Abstract
Fluorescent proteins isolated from coral reef organisms can be roughly grouped into four color classes by emission, cyan, green, yellow, and red. To gain insight into the structural basis for cyan emission, the crystal structure of amFP486 (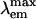 = 486 nm) was determined by molecular replacement, and the model was refined at 1.65-Å resolution. The electron density map reveals a chromophore formed from the tripeptide sequence –K–Y–G– that is indistinguishable from that of GFP (
= 486 nm) was determined by molecular replacement, and the model was refined at 1.65-Å resolution. The electron density map reveals a chromophore formed from the tripeptide sequence –K–Y–G– that is indistinguishable from that of GFP (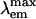 = 509 nm). However, the chromophore environment closely parallels those of the yellow- and red-shifted homologs zFP538, DsRed, and eqFP611. Mutagenesis was performed for Glu-150, Ala-165, His-199, and Glu-217, which are immediately adjacent to the chromophore. His-199 and Ala-165 are key side chains responsible for the blue shift, presumably by localizing chromophore charge density on the phenolate moiety. Furthermore, in the H199T mutant the fluorescence quantum yield is reduced by a factor of ≈110. The crystal structures of H199T (
= 509 nm). However, the chromophore environment closely parallels those of the yellow- and red-shifted homologs zFP538, DsRed, and eqFP611. Mutagenesis was performed for Glu-150, Ala-165, His-199, and Glu-217, which are immediately adjacent to the chromophore. His-199 and Ala-165 are key side chains responsible for the blue shift, presumably by localizing chromophore charge density on the phenolate moiety. Furthermore, in the H199T mutant the fluorescence quantum yield is reduced by a factor of ≈110. The crystal structures of H199T (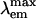 = 515 nm) and E150Q (
= 515 nm) and E150Q (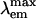 = 506 nm) were determined. Remarkably, the H199T structure reveals that the stacking interaction of His-199 with the chromophore also controls the fluorescence efficiency, because the chromophore is statistically distributed in a 1:1 ratio between cis (fluorescent) and trans (nonfluorescent) conformations.
= 506 nm) were determined. Remarkably, the H199T structure reveals that the stacking interaction of His-199 with the chromophore also controls the fluorescence efficiency, because the chromophore is statistically distributed in a 1:1 ratio between cis (fluorescent) and trans (nonfluorescent) conformations.
Keywords: autocatalysis, fluorescent proteins, protein crystallography
The discovery in Anthozoa of an entire family of fluorescent proteins (FPs) and nonfluorescent chromoproteins distantly related to the green FP from Aequorea victoria (GFP) has provided new tools that can serve as alternatives to or complement the existing uses of GFP (1–5). Based on absorbance and emission spectra, the Anthozoa proteins can be roughly grouped into five classes: cyan, green, yellow, red, and nonfluorescent chromoproteins. The amino acid sequences are closely related, and in all known cases, a tripeptide –X–Y–G–, where X is highly variable, forms the precursor to the chromophore. The large spectral diversity appears to arise from two causes, the most important being differences in the extent of bond conjugation arising from variations in the chemical structure of the chromophore. For the yellow- and red-emitting FPs, the spectral diversity results from oxidation of the polypeptide backbone to form an acylimine linkage or variations thereof [e.g., DsRed (6–8); eqFP611 (9), zFP538 (10), and the “kindling FP” KFP (11, 12); for chromophore structures, see Fig. 6, which is published as supporting information on the PNAS web site]. However, effects arising from the local environment of the chromophore can have a significant influence on absorption and emission maxima. Variations of up to ≈20 nm in either direction have been achieved by means of single-site substitutions in GFP (13, 14) and in DsRed (15).
Three cyan-emitting Anthozoa FPs (CFPs) [amFP486, dsFP483. and cFP484 (1)] have been characterized in some detail. Although the biological role of these proteins is unknown, due to widespread appearance one may safely assume that cyan fluorescence is important for the ecology of reef organisms. amFP486, cloned from Anemonia majano, is a tetrameric protein with subunit molecular mass of 25 kDa. It shows only 26% identity in amino acid sequence to GFP but is much more similar to DsRed and zFP538 (42% and 50% identity, respectively). CFPs typically have broad emission [full width at half maximum ≈ 50 nm (16)] but narrow Stokes shift. In the case of amFP486, maximal excitation and emission occurs at 454 and 486 nm, respectively.
Although cyan or blue emission is not necessarily practical for labeling in living cells, from the fundamental photophysical perspective, it is important to understand how organisms are able to tune the spectroscopic properties of FPs to meet specific requirements. Such knowledge may have practical application in extending the range of FP indicators. Gurskaya et al. (16) investigated the basis for blue-shifted emission of CFPs by comparing sequences of several FPs. They showed that position 165 (amFP486 numbering) is occupied by either an Ala or His in the CFPs but is Ile or Met in most other FPs. The importance of position 165 was tested in amFP486 by mutation of Ala-165 to Ile or Met and in zFP506 (a GFP from Zoanthus) by mutation of Met-167 to Ala or His. The mutations yielded proteins with fluorescence spectra that were intermediate between those of CFPs and GFPs. This result confirmed the importance of position 165 but suggested that other structural features must also contribute to the blue-shift. Based on a homology model, it was proposed that the small side chain of Ala-165 permits a water molecule to interact with the chromophore phenolate, potentially increasing the chromophore polarization and thereby accounting for some of the 23-nm blue-shift relative to GFP (16).
To test the hypotheses of Gurskaya et al. (16), we determined the crystal structure of a representative CFP. Here, we present high-resolution crystal structures of amFP486 from A. majano. Structural comparisons of amFP486 with other Anthozoa FPs provided a starting point for identification of additional features that are responsible for cyan fluorescence. We focused our attention on the following four positions: Ala-165 (see above), Lys-68, His-122, and a conserved salt bridge network associated with His-199. The results of the study suggest that in amFP486, cyan emission arises from a GFP-like chromophore, the charge distribution of which is shifted toward the phenolate end of the chromophore by polar and electrostatic influences of the immediate environment. Furthermore, the crystal structures give insight into the structural features responsible for efficient fluorescence.
Materials and Methods
Protein Expression, Mutagenesis, Purification, and Characterization. A pQE-30 expression system in Escherichia coli (JM-109 DE3) was used to express amFP486 containing an N-terminal MRGS(H)6GS tag (Clontech). Site-directed mutagenesis was carried out by the PCR method (QuikChange, Stratagene) using the following IDT primers: 5′-catgtggtggaaactcgcattgcgaggaccg-3′/5′-cggtcctcgcaatgcgagtttccaccacatg-3′ (H199T), 5′-gggacccatcttttcagaaaatgactgtctgcg-3′/5′-cgcagacagtcattttctgaaaagatgggtccc-3′ (E150Q), and 5′-gtgcaacagcgtgctgcgtcagctgaac-3′/5′-ggcaactgctttgagcaaaaatccacgtttcatgg-3′ (E217Q).
PCR was followed by transformation into JM-109, and cells were grown on expression plates as described in ref. 10. Purification over Ni-NTA agarose columns (Qiagen, Valencia, CA) was followed by dialysis against 50 mM Hepes (pH 7.9) and then buffer exchange (PD-10 Sephadex columns, Amersham Pharmacia) into 20 mM Hepes (pH 7.9) and 2 mM 2-mercaptoethanol (2-ME). Protein concentrations were determined by using theoretical molar extinction coefficients (17) at 280 nm.
Spectroscopy. Absorption and fluorescence spectra for amFP486 and variants were collected at 25°C in 20 mM Hepes (pH 7.9) and 2 mM 2-ME by using a Shimadzu UV-2101PC spectrophotometer and a PerkinElmer LS-55 fluorometer. For the mutant H199T, the absorbance spectra are pH-dependent and were recorded in 100 mM buffers of citrate, acetate, or Tris as appropriate. Fluorescence quantum yields (ΦF) were determined relative to fluorescein in 0.1 M NaOH (18) by adjusting the absorbance of each sample to be equal at the excitation wavelength of 470 nm. Integrated emission curves were corrected for detector response by using the manufacturer-supplied software.
Crystallization and Data Collection. Tetragonal crystals of wild-type (WT) amFP486, H199T, and E150Q were obtained in 1–2 days by hanging drop vapor diffusion. Setups consisted of 4-μl drops containing one part protein solution (A280 = 20) in 20 mM Hepes (pH 7.9), 2 mM 2-ME, and one part well solution. Crystals of WT and H199T protein grew overnight in well solutions of 1.0 M Na/K tartrate/0.16 M Li2SO4/0.1 M 2-(N-cyclohexylamino)ethanesulfonic acid (CHES) (pH 9.3), and 0.6 M Na/K tartrate/40 mM Li2SO4/0.1 M CHES (pH 9.7), respectively. Crystals of the E150Q mutant grew in 2 days in well solution containing 0.74 M Na/K tartrate, 40 mM Li2SO4, and 0.1 M CHES (pH 9.4), with 1 μl of 2-ME per ml of well solution. Single crystals were swept through paratone oil and then flash frozen at 100 K before diffraction data collection. WT and the H199T variant data sets were collected by using a MarCCD 165 detector at the Advanced Photon Source (Argonne, IL) on beamline 14 ID-B. Data for the E150Q mutant was collected by using an R-axis IV image plate detector (Molecular Structure, The Woodlands, TX) on a rotating anode generator.
Structure Determination and Refinement. Diffraction images were indexed and processed by using hkl2000 and the hkl suite (19). The structure of WT amFP486 was solved by molecular replacement with epmr (20) by using the DsRed A chain [Protein Data Bank ID code 1G7K (7)] as the search model. The H199T and E150Q mutant structures were solved by using epmr with the refined WT amFP486 structure. Rigid body refinement at 50–4.0 Å was performed with tnt (21). Model building was carried out by using the o program (22). Positional and B-factor refinement were performed with tnt by using all data (no sigma or F cutoffs) in the resolution range of 6.0 Å or higher. The model of amFP486 was constructed into the experimental electron density map beginning with a polyalanine backbone of DsRed. In the final stages, solvent molecules were modeled into large positive difference electron density peaks that were within hydrogen-bond distance of suitable partners. Before construction of the H199T and E150Q models, solvent molecules were removed, as were the chromophore and side chains of residues His-199, Glu-150, and Glu-217. Model analysis was carried out by using prochek (23), and figures were created with pymol (24). The program platon (25) was used to calculate the out-of-plane angles of the H199T amFP486 chromophores.
Results
Structural Analysis of WT and Mutant amFP486. Crystals of WT amFP486 and the variants are isomorphous and have space group I4122. The asymmetric unit contains one molecule with packing coefficient Vm = 2.4–2.5 Å3/Da (26). Data collection and refinement statistics, which are excellent, are presented in Table 1. The final model of WT amFP486 (R = 0.167 (all data), Rfree = 0.208, 1.65-Å resolution), contains residues 5–224, 170 water molecules, and 1 molecule of 2-ME forming a disulfide link to Cys-120. The 1.72-Å resolution model of H199T contains residues 5–224, 132 water molecules, and 1 2-ME. The E150Q model was refined at 2.0-Å resolution and contains residues 5–224 (except for disordered loop residues 89–91), 73 solvent molecules, and 3 2-ME. prochek analysis of the models indicates excellent stereochemistry. For WT, H199T, and E150Q, 96.1%, 93.9%, and 92% of residues are found in the “most favored” regions, 3.9%, 6.1%, and 8% in are found in “additionally allowed” regions, and none are found in “generously allowed” or disallowed regions of the Ramachandran plot, respectively.
Table 1. Summary of crystallographic statistics.
| Statistic | WT | H199T | E150Q |
|---|---|---|---|
| Total reflections | 1,516,052 | 530,197 | 450,768 |
| Unique reflections | 31,584 | 27,044 | 18,458 |
| Cell dimensions (a, b, c), Å | 112.5, 112.5, 81.4 | 113.0, 113.0, 82.0 | 113.9, 113.9, 82.6 |
| Resolution, Å | 50-1.65 | 50-1.72 | 50.0-2.0 |
| Highest resolution shell, Å | 1.71-1.65 | 1.83-1.72 | 2.07-2.0 |
| Completeness,* % | 99.9 (99.5) | 95.2 (97.7) | 99.8 (100) |
| Average I/σ* | 40.9 (5.8) | 21.9 (2.4) | 34.3 (2.6) |
| Rmerge*† | 0.064 (0.340) | 0.054 (0.288) | 0.077 (0.731) |
| Rwork‡ | 0.165 | 0.169 | 0.18 |
| Rfree | 0.208 | 0.225 | 0.249 |
| R factor (all data) | 0.167 | 0.172 | 0.181 |
| Average B factors, Å2 | 21.4 | 28.7 | 40.4 |
| Protein atoms | 20.4 | 28 | 40.3 |
| Solvent | 32.4 | 38 | 41.5 |
| rmsd bond lengths, Å | 0.01 | 0.008 | 0.006 |
| rmsd bond angles, ° | 2.59 | 2.4 | 2.11 |
Values in parentheses indicate statistics for the highest-resolution shell.
Rmerge = ΣiΣj(Iij — 〈I〉i)/ΣiΣj| 〈I〉i, where Iij is the amplitude of the jth observation of reflection i and 〈I〉i is the mean value of observations Iij.
R factor = Σ∥Fo| — |Fc∥/Σ|Fo| |, where Fo and Fc are the observed and calculated structure amplitudes.
As expected, the fold of amFP486 is the so-called “β-can” motif (27) with an α-helix coaxial with an 11-stranded β-barrel. Crystallographic symmetry operations produce tightly packed tetramers closely resembling those of other Anthozoa FPs (7–10, 15). Within the subunits, superposition of α-carbon positions of amFP486 with DsRed and zFP538 gives rms deviations of 0.67 Å for 191 equivalences and 0.60 Å for 192 equivalences, respectively. The divergence from GFP is much greater, because α-carbons superimpose to 1.46-Å rms for 195 equivalences. amFP486 contains an insertion in the loop between β-strands 3 and 4 (residues 51–52) not found in DsRed or zFP538. The structural studies showed that the effects of point substitutions are generally confined to the immediate region of the modification.
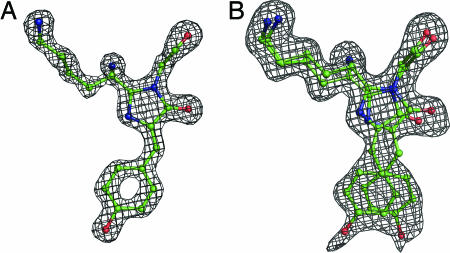
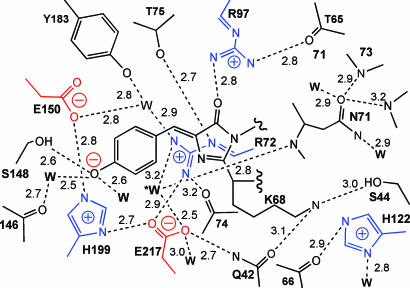
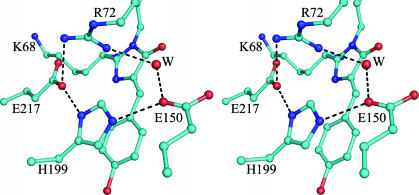
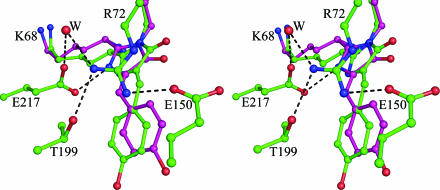
The Chromophore and Environment. The chromophore of WT amFP486 forms from the tripeptide –K68–Y69–G70–. A 1.65-Å (Fo – Fc) omit map (Fig. 1A) reveals the chromophore to be cis-coplanar and, except for Lys instead of Ser at the first position, it appears to be identical to that of GFP. The environment surrounding the amFP486 chromophore (Fig. 2) is very polar as observed in DsRed (7, 8), eqFP611 (9), and zFP538 (10). The similarity of amFP486 with yellow- and red-shifted FPs zFP538 and eqFP611 is quite striking. Residues Arg-72, Glu-150, His-199, Glu-217, and a water molecule form a quadrupole network of salt bridge and hydrogen-bond interactions adjacent to the chromophore in all three proteins (Figs. 2 and 3). Although the overall charge on the network is probably zero, the side chain of the conserved histidine stacks against the chromophore phenolate and is presumably charged, as the imidazole ring interacts with carboxylate oxygens of the conserved Glu residues. From the opposite side of the β-barrel, the essential Arg residue (97 in amFP486) forms hydrogen bonds with the imidazolinone oxygen, Glu-150, and conserved Tyr-183.
Fig. 1.
Representative electron density maps. (A) WT amFP486 Fo – Fc chromophore omit map contoured at 5 σ with superimposed atomic model. (B) Chromophore omit map for H199T amFP486 contoured at 2 σ. The chromophore is represented as a ball-and-stick model with carbon in green, oxygen in red, and nitrogen in blue.
Fig. 2.
Schematic diagram of the amFP486 chromophore environment. Charged side chains are drawn in blue and red. Dashed lines represent hydrogen bonds with distances indicated in angstroms.
Fig. 3.
Stereo drawing of the amFP486 chromophore emphasizing the quadrupole salt bridge network associated with His-199. Hydrogen bonds are represented by dashed lines. Oxygen atoms are drawn as red spheres, and nitrogen is shown in blue. W represents a bound solvent molecule.
As hypothesized by Gurskaya et al. (16), in amFP486 two presumed water molecules are found within hydrogen-bonding distance of the phenolate oxygen. Structural overlays indicate that the small side chain of Ala-165 permits a solvent molecule to occupy the space filled by a bulky hydrophobic group in other FPs. It is interesting that this water is also the first member of an unusual internal channel of water molecules connecting the chromophores, i.e., connecting the phenolate oxygens of protomer A with C and B with D, forming a possible “wire” for proton transfer between chromophores.
Two other potentially charged residues in the chromophore environment (Lys-68 and His-122) are of particular interest because of their demonstrated or suspected importance in other FPs. Lys-68, the first position of the chromophore triplet, autocatalytically forms a second heterocycle in the chromophore of yellow fluorescent zFP538 (10). However, in amFP486, this side chain is fully extended and forms a hydrogen bond with the side chain of Ser-44 (Ile in zFP538). In amFP486, Ser-44 may be important to prevent Lys-68 from undergoing a similar cyclization, although this hypothesis presupposes the prior formation of a transient acylimine linkage (10). Second, among the FPs of known structure, position 122 is either His or Tyr. In zFP538, His-122 forms an internal salt bridge with Asp-69, an interaction that is important, but not essential for development of yellow fluorescence (16). In amFP486, His-122 is rotated away from the corresponding Asn-71. This conformation, which is unique to amFP486, locates the imidazole ring ≈1.0 Å closer to the chromophore, but the ring presumably remains cationic, as it forms hydrogen bonds with a buried solvent molecule and the backbone carbonyl of residue 66.
Based on the structural analysis, we selected several of the above identified sites for mutagenesis. In the following sections, we report the spectroscopic and structural consequences of those alterations.
Spectroscopic Properties of amFP486 Variants. Position 199. The substitution of His-199 with Thr (Thr 203 in GFP), Ser (Ser 197 in DsRed), Ala, Gln, and Asn has dramatic effects on both the structure (see below) and fluorescence emission of amFP486 (see Table 2). Purified samples of each mutant protein appear orange under room lighting, as opposed to the yellow-green appearance of WT amFP486. All His-199 variants exhibit markedly decreased fluorescence quantum yields (ΦF) and significantly red-shifted absorption and emission maxima (see Table 4, which is published as supporting information on the PNAS web site).
Table 2. Spectroscopic properties of amFP486 and mutants.
| Protein | λabs_max, nm | λem_max, nm | λex_max, nm | ΦF |
|---|---|---|---|---|
| WT | 454 | 486 | 450 | 0.71 |
| H199T | 470 | 515 | 500 | 0.006 |
| E150Q | 485 | 506 | 483 | 0.67 |
| E217Q | 469 | 495 | 467 | 0.08 |
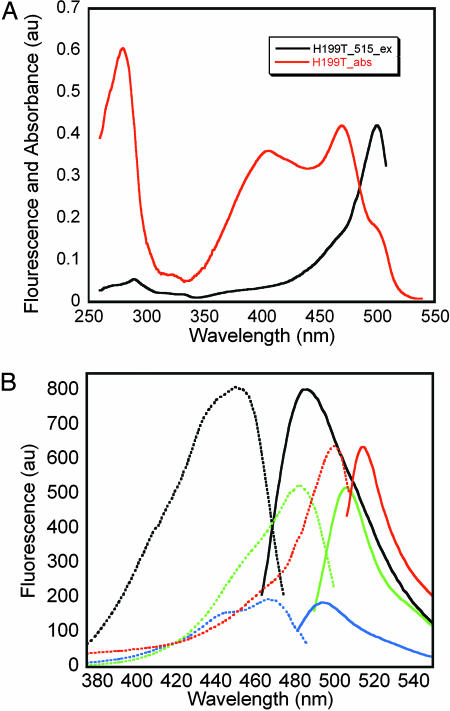
The absorbance spectra typically display two peaks, depending on pH, as follows: “A” in the range 390–405 nm and “B” in the range 470–500 nm, indicative of a mixed population of phenol and phenolate chromophores (see Fig. 7, which is published as supporting information on the PNAS web site). The H199T mutant is of particular interest because its absorbance extends beyond 500 nm (Fig. 4A) and is best modeled, by using least-squares Gaussian decomposition, by a summation of four components with absorbance maxima at ≈398, 420, 473, and 501 nm (Table 3). Unlike WT, the absorbance spectrum of H199T is pH sensitive. At low pH, the peak at ≈398 nm, which presumably corresponds to the neutral chromophore, dominates the absorbance. As the pH is increased, peaks at ≈473 and ≈501 nm, presumably corresponding to the anionic chromophore, become more prominent (see Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 4.
Spectroscopy of amFP486 and variants. (A) Comparison of the amFP486/H199T absorbance spectrum (red) and excitation spectra giving rise to emission at 515 nm (black). (B) Comparison of fluorescence excitation (dashed lines) and emission (solid lines) spectra for WT (black) and variants H199T (red), E150Q (green), and E217Q (blue). Protein concentrations were 8.5 μg/ml for WT, E150Q, and E217Q and 1.7 mg/ml for H199T.
Table 3. Gaussian parameters for peaks in decomposed amFP486/H199T absorbance spectrum.
| Peak | Amplitude | λmax, nm | σ, nm |
|---|---|---|---|
| 1 | 0.067 ± 0.014 | 397.9 ± 2.0 | 22.6 ± 4.3 |
| 2 | 0.308 ± 0.013 | 420.4 ± 2.4 | 68.6 ± 2.9 |
| 3 | 0.205 ± 0.014 | 473.0 ± 0.6 | 18.2 ± 1.3 |
| 4 | 0.125 ± 0.013 | 501.2 ± 0.8 | 10.1 ± 1.3 |
A comparison of the WT and H199T fluorescence spectra is shown in Fig. 4B. The quantum yield of H199T is ≈110-fold less than WT, and the emission maximum is red-shifted by 29 nm to 515 nm. The decrease in ΦF is underscored by the fact that excitation at 280 nm gives rises to readily detectable Trp fluorescence at 337 nm (data not shown). The highly conserved His-199 is thus important for both the blue shift and for efficient fluorescence. Furthermore, the fluorescence excitation spectrum of H199T reveals that only the putative anionic forms of the chromophore are fluorescent, with a surprisingly narrow Stokes shift of 15 nm. Maximal excitation is at 500 nm with a broad shoulder near 475 nm. The 500-nm excitation peak and 475-nm shoulder (Fig. 4A) correlate well with two of the peaks found in the absorbance spectrum.
Positions 150 and 217. Because His-199 is involved in a salt bridge with Glu-150 and Glu-217, the mutations E150Q, E217Q, and the double mutant were made to address the effect of the presumed positive charge on the imidazole ring. Protein yields for E150Q and E217Q were reduced relative to WT, whereas the E150Q/E217Q double mutant yielded nonfluorescent protein. ΦF for E150Q and E217Q is reduced by factors of ≈1.1 and ≈9, and emission maxima are 506 and 495 nm, respectively (Fig. 4B). Assuming that His-199 is uncharged in these mutants, the results support the suspected role of this charge in blue-shifting both the absorbance and emission of WT amFP486.
Positions 68, 122, and 165. Substitution of Lys-68 in the chromophore triplet with Glu, Asp, Gln, Asn, Arg, Gly, Ala, Ser, Thr, or Met resulted in highly fluorescent proteins that expressed well but showed only small changes in absorbance and fluorescence maxima (data not shown). This result is not surprising because Lys-68 is not conserved among CFPs (16). Position 122, which is highly conserved among Anthozoa FPs and is usually His or Tyr, was changed to Gln or Asn. Again, the resulting proteins expressed well and were highly fluorescent; however, their spectra were essentially identical to that of WT (data not shown). This position may be conserved because of its importance for efficiency of maturation, which was not investigated. Finally, substitutions of Ala-165 with Met or Ile resulted in amFP486 variants with emission maxima intermediate between those of WT and GFPs (data not shown), in agreement with Gurskaya et al. (16).
Results of Crystallographic Analysis. Position 199. The most striking structural feature of the weakly fluorescent H199T variant is that the chromophore is statistically distributed among both cis and trans conformational states. This feature is clearly shown in a (Fo – Fc) omit map (Fig. 1B). After careful inspection of difference electron density maps calculated for various partial models, the superposed cis and trans conformations were assigned equal occupancy (0.5) and subjected to further positional refinement. This model proved adequate, as indicated by a relatively featureless final (Fo – Fc) difference electron density map. The (Fo – Fc) omit map also suggested a noncoplanar arrangement of the five- and six-membered rings; so during the last cycles of crystallographic refinement, coplanarity restraints were removed. In the final atomic model of H199T, both the cis and trans chromophores are nonco-planar, with calculated angles between the planes defined by the imidazolinone and p-hydroxybenzyl rings of 22.9° and 30.3° for cis and trans, respectively. These parameters are poorly determined and should be treated with caution.
The H199T substitution also results in dramatic changes in the network of charged residues surrounding the chromophore. Arg-72 moves to a more extended conformation where it partially occupies the space created by the substitution. As shown in Fig. 5, Arg-72 forms a hydrogen bond with the Oγ of Thr-199 and a salt bridge to Glu-150. The salt bridge interaction between Arg-72 and Glu-217 found in WT is replaced by a water-meditated hydrogen bond in the H199T variant. The Glu-217 side chain, inferred by the hydrogen bond pattern to be uncharged, moves in to form a hydrogen bond with N2 of the imidazolinone ring. On the opposite end, the phenol(ate) oxygen of the chromophore (in either configuration) participates in a hydrogen bond with Oγ of Ser-148. The phenol(ate) moiety of the cis chromophore appears to accept hydrogen bonds from two water molecules; by contrast, in the trans chromophore this oxygen has no other hydrogen bond partners and may be protonated.
Fig. 5.
Stereo drawing of the superimposed refined partial models for the amFP486/H199T chromophore. Oxygen atoms are drawn as red spheres and nitrogen as blue. The trans conformation is shown in purple.
Position 150. Comparison of the structure of the E150Q mutant with WT reveals only one significant difference in the vicinity of the chromophore. This difference involves the movement of a single presumed water molecule, which in WT forms hydrogen bonds with the side chains of Glu-150, Tyr-183, and Arg-72 and the imidazolinone oxygen of the chromophore. The substitution appears to force the water molecule away from the side-chain oxygen of residue 150, increasing the hydrogen bond distance from ≈2.7 to ≈3.2 Å. However, the same water now makes a better hydrogen bond (≈2.8 Å compared with ≈3.2 Å in WT) with the carbonyl oxygen of the imidazolinone ring. We think that the changes in the hydrogen bond pattern are less important than the change in the charge distribution, so that the red shift in absorbance and emission can be attributed to the neutralization of residues 150 and 199. The overall hydrogen-bond network remains neutral, but now contains a dipole consisting of the Arg-72–Glu-217 interaction.
Discussion
Structural Basis for Cyan Fluorescence. Gurskyaya et al. (16) used site-directed mutagenesis based on a homology model to demonstrate that the size and polarity of the residue at position 165 could account for a portion of the blue shift observed in CFPs, compared with GFP. The structural studies confirm that the small side chain of Ala-165 allows a water molecule to occupy space that is usually filled by bulky, hydrophobic side chains Ile or Met in other FPs. In turn, this arrangement suggests that the polar interaction causes a shift in electron density toward the phenolate end of the chromophore. However, additional effects attributed to the presence of the presumably cationic His-199 side chain have a more profound influence. Substitution of His-199 with T, S, A, Q, or N results in dramatic red shifts in both absorbance and emission maxima and also decreases the fluorescence efficiency. To test whether the presumed positive charge on His-199 was important, we mutated nearby Glu-150 and Glu-217 to Gln. These isosteric substitutions also resulted in large red shifts in fluorescence emission suggesting neutralization of the His-199 side chain.
The structural and spectroscopic studies of amFP486 variants H199T, E150Q, and E217Q led us to conclude that the positive charge on H199 is the most important determinant of the blue-shifted excitation and emission of the chromophore. One effect, of course, is to stabilize the anionic form of the chromophore. The second effect may be to reduce the extent of charge transfer in the excited state. The results of several theoretical calculations (28–31) performed on the anionic form of the GFP chromophore led to the conclusion that absorption of a photon results in partial charge transfer from the p-hydroxybenzyl ring to the imidazolinone ring. In amFP486, the placement of a positive charge on His-199 near the phenolate moiety is expected to reduce the extent of charge delocalization, thus reducing both excitation and emission wavelengths. These conclusions are well supported by spectroscopic data obtained from model GFP chromophores in dioxane (32). In that study, it was found that placement of an electron accepting group at the position of the phenolate oxygen leads to blue shifts in absorbance and emission maxima. Conversely, placement of an electron donating group leads to red shifts in absorbance and emission (see derivatives I-3, I-19, and I-22 in table 1 of ref. 32). We propose that stabilization of the charge density distribution on the p-hydroxybenzyl ring of the chromophore is a general means by which to achieve blue shifts in absorbance and emission in FPs.
Unexpected Consequences of the H199T Substitution. Substitution of conserved His-199 revealed that this side chain has multiple roles. The substitution had the unanticipated consequence of dramatically lowering the quantum yield of fluorescence (ΦF) and increasing the disorder in the chromophore. There is little doubt that these phenomena are interrelated. The structural and spectroscopic studies of the H199T variant suggest that the chromophore exists as a statistical distribution of four states, protonated, anionic, cis, and trans. In accord with this result, the absorbance spectrum is adequately modeled by superposition of four Gaussian peaks (Table 3). However, the fluorescence excitation spectra of the H199T (Fig. 4A) suggest that only one species (anionic and presumably cis) is primarily responsible for the excitation and emission properties of this variant. We suggest that in WT amFP486, His-199 stabilizes the fluorescent state by “locking” the chromophore into the cis-planar state and lowering the pKa so that the anionic state is predominant. The replacement of His-199 allows the chromophore to occupy both cis and trans noncoplanar conformations. In turn, noncoplanarity presumably results in decreased ΦF due to increased vibrational degrees of freedom and additional opportunities for nonradiative modes of excitation decay.
There are very interesting structural parallels between the weakly fluorescent H199T variant of amFP486 and the nonfluorescent chromoproteins Rtms5 (33) and the kindling FP, KFP-1 (11, 12, 34–36). Superposition of the amFP486 H199T and Rtms5 structures reveal that the guanidinium groups of Arg-72 (as relocated in the H199T variant) and Arg-197 (Rtms5) occupy essentially the same spatial location adjacent to the chromophore, except that the side chains originate from different positions in the backbone. Strikingly, Arg-197 of Rtms5 corresponds to the conserved His found in amFP486 (His-199) and other strongly FPs. This finding suggests that in Rtms5, one function of Arg-197 is to destabilize the planar (and presumably fluorescent) forms of the chromophore. In contrast, KFP-1 contains His-197 (equivalent to His-199 in amFP486), but the chromophore is trans and noncoplanar. This chromoprotein becomes transiently fluorescent (“kindles”) when exposed to intense green light (35, 36). The structure of KFP-1 in the dark state was recently published (11, 12). His-197 was found to be statistically distributed among two different conformers (11), one of which corresponds to the stacking interaction seen in strongly fluorescent proteins such as amFP486. His-197 was proposed to play a key role in the kindling phenomenon and to act as a binary switch to stabilize the fluorescent chromophore state of KFP-1 (11). The structure of the H199T variant of amFP486, reported here, clearly demonstrates the importance of this stacking interaction for efficient fluorescence.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grant MCB-0111053 (to S.J.R.) and National Institutes of Health Training Grant GM-07759 (to the Institute of Molecular Biology). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-Eng-38. Use of BioCARS Sector 14 was supported by the National Institutes of Heath, National Center for Research Resources, under Grant RR07707.
Author contributions: J.N.H. and S.J.R. designed research; J.N.H. performed research; J.N.H. analyzed data; and J.N.H. and S.J.R. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FP, fluorescent protein; CFP, cyan-emitting Anthozoa FP; 2-ME, 2-mercaptoethanol.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2A46, 2A47, and 2A48).
References
- 1.Matz, M. V., Arkady, F. F., Labas, Y. A., Savitsky, A. P., Zaraisky, A. G., Markelov, M. L. & Lukyanov, S. A. (1999) Nat. Biotechnol. 17, 969–973. [DOI] [PubMed] [Google Scholar]
- 2.Labas, Y. A., Gurskaya, N. G., Yanushevich, Y. G., Fradkov, A. F., Lukyanov, K. A., Lukyanov, S. A. & Matz, M. V. (2002) Proc. Natl. Acad. Sci. USA 99, 4256–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gurskaya, N. G., Fradkov, A. F., Terskikh, A., Matz, M. V., Labas, Y. A., Martynov, V. I., Yanushevich, Y. G., Lukyanov, K. A. & Lukyanov, S. A. (2001) FEBS Lett. 507, 16–20. [DOI] [PubMed] [Google Scholar]
- 4.Dove, S. G., Takabayashi, M. & Hoegh-Guldberg, O. (1995) Biol. Bull. 189, 288–297. [DOI] [PubMed] [Google Scholar]
- 5.Dove, S. G., Hoegh-Guldberg, O. & Ranganathan, S. (2001) Coral Reefs 19, 197–204. [Google Scholar]
- 6.Gross, L. A., Baird, G. S., Hoffman, R. C., Baldridge, K. K. & Tsien, R. Y. (2000) Proc. Natl. Acad. Sci. USA 87, 11990–11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yarbrough, D., Wachter, R. M., Kallio, K., Matz, M. V. & Remington, S. J. (2001) Proc. Natl. Acad. Sci. USA 98, 462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wall, M. A., Socolich, M. & Ranganathan, R. (2000) Nat. Struct. Biol. 7, 1133–1138. [DOI] [PubMed] [Google Scholar]
- 9.Petersen, J., Wilmann, P. G., Beddoe, T., Oakley, A. J., Devenish, R. J., Prescott, M. & Rossjohn, J. (2003) J. Biol. Chem. 278, 44626–44631. [DOI] [PubMed] [Google Scholar]
- 10.Remington, S. J., Wachter, R. M., Yarbrough, D. K., Branchaud, B., Anderson, D. C., Kallio, K. & Lukyanov, K. A. (2005) Biochemistry 44, 202–212. [DOI] [PubMed] [Google Scholar]
- 11.Quillin, M. L., Anstrom, D. M., Shu, X., O'Leary, S., Kallio, K., Chudakov, D. M. & Remington, S. J. (2005) Biochemistry 44, 5774–5787. [DOI] [PubMed] [Google Scholar]
- 12.Wilmann, P. G., Petersen, J., Dvenish, R. J., Prescott, M. & Rossjohn, J. (2005) J. Biol. Chem. 280, 2401–2404. [DOI] [PubMed] [Google Scholar]
- 13.Heim, R., Prasher, D. C. & Tsien, R. Y. (1994) Proc. Natl. Acad. Sci. USA 91, 12501–12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ormo, M., Cubitt, A. B., Kallio, K., Gross, L. A., Tsien, R. Y. & Remington, S. J. (1996) Science 273, 1392–1395. [DOI] [PubMed] [Google Scholar]
- 15.Baird, G. S., Zacharias, D. A. & Tsien, R. Y. (2000) Proc. Natl. Acad. Sci. USA 97, 11984–11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurskaya, N. G., Savitsky, A. P., Yanushevich, Y. G., Lukyanov, S. A. & Lukyanov, K. A. (2001) BMC Biochem. 2, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gill, S. C. & von Hippel, P. H. (1989) Anal. Biochem. 182, 319–326. [DOI] [PubMed] [Google Scholar]
- 18.Weber, G. & Teale, F. W. J. (1957) Trans. Faraday Soc. 53, 646–655. [Google Scholar]
- 19.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 20.Kissinger, C. R., Gehlhaar, D. K. & Fogel, D. B. (1999) Acta Crystallogr. D 55, 484–491. [DOI] [PubMed] [Google Scholar]
- 21.Tronrud, D. E., Ten Eyck, L. F. & Matthews, B. W. (1987) Acta Crystallogr. A 43, 489–503. [Google Scholar]
- 22.Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 24.DeLano, W. L. (2002) pymol (DeLano Scientific, San Carlos, CA).
- 25.Spek, A. L. (2005) platon, A Multipurpose Crystallographic Tool (Utrecht University, Utrecht, The Netherlands).
- 26.Matthews, B. W. (1968) J. Mol. Biol. 33, 491–497. [DOI] [PubMed] [Google Scholar]
- 27.Yang, F., Moss, L. G. & Phillips, G. N., Jr. (1996) Nat. Biotechnol. 14, 1246–1251. [DOI] [PubMed] [Google Scholar]
- 28.Voityuk, A. A., Michel-Beyerle, M.-E. & Roesch, N. (1998) Chem. Phys. 196, 269–276. [Google Scholar]
- 29.Helms, V., Winstead, C. & Langhoff, P. W. (2000) J. Mol. Struct. 506, 179–189. [Google Scholar]
- 30.Cinelli, R. A. G., Tozzini, V., Pellegrini, V., Beltram, F., Cerullo, G., Zavelani-Rossi, M., De Silvestri, S., Tyagi, M. & Giacca, M. (2001) Phys. Rev. Lett. 86, 3439–3442. [DOI] [PubMed] [Google Scholar]
- 31.Marques, M. A. L., Lopez, X., Varsano, D., Castro, A. & Rubio, A. (2003) Phys. Rev. Lett. 90, 258101–258104. [DOI] [PubMed] [Google Scholar]
- 32.Follenius-Wund, A., Bourotte, M., Schmitt, M., Iyice, F., Lami, H., Bourguignon, J.-J., Haiech, J. & Pigault, C. (2003) Biophys. J. 85, 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prescott, M., Ling, M., Beddoe, T., Oakley, A. J., Hoegh-Guldberg, O., Devenish, R. J. & Rossjohn, J. (2003) Structure (London) 11, 275–284. [DOI] [PubMed] [Google Scholar]
- 34.Lukyanov, K. A., Fradkov, A. F., Gurskaya, N. G., Matz, M. V., Labas, Y. A., Savitsky, A. P., Markelov, M. L., Zaraisky, A. G., Zhao, X., Fang, Y., et al. (2000) J. Biol. Chem. 275, 25879–25882. [DOI] [PubMed] [Google Scholar]
- 35.Chudakov, D. M., Belousov, V. V., Zaraisky, A. G., Novoselov, V. V., Staroverov, D. B., Zorov, D. B., Lukyanov, S. & Lukyanov, K. A. (2003) Nat. Biotechnol. 21, 191–194. [DOI] [PubMed] [Google Scholar]
- 36.Chudakov, D. M., Foefanov, A. V., Mudrik, N. N., Lukyanov, S. & Lukyanov, K. A. (2003) J. Biol. Chem. 278, 7215–7219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.