protective immune responses of humans.
The exact function of AMA1 is not clear, although ama1 is an essential gene (10, 25), and both domain I and domain III of AMA1 are likely to be functionally critical. Both are bound by inhibitory antibodies in vitro, and the clustering of polymorphisms suggests that they are both targets of protective antibody responses in humans (21, 26–29). Domain I is conserved among Plasmodium species and in other apicomplexan parasites. It presumably has a function common to all apicomplexa that is not specific to a particular host cell type. Domain III is not well conserved in Babesia bovis and Toxoplasma gondii AMA1 sequences, and this region of AMA1 may have a Plasmodium genus-specific function. AMA1 is released from micronemes at the time of invasion, and antibodies that target AMA1 allow merozoites to attach to red blood cells but prevent merozoite reorientation and the establishment of a tight junction (30, 31). Isolated recombinant preparations of AMA1 do not interact with red blood cells, but other studies suggest that an AMA1–red cell interaction may occur (25, 32).
In this study, we determined the structure of the central portion of AMA1 to examine the location of the polymorphisms and to determine the impact that the polymorphisms will have on AMA1 as a potential vaccine. The AMA1 polymorphisms were clustered in two ways. Polymorphic sites where only two amino acids are found (dimorphisms) are concentrated on one face of AMA1, presumably the face that is exposed on the parasite surface. In contrast, highly polymorphic residues surround a hydrophobic pocket that we predict is critical to AMA1 function.
Materials and Methods
Protein Expression, Purification, and Crystallization. AMA1 domains I+II (104-NYMG-PIEVE-438) was expressed in Escherichia coli as a N-terminal His-tagged protein and refolded by dialysis into a buffer containing a redox couple, 2 mM 2-mercaptoethanol and 0.2 mM cystamine, to allow disulfide bond formation. The protein was extensively purified, the tag was removed by using TEV protease, and crystals were grown in 20 mM MES (pH 6), 10 mM MnCl2, and 8% PEG 3350. Crystals were “dehydrated” by placing them in mother liquor over 35% PEG and transferred into a cryo or stabilization solution (88 mM MES, pH 6/44 mM MnCl2/38% PEG 3350). This procedure considerably improved the crystal mosaicity and diffraction resolution. The rational for the choice of the NYMG-PIEVE fragment and the expression, purification, and crystallization procedures are described in detail in ref. 33.
Heavy Atom Derivatization and Data Collection. Heavy atoms that bound to AMA1 in solution were identified by electrophoresis mobility-shift experiments in native gels (34). Successful derivatives resulted from a 20-min incubation in 10 mM K2ReCl6 and an overnight incubation in 2 mM K2IrCl6. K2IrCl6 data sets were collected in-house with crystals maintained at 100 K by using a rotating anode generator and image plate detector. Native and Re data sets were collected at beamline X25 at the National Synchrotron Light Source, Brookhaven National Laboratory. All data sets were collected in 0.5° sections. The K2ReCl6 anomalous data set was collected by using inverse beam geometry every 20°. The native data set was collected in two 240° sweeps, one at long exposure (15 sec) and one at short exposure (3 sec), and the data were merged. Data sets were indexed by using d*trek (35) or hkl2000 (36). Data sets were scaled by using scaleit, and heavy atom sites were identified by using Patterson and difference Fourier maps (37). K2IrCl6 bound to three sites in the asymmetric unit cell, and K2ReCl6 bound to two sites. One of the sites was occupied by both compounds.
Structure Determination. Phases calculated by using mlphare were reasonable to 4 Å resolution (figure of merit = 0.5) but dropped precipitously in quality at higher resolution. The asymmetric unit cell contained two AMA1 molecules with a solvent content of 50%. At low resolution, data sets could be indexed in space group p3121, indicating that the noncrystallographic symmetry (NCS) twofold axis was almost perpendicular to the p31 screw axis, confirmed by the presence of twofold self-rotation peaks identified by using polarrfn (37). Solvent-flattened maps were generated by using dm (37), and the NCS transformation matrix was calculated by using heavy atom sites and by using the bones ncs_op facility in o (38). NCS-averaged, solvent-flattened, and histogram-matched maps were calculated with dm by using the automask option. Phases could be extended to3Åresolution before the electron density started to become disconnected. The resulting map was of sufficient quality to allow ≈75% of the structure to be traced. Iterative cycles of refinement using refmac (39) or cns (40) and model building in sigmaa-weighted maps at increasing resolution using o (38) were used to generate the final refined structure. The same set of test reflections was always used. Refinement statistics are for all data extending from 30–1.9 Å with TLS parameters refined for fixed individual B factors before refinement of individual isotropic B factors. Each molecule in the asymmetric unit cell was defined as a TLS group.
Analysis of Polymorphisms. A set of 129 nonredundant P. falciparum AMA1 sequences was compiled from several studies (21–23, 41). The lowest coverage was at the N terminus of the crystallized region (34 sequences). Polymorphic sites were categorized as “highly polymorphic” if the third most abundant amino acid had a frequency of at least 5% in the nonredundant set. Dimorphic sites were categorized as “high-frequency dimorphisms” if the frequency of the less abundant amino acid was between 15% and 50%. For “low-frequency dimorphisms,” the frequency of the less abundant amino acid was between 2% and 15%. Amino acids below a frequency of 2% were assumed to be neutral mutations or errors.
Results and Discussion
Overview of the Crystal Structure. The fragment of AMA1 that we crystallized includes domains I+II of AMA1 and 20 aa of sequence N-terminal to domain I. Domains I and II encompass the central portion of AMA1 and make up the only portion of the extracellular region of AMA1 that is conserved in the most evolutionarily distant AMA1 or AMA-related sequences: T. gondii AMA1 (10, 42) or MAEBL (43). The structure was solved by using the technique of multiple isomorphous replacement with anomalous scattering. The refined structure (Table 1) contains two molecules in the asymmetric unit and extends from residues 108 to 438 with a refined free R value of 23% and an R value of 19%. The two molecules in the asymmetric unit cell are virtually identical with an rms fit of 0.45 over 305 carbon alpha atoms. Three flexible loops were not observed in the electron density: a 5-aa stretch in domain I (172-GNQYL), a 9-aa stretch in domain I (265-KDESKRNSM), and a 4-aa stretch in domain II (383-GAFK).
Table 1. Crystallographic data and refinement statistics.
| Data sets | Native | K2ReCl6 | K2IrCl6 |
|---|---|---|---|
| Data reduction | |||
| Wave length, Å | 1.100 | 1.176 | 1.541 |
| Space group | p31 | p31 | p31 |
| Unit cell, Å | 54.1, 54.1, 214.1 | 54.2, 54.2, 214.8 | 54.1, 54.1, 214.1 |
| Mosaicity, ° | 0.4 | 0.5 | 0.4 |
| Resolution, Å | 30-1.9 | 30-2.7 | 30-2.4 |
| Rsym, % | 6.2 (30) | 10.6 (44) | 7.9 (38) |
| Completeness, % | 99.2 (100) | 99.9 (100) | 99.9 (100) |
| Redundancy | 9.5 (10.5) | 7.1 (7.2) | 2.7 (2.8) |
| I/σ | 30 (11) | 41 (8) | 15 (3) |
| Heavy-atom refinement (data extending to 3 Å) | |||
| Risomorphous, % | 21 | 8 | |
| Number of sites | 2 | 3 | |
| Rcullis (anomalous) | 0.89 (0.78) | 0.91 | |
| Figure of merit | 0.47 | ||
| Refinement statistics (no data cutoff) | |||
| Reflections (test) | 51,869 (2,629) | ||
| Protein atoms | 4,882 | ||
| Waters | 253 | ||
| Rcryst, % (Rfree) | 19.4 (23.5) | ||
| Average B factor, Å2 | protein 38.5, waters 38.4 | ||
| Rms deviations from ideal | |||
| Bond lengths, Å | 0.015 | ||
| Bond angles, ° | 1.44 | ||
| Ramachandran plot* | 87.4% core, 12.2% allowed, 0.4% generous, 0.0% disallowed | ||
Data reduction statistics values in parentheses are for the outer shell.
Proportion of the molecule within the most favored of disfavored phi/psi regions determined by using procheck (58).
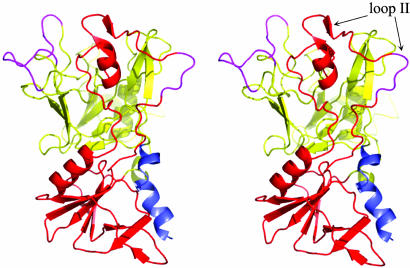
The structure of AMA1 domains I+II can be split into two domains: domain I (yellow, Fig. 1) and domain II (red, Fig. 1). The N-terminal extension forms a distorted helix and packs against domain II. A loop extending from domain II contains an α-helix and a pair of β-strands and packs against one side of domain I (loop II, Fig. 1). Apart from loop II, domain II forms a typical protein domain in that it largely consists of closely packed secondary structure elements that associate mostly by hydrophobic interactions. At the core of domain II is a five-stranded β-sheet that packs against a 12-residue α-helix on one face in addition to interacting with three other β-strands on the opposite face (Fig. 1).
Fig. 1.
Stereo view of the AMA1 domain I+II structure showing the two interconnected domains. The 20 aa from the N-terminal extension are colored blue, domain I is yellow, domain II is red, and loops that are disordered in the structure are violet. This and all other figures depicting the structure were generated by using pymol (www.pymol.org).
The structure of domain I is considerably more irregular. Large sections of domain I consist of loops that are interspersed with secondary structure elements. Most of the loops are well ordered in the structure, even at the protein surface. This is partially the result of fortuitous crystal contacts. The most extensive crystal contact (1,660 Å2) results from the intercalation of three of the domain I loops (data not shown). The loops adopt identical conformations in the two AMA1 molecules in the asymmetric unit cell; however, on the parasite surface these loops may be flexible and adopt alternate conformations. The irregularity of domain I extends into its interior. Approximately 50% of domain I has no secondary structure and consists of “nonrepetitive structure” (44). As is typical of nonrepetitive structure, there are many isolated turns, many buried waters, and a large number of buried hydrophilic residues that form side chain–main chain interactions. However, to one side of domain I, immediately adjacent to domain II, there is a five-stranded β-sheet, packed against a 12-residue helix.
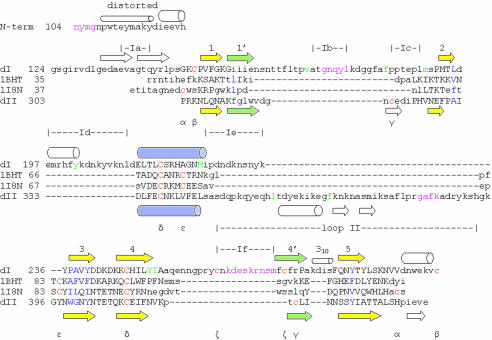
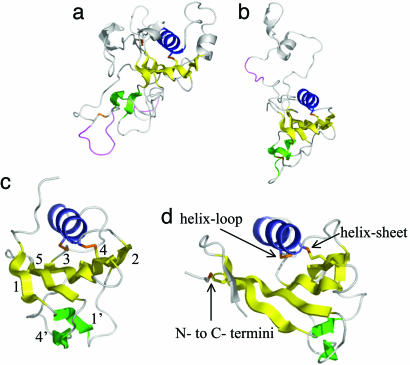
AMA1 Domains I+II Consists of Two Tandem PAN Domains. A three-dimensional alignment of the sheet and helix in domains I and II revealed a close alignment (rms fit of 1.25 Å for 55 carbon alpha atoms with a sequence identity of 13%). AMA1 domains I+II presumably results from an ancient domain duplication that has undergone significant adaptation resulting in the extensive loops in domain I and the long domain II loop. A three-dimensional homology search (45) employing the core domain II structure revealed that AMA1 domains I or II have a PAN or apple domain fold (46). The PAN domain consists of a central five-stranded sheet (yellow) that wraps around a 12-residue helix (blue) (Fig. 3). The helix and five-stranded sheet of AMA1 domains I or II align well with the two published PAN domain crystal structures. Alignments of AMA1 domain II with the N domain of hepatocyte growth factor (Fig. 2c; sequence identity of 5% over 59 atoms) (47) or Leech antiplatelet protein (Fig. 2d; sequence identity 19% over 47 atoms) (48) resulted in rms fits of ≈1.6 Å, reasonable overall alignments.
Fig. 3.
Sequence alignment based on the tertiary alignment of PAN domain crystal structures. Carbon alphas that align within 2.5 Å in the tertiary structures are shown in uppercase. N-term, the N-terminal 20 aa; dI, domain I of AMA1; dII, domain II of AMA1; 1BHT, N domain of hepatocyte growth factor (47); 1I8N, leach anti-platelet protein (48). Central PAN domain sheets are colored yellow, the PAN helix is blue, and the peripheral sheets are green, with secondary structure elements numbered as in Fig. 2. Residues in violet are disordered in the structure. Cysteines are colored orange. The same Greek letter below two cysteines indicates that they are disulfide-bonded. Residues at PAN-domain conserved hydrophobic positions are colored blue. Surface-exposed hydrophobic Plasmodium-conserved residues that line the base of the hydrophobic trough are colored green. The N-terminal “distorted” helix is helical but contains carbonyls that interact with waters, side chains, and main-chain nitrogens in 310 and α-helical connections. The short 310 helix is shown as a small cylinder. α-helices are shown as large cylinders, and β-strands as arrows. Positions of domain I loops, Ia to If, are indicated together with the domain II loop (loop II).
Fig. 2.
The PAN domains of AMA1 domain I (a) and AMA1 domain II (b), N domain of hepatocyte growth factor (c) [1BHT (47)], and the apple domain of leech antiplatelet protein (d) [1I8N (48)]. Disulfide linkages are shown in orange. The numbering of the secondary structure elements is shown in c with the corresponding numbering in the primary sequence in Fig. 3 according to the scheme of Ultsch et al. (47). Characteristic PAN or apple domain disulfides are shown in d. Disordered loops in the AMA1 structure are colored violet.
On the opposite side of the helix the central PAN domain sheet interacts with two loops. The loops connecting β-strands 1 and 3 and β-strands 4 and 5 adopt different conformations in PAN domain structures, and the 1′ and 4′ β-strands they contain are variable in length and position (Fig. 2). A primary sequence alignment based on the tertiary alignment of PAN crystal structures is shown in Fig. 3. Sequence conservation is restricted to one disulfide bond (Fig. 3, δ) and six hydrophobic residues (Fig. 3, blue). AMA1 was not previously recognized as a PAN domain protein because both domain I and II have lost disulfides that are characteristic of PAN domains and have acquired other disulfides that are not characteristic of PAN domains (Figs. 2 and 3).
PAN or apple domains occasionally exist in isolation, such as in leech antiplatelet protein, and can occur as individual domains in multiple domain proteins, such as plasminogen or hepatocyte growth factor. Most frequently, however, PAN domains are present in tandem arrays (46). For example, clotting factor XI consists of a serine protease domain fused to four tandem apple domains, each domain with a specific ligand-binding function (49–51). Similarly, T. gondii MIC-4 apple domains have distinct functions. The sixth apple domain interacts with host cells, whereas the first two apple domains are required for microneme targeting (52, 53). The number of studies in which PAN domain interactions have been examined at a molecular level is limited, but the manner by which PAN domains interact with their ligands does not appear to be conserved. The third apple domain of factor XI interacts with platelets by using side chains on β-strand 4′ or at the C terminus of the domain (51), whereas, hepatocyte growth factor N domain uses residues at the opposite surface of the domain (the helix and β-strand 2) to interact with heparin (54).
AMA1 domains I and II are remarkable in that they are extreme adaptations of the PAN domain fold, with several long loops extending from their PAN domain cores (Fig. 2 a and b). It is likely that the acquisition of the loops during AMA1 evolution has provided a means of evading protective antibody responses. The flexible loops, which would tolerate the acquisition of mutations, may divert antibodies from the functionally critical region of the PAN domain.
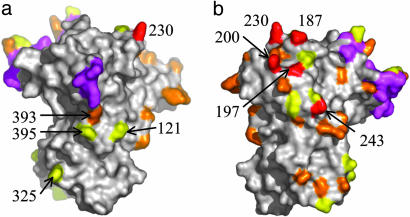
AMA1 Polymorphisms. Within the population of P. falciparum AMA1 sequences there are five “highly polymorphic” residues (positions 187, 197, 200, 230, and 243). These form a cluster at the top left of the molecule as orientated in Fig. 4b. Most AMA1 polymorphic sites are dimorphic, with only two amino acid residues found in the population (Fig. 4: orange, high frequency; yellow, low frequency). These are also clustered and are mostly located on one side of AMA1, “the polymorphic face” as viewed in Fig. 4b. There are four dimorphic residues in the nonpolymorphic face at positions 121, 325, 393, and 395 (Fig. 4a). The dimorphisms at positions 121 and 325 are at a low frequency, 3–5%, and the dimorphism at position 395 is conservative (R or K). This leaves one high-frequency nonconservative dimorphism that lies on the nonpolymorphic face (position 393, orange at center of Fig. 4a). The contrast between the two faces in the number of polymorphisms is striking. One possibility is that only a single face of AMA1 is exposed on the parasite surface.
Fig. 4.
Clustering of polymorphic residues on the surface of AMA1. Side chains 197, 200, 230, and 243, shown in red, are highly polymorphic. High-frequency dimorphisms are shown in orange, with low-frequency dimorphisms in yellow, as described in Materials and Methods. (a) Surface view of the “nonpolymorphic face.” (b) Surface view of the “polymorphic face.” Areas colored violet are disordered in the structure.
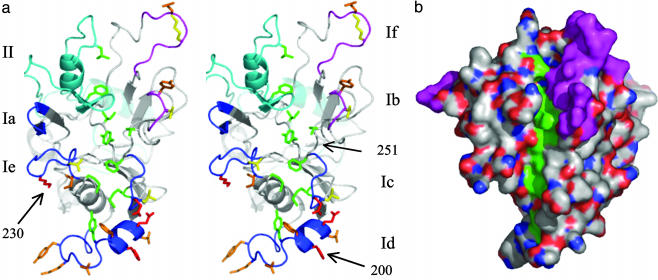
Extended Hydrophobic Pocket Between the AMA1 Loops. The selective acquisition of several loops on AMA1 domains I and II during evolution of the PAN domains suggests that the loops serve a purpose, possibly that of “protecting” a functionally critical portion of the molecule. Examination of the region between the loops revealed the presence of an extended pocket with a base that contains a series of hydrophobic side chains (Fig. 5). This hydrophobic trough consists of nine hydrophobic amino acid side chains that are solvent exposed and hydrophobic in all Plasmodium AMA1 sequences (Figs. 3 and 5). Tyrosine 251, at the center of the trough, rises above the floor of the trough, and is identical in all AMA1 sequences, even those of the more distantly related T. gondii and B. bovis parasites (10, 11). Similarly, V169, F183, L357, and F367 are hydrophobic in all apicomplexan sequences. The overall features of the hydrophobic trough are, therefore, conserved in all AMA1 molecules.
Fig. 5.
AMA1 loops surround a conserved hydrophobic trough. (a) Stereo view with surface-exposed Plasmodium-conserved hydrophobic residues that line the base of the trough shown in green. Tyrosine 251, located in the trough center, is identical in all apicomplexan sequences. Polymorphic sites are shown in red, with K230 and H200 indicated. Dimorphic sites are colored orange (high frequency) and yellow (low frequency). “Ordered” loops are colored blue with the domain I loops Ia, Ic, Id, and Ie in dark blue and the domain II loop in light blue. Disordered loops Ib and If are colored violet. The disordered loops contain four dimorphic sites, and the approximate location of these residues is indicated. (b) Surface view of the hydrophobic trough. Carbon and sulfur atoms that form the base of the hydrophobic trough are colored green. Other carbon and sulfur atoms are colored white. Oxygen atoms are colored red, and nitrogen atoms are blue. Disordered regions are colored violet.
Consistent with the functional importance of the hydrophobic trough is the presence of polymorphic residues on the loops that surround the trough. Of the five highly polymorphic residues in P. falciparum sequences, four (positions 187, 197, 200, and 230) surround one end of the hydrophobic pocket (Fig. 5). In addition, 12 dimorphic residues are distributed throughout all of the loops apart from loop Ia and loop II (Fig. 5). The loops that surround the hydrophobic trough on AMA1 are analogous to the variable loops that hide the CD4 binding site of HIV gp120 (55) or the ring of polymorphisms that surround the sialic acid binding pocket of influenza hemagglutinin (56). Although there is no direct evidence for the functional importance of the hydrophobic pocket on AMA1, these precedents would indicate that this is likely to be a functionally critical region of AMA1 that is a frequent target of protective immunity.
Acknowledgments
We extend our thanks to Janna Wehrle, Steve Soisson, Mario Amzel, Jun Aishima, Kris Tesh, Georgia Allison, Cynthia Wolberger, Sandra Gabelli, Mario Bianchet, Eric Toth, Avi Gnatt, Elizabeth Reisinger, and Derek Piper. Beamline X25 at the National Synchrotron Light Source is supported by the Offices of Biological and Environmental Research and of Basic Energy Sciences of the U.S. Department of Energy, and by the National Center for Research Resources of the National Institutes of Health.
Author contributions: A.H.B. designed research; T.B., M.B., and A.H.B. performed research; P.S., V.J.M., and R.F.A. contributed new reagents/analytic tools; T.B., M.B., P.S., R.F.A., and A.H.B. analyzed data; R.F.A. and A.H.B. wrote the paper; and A.G. determined an improved protein purification protocol critical to the success of the project.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited at the Protein Data Bank, www.pdb.org (PDB ID code 1Z40).
Note. Just before submission of this manuscript, a structure of P. vivax AMA1 was published (57). This protein was expressed in yeast and consisted of full-length AMA1 ectodomain, providing a view of the overall AMA1 structure. However, the structure is less complete, with 15% of the molecule disordered in the crystals. Pizarro et al. report that the binding site of inhibitory monoclonal antibody 4G2 is in the domain II loop that extends into domain I. This loop is disordered in the P. vivax structure, but is observed in the P. falciparum structure reported here (Fig. 1). Interestingly, two of the residues that make up the hydrophobic trough (L357 and F367) are part of the domain II loop. Also, the domain II loop is on the nonpolymorphic face of AMA1. Therefore, AMA1 contains at least one nonpolymorphic epitope that is the target of inhibitory antibodies. It is not clear how immunogenic this epitope will be in humans but it offers the possibility that an AMA1 vaccine will not necessarily be strain specific.
References
- 1.Breman, J. G., Alilio, M. S. & Mills, A. (2004) Am. J. Trop. Med. Hyg. 71, 1–15. [PubMed] [Google Scholar]
- 2.Sachs, J. & Malaney, P. (2002) Nature 415, 680–685. [DOI] [PubMed] [Google Scholar]
- 3.Baird, J. K. (1995) Parasitol. Today 11, 105–111. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, S., McGregor, I. A. & Carrington, S. C. (1961) Nature 192, 733–737. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, S. & Butcher, G. A. (1970) Nature 225, 732–734. [DOI] [PubMed] [Google Scholar]
- 6.Sabchareon, A., Burnouf, T., Ouattara, D., Attanath, P., Bouharoun-Tayoun, H., Chantavanich, P., Foucault, C., Chongsuphajaisiddhi, T. & Druilhe, P. (1991) Am. J. Trop. Med. Hyg. 45, 297–308. [DOI] [PubMed] [Google Scholar]
- 7.Gardner, M. J., Hall, N., Fung, E., White, O., Berriman, M., Hyman, R. W., Carlton, J. M., Pain, A., Nelson, K. E., Bowman, S., et al. (2002) Nature 419, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alonso, P. L., Sacarlal, J., Aponte, J. J., Leach, A., Macete, E., Milman, J., Mandomando, I., Spiessens, B., Guinovart, C., Espasa, M., et al. (2004) Lancet 364, 1411–1420. [DOI] [PubMed] [Google Scholar]
- 9.Saul, A., Lawrence, G., Allworth, A., Elliott, S., Anderson, K., Rzepczyk, C., Martin, L. B., Taylor, D., Eisen, D. P., Irving, D. O., et al. (2005) Vaccine 23, 3076–3083. [DOI] [PubMed] [Google Scholar]
- 10.Hehl, A. B., Lekutis, C., Grigg, M. E., Bradley, P. J., Dubremetz, J. F., Ortega-Barria, E. & Boothroyd, J. C. (2000) Infect. Immun. 68, 7078–7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaffar, F. R., Yatsuda, A. P., Franssen, F. F. & de Vries, E. (2004) Infect. Immun. 72, 2947–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deans, J. A., Knight, A. M., Jean, W. C., Waters, A. P., Cohen, S. & Mitchell, G. H. (1988) Parasite Immunol. 10, 535–552. [DOI] [PubMed] [Google Scholar]
- 13.Collins, W. E., Pye, D., Crewther, P. E., Vandenberg, K. L., Galland, G. G., Sulzer, A. J., Kemp, D. J., Edwards, S. J., Coppel, R. L., Sullivan, J. S., et al. (1994) Am. J. Trop. Med. Hyg. 51, 711–719. [DOI] [PubMed] [Google Scholar]
- 14.Crewther, P. E., Matthew, M. L. S. M., Flegg, R. H. & Anders, R. F. (1996) Infect. Immun. 64, 3310–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narum, D. L., Ogun, S. A., Thomas, A. W. & Holder, A. A. (2000) Infect. Immun. 68, 2899–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stowers, A. W., Kennedy, M. C., Keegan, B. P., Saul, A., Long, C. A. & Miller, L. H. (2002) Infect. Immun. 70, 6961–6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodder, A. N., Crewther, P. E. & Anders, R. F. (2001) Infect. Immun. 69, 3286–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silvie, O., Franetich, J. F., Charrin, S., Mueller, M. S., Siau, A., Bodescot, M., Rubinstein, E., Hannoun, L., Charoenvit, Y., Kocken, C., et al. (2003) J. Biol. Chem. 279, 9490–9496. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy, M. C., Wang, J., Zhang, Y., Miles, A. P., Chitsaz, F., Saul, A., Long, C. A., Miller, L. H. & Stowers, A. W. (2002) Infect. Immun. 70, 6948–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Healer, J., Murphy, V., Masciantonio, R., Anders, R. F., Cowman, A. F. & Batchelor, A. H. (2004) Mol. Microbiol. 52, 199–205. [DOI] [PubMed] [Google Scholar]
- 21.Escalante, A. A., Grebert, H. M., Chaiyaroj, S. C., Magris, M., Biswas, S., Nahlen, B. L. & Lal, A. A. (2001) Mol. Biochem. Parasitol. 113, 279–287. [DOI] [PubMed] [Google Scholar]
- 22.Cortes, A., Mellombo, M., Mueller, I., Benet, A., Reeder, J. C. & Anders, R. F. (2003) Infect. Immun. 71, 1416–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polley, S. D., Chokejindachai, W. & Conway, D. J. (2003) Genetics 165, 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polley, S. D., Mwangi, T., Kocken, C. H., Thomas, A. W., Dutta, S., Lanar, D. E., Remarque, E., Ross, A., Williams, T. N., Mwambingu, G., et al. (2004) Vaccine 23, 718–728. [DOI] [PubMed] [Google Scholar]
- 25.Triglia, T., Healer, J., Caruana, S. R., Hodder, A. N., Anders, R. F., Crabb, B. S. & Cowman, A. F. (2000) Mol. Microbiol. 38, 706–718. [DOI] [PubMed] [Google Scholar]
- 26.Nair, M., Hinds, M. G., Coley, A. M., Hodder, A. N., Foley, M., Anders, R. F. & Norton, R. S. (2002) J. Mol. Biol. 322, 741–753. [DOI] [PubMed] [Google Scholar]
- 27.Kocken, C. H. M., van der Wel, A. M., Dubbeld, M. A., Narum, D. L., van de Rijke, F. M., van Gemert, G.-J., van der Linde, X., Bannister, L., Janse, C., Waters, A. P. & Thomas, A. W. (1998) J. Biol. Chem. 273, 15119–15124. [DOI] [PubMed] [Google Scholar]
- 28.Kocken, C. H. M., Narum, D. L., Massougbodji, A., Ayivi, B., Dubbeld, M. A., van der Wel, A., Conway, D. J., Sanni, A. & Thomas, A. W. (2000) Mol. Biochem. Parasitol. 109, 147–156. [DOI] [PubMed] [Google Scholar]
- 29.Howell, S. A., Withers-Martinez, C., Kocken, C. H. M., Thomas, A. W. & Blackman, M. J. (2001) J. Biol. Chem. 276, 31311–31320. [DOI] [PubMed] [Google Scholar]
- 30.Healer, J., Crawford, S., Ralph, S., McFadden, G. & Cowman, A. F. (2002) Infect. Immun. 70, 5751–5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell, G. H., Thomas, A. W., Margos, G., Dluzewski, A. R. & Bannister, L. H. (2004) Infect. Immun. 72, 154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraser, T. S., Kappe, S. H., Narum, D. L., VanBuskirk, K. M. & Adams, J. H. (2001) Mol. Biochem. Parasitol. 117, 49–59. [DOI] [PubMed] [Google Scholar]
- 33.Gupta, A., Bai, T., Murphy, V. J., Strike, P., Anders, R. F. & Batchelor, A. H. (2005) Protein Expression Purif. 41, 186–198. [DOI] [PubMed] [Google Scholar]
- 34.Boggon, T. J. & Shapiro, L. (2000) Structure Fold. Des. 8, 143–149.10673440 [Google Scholar]
- 35.Pflugrath, J. W. (1999) Acta Crystallogr. D 55, 1718–1725. [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 37.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 38.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 39.Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001) Acta Crystallogr. D 57, 122–133. [DOI] [PubMed] [Google Scholar]
- 40.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 41.Thomas, A. W., Waters, A. P. & Carr, D. (1990) Mol. Biochem. Parasitol. 42, 285–288. [DOI] [PubMed] [Google Scholar]
- 42.Donahue, C. G., Carruthers, V. B., Gilk, S. D. & Ward, G. E. (2000) Mol. Biochem. Parasitol. 111, 15–30. [DOI] [PubMed] [Google Scholar]
- 43.Michon, P., Stevens, J. R., Kaneko, O. & Adams, J. H. (2002) Mol. Biol. Evol. 19, 1128–1142. [DOI] [PubMed] [Google Scholar]
- 44.Richardson, J. S. (1981) Adv. Protein Chem. 34, 167–339. [DOI] [PubMed] [Google Scholar]
- 45.Gibrat, J. F., Madej, T. & Bryant, S. H. (1996) Curr. Opin. Struct. Biol. 6, 377–385. [DOI] [PubMed] [Google Scholar]
- 46.Tordai, H., Banyai, L. & Patthy, L. (1999) FEBS Lett. 461, 63–67. [DOI] [PubMed] [Google Scholar]
- 47.Ultsch, M., Lokker, N. A., Godowski, P. J. & de Vos, A. M. (1998) Structure 6, 1383–1393. [DOI] [PubMed] [Google Scholar]
- 48.Huizinga, E. G., Schouton, A., Connolly, T. M., Kroon, J., Sixma, J. J. & Gros, P. (2001) Acta Crystallogr. D 57, 1071–1078. [DOI] [PubMed] [Google Scholar]
- 49.Baglia, F. A., Jameson, B. A. & Walsh, P. N. (1991) J. Biol. Chem. 266, 24190–24197. [PubMed] [Google Scholar]
- 50.Baglia, F. A., Badellino, K. O., Ho, D. H., Dasari, V. R. & Walsh, P. N. (2000) J. Biol. Chem. 275, 31954–31962. [DOI] [PubMed] [Google Scholar]
- 51.Baglia, F. A., Gailani, D., Lopez, J. A. & Walsh, P. N. (2004) J. Biol. Chem. 279, 45470–45476. [DOI] [PubMed] [Google Scholar]
- 52.Reiss, M., Viebig, N., Brecht, S., Fourmaux, M. N., Soete, M., Di Cristina, M., Dubremetz, J. F. & Soldati, D. (2001) J. Cell Biol. 152, 563–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brecht, S., Carruthers, V. B., Ferguson, D. J., Giddings, O. K., Wang, G., Jakle, U., Harper, J. M., Sibley, L. D. & Soldati, D. (2001) J. Biol. Chem. 276, 4119–4127. [DOI] [PubMed] [Google Scholar]
- 54.Lietha, D., Chirgadze, D. Y., Mulloy, B., Blundell, T. L. & Gherardi, E. (2001) EMBO J. 20, 5543–5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen, B., Vogan, E. M., Gong, H., Skehel, J. J., Wiley, D. C. & Harrison, S. C. (2005) Nature 433, 834–841. [DOI] [PubMed] [Google Scholar]
- 56.Skehel, J. J. & Wiley, D. C. (2000) Annu. Rev. Biochem. 69, 531–569. [DOI] [PubMed] [Google Scholar]
- 57.Pizarro, J. C., Vulliez-Le Normand, B., Chesne-Seck, M. L., Collins, C. R., Withers-Martinez, C., Hackett, F., Blackman, M. J., Faber, B. W., Remarque, E. J., Kocken, C. H., et al. (2005) Science 308, 408–411. [DOI] [PubMed] [Google Scholar]
- 58.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thortnon, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]